Abstract
The phosphatidylinositol 3-kinase (PI3K) signaling axis impacts on cancer cell growth, survival, motility, and metabolism. This pathway is activated by several different mechanisms in cancers, including somatic mutation and amplification of genes encoding key components. In addition, PI3K signaling may serve integral functions for noncancerous cells in the tumor microenvironment. Consequently, therapeutics targeting the PI3K pathway are being developed at a rapid pace, and preclinical and early clinical studies are beginning to suggest specific strategies to effectively use them. However, the central role of PI3K signaling in a large array of diverse biologic processes raises concerns about its use in therapeutics and increases the need to develop sophisticated strategies for its use. In this review, we will discuss how PI3K signaling affects the growth and survival of tumor cells. From this vantage, we will consider how inhibitors of the PI3K signaling cascade, either alone or in combination with other therapeutics, can most effectively be used for the treatment of cancer.
INTRODUCTION
It has been more than 20 years since phosphatidylinositol 3-kinase (PI3K) was first discovered. The transforming ability of viral oncoproteins relied on an association with a PI3K lipid kinase activity.1–4 Over the ensuing years, studies established the central role of PI3K signaling in several cellular processes critical for cancer progression, including metabolism, growth, survival, and motility. Inappropriate co-option of PI3K signaling is one of the most frequent occurrences in human cancer.5,6 Consequently, significant efforts have been made to generate inhibitors of the PI3K pathway to treat cancers. However, it remains unknown which cancers will benefit most from these treatments and how to best use such therapeutics. In addition, the many possible untoward biologic sequelae of PI3K inhibition may limit the potential therapeutic gain of PI3K pathway inhibition. Here we will review data demonstrating the role of PI3K in tumor development and maintenance. We will compare the different potential therapeutic options for inhibiting this pathway and how their efficacy may be affected by the mechanism of PI3K pathway activation in a particular cancer. Finally, we will discuss the emerging data assessing the relative benefits of PI3K pathway inhibitors used as single agents versus combination therapies to treat cancer.
PI3K SIGNALING CASCADE REGULATES CELL GROWTH AND SURVIVAL
There are three classes of PI3Ks grouped according to structure and function. Class IA PI3K is the one most clearly implicated in human cancer.7 Class IA PI3Ks consist of a regulatory subunit and a catalytic subunit. Three mammalian genes, PIK3R1, PIK3R2, and PIK3R3, encode p85α (p85α, p55α, and p50α isoforms), p85β, and p55γ regulatory subunits, respectively, which by convention are referred to collectively as p85.5,7,8 The catalytic isoforms, p110α, p110β, and p110δ, are the products of three genes, PIK3CA, PIK3CB, and PIK3CD.5,8 As will be discussed in greater detail below, both PIK3CA and PIK3R1 are somatically mutated in cancers, and these mutations promote activation of the PI3K pathway.9–12
Class IA PI3Ks are activated by growth factor stimulation through receptor tyrosine kinases (RTKs).13–15 The regulatory subunit, p85, directly binds to phosphotyrosine residues on RTKs and/or adaptors.16 This binding relieves the intermolecular inhibition of the p110 catalytic subunit by p85 and localizes PI3K to the plasma membrane where its substrate, phosphatidylinositol 4,5-bisphosphate (PI[4,5]P2), resides.15,16 PI3K can also be stimulated by activated Ras, which directly binds p110.17 Additionally, the p110β catalytic subunit can be activated by G-protein coupled receptors.8
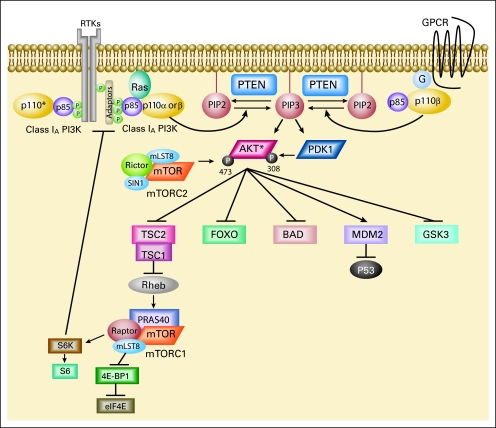
PI3K phosphorylates PIP2 on the 3′OH position to produce PI(3,4,5)P3 (PIP3; Fig 1). The tumor suppressor phosphatase and tensin homolog deleted on chromosome 10 (PTEN) dephosphoryates PIP3 to PIP2, thereby terminating PI3K-dependent signaling. PIP3 propagates intracellular signaling by directly binding pleckstrin homology (PH) domains of various signaling proteins.18 PI(3,4,5)P3 brings two PH domain-containing serine/threonine kinases, phosphoinositide-dependent kinase 1 (PDK1) and AKT, into close proximity. PDK1 activates AKT by phosphorylating AKT at threonine 308.19–21 PI3K-AKT signaling promotes cell growth and survival by several mechanisms. AKT promotes cell survival by inhibiting proapoptotic Bcl-2 family members BAD and BAX.5,18 AKT also impedes negative regulation of the transcription factor NF-κB, leading to increased transcription of antiapoptotic and prosurvival genes.22 Phosphorylation of Mdm2 by AKT antagonizes p53-mediated apoptosis, and AKT negatively regulates forkhead transcription factors, thereby reducing production of cell death-promoting proteins.22 AKT also phosphorylates TSC2, thereby inhibiting the rheb GTPase activity of the TSC1/TSC2 dimer. Activated rheb stimulates the mammalian target of rapamycin (mTOR) –containing protein complex mTORC1, leading to increased p70 S6 kinase activity.5 Activation of mTORC1 results in increased protein synthesis by phosphorylation of eukaryotic initiation factor 4E and the ribosomal S6 protein.5 While mTORC1 relays signals following PI3K-AKT activation, a second mTOR complex, mTORC2, contributes to complete AKT activation by phosphorylating AKT on serine 473.23–25 Of note, activation of the mTORC1 target, S6 kinase, negatively feeds back to diminish PI3K activation. S6 kinase can phosphorylate and inhibit the adaptor protein insulin receptor substrate 1, thereby inhibiting insulin or insulin-like growth factor 1–mediated PI3K activation.26–28
Fig 1.
The phosphatidylinositol 3-kinase (PI3K) signaling cascade. PI3K signaling impacts on cell growth, survival, and metabolism. Arrows represent activation, while bars reflect inhibition. A negative feedback loop has been described from the downstream target S6 kinase (S6K) to the adaptor protein IRS-1. RTK, receptor tyrosine kinase; GPCR, G-protein coupled receptor; P, phosphate; G, G protein; PTEN, phosphatase and tensin homolog; IRS-1, insulin receptor substrate 1; eIF4E, eukaryotic initiation factor 4E; S6, ribosomal S6 protein; PIP2, phosphatidylinositol 4,5-bisphosphate; mTORC2, rapamycin (mTOR) –containing protein complex 2. (*) p110 alpha, beta, or delta.
Inhibitors of PI3K Signaling in Cancer Treatment
Inhibition of PI3K signaling can diminish cell proliferation, and in some circumstances, promote cell death. Consequently, components of this pathway present attractive targets for cancer therapeutics. A number of PI3K pathway inhibitors have been developed and are being evaluated in preclinical studies and in early clinical trials. Rapamycin analogs, such as temsirolimus and everolimus, that specifically inhibit mTORC1 are the most advanced in the clinic, and they have received US Food and Drug Administration approval for the treatment of advanced renal cell carcinoma.29 The role for rapamycin analogs in the treatment of cancer has been extensively reviewed elsewhere and thus will not be discussed further.30 In this review, we will discuss the potential therapeutic roles for other PI3K pathway inhibitors. These include PI3K inhibitors (both pan-PI3K and isoform-specific PI3K inhibitors), dual PI3K-mTOR inhibitors that are catalytic site inhibitors of the p110 isoforms and mTOR (the kinase component of both mTORC1 and mTORC2), mTOR catalytic site inhibitors, and AKT inhibitors. Not only do these agents have the capacity to inhibit cancer cell proliferation and survival signals as described above, but they may also impact tumor angiogenesis, metastasis, and metabolism. Due to space limitations, the impact of PI3K inhibition on tumor angiogenesis and cell motility is discussed in the Appendix (online only).
PI3K and Insulin Signaling: Potential Toxicity and Pharmacodynamic Marker of PI3K Inhibition
PI3K signaling has a central role in mediating the effects of insulin on cellular metabolism that is conserved throughout eurkaryotic evolution.5 Noninsulin-dependent diabetes mellitus, marked by insulin insensitivity, is associated with diminished PI3K response to insulin signaling.5,31 Several transgenic and knockout mice harboring alterations in p85, p110, PTEN, or AKT2 validate the functional significance of this pathway on glucose homeostasis.31–34 These data suggest that insulin resistance may be observed in patients treated with PI3K pathway inhibitors, and indeed this may be used as a pharmacodynamic marker of target inhibition in patients. As will be discussed further below, initial phase I studies with PI3K pathway inhibitors have demonstrated some signs of insulin resistance, but this has not been a dose-limiting toxicity. While both p110α and p110β appear to play specific roles in insulin signaling, studies suggest that glucose homeostasis is predominantly mediated by p110α.35,36 Inhibitors of p110α, but not p110β or p110δ, have been shown to inhibit insulin-stimulated glucose uptake in adipocytes and to block insulin-mediated glucose regulation in mice.36 Consequently, in settings where p110β appears to be the critical PI3K catalytic isoform mediating transformation (eg, some PTEN-deficient tumors, see below), a p110β-specific inhibitor may offer efficacy with decreased risk of insulin resistance compared with a pan-PI3K inhibitor.
ACTIVATION OF PI3K SIGNALING IN CANCER
PI3K signaling is activated in human cancers via several different mechanisms.6,11–13 Increased PI3K signaling is often due to direct mutational activation or amplification of genes encoding key components of the PI3K pathway such as PIK3CA and AKT1, or loss of PTEN.6,9,12,37–41 Genetic alterations in several components of the PI3K signaling pathway have been reported and are summarized in Table 1. PI3K also can be activated by genetic mutation and/or amplification of upstream RTKs, and possibly by mutationally activated Ras.7,17 The mechanism of PI3K activation in an individual cancer may suggest the most effective type of therapeutic to inhibit the pathway.
Table 1.
Genetic Alterations of the PI3K Signaling Pathway in Cancer
| Gene | Alteration | Comments | Tumor Types | References |
|---|---|---|---|---|
| PTEN | LOF mutation | Truncation; loss of phosphatase activity | Bladder, brain, breast, cervical, colorectal, endometrial, gastric, head and neck, kidney | COSMIC, Li et al38 |
| Deletion | Homozygous or hemizygous | Leukemia, liver, lung, lymphoma, melanoma, ovary, prostate, thyroid | ||
| Epigenetic silencing | Transcriptional repression, usually by promoter hypermethylation | Breast, colon, melanoma | Garcia et al,42 Goel et al,43 Berns et al44 | |
| PIK3CA | GOF mutation | Exon 9 (E542K, E545K) helical domain; exon 20 (H1047R) catalytic domain | Breast, colorectal, glioblastoma, endometrial, cervical, esophageal, gastric, head and neck, liver, lung, lymphoma, ovarian, pancreatic, prostate, thyroid | COSMIC, Samuels et al,6 Samuels et al12 |
| Amplification | Increased protein levels and activity | Breast, cervical, gastric, lung, ovarian, prostate | Sun et al,45 Byun et al,46 Campbell et al,47 Ma et al,48 Rácz et al49 | |
| PIK3R1 | GOF mutation | Loss of C-terminal inhibitory domain; constitutive activity | Brain, colon, ovarian | Mizoguchi et al,10 Philp et al11 |
| AKT1 | GOF mutation | Pleckstrin homology domain, membrane localization, and constitutive activation | Breast, colon, endometrial, melanoma, ovarian | Shoji et al,40 Carpten et al,50 Davies et al51 |
| AKT2 | GOF mutation | Kinase domain mutation | Colorectal | Parsons et al52 |
| Amplification | Breast, colon, lymphoma, pancreas | Cheng et al,53 Cheng et al,54 Bellacosa et al55 | ||
| AKT3 | GOF mutation | Pleckstrin homology domain, membrane localization, and constitutive activation | Melanoma | Davies et al51 |
| PDK1 | GOF mutation | Kinase domain mutation | Colorectal | Parsons et al52 |
Abbreviations: LOF, loss of function; COSMIC, Catalogue of Somatic Mutations in Cancer at www.sanger.ac.uk/genetics/CGP/cosmic; GOF, gain of function.
Somatic Alterations of PI3K Pathway Components in Cancer
The most common genetic alteration of the PI3K signaling pathway found in human cancer is inactivation of the PTEN tumor suppressor gene. Inactivation of PTEN leads to loss of its lipid phosphatase activity, causing accumulation of PIP3.56,57 The majority of somatic mutations in PTEN lead to protein truncation. However, missense mutations that typically abrogate PIP3 phosphatase activity are also commonly noted.58 While most PTEN mutations are sporadic, germline mutations in PTEN are noted in hereditary neoplastic disorders, such as Cowden disease.59 Homozygous and hemizygous deletions of PTEN are also observed in human cancers.38,45 Transcriptional repression and epigenetic silencing of PTEN, typically through promoter hypermethylation, is also a mechanism of PTEN inactivation.42,43 Because there are both genetic and epigenetic causes for PTEN loss, accurate assessment of a cancer's PTEN status remains challenging and may ultimately require reliable measurements of protein expression.
More recently, somatic mutations in PIK3CA have been identified in a variety of human tumors, including breast, colon, and endometrial cancers and glioblastomas (see Catalogue of Somatic Mutations in Cancer, http://www.sanger.ac.uk/genetics/CGP/cosmic).6,7,9,12,39 Most of these mutations cluster to two hot spot regions in exons 9 and 20.6,12 Exon 20 encodes the catalytic domain of p110α, and mutations in this domain may constitutively activate its enzymatic activity. Exon 9 encodes the helical domain of p110α, and these mutations de-repress an inhibitory interaction between the N-terminal SH2 domain of p85 and the p110α catalytic subunit.60,61 A smaller cluster of mutations is also found in the N-terminal p85 interacting domain. Interestingly, these mutations increase the lipid kinase activity of p110α but do not appear to alter the interaction between p110α and p85α.9,12
Expression of these PIK3CA mutants leads to increased oncogenic potential in vitro and in vivo.9,37 They cause constitutive signaling along the PI3K pathway in the absence of growth factors and therefore seem to obviate the usual obligate interactions with tyrosine phosphorylated RTKs and/or adapters. Thus, it is intriguing that some studies have suggested that the presence of these mutations confers resistance to therapies targeting RTKs.44,62 Expressing mutated PIK3CA in fibroblasts and mammary epithelial cells results in transformation, growth factor-independent proliferation, and resistance to apoptosis.9,63,64 Additionally, transgenic mice with lung-specific induction of the kinase-domain mutant p110α H1047R develop lung adenocarcinomas.65 In addition to these activating mutations, amplification of PIK3CA is also observed frequently in ovarian cancer and other tumors, but how amplification affects PI3K activation is less clear.39
Mutations in the p85 regulatory subunit PIK3R1 are also observed in a variety of human cancers, including glioblastomas, ovarian cancers, and colorectal cancers.10,11 Mutations in PIK3R1 generally produce either truncations or in-frame deletions that often localize to the inter-SH2 domain of p85α. Structural analyses suggest that the iSH2 domain of p85 interacts with the C2 domain of p110.60 Thus, it seems likely that these p85α mutations also activate PI3K signaling by relieving the inhibitory effect of p85 on p110.11,66 Laboratory studies suggest that these mutations also lead to constitutive PI3K signaling.11,66
Mutations in AKT family genes encoding for AKT1, AKT2, and AKT3 have also been identified in human cancers. A single amino acid substitution, E17K, in the lipid-binding PH domain of AKT1 has been identified in breast, colorectal, endometrial, and ovarian cancers, as well as melanoma.40,50,51 This amino acid change alters AKT1 lipid binding, presumably leading to constitutive membrane localization in the absence of PIP3. However, although phosphorylation on Ser473 was constitutive in this mutant, T308 phosphorylation was still responsive to PI3K activation.50 Thus, it is unclear if PI3K inhibitors will effectively decrease AKT signaling in cancers with these mutations. The E17K mutation has also been identified in AKT3 in some melanomas.51 In addition, mutations affecting the kinase domain of AKT2 have been found in colorectal cancers; however, the functional consequences of these mutations have not been assessed.52 Amplification of AKT2 has also been reported in human tumors.53,54
PI3K Activation by Receptor Tyrosine Kinases and Ras
In normal epithelial cells, PI3K is often activated downstream of RTK signaling. In cancers, these RTKs are often mutated, amplified, or overexpressed, causing aberrant PI3K activation. When therapies targeting RTKs are effective, they invariably lead to loss of PI3K signaling.67 For example, PI3K is activated by epithelial growth factor receptor (EGFR) in lung cancers harboring somatic activating mutations in EGFR, and by human epidermal growth factor receptor 2 (HER2) in breast cancers with HER2 amplification.65,68–70 In these cancers, EGFR or HER2 phosphorylates the kinase-dead ErbB3 that, in turn, directly binds and activates PI3K.65,68–70 Thus, when these cancers are successfully treated with EGFR- and HER2-targeted therapies, respectively, PI3K signaling is turned off and the cells undergo cell death. Similarly, glioblastomas frequently exhibit PI3K activation, either through integration of signaling from multiple activated RTKs, such as the constitutively active EGFRvIII mutant, or through the combined activation of RTKs and loss of PTEN.71,72
The small GTPase Ras is also frequently mutated in human cancers, and PI3K is an effector of Ras-mediated oncogenic signaling.73 Early studies showed that Ras directly bound p110, and provided a direct link between Ras and PI3K.74 In addition, functional studies demonstrated that PI3K activation appears to be crucial for tumor initiation. For example, expression of a dominant-negative p85α lacking the p110-binding domain inhibited Ras-mediated transformation.8,75 In addition, expression of a p110α mutant that does not directly bind Ras inhibited K-Ras–induced lung adenocarcinomas in genetically engineered mouse models.76 Similarly, deletion of Pik3r1 and Pik3r2 abrogated K-Ras G12D-induced lung tumorigenesis.65 Although PI3K activation may be necessary for K-Ras–induced tumorigenesis, preliminary studies suggest that inhibition of PI3K signaling alone may not be sufficient to shrink established tumors in vivo or effectively treat K-Ras–mutated cancer cell lines in vitro.65,77 These findings underscore the difference between killing established cancers and blocking tumorigenesis and cell transformation. Furthermore, these studies suggest that established cancers with KRAS mutations may not be sensitive to single-agent PI3K pathway inhibitors.
Potential Roles for p110β, p110δ, and p110γ in Transformation
While activating mutations in PIK3CA are frequently identified in human cancers, no oncogenic mutations have been verified in p110β, p110δ, or the class IB catalytic isoform p110γ. Although rare somatic single-residue substitutions have been found in p110β and p110γ (www.sanger.ac.uk/perl/genetics/CGP/cosmic), the function of these substitutions is unknown. Despite the lack of evidence for activating mutations in these other p110 catalytic isoforms, recent work has demonstrated the oncogenic potential of p110β, p110δ, and p110γ. Interestingly, unlike p110α, overexpression of wild-type p110β, p110δ, or p110γ is transforming in cell culture.13 Although expression of the γ and δ isoforms is normally restricted to leukocytes, increased p110δ (as well as p110β) has been identified in some colon and bladder cancers, as well as in glioblastomas.13 p110δ appears to provide the critical PI3K activity in acute myelogenous leukemia, while p110γ is upregulated by the Bcr-Abl oncogene in chronic myelogenous leukemia.78,79
Recent data also suggest a prominent role for p110β in PTEN-deficient tumors. Targeted deletion of pten in the mouse prostate results in prostatic intraepithelial neoplasia and carcinoma.80 Concomitant ablation of p110β, but not p110α, decreased PI3K proliferation signaling and prevented prostate tumorigenesis.35 Similarly, inducible depletion of p110β, but not p110α, using short hairpin RNA in PTEN-deficient human cancer cell lines extinguished PI3K-mediated signaling and inhibited growth in vitro and in vivo.81 Deletion of p110β also abrogated transformation of mouse embryo fibroblasts by activated Ras or EGFR mutants to a more pronounced extent than did p110α loss.35 These studies suggest that although cancers driven by PIK3CA mutations are candidates for treatment with p110α-specific inhibitors, treatment of PTEN-deficient cancers may require agents with activity against p110β.
PI3K PATHWAY INHIBITORS ENTERING THE CLINIC: PRECLINICAL AND EARLY CLINICAL DATA
A number of potential therapeutics targeting the PI3K signaling cascade have been generated. We will consider four different classes of PI3K pathway inhibitors: dual PI3K-mTOR inhibitors, PI3K inhibitors (that do not inhibit mTOR), AKT inhibitors, and mTOR catalytic site inhibitors. Table 2 summarizes PI3K pathway inhibitors in clinical trials.
Table 2.
PI3K Pathway Inhibitors in Clinical Trials
| Targets | Compound (Company) | Study Population | Reported Efficacy/Responses | Reported Toxicities | References |
|---|---|---|---|---|---|
| PI3K/mTOR | SF-1126 (Semafore Pharmaceuticlas) | Phase I: advanced solid tumors | SD in 11 of 28 pts | DLT: grade 3 diarrhea (one pt) | Chiorean et al82 |
| NVP-BEZ235 (Novartis) | Phase I/II: advanced solid tumors (breast cancer-enriched) | N/A | N/A | ClinicalTrials.gov | |
| NVP-BGT226 (Novartis) | Phase I/II: advanced solid tumors (including breast cancer) | N/A | N/A | ClinicalTrials.gov | |
| XL765 (Exelixis) | Phase I: refractory solid tumors | SD in five of 36 pts | Most frequent (> 10%) AEs: elevated liver enzymes, nausea, diarrhea | LoRusso et al83 | |
| Other AEs: anorexia/hypophosphatemia, rash, vomiting, and neurologic complaints | |||||
| PI3K | PX-866 (Oncothyreon) | Phase I: advanced solid tumors | SD in two of six pts (initial dosing cohorts) | Possible AEs: abdominal discomfort, mild diarrhea | Jimeno et al84 |
| No DLTs in early dosing cohorts | |||||
| XL147 (Exelixis) | Phase I: advanced solid tumors | SD (> 6 months) in six of 39 pts; one pt with HR CaP with normalized PSA | AEs: grade 3 rash (DLT in two of three pts at highest tested dose); grade 3 arterial thrombosis (one pt); grade 2 transaminitis (one pt); grade 1 hyperglycemia (four pts) | Shapiro et al85 | |
| NVP-BKM120 (Novartis) | Phase I: solid tumors | N/A | N/A | Markman et al86 | |
| GDC-0941 (Genentech/Piramed) | Phase I: advanced solid tumors | Evidence for potential antitumor activity in three of 19 pts | Most frequent AEs: Grade 1 to 2 nausea, fatigue, diarrhea, dysgeusia, and peripheral edema | Wagner et al87 | |
| CAL-101 (Calistoga Pharmaceuticals) | Phase I: relapsed/refractory CLL or B-cell NHLs | PR in two of six pts, SD in four of six pts in early dosing cohorts | No AEs grade > 1 in first two dosing cohorts | Flinn et al88 | |
| Akt (allosteric) | MK-2206 (Merck) | Phase I: advanced solid tumors | SD in six of 19 patients | Most frequent AEs: skin (47.1%) and GI (41.2%) toxicities; DLTs: grade 3 to 4 rash, grade 3 mucositis | Tolcher et al89 |
Abbreviations: SD, stable disease; DLT, dose-limiting toxicity; Pt, patient; N/A, results from patient treatment in clinical trials have not been reported to date; AE, adverse event; HR CaP, hormone-refractory prostate cancer; PSA, prostate-specific antigen; CLL, chronic lymphocytic leukemia; NHL, non-Hodgkin's lymphoma; PR, partial response.
Dual PI3K-mTOR Inhibitors
The catalytic domains of the p110 subunits and mTOR are structurally similar, because they all belong to the phosphatidylinositol kinase–related kinase family of kinases. Many chemical inhibitors under development inhibit both mTOR and the p110 catalytic subunits. These are termed dual PI3K-mTOR inhibitors. When compared with the other types of PI3K pathway inhibitors, dual PI3K-mTOR inhibitors have the possible advantage of inhibiting all PI3K catalytic isoforms, mTORC1, and mTORC2. Thus, they should effectively turn off this pathway completely and overcome feedback inhibition normally observed with mTORC1 inhibitors (ie, rapamycin analogs) that may limit their efficacy (Fig 1).28 However, it remains unknown if dual PI3K-mTOR inhibitors will be tolerable at doses that effectively inhibit all p110 isoforms and mTOR, or if their use will necessitate sacrificing complete inhibition of one or more of the potential targets.
For many years, the PI3K inhibitor LY294002, a dual PI3K-mTOR inhibitor, has been extensively used in preclinical studies. Although LY294002 is unsuitable for patient use, the backbone structure of this compound has been exploited in the design of novel PI3K inhibitors.7,90 SF-1126 (Semafore, Indianapolis, IN) is a prodrug of LY294002 that is conjugated to a tetra-peptide designed to target tumor vasculature, and this compound has demonstrated efficacy in solid tumor xenograft models.7,90,91 In a phase I study of SF-1126, mTORC1 inhibition in cancers was demonstrated by decreased S6 phosphorylation.82 No responses were observed, but stable disease was achieved in 11 of 28 patients below the maximum tolerated dose, without consistent effects on blood glucose.82
Other dual PI3K-mTOR inhibitors, such as NVP-BEZ235 and NVP-BGT226 (Novartis, Basel, Switzerland) and XL765 (Exelixis, San Francisco, CA) have entered phase I testing in clinical trials.30,90,92 There have been several preclinical evaluations of NVP-BEZ235. NVP-BEZ235 slowed the growth of PTEN-deficient human cancer cell line xenografts in mice, and it was well tolerated with no significant changes in body weight.92 Additionally, breast cancer cell lines with HER2 amplification and/or PIK3CA mutations appeared to be particularly sensitive to these agents; however, it should be noted that only tumor stasis, and not tumor regression, was observed in vivo.93 NVP-BEZ235 was further shown to induce apoptosis in estrogen-deprived estrogen receptor–positive breast cancer cells harboring either PIK3CA mutations or PIK3CB amplification.94 In this study, RNA interference was also used to knock down p110α, p110β, or both in these cells. In some estrogen receptor–positive breast cancer cell lines harboring a PIK3CA mutation, dual knockdown of both p110α and p110β led to greater apoptosis following estrogen deprivation compared with knockdown of either isoform alone.94 These results underscore a potential benefit of inhibiting both p110α and p110β, even in cancers that harbor specific genetic activation of one isoform.94 A study with genetically engineered mice also demonstrated that NVP-BEZ235 was highly effective at shrinking murine lung tumors driven by a p110α H1047R transgene.65 Recently, phase I results for the dual PI3K-mTOR inhibitor, XL765 were reported at the 45th American Society of Clinical Oncology (ASCO) annual meeting (2009). There were no responses, but stable disease was noted in five of 36 patients.83 There was evidence of 50% to 80% pathway inhibition in surrogate tissue. However, it is unclear whether this level of inhibition will be sufficient to induce shrinkage in potentially responsive tumors, or whether more complete inhibition will be required. No significant changes in serum glucose were noted, although an augmentation in food-induced plasma insulin increases was observed.83
PI3K Inhibitors
The PI3K inhibitors can be divided into isoform-specific inhibitors or pan-PI3K inhibitors. Pan-PI3K inhibitors target all class IA PI3K in the cancer. These include wortmannin derivatives such as PX-886 or wortmannin prodrugs such as the self-activating viridans modified by dextran linker moieties that are designed to increase permeability and extend serum half-life.95–97 These agents exhibit cytostatic antitumor effects in vivo.95–97 The presence of PIK3CA mutations appears to predict for sensitivity to PX-866 across an array of cancer cell lines derived from different tissues of origin.77 Interestingly, PTEN loss also appears to predict for PX-866 sensitivity, despite its relatively low efficacy toward p110β.77 Animals treated with PX-866 experienced hyperglycemia with decreased glucose tolerance as a major toxicity of PI3K inhibition, but this could be overcome with the oral antidiabetic agent pioglitozone.98 Phase I clinical trial results for other pan-PI3K inhibitors have also been reported. Of 19 patients with solid tumors treated with GDC-0941 (Piramed/Genentech, Slough, United Kingdom/South San Francisco, CA), three demonstrated potential signs of antitumor activity.87 Another pan-PI3K inhibitor, XL147, produced durable disease control in six of 39 treated patients. As observed with the dual PI3K-mTOR inhibitor XL765, plasma glucose levels were minimally affected by XL147, although an augmentation of food-induced plasma insulin increases was noted.85
A possible advantage of isoform-specific PI3K inhibitors is that they may be tolerated at doses resulting in more complete target inhibition with fewer adverse effects. Isoform-specific inhibitors that selectively inhibit p110α, β, δ, or γ catalytic subunits are under investigation in preclinical studies.99,100 Indeed, a p110δ-specific inhibitor tested for refractory non-Hodgkin's lymphoma and chronic lymphocytic leukemia induced responses in 6 out of 12 patients (presented at the 45th Annual Meeting of ASCO in 2009).80
AKT Inhibitors
Both adenosine triphosphate (ATP) mimetics and noncatalytic-site AKT inhibitors are under active clinical development.90,101 Cancers with AKT1 mutations and AKT1 and AKT2 amplifications may be expected to be among the more sensitive to AKT inhibitors. However, this class of inhibitors will not block the non-AKT effectors of PI3K signaling and, paradoxically, could actually increase PI3K-dependent activation of those effectors via loss of negative feedbacks. This is especially important in light of the recent findings that the PDK1 substrate SGK3, and not AKT, may play a more prominent role in promoting PI3K-dependent viability in some cancers harboring PIK3CA mutations.102 Despite these findings, a recent study demonstrated that a noncatalytic site AKT1/AKT2 inhibitor was effective against breast cancer cell lines with PIK3CA mutations and HER2 amplifications.101 Phase I results for the allosteric pan-AKT inhibitor MK-2206 (Merck, Whitehouse Station, NJ) showed stable disease in six of 19 patients and decreases in CA125 in patients with ovarian cancer. Adverse effects included rash and hyperglycemia.89
mTOR Catalytic Site Inhibitors
Rapamycin interacts with FKBP12 in mammalian cells to form a complex that directly binds to the FKBP12-rapamycin-binding domain of mTOR in mTORC1, but not in mTORC2.103,104 Conversely, ATP-competitive mTOR inhibitors target the kinase domain of mTOR to impede the activity of both mTORC1 and mTORC2. Inhibiting mTORC2 would provide the theoretical advantage of blocking AKT activation. An ATP-competitive mTOR inhibitor might be more effective than rapamycin because, by blocking AKT activation, it would mitigate the activation of PI3K that often accompanies mTORC1 inhibition (ie, de-repression of the negative feedback, Fig 1). Intriguing preclinical data are emerging from studies of these compounds that shed new light on the potential limitations of rapamycin analogs. Feldman et al105 demonstrated that two mTOR kinase domain inhibitors, PP242 and PP30, inhibit both mTORC1 and mTORC2. Unlike acute rapamycin treatment, which activates AKT, PP242 administration to mice suppressed AKT activation in tissues. PP242 was also a more effective inhibitor of proliferation than rapamycin.105 Surprisingly, the improved efficacy of PP242 appeared to be due to more effective mTORC1 inhibition, rather than through its additional inhibition of mTORC2.105 Similarly, the ATP-competitive mTOR inhibitor Torin1 impeded cell proliferation predominantly via its effects on mTORC1, not mTORC2.106 Both PP242 and Torin1 were more effective inhibitors of 4E-BP1 phosphorylation and cap-dependent RNA translation than rapamycin.103,105,106 Three additional ATP-competitive mTOR inhibitors—WAY-600, WYE-687, and WYE-354—have been shown to inhibit proliferation of a variety of cancer cell lines more effectively than rapamycin, causing G1 cell cycle arrest, and in some cases, apoptosis.104
Although the clinical results with PI3K pathway inhibitors are preliminary, their efficacy has modest at best. For their effective development, it will be imperative to understand why these drugs fail to produce a response when they do. Is the lack of activity due to inadequate inhibition of the target, or because complete inhibition of the target is not sufficient to produce antitumor activity? Indeed, most of the studies to date have not assessed this issue systematically. To answer this question, future studies with quantitative pharmacodynamic assessments will be required to determine the degree of target inhibition. For example, a study with even a small number of patients with favorable genotypes (eg, PIK3CA mutants or HER2 amplified) that correlates pharmacodynamic responses of PI3K pathway inhibition with outcomes may prove invaluable in identifying the reasons for lack of efficacy.
POTENTIAL CLINICAL USES FOR PI3K PATHWAY INHIBITORS
Thus far, preclinical studies have shown that PI3K pathway inhibitors may have significant single-agent activity in a few types of genetically defined cancers: HER2-amplified breast cancers, cancers with PIK3CA mutations, and PTEN-deficient cancers.65,77,92,93,101 To this point, data suggest that cancers with KRAS mutations may be fairly resistant to PI3K pathway inhibitors.65,77 Consequently, it seems likely that the presence of KRAS mutations will limit the efficacy of single-agent PI3K pathway inhibitors in cancers harboring both KRAS and PIK3CA mutations, such as many colon cancers.
In addition to these genetically defined settings, there may be other opportunities to target the PI3K pathway. For example, PI3K pathway inhibitors may be effective agents in the treatment of certain cancers that acquire resistance to RTK inhibitors. Cancers that are sensitive to receptor tyrosine kinase inhibitors (TKIs) have PI3K under the exclusive control of that RTK.67 When a TKI works, it leads to downregulation of PI3K activity. For example in HER2-amplified breast cancers, trastuzumab disrupts the interaction between ErbB2 and ErbB3, resulting in ErbB3 dephosphorylation and loss of interaction with PI3K.107 Furthermore, the presence of an activating PIK3CA mutation or depletion of PTEN correlates with a poor response to trastuzumab, presumably because these cancers fail to downregulate PI3K signaling in response to the anti-HER2 therapy. Furthermore, when cancers that were initially sensitive to TKIs subsequently develop resistance, they invariably find a way to maintain PI3K signaling in the presence of the TKI.67 These cancers therefore may be susceptible to addition of a PI3K pathway inhibitor to the TKI to re-induce remissions. This is an attractive approach in HER2-amplified breast cancers, because they appear to be sensitive to PI3K pathway inhibitors even before they develop resistance to anti-HER2 –based therapies. Indeed, the HER2-amplified breast cancer cell lines with PIK3CA mutations are resistant to trastuzumab, and this can be overcome with treatment with GDC-0941.44,107 Similarly, PTEN loss, or activating mutations in PIK3CA, confers resistance to lapatinib, which can be overcome by treatment with NVP-BEZ235.68
Potential of Combining PI3K With MEK Pathway Inhibitors
When cancers are sensitive to TKIs (ie, oncogene-addicted to an RTK), the TKI usually leads to downregulation of PI3K and other pathways, including the MEK-MAPK pathway. Thus, it remains unclear whether single-agent PI3K pathway inhibitors will promote dramatic responses (comparable to gefitinib in EGFR-mutant lung cancers or imatinib in chronic myelogenous leukemia), even in sensitive cancers. Most models of cancers that are sensitive to single-agent PI3K pathway inhibitors have demonstrated tumor stasis in vivo rather than frank tumor regressions.91–93,95–97 Consequently, it may be necessary to combine PI3K pathway inhibitors with other agents to induce dramatic responses. Furthermore, there may be cancers that show no response to single-agent PI3K pathway inhibitors that will respond to PI3K pathway inhibitors combined with other therapies. For example, inhibition of PI3K signaling with NVP-BEZ235 failed to shrink established Kras G12D-driven lung tumors.65 However, combined PI3K and MAPK pathway inhibition by treatment with NVP-BEZ235 and the MEK inhibitor ARRY-142886 led to marked tumor regression in this Kras lung cancer model.65 Similarly, combined PI3K and MEK inhibition was required to effectively shrink EGFR-mutant lung cancers in genetically engineered mouse models.108 Findings such as these are spurring the biotechnology and pharmaceutical industries to combine therapeutic inhibitors of these two pathways.
SUMMARY
Great strides are being made in our understanding of the diverse roles that PI3K signaling plays in cancer initiation, progression, and maintenance. Novel therapeutics targeting different components of this pathway are demonstrating efficacy in an array of human cancer types in preclinical studies, and these drugs are being carried forward into clinical trials. There is growing preclinical evidence that some genetically defined cancer subtypes may be the most sensitive to single-agent PI3K pathway inhibitors. These include cancers with PIK3CA activating mutations, loss of PTEN, and breast cancers with HER2 amplification. However, it remains to be determined whether these sensitive cancers will demonstrate stable disease or tumor shrinkage in response to single-agent therapeutics. Conversely, cancers harboring activated Ras mutants appear to be insensitive to PI3K pathway inhibition alone. In such cases, effective treatment with PI3K inhibitors may require concomitant therapies targeting MAPK signaling.
Acknowledgment
We apologize to the many authors whose work we could not cite directly because of space limitations. We are grateful to Lew Cantley for his mentorship and for thoughtful discussions and insights regarding PI3K signaling.
Appendix
Inhibition of PI3K Signaling Impedes Tumor Angiogenesis
In addition to their potential direct effects on cancer cells, PI3K pathway inhibitors may have profound effects on the tumor microenvironment. Recent studies have demonstrated that endothelial cell survival, vascular development, and tumor neovascularization require intact PI3K signaling (Graupera M: Nature 453:662-666, 2008; Yuan TL: Proc Natl Acad Sci U S A 105:9739-9744, 2008). PI3K is a component of a complex that includes vascular endothelial cadherin (VE-cadherin), β-catenin, and vascular endothelial growth factor receptor-2 (VEGFR-2) in endothelial cells. Deletion or truncation of VE-cadherin disrupts this complex in mice, resulting in endothelial cell apoptosis and embryonic lethality due to impaired survival signaling from VEGF-A to PI3K-AKT (Carmeliet P: Cell 98:147-157, 1999).8 Direct chemical inhibition of PI3K also inhibits VEGF-A–mediated endothelial cell survival (Carmeliet P: Cell 98:147-157, 1999).8 Deletion of the p85 regulatory subunits of Class IA PI3K in endothelial cells of developing mice results in hemorrhage and embryonic lethality (Yuan TL: Proc Natl Acad Sci U S A 105:9739-9744, 2008). Mice that retain one allele of p85α are viable but show impaired tumor growth using transplanted allografts with alterations in tumor vessel integrity, including blood vessel leakage and decreased number of large vessels (Yuan TL: Proc Natl Acad Sci U S A 105:9739-9744, 2008). An elegant series of studies that evaluated specific loss of p110α function in endothelial cells revealed that p110α, but not p110β or p110δ, appear to be critical for proper vascular development (Graupera M: Nature 453:662-666, 2008). These findings suggest that a pan-PI3K inhibitor, or perhaps an inhibitor with specificity toward the p110α catalytic subunit, might be effective at impeding tumor angiogenesis independent of the effects of PI3K inhibition within the tumor cells themselves. Indeed, Yuan et al (Yuan TL: Proc Natl Acad Sci U S A 105:9739-9744, 2008) demonstrated that a dual PI3K-mTOR inhibitor, NVP-BEZ235 (Novartis, Basel, Switzerland), significantly impaired vascularization in response to VEGF. Similarly, NVP-BEZ235 was shown to reduce tumor microvessel permeability and to decrease tumor interstitial fluid pressure in orthotopic rat mammary tumor models (Schnell CR: Cancer Res 68:6598-6607, 2008). Because drugs that affect vessel growth and integrity (eg, bevacuzimab) have shown efficacy in a number of cancer types, there is a possibility that PI3K pathway inhibitors may also have therapeutic efficacy via effects on the vasculature.
The Role of PI3K Signaling in Cell Motility: Potential Contributions to Tumor Metastasis
In addition to its effects on vascular development and tumor angiogenesis, the PI3K pathway may be integral to promoting tumor metastasis. Laboratory studies have repeatedly demonstrated that the PI3K pathway contributes to cytoskeletal reorganization following growth factor stimulation or Ras activation (Hawkins PT: Curr Biol 5:393-403, 1995).8,75 Platelet-derived growth factor (PDGF)–mediated PI3K stimulation results in Rac activation and formation of lamellipodia (Hawkins PT: Curr Biol 5:393-403, 1995). Indeed, cells expressing a mutant PDGF receptor-β incapable of binding to PI3K have impaired chemotaxis in response to PDGF-BB (Heuchel R: Proc Natl Acad Sci U S A 96:11410-11415, 1999). PI3K signaling also can be activated by the G-protein-coupled chemokine receptor CXCR4, which appears to be the principal chemokine receptor expressed on cancer cells (Pan J: Mol Cancer 5:56, 2006; Staller P: Nature 425:307-311, 2003). The interaction between chemokine receptors expressed on cancer cells and their ligands expressed at potential sites of metastasis may govern organ-specific metastasis, and thus chemokine-induced activation of PI3K may allow for enhanced survival of cancer cells at particular potential sites of metastases (Pan J: Mol Cancer 5:56, 2006; Staller P: Nature 425:307-311, 2003).
Interestingly, emerging data suggest that AKT1 and AKT2 may have isoform-specific roles in cancer development and metastasis. In transgenic mouse models of breast cancer, expression of activated AKT1 accelerated tumor development without affecting metastatic potential, while activated AKT2 expression increased metastases without altering tumor latency (Dillon RL: Cancer Res 69:5057-5064, 2009). Similar results suggesting a central role for AKT2, but not AKT1 in cancer invasion and metastases, were observed in mammary epithelial cells in vitro and human colorectal cancer cell lines in vivo (Irie HY: J Cell Biol 171:1023-1034, 2005; Rychahou PG: Proc Natl Acad Sci U S A 105:20315-20320, 2008). Interestingly, loss of AKT1 appeared to enhance cellular invasiveness and metastasis, presumably by increasing signaling through AKT2. This raises theoretical concerns about potential deleterious effects from AKT1-isoform specific inhibitors.
Footnotes
Supported by an American Society of Clinical Oncology Young Investigator Award and a Genentech Dana-Farber/Harvard Cancer Center (DF/HCC) Kidney Cancer Career Development Award (K.D.C.), by National Institutes of Health K08 Grants No. CA120060, No. R01CA140594, and No. R01CA137008, by National Cancer Institute Lung Special Program of Research Excellence (SPORE) Grant No. P50CA090578, DF/HCC Gastrointestinal Cancer SPORE Grant No. P50 CA127003, the Ellison Foundation Scholar award (J.A.E.), and the American Association for Cancer Research Stand Up To Cancer program.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Jeffrey A. Engelman, Novartis (C), AstraZeneca (C), Millennium Pharmaceuticals (C), Bristol-Myers Squibb (C), Hoffman-La Roche (C), OSI Pharmaceuticals (C), Schering-Plough (C), MedImmune (C), Daiichi Sankyo (C) Stock Ownership: None Honoraria: None Research Funding: Kevin D. Courtney, Genentech; Jeffrey A. Engelman, Novartis Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Kevin D. Courtney, Ryan B. Corcoran, Jeffrey A. Engelman
Manuscript writing: Kevin D. Courtney, Ryan B. Corcoran, Jeffrey A. Engelman
Final approval of manuscript: Kevin D. Courtney, Ryan B. Corcoran, Jeffrey A. Engelman
REFERENCES
- 1.Jia S, Roberts TM, Zhao JJ. Should individual PI3 kinase isoforms be targeted in cancer? Curr Opin Cell Biol. 2009;21:199–208. doi: 10.1016/j.ceb.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan DR, Whitman M, Schaffhausen B, et al. Common elements in growth factor stimulation and oncogenic transformation: 85 kd phosphoprotein and phosphatidylinositol kinase activity. Cell. 1987;50:1021–1029. doi: 10.1016/0092-8674(87)90168-1. [DOI] [PubMed] [Google Scholar]
- 3.Sugimoto Y, Whitman M, Cantley LC, et al. Evidence that the Rous sarcoma virus transforming gene product phosphorylates phosphatidylinositol and diacylglycerol. Proc Natl Acad Sci U S A. 1984;81:2117–2121. doi: 10.1073/pnas.81.7.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitman M, Kaplan DR, Schaffhausen B, et al. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 5.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 6.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 7.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: Variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katso R, Okkenhaug K, Ahmadi K, et al. Cellular function of phosphoinositide 3-kinases: Implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 9.Ikenoue T, Kanai F, Hikiba Y, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 10.Mizoguchi M, Nutt CL, Mohapatra G, et al. Genetic alterations of phosphoinositide 3-kinase subunit genes in human glioblastomas. Brain Pathol. 2004;14:372–377. doi: 10.1111/j.1750-3639.2004.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philp AJ, Campbell IG, Leet C, et al. The phosphatidylinositol 3′-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61:7426–7429. [PubMed] [Google Scholar]
- 12.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 13.Kang S, Denley A, Vanhaesebroeck B, et al. Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skolnik EY, Margolis B, Mohammadi M, et al. Cloning of PI3 kinase-associated p85 utilizing a novel method for expression/cloning of target proteins for receptor tyrosine kinases. Cell. 1991;65:83–90. doi: 10.1016/0092-8674(91)90410-z. [DOI] [PubMed] [Google Scholar]
- 15.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpenter CL, Auger KR, Chanudhuri M, et al. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J Biol Chem. 1993;268:9478–9483. [PubMed] [Google Scholar]
- 17.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 18.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 19.Alessi DR, James SR, Downes CP, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 20.Currie RA, Walker KS, Gray A, et al. Role of phosphatidylinositol 3,4,5-trisphosphate in regulating the activity and localization of 3-phosphoinositide-dependent protein kinase-1. Biochem J. 1999;337:575–583. [PMC free article] [PubMed] [Google Scholar]
- 21.Majumder PK, Sellers WR. Akt-regulated pathways in prostate cancer. Oncogene. 2005;24:7465–7474. doi: 10.1038/sj.onc.1209096. [DOI] [PubMed] [Google Scholar]
- 22.Duronio V. The life of a cell: Apoptosis regulation by the PI3K/PKB pathway. Biochem J. 2008;415:333–344. doi: 10.1042/BJ20081056. [DOI] [PubMed] [Google Scholar]
- 23.Hresko RC, Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- 24.Sarbassov DD, Ali SM, Sengupta S, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 25.Sarbassov DD, Guertin DA, Ali SM, et al. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 26.Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: Of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- 27.Harrington LS, Findlay GM, Gray A, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Reilly KE, Rojo F, She QB, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma WW, Adjei AA. Novel agents on the horizon for cancer therapy. CA Cancer J Clin. 2009;59:111–137. doi: 10.3322/caac.20003. [DOI] [PubMed] [Google Scholar]
- 30.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo J, Sobkiw CL, Hirshman MF, et al. Loss of class IA PI3K signaling in muscle leads to impaired muscle growth, insulin response, and hyperlipidemia. Cell Metab. 2006;3:355–366. doi: 10.1016/j.cmet.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Cho H, Mu J, Kim JK, et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi CM, Tran TT, Kondo T, et al. Phosphoinositide 3-kinase regulatory subunit p85alpha suppresses insulin action via positive regulation of PTEN. Proc Natl Acad Sci U S A. 2006;103:12093–12097. doi: 10.1073/pnas.0604628103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueki K, Yballe CM, Brachmann SM, et al. Increased insulin sensitivity in mice lacking p85beta subunit of phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2002;99:419–424. doi: 10.1073/pnas.012581799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia S, Liu Z, Zhang S, et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight ZA, Gonzalez B, Feldman ME, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 39.Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 40.Shoji K, Oda K, Nakagawa S, et al. The oncogenic mutation in the pleckstrin homology domain of AKT1 in endometrial carcinomas. Br J Cancer. 2009;101:145–148. doi: 10.1038/sj.bjc.6605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 42.García JM, Silva J, Peña C, et al. Promoter methylation of the PTEN gene is a common molecular change in breast cancer. Genes Chromosomes Cancer. 2004;41:117–124. doi: 10.1002/gcc.20062. [DOI] [PubMed] [Google Scholar]
- 43.Goel A, Arnold CN, Niedzwiecki D, et al. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res. 2004;64:3014–3021. doi: 10.1158/0008-5472.can-2401-2. [DOI] [PubMed] [Google Scholar]
- 44.Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 45.Sun X, Huang J, Homma T, et al. Genetic alterations in the PI3K pathway in prostate cancer. Anticancer Res. 2009;29:1739–1743. [PubMed] [Google Scholar]
- 46.Byun DS, Cho K, Ryu BK, et al. Frequent monoallelic deletion of PTEN and its reciprocal association with PIK3CA amplification in gastric carcinoma. Int J Cancer. 2003;104:318–327. doi: 10.1002/ijc.10962. [DOI] [PubMed] [Google Scholar]
- 47.Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 48.Ma YY, Wei SJ, Lin YC, et al. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000;19:2739–2744. doi: 10.1038/sj.onc.1203597. [DOI] [PubMed] [Google Scholar]
- 49.Rácz A, Brass N, Heckel D, et al. Expression analysis of genes at 3q26–q27 involved in frequent amplification in squamous cell lung carcinoma. Eur J Cancer. 1999;35:641–646. doi: 10.1016/s0959-8049(98)00419-5. [DOI] [PubMed] [Google Scholar]
- 50.Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 51.Davies MA, Stemke-Hale K, Tellez C, et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer. 2008;99:1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsons DW, Wang TL, Samuels Y, et al. Colorectal cancer: Mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 53.Cheng JQ, Godwin AK, Bellacosa A, et al. AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc Natl Acad Sci U S A. 1992;89:9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng JQ, Ruggeri B, Klein WM, et al. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci U S A. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bellacosa A, de Feo D, Godwin AK, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 56.Haas-Kogan D, Shalev N, Wong M, et al. Protein kinase B (PKB/Akt) activity is elevated in glioblastoma cells due to mutation of the tumor suppressor PTEN/MMAC. Curr Biol. 1998;8:1195–1198. doi: 10.1016/s0960-9822(07)00493-9. [DOI] [PubMed] [Google Scholar]
- 57.Myers MP, Pass I, Batty IH, et al. The lipid phosphatase activity of PTEN is critical for its tumor suppressor function. Proc Natl Acad Sci U S A. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han SY, Kato H, Kato S, et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 2000;60:3147–3151. [PubMed] [Google Scholar]
- 59.Liaw D, Marsh DJ, Li J, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 60.Huang CH, Mandelker D, Schmidt-Kittler O, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 61.Miled N, Yan Y, Hon WC, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 62.Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Isakoff SJ, Engelman JA, Irie HY, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 64.Zhao JJ, Liu Z, Wang L, et al. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shekar SC, Wu H, Fu Z, et al. Mechanism of constitutive phosphoinositide 3-kinase activation by oncogenic mutants of the p85 regulatory subunit. J Biol Chem. 2005;280:27850–27855. doi: 10.1074/jbc.M506005200. [DOI] [PubMed] [Google Scholar]
- 67.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–79. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 68.Eichhorn PJ, Gili M, Scaltriti M, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holbro T, Beerli RR, Maurer F, et al. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci U S A. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moasser MM, Basso A, Averbuch SD, et al. The tyrosine kinase inhibitor ZD1839 (“Iressa”) inhibits HER2-driven signaling and suppresses the growth of HER2-overexpressing tumor cells. Cancer Res. 2001;61:7184–7188. [PubMed] [Google Scholar]
- 71.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 72.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 73.Lim KH, Counter CM. Reduction in the requirement of oncogenic Ras signaling to activation of PI3K/AKT pathway during tumor maintenance. Cancer Cell. 2005;8:381–392. doi: 10.1016/j.ccr.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 74.Pacold ME, Suire S, Perisic O, et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 75.Rodriguez-Viciana P, Warne PH, Khwaja A, et al. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 76.Gupta S, Ramjaun AR, Haiko P, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 77.Ihle NT, Lemos R, Jr, Wipf P, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res. 2009;69:143–150. doi: 10.1158/0008-5472.CAN-07-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hickey FB, Cotter TG. BCR-ABL regulates phosphatidylinositol 3-kinase-p110gamma transcription and activation and is required for proliferation and drug resistance. J Biol Chem. 2006;281:2441–2450. doi: 10.1074/jbc.M511173200. [DOI] [PubMed] [Google Scholar]
- 79.Sujobert P, Bardet V, Cornillet-Lefebvre P, et al. Essential role for the p110delta isoform in phosphoinositide 3-kinase activation and cell proliferation in acute myeloid leukemia. Blood. 2005;106:1063–1066. doi: 10.1182/blood-2004-08-3225. [DOI] [PubMed] [Google Scholar]
- 80.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 81.Wee S, Wiederschain D, Maira SM, et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chiorean EG, Mahadevan D, Harris WB, et al. Phase I evaluation of SF1126, a vascular targeted PI3K inhibitor, administered twice weekly IV in patients with refractory solid tumors. J Clin Oncol. 2009;27(suppl):122s. abstr 2558. [Google Scholar]
- 83.LoRusso P, Markman B, Tabernero J, et al. A phase I dose-escalation study of the safety, pharmacokinetics (PK), and pharmacodynamics of XL765, a PI3K/TORC1/TORC2 inhibitor administered orally to patients (pts) with advanced solid tumors. J Clin Oncol. 2009;27(suppl):146s. abstr 3502. [Google Scholar]
- 84.Jimeno A, Hong DS, Hecker S, et al. Phase I trial of PX-866, a novel phosphoinositide-3-kinase (PI-3K) inhibitor. J Clin Oncol. 2009;27(suppl):156s. abstr 3542. [Google Scholar]
- 85.Shapiro G, Kwak E, Baselga J, et al. Phase I dose-escalation study of XL147, a PI3K inhibitor administered orally to patients with solid tumors. J Clin Oncol. 2009;27(suppl):146s. abstr 3500. [Google Scholar]
- 86.Markman B, Atzori F, Pérez-Garcia J, et al. Status of PI3K inhibition and biomarker development in cancer therapeutics. Ann Oncol. doi: 10.1093/annonc/mdp347. doi: 10.1093/annonc/mdp347 [epub ahead of print on August 27, 2009] [DOI] [PubMed] [Google Scholar]
- 87.Wagner AJ, Von Hoff DH, LoRusso PM, et al. A first-in-human phase I study to evaluate the pan-PI3K inhibitor GDC-0941 administered QD or BID in patients with advanced solid tumors. J Clin Oncol. 2009;27(suppl):146s. abstr 3501. [Google Scholar]
- 88.Flinn IW, Byrd JC, Furman RR, et al. Preliminary evidence of clinical activity in a phase I study of CAL-101, a selective inhibitor of the p1108 isoform of phosphatidylinositol 3-kinase (PI3K), in patients with select hematologic malignancies. J Clin Oncol. 2009;27(suppl):156s. abstr 3543. [Google Scholar]
- 89.Tolcher AW, Yap TA, Fearen I, et al. A phase I study of MK-2206, an oral potent allosteric Akt inhibitor (Akti), in patients (pts) with advanced solid tumor (ST) J Clin Oncol. 2009;27(suppl):146s. abstr 3503. [Google Scholar]
- 90.Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene. 2008;27:5511–5526. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]
- 91.Garlich JR, De P, Dey N, et al. A vascular targeted pan phosphoinositide 3-kinase inhibitor prodrug, SF1126, with antitumor and antiangiogenic activity. Cancer Res. 2008;68:206–215. doi: 10.1158/0008-5472.CAN-07-0669. [DOI] [PubMed] [Google Scholar]
- 92.Maira SM, Stauffer F, Brueggen J, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 93.Serra V, Markman B, Scaltriti M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 94.Crowder RJ, Phommaly C, Tao Y, et al. PIK3CA and PIK3CB inhibition produce synthetic lethality when combined with estrogen deprivation in estrogen receptor-positive breast cancer. Cancer Res. 2009;69:3955–3962. doi: 10.1158/0008-5472.CAN-08-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blois J, Yuan H, Smith A, et al. Slow self-activation enhances the potency of viridin prodrugs. J Med Chem. 2008;51:4699–4707. doi: 10.1021/jm800374f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Howes AL, Chiang GG, Lang ES, et al. The phosphatidylinositol 3-kinase inhibitor, PX-866, is a potent inhibitor of cancer cell motility and growth in three-dimensional cultures. Mol Cancer Ther. 2007;6:2505–2514. doi: 10.1158/1535-7163.MCT-06-0698. [DOI] [PubMed] [Google Scholar]
- 97.Smith A, Blois J, Yuan H, et al. The antiproliferative cytostatic effects of a self-activating viridin prodrug. Mol Cancer Ther. 2009;8:1666–1675. doi: 10.1158/1535-7163.MCT-08-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ihle NT, Paine-Murrieta G, Berggren MI, et al. The phosphatidylinositol-3-kinase inhibitor PX-866 overcomes resistance to the epidermal growth factor receptor inhibitor gefitinib in A-549 human non-small cell lung cancer xenografts. Mol Cancer Ther. 2005;4:1349–1357. doi: 10.1158/1535-7163.MCT-05-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen JS, Zhou LJ, Entin-Meer M, et al. Characterization of structurally distinct, isoform-selective phosphoinositide 3′-kinase inhibitors in combination with radiation in the treatment of glioblastoma. Mol Cancer Ther. 2008;7:841–850. doi: 10.1158/1535-7163.MCT-07-0393. [DOI] [PubMed] [Google Scholar]
- 100.Torbett NE, Luna-Moran A, Knight ZA, et al. A chemical screen in diverse breast cancer cell lines reveals genetic enhancers and suppressors of sensitivity to PI3K isoform-selective inhibition. Biochem J. 2008;415:97–110. doi: 10.1042/BJ20080639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.She QB, Chandarlapaty S, Ye Q, et al. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS One. 2008;3:e3065. doi: 10.1371/journal.pone.0003065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102.Vasudevan KM, Barbie DA, Davies MA, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2:pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 104.Yu K, Toral-Barza L, Shi C, et al. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 2009;69:6232–6240. doi: 10.1158/0008-5472.CAN-09-0299. [DOI] [PubMed] [Google Scholar]
- 105.Feldman ME, Apsel B, Uotila A, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thoreen CC, Kang SA, Chang JW, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Junttila TT, Akita RW, Parsons K, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 108.Faber AC, Li D, Song Y, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Natl Acad Sci U S A. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]