Abstract
Purpose
Cisplatin is a chemotherapeutic agent not used routinely for breast cancer treatment. As a DNA cross-linking agent, cisplatin may be effective treatment for hereditary BRCA1-mutated breast cancers. Because sporadic triple-negative breast cancer (TNBC) and BRCA1-associated breast cancer share features suggesting common pathogenesis, we conducted a neoadjuvant trial of cisplatin in TNBC and explored specific biomarkers to identify predictors of response.
Patients and Methods
Twenty-eight women with stage II or III breast cancers lacking estrogen and progesterone receptors and HER2/Neu (TNBC) were enrolled and treated with four cycles of cisplatin at 75 mg/m2 every 21 days. After definitive surgery, patients received standard adjuvant chemotherapy and radiation therapy per their treating physicians. Clinical and pathologic treatment response were assessed, and pretreatment tumor samples were evaluated for selected biomarkers.
Results
Six (22%) of 28 patients achieved pathologic complete responses, including both patients with BRCA1 germline mutations;18 (64%) patients had a clinical complete or partial response. Fourteen (50%) patients showed good pathologic responses (Miller-Payne score of 3, 4, or 5), 10 had minor responses (Miller-Payne score of 1 or 2), and four (14%) progressed. All TNBCs clustered with reference basal-like tumors by hierarchical clustering. Factors associated with good cisplatin response include young age (P = .001), low BRCA1 mRNA expression (P = .03), BRCA1 promoter methylation (P = .04), p53 nonsense or frameshift mutations (P = .01), and a gene expression signature of E2F3 activation (P = .03).
Conclusion
Single-agent cisplatin induced response in a subset of patients with TNBC. Decreased BRCA1 expression may identify subsets of TNBCs that are cisplatin sensitive. Other biomarkers show promise in predicting cisplatin response.
INTRODUCTION
Triple-negative breast cancers (TNBCs), those that do not express estrogen or progesterone receptors or contain an amplified HER2/Neu gene, often demonstrate sensitivity to cytotoxic neoadjuvant treatment regimens1–4; however, no specific molecular targets or chemotherapeutic vulnerabilities have been identified. TNBCs comprise 15% to 20% of breast cancers in Western countries, and the vast majority are sporadic.5 Approximately 70% of breast cancers in individuals carrying a germline BRCA1 mutation are triple negative; BRCA1-associated and sporadic TNBCs share many histopathologic features. They are almost always high grade,6 with common histologic7–11 and cytokeratin expression patterns,12,13 and share molecular features, including frequent p53 mutation14 and abnormalities of the inactivated X chromosome.15 Both BRCA1-associated and sporadic TNBCs are typically basal-like by hierarchical clustering of transcriptional profiles.16,17 Further, these tumors share a pattern of genomic instability characterized by allelic loss.6,15 These similarities have led to speculation that BRCA1-associated and at least a subset of sporadic TNBCs may share defects in a BRCA1-associated pathway; DNA repair has received the most attention.18
BRCA1-deficient cells are particularly susceptible to the interstrand cross-linking agents mitomycin and cisplatin. A cell line from a BRCA1-associated breast tumor was shown to be defective in DNA double-strand break repair19 and was also cisplatin sensitive20; these properties were reversed by adding wild-type BRCA1.19,20 BRCA1-deficient tumors in mouse models also demonstrated cisplatin sensitivity.21 Some cell lines representing sporadic TNBCs show cisplatin and mitomycin sensitivity (D. Silver and D.M. Livingston, unpublished data), suggesting that these tumors may have defects in the BRCA1 pathway.
Given these observations, we conducted a neoadjuvant trial of four cycles of cisplatin in TNBC. The trial end point was pathologic response. We analyzed pretreatment specimens for predictors of response to cisplatin, including BRCA1 expression levels, and BRCA1 promoter methylation. Other features that may predispose to cisplatin sensitivity, including gene expression patterns, p53 mutation, and the presence of a cisplatin-specific apoptosis pathway involving the p53 family members p63 and p73, were also explored.
PATIENTS AND METHODS
Patients
Newly diagnosed patients with T1, N1-3, M0 or T2-4, N0-3, M0 breast cancers (with tumors ≥ 1.5 cm), negative for estrogen and progesterone receptors defined as < 1% nuclear staining by immunohistochemistry, HER2/Neu 0 or 1+ by immunohistochemistry, or HER2 nonamplified by fluorescent in situ hybridization were eligible for this trial.
Study Design and Treatment Plan
A core biopsy was performed to obtain tumor tissue for study, and a radio-opaque clip was placed in the tumor bed; four treatments of cisplatin at 75 mg/m2 every 21 days were administered. Patients then received definitive surgery, including an axillary lymph node dissection in patients with positive sentinel lymph node biopsy. The specimen was evaluated for chemotherapy response, with focused sampling of the tumor bed marked by the radio-opaque clip. The Miller-Payne scoring system21a was used to assess tumor response. A score of 3, 4, or 5, with 5 being a pathologic complete response (pCR), is hereafter termed a good response.
For details of the patients on trial, study design and treatment plan, specimen analysis, Exon Grouping Analysis genotyping, gene array analysis, quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), and microarray data analysis, see Supplemental Methods (online only).
RESULTS
Patient Characteristics
Twenty-nine women were enrolled on the study: one was ineligible and never received protocol treatment. The last two patients entered signed consent forms concurrently, so a total of 28 instead of 27 women were treated on study. Patient age ranged from 29 to 69 years at diagnosis (Table 1). The median pretreatment tumor size, determined by magnetic resonance imaging, was 3.7 cm (range, 2.0 to 7.0 cm); 13 patients had axillary metastases. Research genotyping detected only the two BRCA1 germline mutations previously identified through clinical testing. Research biopsies of primary tumor provided adequate material for most molecular analyses from 24 of the 28 patients.
Table 1.
Clinical Characteristics and Response to Treatment of the Study Population (N = 28)
| Patient No. | Age (years) | BRCA1 Germline* | Baseline Tumor Diameter by MRI (cm) | Baseline Nodal Status† | Clinical Response | Pathologic Response (Miller-Payne scale)‡ |
|---|---|---|---|---|---|---|
| 15 | 57 | wt | 6.5 | − | SD | Progression |
| 21 | 59 | wt | 2.4 | − | PD | Progression |
| 26 | 39 | wt | 3.2 | − | PD | Progression |
| 27 | 63 | wt | 3.3 | + | PD | Progression |
| 4 | 68 | wt | 2.5 | + | SD | 1 |
| 6 | 62 | wt | Not done | + | cPR | 1 |
| 12 | 53 | wt | 2.7 | − | SD | 1 |
| 13 | 56 | wt | 3.2 | − | cPR | 1 |
| 16 | 45 | wt | 7.0 | + | SD | 1 |
| 14 | 43 | wt | 2.3 | − | cPR | 2 |
| 20 | 69 | wt | 5.0 | + | SD | 2 |
| 22 | 67 | wt | 4.7 | + | cPR | 2 |
| 24 | 50 | wt | 3.3 | − | cPR | 2 |
| 28 | 60 | wt | 3.7 | + | cPR | 2 |
| 1 | 59 | wt | 3.0 | − | cPR | 3 |
| 11 | 41 | wt | 4.0 | + | SD | 3 |
| 23 | 29 | wt | 4.5 | − | cPR | 3 |
| 25 | 40 | wt | 2.4 | − | cPR | 3 |
| 2 | 49 | wt | 4.0 | + | SD | 4 |
| 7 | 39 | wt | 3.7 | + | cCR | 4 |
| 8 | 51 | wt | 4.2 | − | cPR | 4 |
| 10 | 43 | wt | 2.5 | − | cCR | 4 |
| 3 | 39 | wt | 4.5 | + | cPR | 5 |
| 5 | 44 | mut | 5.8 | − | cPR | 5 |
| 9 | 31 | wt | 4.0 | − | cPR | 5 |
| 17 | 52 | wt | 2.0 | + | cCR | 5 |
| 18 | 48 | mut | 2.8 | + | cCR | 5 |
| 29 | 44 | wt | 6.3 | − | cPR | 5 |
Abbreviations: MRI, magnetic resonance imaging; wt, wild type, no germline mutation; SD, stable disease; PD, progressive disease; cPR, clinical partial response; cCR, clinical complete response; mut, presence of a pathogenic germline BRCA1 mutation.
BRCA1 germline genotype determined as in Patients and Methods section.
Axillary lymph node status determine at baseline; + indicates presence of lymph node metastasis determined by sentinel node biopsy or fine-needle aspiration; − indicates sentinel node biopsy negative for metastasis.
Pathologic assessment of response to treatment using Miller-Payne method and grading scale: progression = off study prior to surgery for clinical progression or additional nonprotocol therapy, 1 = no or minimal reduction in tumor, 2 = up to 30% reduction, 3 = 30% to 90% reduction, 4 = > 90% reduction but with some residual invasive (or axillary) disease, and 5 = no residual invasive carcinoma or axillary metastasis (pathologic complete response).
Treatment Response
Eighteen patients had a clinical response (Table 1, 14 had a partial response and four had a complete response), for an estimated response rate of 64% (95% conditional CI, 44% to 81%). The Miller-Payne score for responses to four cycles of cisplatin are listed in Table 1. Six patients had pCR (21%; 95% conditional CI, 9% to 43%), and eight additional patients had significant pathologic partial responses defined as Miller-Payne 3 or 4 (29%, for an overall good response rate of 50%; 95% conditional CI, 31% to 70%). Four patients had clinical progression while on cisplatin.
Table 2 lists the distribution of various clinical and pathologic characteristics by the response outcomes (Miller-Payne 3, 4, and 5 responses, pCR, and overall clinical response). Neither tumor size (by magnetic resonance imaging) nor axillary lymph node positivity were significantly related to any of the three response outcomes (P ≥ .48 and P ≥ .43, respectively).
Table 2.
Covariates and Response Variables
| Covariate | Miller-Payne 3,4,5 Responses |
pCR |
Clinical Response (CR and PR) |
||||
|---|---|---|---|---|---|---|---|
| No. of Patients | Observed (%) | P* | Observed (%) | P* | Observed (%) | P* | |
| All | 28 | 50 | — | 21 | — | 64 | — |
| Age, years | .001† | .13† | .46† | ||||
| 29-41 (Q1) | 7 | 86 | 29 | 71 | |||
| 42-49 (Q2) | 7 | 71 | 43 | 71 | |||
| 50-59 (Q3) | 8 | 38 | 13 | 63 | |||
| 60-69 (Q4) | 6 | 0 | 0 | 50 | |||
| Tumor size, cm (by MRI) | .75† | .48† | .83† | ||||
| Unknown | 1 | — | — | — | |||
| 2.0-2.7 (Q1) | 6 | 50 | 17 | 67 | |||
| 2.8-3.7 (Q2) | 7 | 29 | 14 | 57 | |||
| 3.8-4.5 (Q3) | 8 | 88 | 25 | 75 | |||
| 4.6-7.0 (Q4) | 6 | 33 | 33 | 50 | |||
| Lymph nodes | 1.00‡ | 1.00‡ | .43‡ | ||||
| Negative | 15 | 53 | 20 | 73 | |||
| Positive | 13 | 46 | 23 | 54 | |||
| BRCA1 mRNA levels, arbitrary relative units | .03† | .79† | .65† | ||||
| Unknown/NA | 7 | 57 | 42 | 71 | |||
| 0.00-0.03 (Q1) | 5 | 100 | 0 | 80 | |||
| 0.04-0.23 (Q2) | 6 | 33 | 33 | 50 | |||
| 0.25-0.44 (Q3) | 5 | 40 | 0 | 60 | |||
| 0.57-3.69 (Q4) | 5 | 20 | 20 | 60 | |||
| BRCA1 methylation | .04‡ | 1.00‡ | .40‡ | ||||
| Unknown/NA | 5 | 80 | 60 | 80 | |||
| Negative | 15 | 27 | 13 | 53 | |||
| Positive | 8 | 75 | 13 | 75 | |||
| ΔNp63/TAp73 ratio | .39‡ | .26‡ | .66‡ | ||||
| Unknown | 6 | 50 | 33 | 67 | |||
| > 2 | 9 | 67 | 33 | 56 | |||
| < 2 | 13 | 38 | 8 | 69 | |||
| Type of p53 mutation | .03‡ | .78‡ | .09‡ | ||||
| Unknown | 6 | 50 | 33 | 67 | |||
| MSM | 10 | 30 | 10 | 60 | |||
| NSM | 6 | 100 | .01§ | 33 | 1.00§ | 100 | .23§ |
| wt | 6 | 33 | .64¶ | 17 | 1.00¶ | 33 | .14¶ |
Abbreviations: pCR, pathologic complete response; CR, complete response; PR, partial response; Q1/Q2/Q3/Q4, first, second, third, and fourth quartiles, respectively; MRI, magnetic resonance imaging; NA, not assessed because of BRCA1 mutation; MSM, missense mutation; NSM, nonsense or frameshift mutation; wt, wild type (no mutation).
P values when patients with unknown values were omitted.
Fisher's exact test on four ordered categories of the covariate (equivalent to a Wilcoxon rank sum test using the quartile number as the observation and comparing responders to nonresponders).
Fisher's exact test on unordered categories of the covariate.
Fisher's exact test on NSM v MSM.
Fisher's exact test on wt v MSM + NSM.
Toxicity
Severe toxicity was uncommon. One patient had a grade 4 elevation of AST/AST. There were nine grade 3 toxicities reported: tinnitus, neutropenia, fatigue, hyperkalemia, elevation of ALT/ALT, nausea, myalgia, skin toxicity, and GI toxicity.
Predictors of Response
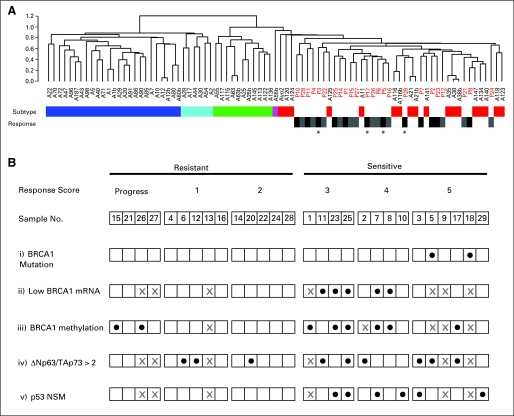
The TNBCs tend to be classified as basal-like in gene expression array hierarchical cluster analysis using the intrinsic genes.12,22 We determined the intrinsic subtype of all cases with adequate material (n = 24) by co-clustering these cases with a reference set of tumors for which intrinsic subtype had been determined independently (Supplemental Fig 1, Supplemental Methods). All of the trial TNBCs co-clustered with the reference basal-like tumors (Fig 1A). Hierarchical clustering with the intrinsic genes did not reveal distinct subclusters of cisplatin-resistant or -sensitive tumors.
Fig 1.
Predictors of response to cisplatin therapy in triple-negative basal-like tumors. (A) The sample dendrogram of gene expression hierarchical cluster analysis with the intrinsic genes47 is shown. Cisplatin pretreatment samples (sample numbers in red) are co-clustered with a reference set of breast tumors (sample numbers in black). Intrinsic subtype of the reference cases, determined by an independent hierarchical cluster analysis, is indicated by the color bar below the dendrogram as follows: luminal A, dark blue; luminal B, light blue; ErbB2, green; normal-like, purple; basal-like, red. Cisplatin response of the trial patients is indicated on the lower row as follows: resistant (progression, Miller-Payne score of 1 or 2) in gray; sensitive (Miller-Payne score of 3, 4, or 5) in black. (*) Trial cases with pathologic complete response (pCR; Miller-Payne score of 5). (B) Relationship of BRCA1 biomarkers and p53 family biomarkers to cisplatin sensitivity. Each trial patient is indicated by sample number, and patients are arranged according to relative response to cisplatin chemotherapy. Progression or Miller-Payne response scores are indicated above each sample. Predictive biomarker positivity is indicated with solid circles as follows: i = the presence of a BRCA1 germline mutation, ii = the lowest quartile of BRCA1 mRNA expression measured by quantitative reverse transcriptase polymerase chain reaction, iii = the presence of BRCA1 promoter methylation, iv = the ratio of mRNA expression levels of ΔNp63/TAp73 measured by quantitative reverse transcriptase polymerase chain reaction > 2, and v = the presence of p53 protein-truncating mutations. For each biomarker, samples with no data are indicated by a gray X; in addition, for BRCA1 mRNA expression and promoter methylation, a gray X indicates “not applicable” for the two cases with known BRCA1 germline mutation. NSM, nonsense or frameshift mutations.
Age
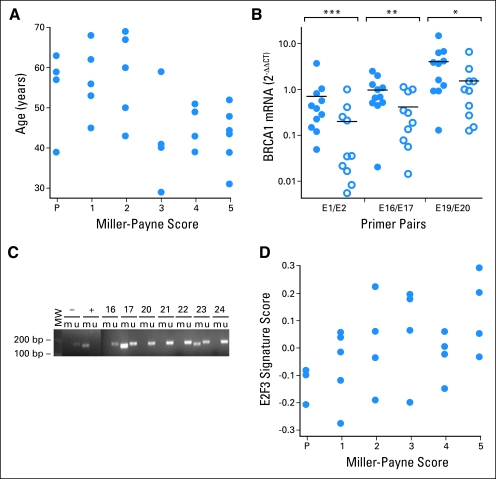
There was a strong association between younger age and good response (P = .001 based on quartiles of age, significant even after Bonferroni adjustment for multiple comparisons; when the two BRCA1 mutations carriers were excluded, P = .001 and the Miller-Payne response rate in patients age 42 to 49 years decreased to 60%; Table 2; Fig 2A). This effect was not attributable to decreased dose or dose delays in older patients (data not shown). Age was not significantly associated with pCR (P = .13) or clinical response (P = .46).
Fig 2.
Relationship of cisplatin treatment response to clinical and molecular features. (A) The patient age in years (y-axis) and Miller-Payne pathologic response score to neoadjuvant cisplatin therapy (x-axis) are plotted for each patient in the cohort as indicated by solid circles (P = .001 based on ordered quartiles of age). (B) Relative BRCA1 mRNA level measured by quantitative reverse transcriptase polymerase chain reaction (PCR; 2−ΔΔCT)48 is plotted for resistant tumors (solid circles) and sensitive tumors (open circles). The average mRNA level of each group is indicated by a black horizontal line. Measurements were performed using PCR primer pairs encompassing exons 1 and 2 (E1/E2), exons 16 and 17 (E16/E17), and exons 19 and 20 (E19/E20) as indicated along the bottom of the plot. The Wilcoxon P values for difference between sensitive and resistant tumors are indicated above each primer pair as follows: (***) P = .020; (**) P = .048; (*) P = .098. (C) Electrophoresis of PCR products spanning the BRCA1 promoter from bisulfite-treated DNA. Each lane contains products generated from separate PCR reactions using primers specific for methylated (m) or unmethylated (u) DNA template. Bacterial methylase-treated lymphocyte DNA was used for the positive control (+). DNA from normal lymphocytes was used as a negative control (−). The lane marked MW indicates molecular weight markers measured in base pairs (bp). Paired methylated- and unmethylated-specific primer reactions are marked by a line over the paired lanes and labeled corresponding to the template DNA used in the reaction (positive control, negative control, and patient No.). Patients 17 and 23 demonstrate bands in both the unmethylated (u) and methylated (m) lanes indicating the presence of BRCA1 promoter methylation. Patients 16, 20, 21, 22, and 24 lack bands with the methylated primer pair, signifying the absence of BRCA1 promoter methylation. (D) The E2F3 signature score (y-axis) and Miller-Payne pathologic response scores (x-axis) plotted (solid circles) for each patient in the cohort with available gene expression array data (Pearson correlation, 0.46; P = .025).
BRCA1 Genotype
The two BRCA1 mutation carriers achieved pCR; the other four patients with pCR were germline BRCA1 wild type (Table 1, Fig 1Bi). Without the two BRCA1 mutation carriers, the overall clinical response rate was 16 (62%) of 26, the good response rate was 12 (46%) of 26, and the pCR rate was four (15%) of 26.
BRCA1 mRNA Expression
Lower BRCA1 mRNA expression (measured by qRT-PCR) was significantly associated with a larger percent of patients having good response (P = .03; Table 2 and Figs 1Bii and 2B). All five patients with the lowest quartile of BRCA1 expression had a good response while only five of the 16 other patients with BRCA1 expression data had good responses. Multiple pairs of primers gave similar results. Excluding the tumors from BRCA1 carriers, the remaining three evaluable tumors showing pCR did not express BRCA1 at low levels. However, five of seven evaluable tumors in the Miller-Payne 3 and 4 groups had BRCA1 expression levels in the lowest quartile. The levels of the BRCA1 transcriptional repressor, ID4, have been shown to correlate inversely with BRCA1 expression levels23; we did not see an inverse correlation between array-measured ID4 expression and BRCA1 expression measured by qRT-PCR (Spearman correlation −0.05; P = .84; data not shown).
BRCA1 Promoter Methylation
BRCA1 promoter methylation was analyzed by methylation-specific PCR, a method sensitive to low levels of methylation (Fig 1Biii and Fig 2C). BRCA1 expression levels were lower in tumors with BRCA1 promoter methylation compared with tumors without (medians of 0.025 and 0.33, respectively; Wilcoxon rank sum test P = .06; Supplemental Fig 2). Tumors with BRCA1 promoter methylation were more likely to have good response, but not pCR or clinical response, than tumors without methylation (75% v 27%; P = .04).
ΔNp63/TAp73 Ratio
The transcription factor ΔNp63α promotes survival in a subset of tumors through its ability to repress the proapoptotic activity of the related p53 family member TAp73.24–26 Phosphorylation of TAp73 after cisplatin treatment causes TAp73 release from ΔNp63α, enabling TAp73 to activate a program of apoptosis.25 Tumors with this pathway intact would be predicted to be platinum sensitive and to have high levels of ΔNp63 relative to TAp73 to repress TAp73 activity in the absence of cisplatin. For this reason, levels of ΔNp63 and TAp73 were measured by qRT-PCR using RNA from microdissected primary tumor samples. Nine (41%) of the 22 evaluable tumors had a ΔNp63/TAp73 ratio > 2, the predetermined cutoff value suggesting active repression of TAp73 by ΔNp6325 (data not shown). Of these nine tumors, six (67%) had good responses to cisplatin and three (33%) had a pCR. Of 13 tumors with ΔNp63/TAp73 < 2, five (38%) had good responses and only one (8%) had a pCR (Fig 1Biv; P = .39 for good response; P = .26 for pCR; P = .66 for clinical response). Of the complete responders with material available, three of four patients had a ΔNp63/TAp73 > 2. These results provide preliminary evidence that a ΔNp63/TAp73 ratio > 2 associates with a greater chance of response to cisplatin.
p53 Mutation
The sequence of the p53 gene in tumor DNA was determined in 22 patients. Six tumors had nonsense or frameshift mutations (NSM), 10 had missense mutations (MSM), and six were wild type (wt), confirming the high frequency of p53 mutation in TNBC. There was no significant association of good response with the presence of a p53 mutation compared with wt (P = .64). However, the tumors with NSM tended to have a higher good response percent than those in the other two groups (100% v 30% v 33% in NSM, MSM, and wt groups, respectively; P = .03), and the difference in response rate between tumors with NSM and MSM was significant (P = .01). These tumors also had a higher rate of pCR (33% v 10% and 17%) and clinical response (100% v 60% and 33%) but these differences were not significant (P = .30 and P = .11).
Several Predictors of Miller-Payne Response Used Together
In an exploratory analysis of whether any of the other variables might add to the prediction based on age, we did step-up logistic regression for the outcome of good response, omitting the oldest age quartile. Using this approach, BRCA1 mRNA added significantly to predictions based on age alone (for details, see Supplemental Data).
Array Mining
We used several approaches to search for genes or gene signatures associated with cisplatin response. First, we identified candidate genes reported to have association with cisplatin response (eg, ERCC1, BIRC5; see Supplemental Data 1) and genes associated with subsets within TNBC (eg, basal keratins, EGFR, alpha B-crystallin). We evaluated the Pearson correlation between the Miller-Payne score and gene expression array level of these 114 candidate genes. Only a single gene correlation, AIFM1, remained significant after correction for multiple hypothesis testing (corrected P = .041). However, the expression level and standard deviation were low for this probe, suggesting random fluctuation. A complete list of candidate genes tested and P values are listed in Supplemental Data 1.
In addition, we evaluated published gene signatures consisting of a set of co-regulated gene expression changes in response to a specific oncogene pathway activation27 or specific biologic processes, including cell cycle,28 chromosome instability,29 and core serum response.30 We also tested an immune response classifier reported to have prognostic value in estrogen receptor–negative breast cancers.31 For each signature, we calculated a score estimating the relative level of the gene signature present in the array data from each tumor. We tested this score for association with response using Pearson correlation (see Supplemental Data 2). No signature was statistically significant after Bonferroni correction for multiple hypothesis testing. The strongest association was with a signature of E2F3 oncogenic pathway activation (Fig 2D; r = 0.46; P = .025), which has been associated with general chemotherapy responsiveness.27
Finally, we identified all genes with correlation to the Miller-Payne response score, a standard deviation > 0.5, and a P value < .01. No correlation was significant after Bonferroni correction (see Supplemental Data 3). The highest correlations were with inhibitor of growth family member 3 (ING3; r = 0.69; P = .0002) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a non-protein coding gene (r = .62; P = .001). The reproducibility of these potential candidate biomarkers will have to be determined by analysis of independent cohorts of similarly treated patients.
DISCUSSION
Six (21%) of 28 patients with TNBC achieved pCR with single-agent neoadjuvant cisplatin. Among the 28 patients in this trial were two BRCA1 carriers, both of whom achieved pCR; four (15%) of the 26 women with sporadic TNBC also achieved pCR to cisplatin. Overall, 50% of the patients had a good response to cisplatin defined by a Miller-Payne score of 3, 4, or 5. These results are consistent with preclinical and recent clinical data32 showing that BRCA1-associated tumors are responsive to cisplatin, and data that suggest a subset of basal-like breast cancers with intact BRCA1 share some fundamental molecular defects with BRCA1-deficient tumors.
The effect of increasing dose, intensity, and/or duration of cisplatin on response rates is unknown. Sporadic TNBCs show heterogeneity in response to other cytotoxic chemotherapies, with reported pCR rates of TNBC ranging from 12% for single-agent taxane regimens to 27% to 45% in multiagent neoadjuvant trials.1,3,4,33 Our data also suggest heterogeneity among these patients on the basis of their response to cisplatin. Clinical progression on cisplatin was also observed: other neoadjuvant studies of TNBC either have used different criteria or have not reported on progression, making it difficult to make meaningful comparisons of progression rates.3,4 We studied a variety of biomarkers to try to discover those that would distinguish patients likely to respond from patients unlikely to respond to cisplatin.
In our hands, triple negativity using a strict criteria of < 1% nuclear staining for estrogen and progesterone receptor immunohistochemistry, criteria for HER2 negativity of 0 or 1+ by immunohistochemistry, or HER2 nonamplification by fluorescent in situ hybridization reliably predicted classification into the basal-like subtype by hierarchical cluster analysis of the intrinsic genes.
Age correlated with response whether or not the two patients with BRCA1 mutations were included, with younger patients more likely to respond. The biologic explanation for this finding is unclear. It does not seem related to cisplatin dosing or dose intensity because these did not vary with age. Younger patients may develop subtypes of TNBC that are more responsive to chemotherapy in general or cisplatin in particular. Of note, younger age was a predictor of breast cancer sensitivity to neoadjuvant therapy with paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide in a recent study.34
We tested the hypothesis that low BRCA1 expression is an explanation of the phenotypic similarity of sporadic TNBC and BRCA1-related breast cancer. BRCA1 mRNA levels were lower in patients with cisplatin sensitivity, consistent with the suggestion that the “BRCA-ness” of these tumors may be related to decreased BRCA1 expression.18,23 However, the lowest BRCA1-expressing tumors were not those with pCR (excluding the BRCA1 germline mutation tumors), but rather were those with moderate responses to cisplatin (Miller-Payne score of 3 or 4). BRCA1 promoter methylation, assessed by a sensitive methylation-specific PCR assay, was also statistically significantly correlated with response to cisplatin (P = .04) and was inversely correlated to BRCA1 mRNA levels (P = .06). These data suggest that a subset of TNBCs may be sensitive to cisplatin on the basis of low BRCA1 expression levels. The rarity of low BRCA1 levels among tumors with pCR to cisplatin may be a reflection of the small number of patients analyzed and emphasizes the need to validate results of this exploratory trial in an independent cohort. In this regard, it is interesting that low levels of BRCA1 expression in ovarian tumors correlate with better survival in patients treated with cisplatin-containing regimens.35
Our trial and a recent report32 of a neoadjuvant trial using the same cisplatin regimen in women with germline BRCA1 mutations suggest that tumors from women with hereditary BRCA1 mutations have a high rate of response to cisplatin. Whether tumors from BRCA1 mutation carriers truly represent a homogeneous group with respect to cisplatin response or to other cytotoxic agents will require further experience. The small number of BRCA1 mutation carriers treated to date does not provide sufficient data for clinical use of neoadjuvant cisplatin outside of a trial.
p53 mutation status has been investigated as a potential predictor of chemotherapy responsiveness in solid tumors36–40: Nonsense and truncating p53 mutations have been shown to be common in BRCA1-mutated breast cancer.41 We found a significant association of tumor p53 protein-truncating mutations (NSM) with cisplatin response.
A pathway of apoptosis activated by cisplatin involving the p53 family members TAp73 and ΔNp63α may play a role in cisplatin sensitivity.25 We measured the ratio of the mRNA of these two genes as a marker of the potential integrity of this pathway. Although not statistically significant, three (75%) of four patients with pCR related to cisplatin were positive for this biomarker. This finding is consistent with a recent retrospective analysis of response following neoadjuvant cisplatin-based chemotherapy.42
In exploratory analyses (given the small sample size and large number of measured genes), no single gene on transcriptional arrays convincingly segregated responders from nonresponders. None of a smaller set of candidate genes reported to have some relationship to cisplatin response showed a strong association with response. Notable negatives include ERCC1, reported prognostic in cisplatin-treated lung cancer,43 and alpha B-crystallin.44 Of several signatures consisting of responses of many genes to a perturbation relevant for tumorigenesis, only one stood out in this analysis, a signature derived by overexpression of the transcription factor E2F3.27 A proportion of TNBCs have copy number gain on chromosome 6p at the E2F3 locus (Z.C. Wang, unpublished data), and there is evidence for inactivation of the Rb pathway in a proportion of TNBCs,45,46 which leads to E2F3 overexpression. Furthermore, the E2F3 pathway signature is associated with response of TNBC to neoadjuvant chemotherapy using drugs other than cisplatin.40 It is unclear whether any of the biomarkers correlated with cisplatin response reported here represent specific markers of cisplatin response; they may indicate general chemotherapy responsiveness. However, in cell culture experiments and in a retrospective clinical analysis, the p63/p73 pathway is related specifically to cisplatin response and not to response to a variety of other chemotherapeutic agents.25,26
We emphasize that patients in this neoadjuvant trial received standard therapy after surgery, and the relatively low pCR rate to single-agent cisplatin (21% for all patients, 15% excluding the BRCA1 mutation carriers) argues against administration of single-agent cisplatin as adjuvant or neoadjuvant therapy for unselected TNBCs. Multiagent neoadjuvant therapy has achieved higher pCR rates: 45% to 24 weeks of paclitaxel/fluorouracil, doxorubicin, and cyclophosphamide (T/FAC),4 and 24% to doxorubicin and cyclophosphamide (AC) often followed by a taxane.3 In this study, we could not assess whether the tumors that responded to cisplatin were the same as or different from tumors that would have responded to established multiagent regimens. We believe that these results justify exploring therapeutic combinations including platinum agents in TNBC and invite additional efforts at discovering biomarkers predictive of response.
Supplementary Material
Glossary Terms
- Promoter methylation:
Methylation of DNA sequences within the promoters of genes; this often occurs on the cytosine residue of CpG dinucleotides and is correlated with decreased expression of the adjacent gene.
- BRCA1 expression:
A tumor suppressor gene, the breast cancer 1 susceptibility gene is known to play a role in repairing DNA breaks. Mutations in this gene are associated with increased risks of developing breast or ovarian cancer.
- Hierarchical cluster:
An analytical tool used to find the closest associations among gene profiles and specimens under evaluation.
- PCR (polymerase chain reaction):
PCR is a method that allows exponential amplification of short DNA sequences within a longer DNA molecule.
- Double-strand break repair:
Any of several DNA repair processes used by organisms to repair breaks in DNA that span both strands of DNA at a single location.
- BRCA1:
A tumor suppressor gene that prevents ovarian and breast cancer.
- Gene signature:
The coordinated response of many genes to a particular stimulus; for example, the “myc oncogene signature” is the response of many genes to the forced overexpression of the myc oncogene.
- Intrinsic genes:
A set of genes whose level of expression is used to sort breast cancers into subtypes.
- Methylation-specific PCR:
A molecular assay that detects methylation of a particular stretch of DNA.
- Gene array:
A microchip on which DNA sequences for many genes are embedded; used to measure gene expression of many genes simultaneously.
- BRCA-ness:
A term applied to breast cancers, referring to the degree of relationship of a given breast cancer to one deficient in the BRCA1 gene.
Footnotes
Supported by Grants No. CA089393 from the National Cancer Institute Program of Research Excellence (SPORE) in Breast Cancer at the Dana-Farber/Harvard Cancer Center and No. R21LM008823-01A1 from the National Institutes of Health, and by the Breast Cancer Research Foundation, Sidney Kimmel Foundation, Avon supplement to the Dana-Farber/Harvard Cancer Center support grant, and Susan G. Komen for the Cure.
Presented in part at the San Antonio Breast Cancer Conference, December 14-17, 2006, San Antonio, TX, and the National Cancer Institute Translational Science Meeting, November 7-9, 2008, Washington, DC.
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Clinical trial information can be found for the following: NCT00148694.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: None Stock Ownership: None Honoraria: None Research Funding: Judy E. Garber, AstraZeneca Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Daniel P. Silver, Andrea L. Richardson, Rebecca S. Gelman, Paula D. Ryan, Shridar Ganesan, Leif W. Ellisen, Eric P. Winer, Judy E. Garber
Financial support: Daniel P. Silver, Andrea L. Richardson, Leif W. Ellisen, Eric P. Winer, Judy E. Garber
Administrative support: Daniel P. Silver, Andrea L. Richardson, Eric P. Winer, Judy E. Garber
Provision of study materials or patients: Daniel P. Silver, Andrea L. Richardson, Diana Calogrias, Paula D. Ryan, Nadine M. Tung, Dennis C. Sgroi, Eric P. Winer, Judy E. Garber
Collection and assembly of data: Daniel P. Silver, Andrea L. Richardson, Zhigang C. Wang, Chee-Onn Leong, Diana Calogrias, Ayodele Buraimoh, Aquila Fatima, Arcangela De Nicolo, Shridar Ganesan, Alexander Miron, Christian Colin, Dennis C. Sgroi, Leif W. Ellisen, Eric P. Winer, Judy E. Garber
Data analysis and interpretation: Daniel P. Silver, Andrea L. Richardson, Aron C. Eklund, Zhigang C. Wang, Zoltan Szallasi, Qiyuan Li, Nicolai Juul, Chee-Onn Leong, Rebecca S. Gelman, Arcangela De Nicolo, Shridar Ganesan, Alexander Miron, Christian Colin, Dennis C. Sgroi, Leif W. Ellisen, Eric P. Winer, Judy E. Garber
Manuscript writing: Daniel P. Silver, Andrea L. Richardson, Zoltan Szallasi, Rebecca S. Gelman, Nadine M. Tung, Christian Colin, Eric P. Winer, Judy E. Garber
Final approval of manuscript: Daniel P. Silver, Andrea L. Richardson, Aron C. Eklund, Zhigang C. Wang, Zoltan Szallasi, Qiyuan Li, Nicolai Juul, Chee-Onn Leong, Diana Calogrias, Ayodele Buraimoh, Aquila Fatima, Rebecca S. Gelman, Paula D. Ryan, Nadine M. Tung, Arcangela De Nicolo, Shridar Ganesan, Alexander Miron, Christian Colin, Dennis C. Sgroi, Leif W. Ellisen, Eric P. Winer, Judy E. Garber
REFERENCES
- 1.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 2.Berry DA, Cirrincione C, Henderson IC, et al. Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer. JAMA. 2006;295:1658–1667. doi: 10.1001/jama.295.14.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 4.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 5.Foulkes WD, Stefansson IM, Chappuis PO, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 6.Wang ZC, Lin M, Wei LJ, et al. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Res. 2004;64:64–71. doi: 10.1158/0008-5472.can-03-2570. [DOI] [PubMed] [Google Scholar]
- 7.Eisinger F, Jacquemier J, Charpin C, et al. Mutations at BRCA1: The medullary breast carcinoma revisited. Cancer Res. 1998;58:1588–1592. [PubMed] [Google Scholar]
- 8.Marcus JN, Watson P, Page DL, et al. Hereditary breast cancer: Pathobiology, prognosis, and BRCA1 and BRCA2 gene linkage. Cancer. 1996;77:697–709. doi: 10.1002/(sici)1097-0142(19960215)77:4<697::aid-cncr16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 9.Karp SE, Tonin PN, Begin LR, et al. Influence of BRCA1 mutations on nuclear grade and estrogen receptor status of breast carcinoma in Ashkenazi Jewish women. Cancer. 1997;80:435–441. doi: 10.1002/(sici)1097-0142(19970801)80:3<435::aid-cncr11>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 10.Verhoog LC, Brekelmans CT, Seynaeve C, et al. Survival and tumour characteristics of breast-cancer patients with germline mutations of BRCA1. Lancet. 1998;351:316–321. doi: 10.1016/s0140-6736(97)07065-7. [DOI] [PubMed] [Google Scholar]
- 11.Silver DP. HIN-1 and the nosology of breast cancer. Cancer Biol Ther. 2003;2:564–565. doi: 10.4161/cbt.2.5.512. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 13.Lakhani SR, Reis-Filho JS, Fulford L, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 14.Feki A, Irminger-Finger I. Mutational spectrum of p53 mutations in primary breast and ovarian tumors. Crit Rev Oncol Hematol. 2004;52:103–116. doi: 10.1016/j.critrevonc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 17.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 19.Scully R, Ganesan S, Vlasakova K, et al. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 20.Tassone P, Tagliaferri P, Perricelli A, et al. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br J Cancer. 2003;88:1285–1291. doi: 10.1038/sj.bjc.6600859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rottenberg S, Nygren AO, Pajic M, et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc Natl Acad Sci U S A. 2007;104:12117–12122. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Ogston KN, Miller ID, Payne S, et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: Prognostic significance and survival. Breast (Edinburgh, Scotland) 2003;12:320–327. doi: 10.1016/s0960-9776(03)00106-1. [DOI] [PubMed] [Google Scholar]
- 22.Kreike B, van Kouwenhove M, Horlings H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner NC, Reis-Filho JS, Russell AM, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- 24.DeYoung MP, Johannessen CM, Leong CO, et al. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006;66:9362–9368. doi: 10.1158/0008-5472.CAN-06-1619. [DOI] [PubMed] [Google Scholar]
- 25.Leong CO, Vidnovic N, DeYoung MP, et al. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest. 2007;117:1370–1380. doi: 10.1172/JCI30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocco JW, Leong CO, Kuperwasser N, et al. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 28.Whitfield ML, Sherlock G, Saldanha AJ, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter SL, Eklund AC, Kohane IS, et al. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 30.Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teschendorff AE, Caldas C. A robust classifier of high predictive value to identify good prognosis patients in ER-negative breast cancer. Breast Cancer Res. 2008;10:R73. doi: 10.1186/bcr2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byrski T, Huzarski T, Dent R, et al. Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat, 2009;115:359–363. doi: 10.1007/s10549-008-0128-9. [DOI] [PubMed] [Google Scholar]
- 33.Hayes DF, Thor AD, Dressler LG, et al. HER2 and response to paclitaxel in node-positive breast cancer. N Engl J Med. 2007;357:1496–1506. doi: 10.1056/NEJMoa071167. [DOI] [PubMed] [Google Scholar]
- 34.Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 35.Quinn JE, James CR, Stewart GE, et al. BRCA1 mRNA expression levels predict for overall survival in ovarian cancer after chemotherapy. Clin Cancer Res. 2007;13:7413–7420. doi: 10.1158/1078-0432.CCR-07-1083. [DOI] [PubMed] [Google Scholar]
- 36.Lowe SW, Ruley HE, Jacks T, et al. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 37.Di Leo A, Tanner M, Desmedt C, et al. p-53 gene mutations as a predictive marker in a population of advanced breast cancer patients randomly treated with doxorubicin or docetaxel in the context of a phase III clinical trial. Ann Oncol. 2007;18:997–1003. doi: 10.1093/annonc/mdm075. [DOI] [PubMed] [Google Scholar]
- 38.Aas T, Børresen AL, Geisler S, et al. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med. 1996;2:811–814. doi: 10.1038/nm0796-811. [DOI] [PubMed] [Google Scholar]
- 39.Bertheau P, Turpin E, Rickman DS, et al. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4:e90. doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tordai A, Wang J, Andre F, et al. Evaluation of biological pathways involved in chemotherapy response in breast cancer. Breast Cancer Res. 2008;10:R37. doi: 10.1186/bcr2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holstege H, Joosse SA, van Oostrom CT, et al. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res. 2009;69:3625–3633. doi: 10.1158/0008-5472.CAN-08-3426. [DOI] [PubMed] [Google Scholar]
- 42.Rocca A, Viale G, Gelber RD, et al. Pathologic complete remission rate after cisplatin-based primary chemotherapy in breast cancer: Correlation with p63 expression. Cancer Chemother Pharmacol. 2008;61:965–971. doi: 10.1007/s00280-007-0551-3. [DOI] [PubMed] [Google Scholar]
- 43.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 44.Ivanov O, Chen F, Wiley EL, et al. alphaB-crystallin is a novel predictor of resistance to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2008;111:411–417. doi: 10.1007/s10549-007-9796-0. [DOI] [PubMed] [Google Scholar]
- 45.Gauthier ML, Berman HK, Miller C, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herschkowitz JI, He X, Fan C, et al. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10:R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.