Abstract
Purpose
To determine whether plasma estradiol (E2) levels are related to gene expression in estrogen receptor (ER)–positive breast cancers in postmenopausal women.
Materials and Methods
Genome-wide RNA profiles were obtained from pretreatment core-cut tumor biopsies from 104 postmenopausal patients with primary ER-positive breast cancer treated with neoadjuvant anastrozole. Pretreatment plasma E2 levels were determined by highly sensitive radioimmunoassay. Genes were identified for which expression was correlated with pretreatment plasma E2 levels. Validation was performed in an independent set of 73 ER-positive breast cancers.
Results
The expression of many known estrogen-responsive genes and gene sets was highly significantly associated with plasma E2 levels (eg, TFF1/pS2, GREB1, PDZK1 and PGR; P < .005). Plasma E2 explained 27% of the average expression of these four average estrogen-responsive genes (ie, AvERG; r = 0.51; P < .0001), and a standardized mean of plasma E2 levels and ER transcript levels explained 37% (r, 0.61). These observations were validated in an independent set of 73 ER-positive tumors. Exploratory analysis suggested that addition of the nuclear coregulators in a multivariable analysis with ER and E2 levels might additionally improve the relationship with the AvERG. Plasma E2 and the standardized mean of E2 and ER were both significantly correlated with 2-week Ki67, a surrogate marker of clinical outcome (r = −0.179; P = .05; and r = −0.389; P = .0005, respectively).
Conclusion
Plasma E2 levels are significantly associated with gene expression of ER-positive breast cancers and should be considered in future genomic studies of ER-positive breast cancer. The AvERG is a new experimental tool for the study of putative estrogenic stimuli of breast cancer.
INTRODUCTION
Approximately 80% of invasive breast cancers present in developed countries as estrogen receptor (ER)–positive and are usually treated with endocrine therapy in the form of antiestrogens or estrogen suppressants—aromatase inhibitors (AIs) in postmenopausal women. ER-positive tumors, although highly heterogeneous, consists largely of tumors described as luminal1 and are now recognized as being genetically distinct from ER-negative tumors. There is considerable clinical interest in identifying molecular factors that reflect the variable dependence of ER-positive tumors on estrogenic stimulation and, by extension, the likelihood of these tumors benefiting from endocrine therapy2; however, few genomic studies have focused specifically on the ER-positive subgroup.
Although expression of progesterone receptor (PgR), which is strongly influenced by estrogens in vitro3 and in vivo,4–6 has been evaluated in this manner, it is clear that, as a single factor, PgR does not fully reflect estrogen dependence: many PgR-negative tumors respond to tamoxifen or AIs.7,8 Trefoil factor 1 (TFF1), originally known as pS2, is used often as a marker in laboratory studies9 but infrequently for clinical studies. Other studies of ER-positive breast cancer cells in vitro have identified additional genes for which expression is dependent on estrogenic stimulation,10 but most have not been translated to clinical utility.
Plasma levels of estradiol (E2) vary markedly between postmenopausal women and are significantly correlated with risk of development of breast cancer.11 Exogenous estrogens increase risk, particularly when administered with progestins.12 Plasma levels of endogenous E2 are, however, not known to influence breast cancer biology significantly; conversely, it is widely considered that intratumoral synthesis of estrogens provides the predominant estrogenic influence.13
The primary objective of this study was to determine whether plasma E2 levels correlated with gene expression in ER-positive breast cancers and, if so, whether associated genes were known to be estrogen regulated. Secondary objectives were to determine the relative contribution of plasma E2 and ESR1 expression levels to any associations with estrogen-regulated genes and to assess the relationship of these genes with clinical and biologic measures of clinical efficacy of estrogen deprivation (ie, aromatase inhibition). Exploratory analyses included an assessment of the relationship of steroid receptor coregulators with expression of estrogen-regulated genes.
MATERIALS AND METHODS
Patient Samples
Core-cut tumor biopsies (14-gauge) were obtained from 112 postmenopausal women with stage I to IIIB, ER-positive, early breast cancer before and after 2 weeks of anastrozole treatment in a neoadjuvant trial.14 Patients who received gefitinib during the first 2 weeks were excluded. Tissue was later stored in RNA (Ambion, Austin, TX) at −20°C. Two 4-μm sections from the core were stained with hematoxylin and eosin to confirm the presence of cancerous tissue and the histopathology. Total RNA was extracted by using RNeasy (Qiagen, Sussex, United Kingdom). RNA quality was checked by using an Agilent Bioanalyser (Agilent Technologies, Santa Clara, CA): samples with RNA integrity values less than 5 were excluded from additional analysis. ER status and Ki67values by immunohistochemistry were already available.14 The validation set was derived from baseline frozen biopsy samples obtained from 73 patients receiving neoadjuvant letrozole therapy in a phase II clinical trial.15
Gene Expression Analysis and Data Preprocessing
RNA amplification, labeling, and hybridization on HumanWG-6 v2 Expression BeadChips (Illumina, San Diego, CA) were performed according to the manufacturer's instructions at a single Illumina BeadStation facility. Tumor RNA of sufficient quality and quantity was available to generate expression data from 104 pretreatment biopsies. Data was extracted by using BeadStudio (Illumina) software and was normalized with variance-stabilizing transformation (VST) and Robust Spline Normalization method in the Lumi package.16 Probes that were not detected in any samples (detection P > 1%) were discarded from additional analysis. The gene expression approach (Agilent Technologies) and gene expression data sets for the validation study are described in Crowder et al.17
Data Analysis
Primary analysis.
Multiple correlation analysis was performed in BRB-Array Tools (http://linus.nci.nih.gov/BRB-ArrayTools.html). A statistical significance level for each gene for testing the hypothesis that the Spearman correlation between gene expression and plasma estradiol was zero was calculated, and P values then were used in a multivariate permutation test,18 from which false discovery rates were computed. Gene set enrichment analysis (GSEA) was carried out on the ranked lists of correlated genes by using the GSEA preranked function of the software across the curated gene sets of the Broad Institute Molecular Signatures Database (MSigDB).19 Pathway analysis by using Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) was conducted on the list of genes correlated with P ≤ .005. Other statistical analyses were performed in SPSS for Windows (SPSS, Chicago, IL), S-Plus (TIBCO Software, Palo Alto, CA), and Graphpad Prism (Graphpad Software, La Jolla, CA).
Intrinsic subtypes were assigned with a 50-gene subtype predictor.20 Data from the exclusively ER-positive tumors in this study were normalized before subtyping analysis with data from an additional 64 tumors, including 23 ER-negative cancers, analyzed concurrently on Illumina arrays.
Secondary and exploratory analyses.
Multivariable analysis was performed in a forward stepwise fashion, and the most significant additional variable (satisfying P < .05) was added at each stage. Occurrences with missing values for any of the variables in the model were excluded from analysis (Appendix Table A1). Statistically significant results were assessed when data availability allowed in the validation set.
Blood Samples and Hormone Measurement
Baseline plasma E2 levels before anastrozole or letrozole treatment were determined by a highly sensitive radioimmunoassay after ether extraction21 by using rabbit antiserum against an estradiol-6-carboxymethyloxime-BSA conjugate (EIR, Wurenlingen, Switzerland) and estradiol iodine 125 reagent DSL 4420 from the estradiol RIA kit DSL 4800 (Beckman Coulter [formerly known as DSL], Fullerton, CA). Sensitivity was 3 pmol/L by calculation from the 95% CIs of the zero standard. Plasma samples from both the anastrozole and letrozole cohorts were analyzed in the same laboratory.
RESULTS
Relationship of Plasma E2 With Tumor Gene Expression
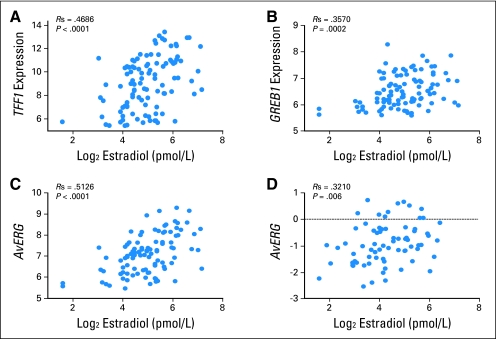
Quantitative trait analysis (QTA) by Spearman correlation was used to identify 226 genes from the 25,386 filtered genes for which expression in tumor biopsies correlated with plasma E2 levels at a P < .005 (Table 1). Genes previously known to be associated with a transcriptional response to estrogen appeared to be overrepresented among the genes closely correlated with plasma E2. TFF1 expression correlated most highly with pretreatment plasma E2 levels (Fig 1A). GREB1 (gene regulated by estrogen in breast cancer), an early response gene in the estrogen receptor-regulated pathway,22,24 (Fig 1B) was the fifth-most positively correlated gene, whereas expression of the PGR, the gene encoding the progesterone receptor, was also significantly correlated (P = .003). Plasma E2 levels were significantly higher in patients with immunohistochemical (IHC) PgR-positive tumors than in those with PgR-negative tumors (medians, 35 and 23 pmol/L, respectively; P = .0002). There was no significant relationship between plasma E2 and ER IHC levels. Plasma E2 did not correlate with either nodal status or tumor size (P = .55 and 0.52, respectively).
Table 1.
Genes Positively Correlated With Plasma Estradiol Concentration
| Spearman Rank | Gene Symbol | Genbank Accession No. | Spearman Correlation Coefficient | Parametric P | Known Estrogenic Function |
|---|---|---|---|---|---|
| 1 | TFF1 | NM_003225 | 0.469 | 7.00E-07 | Estradiol-responsive gene in vitro and in vivo10,22,23 |
| 2 | CAPN2 | NM_001748 | 0.430 | 6.40E-06 | |
| 3 | CAPN9 | NM_016452 | 0.378 | .73E-05 | |
| 4 | UNG2 | NM_021147 | 0.374 | .0001027 | |
| 5 | GREB1 | NM_148903 | 0.360 | .0001955 | Estradiol-responsive gene in vitro and in vivo10,22–24 |
| 6 | RAGE | NM_014226 | 0.357 | .0002178 | Estradiol-responsive gene in vitro25 |
| 7 | BTD | NM_000060 | 0.346 | .000348 | |
| 8 | VN1R2 | NM_173856 | 0.344 | .000378 | |
| 9 | TLE1 | NM_005077 | 0.337 | .0005016 | |
| 10 | SPG3A | NM_181598 | 0.336 | .0005227 | |
| 14 | AZGP1 | NM_001185 | 0.329 | .0006986 | Estradiol-responsive gene in vivo22 |
| 31 | PDZK1 | XM_943050 | 0.310 | .0014502 | Estradiol-responsive gene in vitro and in vivo10,22,23 |
| 57 | SERPINA3 | NM_001085 | 0.298 | .0022427 | Estradiol-responsive gene in vivo23 |
| 59 | IRS1 | NM_005544 | 0.297 | .0022581 | Estradiol-responsive gene in vitro26 |
| 75 | TFF2 | NM_005423 | 0.290 | .0029109 | Estradiol-responsive gene in GI tract27 |
| 85 | PGR | NM_000926 | 0.286 | .0034066 | Established measure of estrogenicity3 |
| 119 | TFF3 | NM_003226 | 0.276 | .0047773 | Estradiol-responsive gene in vivo23 |
NOTE. Ranked according to Spearman correlation. The 10 most strongly correlated genes are shown together with genes known to be associated with estrogen signaling with P < .005.
Fig 1.
Correlation of plasma estradiol and estrogen-responsive gene expression. (A) Plasma estradiol versus TFF1 expression in discovery data set. (B) Plasma estradiol versus GREB1 expression in discovery data set. (C) Plasma estradiol versus AvERG (average estrogen-responsive genes; ie, mean of TFF1, GREB1, PDZK1, and PGR) expression in discovery data set. (D) Plasma estradiol versus AvERG expression in independent validation data set.
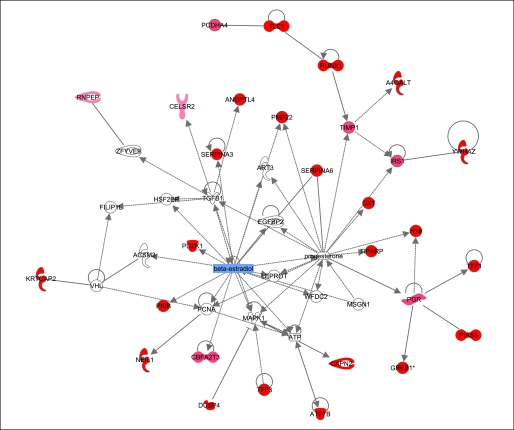
GSEA19 revealed that gene sets associated with the transcriptional response to estrogen deprivation in other published data sets were significantly enriched in the genes most tightly correlated with pretreatment plasma E2 levels. In particular, the genes identified as regulated in vitro by E2 through ER-α and ER-β28 were statistically significantly enriched (q value, < 0.2; Table 2). Pathway analysis by using Ingenuity software on the 98 annotated genes positively correlated with plasma E2 levels (P < .005) and revealed a network showing a significant relationship between 29 of these genes (Fig 2).
Table 2.
Gene Set Enrichment Analysis of Genes Positively Correlated With Plasma Estradiol at FDR q < 0.3
| Gene Set | Size of Gene Set | Normalised Enrichment Score | Nominal P | FDR q |
|---|---|---|---|---|
| Stossi_ER_up* | 25 | 1.723829 | 0 | 0.086808 |
| Breast_cancer_estrogen_signaling* | 54 | 1.678478 | 0 | 0.102272 |
| Falt_bcll_Ig_mutated_v_wt_up | 20 | 1.652155 | .002028 | 0.112083 |
| Creb_brain_8weeks_dn | 24 | 1.588765 | .002006 | 0.238964 |
| Lee_cip_dn | 23 | 1.57291 | .003009 | 0.246045 |
| Uvb_nhek1_c1 | 20 | 1.555116 | .00402 | 0.268565 |
| Ros_mouse_aorta_dn | 43 | 1.545233 | 0 | 0.268399 |
| BRCA_er_pos* | 291 | 1.533465 | 0 | 0.278001 |
| Nab_lung_dn | 26 | 1.532062 | .007 | 0.251437 |
| Basso_germinal_center_cd40_dn | 28 | 1.511978 | .003006 | 0.296837 |
| Frasor_ER_dn* | 27 | 1.509353 | .006006 | 0.279102 |
| Aged_mouse_cerebellum_up | 31 | 1.496655 | .006018 | 0.280914 |
| Basso_germinal_center_cd40_dn | 28 | 1.511978 | .003006 | 0.296837 |
Abbreviation: FDR, false discovery rate.
Estradiol-related pathway.
Fig 2.
Map showing relationship between genes positively correlated with plasma estradiol superimposed on Ingenuity map. Pathway analysis was carried out on 98 positively correlated genes (P < .005), and functional annotation revealed a network of 29 genes related through known estradiol- and progesterone-related signaling pathways. Genes correlated with plasma estradiol are shown in red, and the intensity of red coloration reflects the degree of correlation.
For additional analysis, an index of estradiol-responsive genes was generated by averaging the expression of four key genes, which was denoted AvERG (ie, average estrogen-responsive genes). TFF1, GREB1, and PDZK1 were selected for this index as the highest ranked genes in our analysis that had been responsive to estrogen deprivation with AIs in the two published clinical studies available to date22,23 and in the key published in vitro study of estradiol-stimulated genes in breast cancer cells.10 PGR was included, as it has been identified as a marker of estrogenicity in numerous studies spanning 3 decades3 and is in routine clinical use. No other combination of genes was tested. The resultant index correlated with plasma E2 levels with r = 0.51 (P < .0001; Fig 1C).
Validation of Association Between Plasma E2 and AvERG Expression
To determine the validity of these observations, we tested the four key estradiol-dependent genes in an independent group of ER-positive breast cancer samples from postmenopausal patients before treatment in a phase II trial of neoadjuvant letrozole.15 Plasma taken at diagnosis and Agilent microarray measurements from tumors were available from 73 patients. Of the four classical estrogen-dependent genes identified in our discovery set (ie, TFF1, GREB1, PDZK1, and PGR) only PGR was not individually significantly correlated with plasma E2 (P ≤ .05), whereas the AvERG correlated overall with r = 0.32 (P = .006; Fig 1D). The CIs for the correlation coefficients for PGR in the two sample sets overlapped markedly, which indicated no significant heterogeneity despite the lack of statistical significance in the validation set (initial samples: r = 0.25; 95% CI, 0.053 to 0.44; validation samples: r = 0.17; 95% CI = −0.072 to 0.39). Similarly, plasma E2 levels were higher in PgR-positive (by IHC) than PgR-negative occurrences in the validation set, but this was not statistically significant (P = .17).
Contribution of ESR1 Levels to AvERG Gene Expression
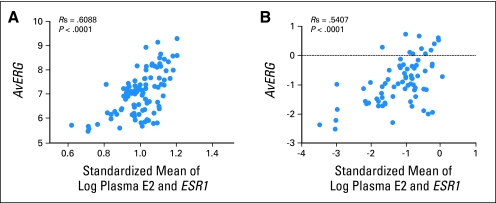
As E2 induces transcription primarily through ER-α (ESR1) in breast carcinomas, we investigated the effect of ESR1 levels, as measured by cDNA microarray analysis, on AvERG. ESR1 expression was correlated with AvERG in our original data set (r = 0.27; P = .006) and not with plasma E2 levels (r = 0.01; P = not significant). On multivariable analysis, variation in plasma E2 and ESR1 were found to account for 27% and 10% of the variation in AvERG, respectively. The correlation of the AvERG with a normalized mean of plasma E2 and ESR1 was 0.61 (P < .0001; Fig 3A). Equivalent analysis of the validation samples found plasma E2 and ESR1 levels accounted for 18% and 15% of the variation in AvERG, respectively, with analysis that included ESR1 levels producing a correlation of r = 0.54 (P < .0001; Fig 3B).
Fig 3.
(A) Standardized mean of plasma estradiol and ESR1 expression versus AvERG (average estrogen-responsive genes; ie, mean of TFF1, GREB1, PDZK1, and PGR) expression in discovery data set. (B) Standardized mean of plasma estradiol and ESR1 expression versus AvERG expression in independent validation data set.
Contribution of Cofactor Levels to AvERG Gene Expression
Coactivators and corepressors interact with the bound ER to modulate gene transcription.29 To assess the role that levels of the primary coactivators and corepressors have on estrogen-dependent gene expression, univariate analysis was undertaken with AvERG expression as the dependent variable and the following coregulators: NCOR1, NCOR2, NCOA1, NCOA2, and NCOA3 as potentially independent covariables. Significant associations were found with NCOR1 (P = .002), NCOA1 (P = .03), and NCOA3 (P = .01). Inclusion of these three factors with plasma E2 and ESR1 in a multivariable analysis revealed log plasma E2 (P < .001), ESR1 (P = .001), and NCOR1 (P = .007) to be independently significant with these factors, which explained 41% of the variation in AvERG expression. None of the cofactors were found to be significant on equivalent analysis of the independent data set, but the Agilent platform provided a lower dynamic range of expression values.
Relationship of Pretreatment Plasma E2 and AvERG With Intrinsic Subtype Status
Intrinsic subtype was ascribable to 69 tumors: 35 luminal A, 17 luminal B, 12 HER2-like, and five basal-like tumors. AvERG and plasma E2 were significantly higher in luminal A tumors compared with the other sets, when considered together (P = .008 and 0.011, respectively). Differences of similar magnitude were present between luminal A and B tumors when considered separately, but the significance levels were lower because of fewer numbers (P = .097 and 0.040, respectively).
Relationship of Pretreatment Plasma E2 With Ki67 Staining After 2 Weeks of Treatment With Anastrozole
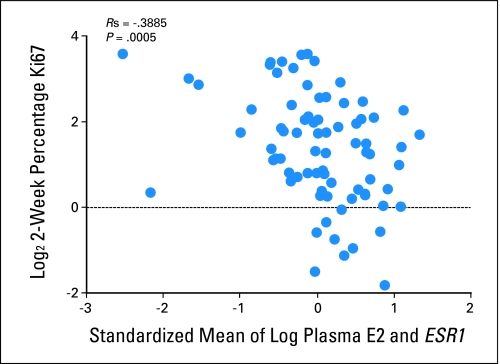
We have previously reported that higher Ki67 expression after 2 weeks of endocrine therapy is significantly associated with lower recurrence-free survival.30 To determine whether pretreatment plasma estradiol and/or ESR1 levels affect this potential surrogate marker for outcome, we investigated the relationship between plasma E2 and the standardized mean of plasma E2 and ESR1 with the level of tumor Ki67 expression after 2 weeks of treatment. Plasma E2 and the standardized mean were both significantly correlated with 2-week Ki67, although this was borderline for the former (r = −0.179; P = .05; and r = −0.389, P = .0005, respectively; Fig 4).
Fig 4.
Standardized mean of plasma estradiol and ESR1 expression versus Ki67 percentage staining 2 weeks after anastrozole treatment.
Relationship of Pretreatment Plasma E2 and AvERG With Clinical Response
We conducted an exploratory analysis to determine whether pretreatment plasma E2 and/or AvERG correlated with clinical response in the approximately 50% of the patients that received anastrozole alone for their complete courses of neoadjuvant therapy. In each occurrence, there was a trend toward higher levels in the complete and partial responders versus in those who showed stable disease or progression. However, in neither instance was this close to statistical significance (P = .16 for n = 50; and P = 0.17 for n = 51, respectively).
DISCUSSION
This study is the first to show a relationship between the expression of estrogen-dependent genes in ER-positive breast carcinomas and basal levels of estrogen. Although such results may not appear surprising, findings that intratumoral E2 levels are 10 to 20 times higher than those in plasma, and the absence of a reported correlation between levels in the two compartments, have led to the view that intratumoral estrogen synthesis has a greater influence on estrogen signaling than uptake from the circulation.13 The data presented in this article challenge that view and strongly suggest that differences in plasma E2 levels between patients have a significant influence on breast tumors. In particular, this influence may be important to consider in future genomic studies of ER-positive breast cancer, and this may extend to premenopausal women in whom the within-patient changes in E2 levels through the menstrual cycle are marked. Our data relating to Ki67 also suggest that E2 levels merit additional investigation in relation to their possible influence on disease outcome on AIs.
The strength of the relationship between AvERG and plasma E2 levels in this heterogeneous group of ER-positive tumors is remarkable. The relationship was replicated in an independent sample set with good statistical confidence, despite a smaller sample size. The low plasma levels of E2 in postmenopausal women measured in this study would be unquantifiable by most routinely used assays31 and may be one reason that this relationship has not been previously observed. Additional exploration should be conducted only with the use of such specialist assays. In a model system, proportional uptake of E2 from plasma was inversely related to plasma E2 concentrations.32 Our data do not allow us to determine whether this inverse relationship extends to the expression of estrogen-related genes.
The inclusion of ESR1 mRNA expression as a variable along with plasma E2 explained 37% of the variability in the AvERG. There is sufficient unexplained variability for intratumoral synthesis to make a meaningful contribution, but analytic imprecision and other factors also may be influential. One such potential influence examined in this study is the expression of nuclear coregulators.29 Additional exploratory modeling that allowed for an interaction between the five coregulators and ER explained 54% of the variability in AvERG (data not shown). These relationships were not confirmed in the validation set, possibly because of the lower dynamic range on the Agilent platform. These apparent relationships need additional study before they could be considered reliable.
The absence of a relationship between plasma E2 and ESR1 expression and the known effect of estrogen deprivation on the index genes suggest that the relationship between plasma E2 and these estrogen index genes is due to signaling influences rather than to the evolution of particularly estrogen-sensitive tumors as a result of higher E2 levels. As well as providing evidence for the importance of basal levels of E2 on breast cancer biology, the creation of AvERG provides a new index for understanding the clinical and biologic importance of other putative estrogenic influences. For example, a number of steroids thought of largely as androgens also have been found to have significant estrogenic activity in vitro,33,34 and it has been speculated that these may be particularly important in patients under treatment with AIs. They would not be expected to be influenced by such treatment; as a result, they may provide a possible resistance mechanism.33 Assessment of the AvERG in relation to plasma levels of these androgens in AI-treated patients would provide evidence for or against the clinical importance of their estrogenic activity. The AvERG also may be useful as an end point in characterizing the pathologic impact of polymorphic genetic differences in components of the estrogen response mechanism (eg, ESR1, coregulators) in breast tumors.
Our selection of the four index genes to create the AvERG was based on the well-established estrogen sensitivity of these genes as recorded in numerous publications and, therefore, minimized the potential for false discovery in their selection from tens of thousands of genes. It is likely that, as a result, the current AvERG will not be the most sensitive possible marker of estrogenic activity and that an optimally selected gene set could lead to an improved AvERG.
There are few data assessing the relationship of plasma estrogen levels and outcome of therapy,35 but the weak, significant relationship of baseline plasma E2 level with the value of Ki67 after 2 weeks of aromatase inhibition, a marker we have previously found related to recurrence-free survival,30 suggests that this merits evaluation in large cohorts of patients. The lack of a significant correlation between either plasma E2 or AvERG with clinical response was not surprising given the small patient cohort available. In addition, we have previously found that Ki67 at 2 weeks is a better predictor of long-term outcome on endocrine therapy than clinical response.30 The higher levels of plasma E2 and AvERG in luminal A tumors compared with the other subtypes is consistent with the expression in this group of many ER-related genes.1
Plasma estrogen and androgen levels are significantly correlated with one another. For example, in published data from our laboratory,36 log plasma E2 and log plasma testosterone showed r2 = 0.31 in greater than 2,000 samples from postmenopausal women (correlation data unpublished). This raises the possibility that the correlation between plasma E2 and AvERG could be due in part to plasma testosterone action as a substrate for intratumoral aromatization. Plasma samples from this study, unfortunately, were lost in a fire, which prevented measurements of testosterone to assess this possibility.
In conclusion, this assessment of plasma E2 levels in association with genome-wide expression studies has revealed new relationships that are likely to be important for additional genomic studies of ER-positive breast cancer, for the assessment of multiple putative estrogenic influences on breast cancer, and possibly for clinical outcome after estrogen deprivation therapy.
Appendix
Table A1.
Patients Contributing to Cofactor Analyses
| Analyses | No. of Patients |
|---|---|
| Study data set | |
| AvERG, plasma E2, ESR1, and any cofactor | 97 |
| AvERG pre and post (difference), plasma E2, ESR1 | 77 |
| AvERG, plasma E2, Ki67 2 week | 65 |
| AvERG, ESR1, Ki67 2 week | 77 |
| AvERG, plasma E2,ESR1, Ki67 2 week | 63 |
| Independent data set | |
| AvERG, plasma E2, ESR1, and any cofactor | 72 |
NOTE. Unless otherwise stated, analyses with subsets of the variables had the same number of patients as the analyses involving all the variables (eg, analysis in the study set of AvERG, plasma E2, and ESR1 only would have involved 97 patients).
Abbreviation: AvERG, average estrogen responsive genes (ie, mean of TFF1, GREB1, PDZK1, and PGR).
Footnotes
Supported by the Mary-Jean Mitchell Green Foundation and Breakthrough Breast Cancer; by National Institutes of Health Grant Nos. CA095614 and CA114722; by the Breast Cancer Research Foundation; by a Clinical Research Affiliate Funding Trials grant from the St Louis Affiliate of the Susan G. Komen for the Cure; and the National Health Service provided funding to the NIHR Biomedical Research Centre.
Presented in part at the 31st San Antonio Breast Cancer Symposium, December, 10-14, 2008, San Antonio, TX.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Mitch Dowsett, AstraZeneca (C) Stock Ownership: Helen Anderson, AstraZeneca; Charles M. Perou, University Genomics; Matthew J. Ellis, University Genomics Honoraria: Matthew J. Ellis, Novartis, AstraZeneca, Pfizer Research Funding: Mitch Dowsett, AstraZeneca Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Anita K. Dunbier, Helen Anderson, Mitch Dowsett
Financial support: Mitch Dowsett
Provision of study materials or patients: Robert J. Crowder, Ian E. Smith, Matthew J. Ellis, Mitch Dowsett
Collection and assembly of data: Anita K. Dunbier, Helen Anderson, Elizabeth J. Folkerd, Robert J. Crowder, Jeremy Hoog, Peter Osin, Ashutosh Nerurkar, Matthew J. Ellis
Data analysis and interpretation: Anita K. Dunbier, Helen Anderson, Zara Ghazoui, Roger A'Hern, Jeremy Hoog, Ian E. Smith, Joel S. Parker, Charles M. Perou, Matthew J. Ellis, Mitch Dowsett
Manuscript writing: Anita K. Dunbier, Helen Anderson, Matthew J. Ellis, Mitch Dowsett
Final approval of manuscript: Anita K. Dunbier, Helen Anderson, Zara Ghazoui, Elizabeth J. Folkerd, Roger A'Hern, Robert J. Crowder, Jeremy Hoog, Ian E. Smith, Peter Osin, Ashutosh Nerurkar, Joel S. Parker, Charles M. Perou, Matthew J. Ellis, Mitch Dowsett
REFERENCES
- 1.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller WR, Larionov A, Renshaw L, et al. Gene expression profiles differentiating between breast cancers clinically responsive or resistant to letrozole. J Clin Oncol. 2009;27:1382–1387. doi: 10.1200/JCO.2008.16.8849. [DOI] [PubMed] [Google Scholar]
- 3.Horwitz KB, Koseki Y, McGuire WL. Estrogen control of progesterone receptor in human breast cancer: Role of estradiol and antiestrogen. Endocrinology. 1978;103:1742–1751. doi: 10.1210/endo-103-5-1742. [DOI] [PubMed] [Google Scholar]
- 4.Miller WR, Dixon JM, Macfarlane L, et al. Pathological features of breast cancer response following neoadjuvant treatment with either letrozole or tamoxifen. Eur J Cancer. 2003;39:462–468. doi: 10.1016/s0959-8049(02)00600-7. [DOI] [PubMed] [Google Scholar]
- 5.Ellis MJ, Coop A, Singh B, et al. Letrozole inhibits tumor proliferation more effectively than tamoxifen independent of HER1/2 expression status. Cancer Res. 2003;63:6523–6531. [PubMed] [Google Scholar]
- 6.Dowsett M, Ebbs SR, Dixon JM, et al. Biomarker changes during neoadjuvant anastrozole, tamoxifen, or the combination: Influence of hormonal status and HER-2 in breast cancer—A study from the IMPACT trialists. J Clin Oncol. 2005;23:2477–2492. doi: 10.1200/JCO.2005.07.559. [DOI] [PubMed] [Google Scholar]
- 7.Anderson H, Bulun S, Smith I, et al. Predictors of response to aromatase inhibitors. J Steroid Biochem Mol Biol. 2007;106:49–54. doi: 10.1016/j.jsbmb.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 8.Elledge RM, Green S, Pugh R, et al. Estrogen receptor (ER) and progesterone receptor (PgR), by ligand-binding assay compared with ER, PgR and pS2, by immuno-histochemistry in predicting response to tamoxifen in metastatic breast cancer: A Southwest Oncology Group Study. Int J Cancer. 2000;89:111–117. [PubMed] [Google Scholar]
- 9.Brown AM, Jeltsch JM, Roberts M, et al. Activation of pS2 gene transcription is a primary response to estrogen in the human breast cancer cell line MCF-7. Proc Natl Acad Sci U S A. 1984;81:6344–6348. doi: 10.1073/pnas.81.20.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frasor J, Stossi F, Danes JM, et al. Selective estrogen receptor modulators: Discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64:1522–1533. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- 11.Key T, Appleby P, Barnes I, et al. Endogenous sex hormones and breast cancer in postmenopausal women: Reanalysis of nine prospective studies. J Natl Cancer Inst. 2002;94:606–616. doi: 10.1093/jnci/94.8.606. [DOI] [PubMed] [Google Scholar]
- 12.Chlebowski RT, Hendrix SL, Langer RD, et al. Influence of estrogen plus progestin on breast cancer and mammography in healthy postmenopausal women: The Women's Health Initiative randomized trial. JAMA. 2003;289:3243–3253. doi: 10.1001/jama.289.24.3243. [DOI] [PubMed] [Google Scholar]
- 13.Simpson ER, Dowsett M. Aromatase and its inhibitors: Significance for breast cancer therapy. Recent Prog Horm Res. 2002;57:317–338. doi: 10.1210/rp.57.1.317. [DOI] [PubMed] [Google Scholar]
- 14.Smith IE, Walsh G, Skene A, et al. A phase II placebo-controlled trial of neoadjuvant anastrozole alone or with gefitinib in early breast cancer. J Clin Oncol. 2007;25:3816–3822. doi: 10.1200/JCO.2006.09.6578. [DOI] [PubMed] [Google Scholar]
- 15.Olson JA, Jr, Budd GT, Carey LA, et al. Improved surgical outcomes for breast cancer patients receiving neoadjuvant aromatase inhibitor therapy: Results from a multicenter phase II trial. J Am Coll Surg. 2009;208:906–914. doi: 10.1016/j.jamcollsurg.2009.01.035. discussion 915-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du P, Kibbe WA, Lin SM. Lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 17.Crowder RJ, Phommaly C, Tao Y, et al. PIK3CA and PIK3CB inhibition produce synthetic lethality when combined with estrogen deprivation in estrogen receptor-positive breast cancer. Cancer Res. 2009;69:3955–3962. doi: 10.1158/0008-5472.CAN-08-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korn EL, Troendle JF, McShane LM, et al. Controlling the number of false discoveries: Application to high-dimensional genomic data. J Stat Plan Interfer. 2004;124:379–398. [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowsett M, Goss PE, Powles TJ, et al. Use of the aromatase inhibitor 4-hydroxyandrostenedione in postmenopausal breast cancer: Optimization of therapeutic dose and route. Cancer Res. 1987;47:1957–1961. [PubMed] [Google Scholar]
- 22.Mackay A, Urruticoechea A, Dixon JM, et al. Molecular response to aromatase inhibitor treatment in primary breast cancer. Breast Cancer Res. 2007;9:R37. doi: 10.1186/bcr1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller WR, Larionov AA, Renshaw L, et al. Changes in breast cancer transcriptional profiles after treatment with the aromatase inhibitor, letrozole. Pharmacogenet Genomics. 2007;17:813–826. doi: 10.1097/FPC.0b013e32820b853a. [DOI] [PubMed] [Google Scholar]
- 24.Rae JM, Johnson MD, Scheys JO, et al. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka N, Yonekura H, Yamagishi S, et al. The receptor for advanced glycation end products is induced by the glycation products themselves and tumor necrosis factor-alpha through nuclear factor-kappa B, and by 17beta-estradiol through Sp-1 in human vascular endothelial cells. J Biol Chem. 2000;275:25781–25790. doi: 10.1074/jbc.M001235200. [DOI] [PubMed] [Google Scholar]
- 26.Molloy CA, May FE, Westley BR. Insulin receptor substrate-1 expression is regulated by estrogen in the MCF-7 human breast cancer cell line. J Biol Chem. 2000;275:12565–12571. doi: 10.1074/jbc.275.17.12565. [DOI] [PubMed] [Google Scholar]
- 27.Campbell-Thompson ML. Estrogen receptor alpha and beta expression in upper gastrointestinal tract with regulation of trefoil factor family 2 mRNA levels in ovariectomized rats. Biochem Biophys Res Commun. 1997;240:478–483. doi: 10.1006/bbrc.1997.7683. [DOI] [PubMed] [Google Scholar]
- 28.Stossi F, Barnett DH, Frasor J, et al. Transcriptional profiling of estrogen-regulated gene expression via estrogen receptor (ER) alpha or ERbeta in human osteosarcoma cells: Distinct and common target genes for these receptors. Endocrinology. 2004;145:3473–3486. doi: 10.1210/en.2003-1682. [DOI] [PubMed] [Google Scholar]
- 29.Shang Y, Hu X, DiRenzo J, et al. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 30.Dowsett M, Smith IE, Ebbs SR, et al. Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst. 2007;99:167–170. doi: 10.1093/jnci/djk020. [DOI] [PubMed] [Google Scholar]
- 31.Dowsett M, Folkerd E. Deficits in plasma oestradiol measurement in studies and management of breast cancer. Breast Cancer Res. 2005;7:1–4. doi: 10.1186/bcr960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue W, Santner SJ, Masamura S, et al. Determinants of tissue estradiol levels and biologic responsiveness in breast tumors. Breast Cancer Res Treat. 1998;49(suppl 1):S1–S7. doi: 10.1023/a:1006026732129. discussion S33-S37. [DOI] [PubMed] [Google Scholar]
- 33.Sikora MJ, Cordero KE, Larios JM, et al. The androgen metabolite 5alpha-androstane-3beta, 17beta-diol (3betaAdiol) induces breast cancer growth via estrogen receptor: Implications for aromatase inhibitor resistance. Breast Cancer Res Treat. 2009;115:289–296. doi: 10.1007/s10549-008-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorgan JF, Stanczyk FZ, Longcope C, et al. Relationship of serum dehydroepiandrosterone (DHEA), DHEA sulfate, and 5-androstene-3 beta, 17 beta-diol to risk of breast cancer in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 1997;6:177–181. [PubMed] [Google Scholar]
- 35.Lønning PE, Helle SI, Johannessen DC, et al. Influence of plasma estrogen levels on the length of the disease-free interval in postmenopausal women with breast cancer. Breast Cancer Res Treat. 1996;39:335–341. doi: 10.1007/BF01806162. [DOI] [PubMed] [Google Scholar]
- 36.Dunning AM, Dowsett M, Healey CS, et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst. 2004;96:936–945. doi: 10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]