Abstract
Purpose
The Gastrointestinal Stromal Tumor Meta-Analysis Group (MetaGIST) project aims to additionally explore the data of the two large, randomized, cooperative-group studies comparing two doses of imatinib (400 mg daily v twice daily) in 1,640 patients with advanced GIST.
Methods
End points were progression-free survival (PFS) and overall survival (OS). Investigated cofactors included age, sex, performance status (PS), primary tumor site, time from diagnosis, prior therapies, baseline biology, and KIT/PDGFRα mutations for a subset of 772 patients. Univariate and multivariate models were used for the analysis.
Results
At a median follow-up of 45 months, a small but significant PFS advantage was documented for the high-dose arm. OS was identical in the two arms. The multivariate prognostic models included the following adverse factors: male sex, poor PS, and high baseline neutrophils counts (PFS and OS); low hemoglobin and GIST from small bowel origin (PFS); and advanced age, large tumor size, low albumin level, and prior chemotherapy (OS). In patients analyzed for mutations, patients with wild type, patients with KIT exon 9 mutations, and patients with other mutations had worse prognoses than patients with KIT exon 11 mutations for both end points. The mutation status was the only predictive factor for the PFS benefit attributed to high-dose treatment that resulted in significantly longer PFS (and higher objective response rate) for patients with KIT exon 9 mutations.
Conclusion
This analysis confirms a small PFS advantage of high-dose imatinib, essentially among patients with KIT exon 9 mutations, but no OS advantage.
INTRODUCTION
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal neoplasms of the gastrointestinal tract, with an incidence of at least 10 new occurrences per million.1 The treatment of localized forms is primarily surgery. These tumors have proven to be insensitive to chemotherapy and radiotherapy.2
Imatinib is a small-molecule tyrosine kinase inhibitor active against BCR-ABL, KIT, and PDGFRα. KIT is expressed in the vast majority of GISTs and is frequently mutated, which leads to constitutive activation in these tumors. An EORTC (European Organisation for Research and Treatment of Cancer) phase I study3 identified the highest feasible dose of imatinib to be 400 mg twice daily and indicated extensive activity in GIST. Phase II studies showed activity at all doses tested (ie, 400 to 800 mg).4,5
Two large, randomized, phase III studies were subsequently conducted to compare the outcome of patients treated with a daily dose of 400 mg once daily (ie, standard dose) and of 400 mg twice daily (ie, high dose) for unresectable or metastatic GIST. One study was conducted jointly in Europe and Australia by EORTC, Italian Sarcoma Group (ISG), and Australasian Gastro-Intestinal Trials Group (AGITG; trial 62005, referred to as EU-AUS); the other study was conducted in the United States and Canada by Southwest Oncology Group (SWOG), Cancer and Leukemia Group B (CALGB), National Cancer Institute of Canada (NCI-C), and Eastern Cooperative Oncology Group (ECOG; trial S0033, referred to as US-CDN). Selection criteria, protocol treatment, and follow-up were purposely identical in the two protocols to enable a meta-analysis and have been described elsewhere.6,7 The primary end point differed between trials: EU-AUS used progression-free survival (PFS), which required enrollment of 600 patients with data analysis after 344 events (ie, progression or death); US-CDN used overall survival (OS), which required 600 patients and observation of 321 deaths. The trials recruited 946 and 746 patients, respectively, both within 1 year.
In EU-AUS, there initially was a significant PFS improvement in favor of the high-dose arm (median follow-up, 25 months; 498 observed events),6 which was no longer demonstrated with longer follow-up (median follow-up, 40 months; 657 observed events) and which did not translate into an OS improvement (412 observed deaths).8 US-CDN did not demonstrate any significant difference in either PFS or OS between the two doses (median follow-up, 54 months; 342 observed deaths; 545 observed progressions or death).7
Patients who experienced disease progression in the standard-dose arm were allowed to cross over to the high-dose regimen. Both trials reported additional disease stabilization after cross over (29% and 28%) including a few objective responses (2% and 3%).9,10
In both trials, a KIT and PDGFRα mutation analysis were performed on paraffin-embedded tumor samples for a subset of patients. In EU-AUS, the mutation analysis, which was performed in 377 patients with externally confirmed GIST, showed superior PFS and OS in patients who had KIT exon 11 mutations when compared with patients who had KIT exon 9 mutations and wild types, whereas patients with KIT exon 9 mutations benefitted most from high-dose therapy in terms of PFS as opposed to other genomic subpopulations.11 In US-CDN, the mutation analysis performed in 397 patients with CD117-immunopositive GIST confirmed the EU-AUS data on prognosis, but the high-dose therapy did not result in significantly superior PFS or OS in patients with KIT exon 9 mutations, although it induced a higher response rate.12
EU-AUS also demonstrated that early resistance was increased in patients with low baseline hemoglobin level, whereas late resistance was increased in patients with either high baseline granulocyte count, large lesions, or nongastric primaries. High granulocyte count and disease origin within the GI tract, but outside the stomach and small bowel, also were predictive factors for the benefit of high-dose therapy. This exploratory study has not been externally validated.13
The objectives of the MetaGIST project were to confirm the already available results of the two studies, to explore patient subsets, to attempt to validate previously suggested prognostic and predictive factors, and to compare the baseline characteristics of the patients that could explain eventual differences between results of the two studies.
METHODS
A common data set that included data of both trials was assembled for the purpose of this work, according to prospectively agreed data specifications. The common data set included the data used for previous publications (median follow-up, 54 and 42 months, respectively, for the US-CDN and the EU-AUS series).7,14 Ineligible patients had been excluded for the US-CDN trial only, but this was not expected to affect the results of the joint analysis. Study protocols were approved by competent, local Human Investigations Committees, and all patients gave written informed consent.
Two primary end points were analyzed: OS, defined from the date of random assignment to the date of death, and PFS, defined from the date of random assignment to the date of documented progression or death, whichever occurred first. Critical significance levels were not adjusted for multiplicity. Patients alive and progression free were censored for both end points at the date of the last reported visit. Best overall response (according to Response Evaluation Criteria in Solid Tumors [RECIST]) and time to imatinib failure (defined as time to second progression or death for patients who crossed over to high-dose imatinib and as time to first progression or death for the other patients) were used as explanatory end points.
All cofactors recorded in the common database were investigated as potential prognostic and predictive factors: randomized daily dose; age; sex; performance status at entry; site of disease origin; time from initial diagnosis of GIST; maximum diameter of the largest lesion at entry; prior surgery, radiotherapy, and chemotherapy; baseline WBCs, neutrophils, platelets, hemoglobin, creatinine, bilirubin, and albumin; and mutation status when available.
When recoding of continuous variables was needed, categories did adhere as far as possible to previous publications, to the (rounded) median (binary variables), or to (rounded) quartiles. Mutations were classified as KIT or PDGFRα mutations, and KIT mutations were subclassified according to the involved exon.
Categoric variables were tabulated by study, and the distributions were compared with the χ2 test (binary and nonordered categories) or with the Cochran-Armitage test for trend (ordered categories). Continuous variables were summarized by using the median and range in each study, and the distributions were compared with the Wilcoxon test.
Best overall response was analyzed as an ordered categoric variable. For the predictive factor analysis, this variable was dichotomized (complete or partial response v no change or progression), and a logistic multivariate model was used.
PFS and OS were estimated by the Kaplan-Meier method. The Wald test (from the Cox model stratified by study) was used for the primary treatment comparison, whereas the log-rank test (unstratified) and the Wilcoxon test were used for sensitivity analyses. The baseline hazard function also was estimated by the life-table method for six time intervals (ie, 0 to 3, 3 to 6, 6 to 12, 12 to 24, 24 to 36, and 36 to 60 months).
Step-down Cox regression was used for the prognostic factors analysis. The validity of final models (ie, stability of included cofactors for different patient selection) was evaluated by using the bootstrap resampling method,15 and the discrimination of final models (ie, ability to correctly classify patients) was evaluated by using the C-index of Harrell, which varies from 0.5 (no discrimination) to 1.0 (perfect discrimination).16
The analysis of predictive factors used Cox multivariate models, including the randomized treatment, the investigated factor, and an interaction test. For this analysis, all variables were dichotomized. Forest plots of hazard ratios were used to illustrate the results. All models were stratified by study.
RESULTS
Comparison of the Study Populations
Distribution of the cofactors differed significantly between studies (Appendix Table A1, online only). US-CDN recruited older patients with a worse performance status; lower baseline WBC, neutrophils, and platelets count; and less recent diagnosis and with more radiotherapy but less chemotherapy than in EU-AUS. There also were proportionally fewer patients with KIT exon 9 mutations (8% v 16%) or with PDGFRα mutations (1.3% v 2.9%), and there were more patients with KIT exon 11 mutations (71% v 66%) or KIT/PDGFRα wild types (17% v 13%).
The distribution of tumor sites (stomach v small bowel v other) and of the size of lesion or lesions at trial entry did not differ between the two trials. All cofactors were evenly distributed between patients with and without mutation data, with the exception of prior surgery (respectively, 89.4% and 80.5%).
Overall Results
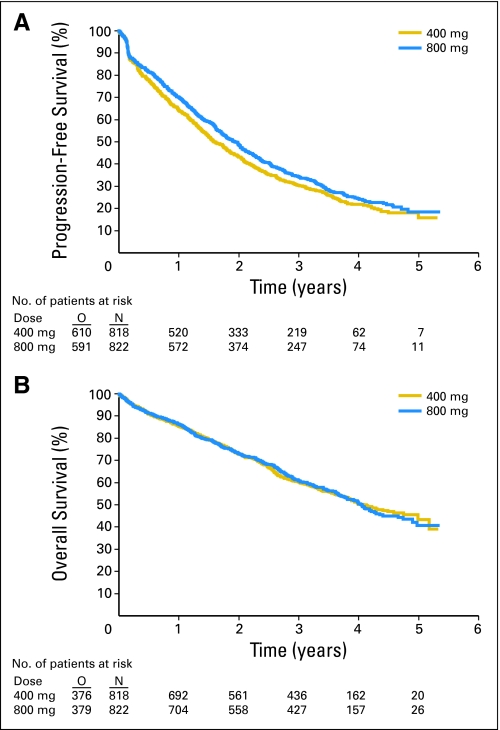
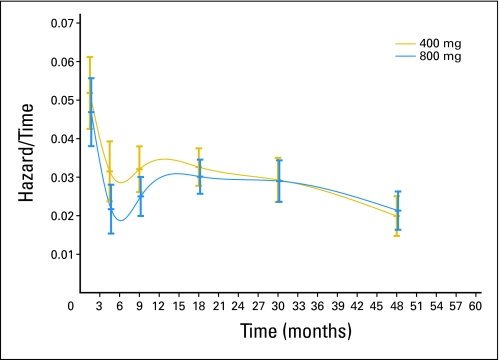
In the pooled data set, a small (estimated hazard ratio [HR], 0.89; 95% CI, 0.79 to 1.00) but significant (P = .04, Wald test) PFS advantage was observed for the high-dose arm (Fig 1). This advantage was consistent across trials (HR, 0.89 in both trials; Table 1). Results were similar for the unadjusted log-rank test but were more significant for the Wilcoxon test that is more sensitive to early differences. The hazard function (which represented the immediate risk of relapse) was consistently higher in the standard-dose arm up to approximately 2 years, but it was the same in both arms afterwards (Appendix Fig A1, online only).
Fig 1.
Progression-free and overall survival in the high-dose arm. N, number of patients; O, events.
Table 1.
Overall Results
| Treatment by Group | No. of Patients | No. of Events | HR | 95% CI | P | Median Time (years) | 95% CI | % at 3 Years | 95% CI |
|---|---|---|---|---|---|---|---|---|---|
| Progression-free survival | |||||||||
| All patients | |||||||||
| Imatinib 400 mg | 818 | 610 | 1.00 | .0412 Wald; .0422 log-rank; .0195 Wilcox | 1.58 | 1.46 to 1.80 | 30.30 | 27.11 to 33.55 | |
| Imatinib 800 mg | 822 | 591 | 0.89 | 0.79 to 1.00 | 1.95 | 1.74 to 2.08 | 34.36 | 31.05 to 37.70 | |
| EU-AUS | |||||||||
| Imatinib 400 mg | 473 | 332 | 1.00 | 0.1232 log-rank; .0560 Wilcox | 1.74 | 1.52 to 1.98 | 31.41 | 27.13 to 35.78 | |
| Imatinib 800 mg | 473 | 324 | 0.89 | 0.76 to 1.03 | 2.02 | 1.80 to 2.24 | 34.95 | 30.53 to 39.40 | |
| US-CDN | |||||||||
| Imatinib 400 mg | 345 | 278 | 1.00 | .1806 log-rank; .1625 Wilcox | 1.46 | 1.30 to 1.79 | 28.82 | 24.10 to 33.69 | |
| Imatinib 800 mg | 349 | 267 | 0.89 | 0.75 to 1.05 | 1.64 | 1.48 to 2.08 | 33.47 | 28.51 to 38.50 | |
| Overall survival | |||||||||
| All patients | |||||||||
| Imatinib 400 mg | 818 | 376 | 1.00 | .9738 Wald; .9635 log-rank; .8621 Wilcox | 4.08 | 3.74 to 4.76 | 60.40 | 56.86 to 63.75 | |
| Imatinib 800 mg | 822 | 379 | 1.00 | 0.86 to 1.15 | 4.05 | 3.75 to 4.36 | 60.96 | 57.43 to 64.30 | |
| EU-AUS | |||||||||
| Imatinib 400 mg | 473 | 208 | 1.00 | .5853 log-rank; .4729 Wilcox | 3.74 | 3.33 to NR | 57.62 | 52.83 to 62.12 | |
| Imatinib 800 mg | 473 | 205 | 0.95 | 0.78 to 1.15 | 3.78 | 3.42 to 4.01 | 60.03 | 55.24 to 64.47 | |
| US-CDN | |||||||||
| Imatinib 400 mg | 345 | 168 | 1.00 | .5819 log-rank; .5760 Wilcox | 4.59 | 3.94 to 5.18 | 63.99 | 58.64 to 68.83 | |
| Imatinib 800 mg | 349 | 174 | 1.06 | 0.86 to 1.31 | 4.27 | 3.82 to 4.98 | 62.05 | 56.67 to 66.96 |
Abbreviations: HR, hazard ratio; EU-AUS, joint studies in Europe and Australia; US-CDN, joint studies in United States and Canada; NR, not reached.
No difference was observed between the two arms in terms of OS (HR, 1.00; P = .97; Fig 1), and this again was consistent across trials (EU-AUS HR, 0.95; US-CDN HR, 1.06; Table 1). No significant difference was observed between the two arms in terms of best overall response (51% v 54%; P = .08), although a borderline advantage of the high-dose arm was observed in the EU-AUS data (50% v 55%; P = .05).
Analysis of Prognostic Factors
The final Cox multivariate model for PFS included five significant, adverse prognostic factors: poor performance status, high neutrophil counts, low hemoglobin level, male sex, and GIST from small bowel origin; randomly assigned dose was of borderline significance when added to this model (P = .06).
In patients with available mutation data, the mutation status was added to the prognostic model with a high significance level (P < .0001); patients with KIT exon 11 mutations had a more favorable prognosis than patients with KIT exon 9 mutations, patients with other mutations, and patients with wild-type disease. The prognostic value of the site of origin substantially decreased and became of borderline significance.
The final Cox multivariate model for OS included seven significant adverse prognostic factors: high neutrophil counts, poor performance status, advanced age, low albumin level, prior chemotherapy, large tumor size, and male sex. Site of tumor origin did not have any significant prognostic value in the univariate or in the multivariate models.
In patients with available mutation data, the mutation status also was added to the prognostic model with a high significance level (P < .0001), whereas the factor of prior chemotherapy lost its significance. All Cox models are detailed in Appendix Table A2 (online only).
The internal validation showed that the survival model was rather stable: the seven significant factors were included in the bootstrap models with the highest frequencies, which ranged from 99% to 80%. This was not the case for the PFS model: only performance status and neutrophil count were included in bootstrap models with a high frequency (90%), as opposed to tumor site (65%), sex (53%), and hemoglobin level (44%). Two other factors were included with a high frequency: creatinine level (56%) and prior chemotherapy (46%). The C-index of Harrell was 66.5% for the survival model but was only 58.3% for the PFS model.
Analysis of Predictive Factors
For PFS, the presence of KIT exon 9 mutation was the only significant predictive factor for the benefit of high-dose therapy (P = .012, interaction test; Appendix Table A3, online only). Within patients with KIT exon 9 mutations, PFS was significantly longer for patients treated with the high-dose arm (P = .017, Wald test). For patients without such mutations, no difference was observed between treatment arms (Fig 2).
Fig 2.
Significant predictive factors for the benefit of high-dose therapy with regard to progression-free survival and overall survival. ex9, exon 9; N, number of patients; O, events.
For OS, none of the investigated cofactors showed any predictive value, and no significant advantage of high-dose therapy (P = .15, Wald test) was documented in patients with KIT exon 9 mutations (Fig 2). Results of treatment comparison in all mutation subgroups are summarized on Forest plots (Fig 3).
Fig 3.
Forest plots of results of treatment comparison in all mutation subgroups. N, number of patients; O, events; E, expected; HR, hazard ratio; SD, standard deviation.
Two explanatory end points were analyzed additionally in patients with KIT exon 9 mutations. Best overall response rate was significantly higher in the high-dose arm (47% v 21%, P = .0037, Cochrane Armitage test for trend) as opposed to other subgroups, for example patients with KIT exon 11 mutations (64% in both arms). Time to imatinib failure did not differ between randomized treatment arms (data not shown).
DISCUSSION
Despite identical eligibility criteria, the populations entered in the two studies differed significantly; some factors had favorable prognosis impacts, and others had adverse prognosis impacts on the US-CDN study as compared with the EU-AUS study. This observation stresses that epidemiologic differences could affect the conclusions of randomized trials, and it justifies the stratification of all analyses by study.
A borderline PFS advantage was documented for the high-dose arm. The reduction of the risk of relapse, however, seems to be limited to the first 2 years of imatinib treatment, which explains the higher significance level of the Wilcoxon test (P = .02) when compared with the Wald and log-rank tests (P = .04). This heterogeneity explains apparent discrepancies between previously published results. EU-AUS initially was reported with a large number of events but with a short follow-up: the positive results translated the early impact of high-dose therapy. US-CDN was published with a longer follow-up, and the early advantage was outweighed by later relapses in the analysis. Currently, the estimated overall HR is the same in the two trials. The short-lasting advantage of high-dose therapy could be linked to frequent dose reductions (44% and 49% at 6 months) reported in this arm.14,17 Also, we know that progression may be induced by several biomolecular mechanisms of resistance,18–20 and we cannot exclude the predominance of a dose-dependent mechanism in the first 2 years and of an independent mechanism later on. It has already been demonstrated that early resistance (ie, progression within 3 months) is not predicted by the same factors as late resistance (ie, progression after 3 months), and factors affecting very late resistance (ie, progression after 18 or 24 months) may be different again. The direct applicability to clinical practice is questionable, as both doses lead to an identical OS, probably because of the efficacy of cross over to high-dose after progression in the standard-dose arm9,10 and because of availability of new drugs as salvage therapy.
Best overall response to therapy was analyzed as an explanatory end point. Despite limitations of RECIST in GIST, the expected rate of objective responses may guide physicians in appropriate choice in patients with locally advanced disease. Because responses rates were similar in both arms, and because we know that the impact of the drug on the size of lesions may be relatively slow (because onset of responses has been observed after years of therapy), systematic use of high-dose neoadjuvant therapy does not seem justified, but it should be explored additionally in patients with KIT exon 9 mutations in whom high-dose therapy did induce a higher response rate (as compared with standard dose).
Prognostic factors identified in the pooled data set are consistent between the two trials and confirm previous findings.7,11–13 Poor performance status and elevated neutrophil count at baseline have a strong adverse prognostic value for PFS and OS, all confirmed by internal validation. For the PFS model, other factors were unstable, probably because of the correlation among sex, hemoglobin, and creatinine levels (data not shown); moreover, the model had a poor discrimination. The OS model was more stable and had a better discrimination. In patients with mutation data, mutation site adds strong prognostic information to the two models; however, as previously reported,11 the site of origin loses part of its significance in the PFS model, presumably because of the predominance of exon 9 mutation in tumors of small bowel origin; prior chemotherapy also steps out of the OS model.
The only identified predictive factor was the presence of KIT exon 9 mutations. The estimated risk of progression or death was reduced by 42% in the high-dose arm (compared with the standard-dose arm) in patients with KIT exon 9 mutations and was increased by 2% in patients with other types of mutations or with no mutations. The risk reduction was significant in the patients with KIT exon 9 mutations (P = .017, Wald test). A nonsignificant risk reduction of 31% was also observed in OS. Those results were not consistent between trials and were essentially observed in the EU-AUS data, which contained more patients who had KIT exon 9 mutations (15.6% v 8.1%), which may have represented a different biologic subset.
This series of patients with KIT exon 9 mutations who were randomly assigned for the initial dose of imatinib was and will probably remain unique, and it also can be considered mature, with a censoring rate of 10% for progression and 33% for survival. Therefore, we carried out a few more exploratory analysis of this patient subset. Time to imatinib failure was identical in the two randomized arms. However, this analysis is biased in favor of the standard-dose arm: in the high-dose arm (and in patients who did not cross over), imatinib failure occurred after a 20% increase of the tumor load (or development of new lesions); in patients who crossed over to the high-dose level, imatinib failure occurred after two successive increases of at least 20% in the tumor load (ie, a total increase of at least 44%, or development of new lesions). Therefore, we cannot exclude an advantage of upfront high-dose therapy on those grounds.
This meta-analysis that was based on 1,640 patients confirmed that initial treatment with a daily dose of imatinib 800 mg, compared with the registered daily dose of 400 mg, does not have any advantage for most patients. As toxicity of the drug is clearly dose dependent,14 400 mg should remain the standard starting dose. The only exception may be patients who harbor KIT exon 9 mutations, for whom high-dose therapy would potentially delay the first occurrence of disease progression and increase objective response rate. Therefore, imatinib 800 mg could be explored as neoadjuvant treatment for patients with locally advanced disease and KIT exon 9 mutations to decrease tumor burden before surgery. Because there was no impact on overall survival or time to imatinib failure in our study, it remains unclear whether an initial daily dose of imatinib 400 mg followed by dose escalation to 800 mg at the time of first progression would be equivalent as systemic treatment for patients with metastatic disease who harbor KIT exon 9 mutations.
Acknowledgment
Supported in part by Grants No. 5U10 CA11488-30 to 5U10 CA011488-39 (to the EORTC Data Center), CA38926, CA32102, CA27057, CA68183, CA46441, CA04919, CA20319, CA45377, CA35192, CA14028, CA35261, CA63844, CA63850, CA35119, CA76447, CA13612, CA46282, CA58861, CA35178, CA46113, CA42777, CA35090, CA45560, CA45807, CA35176, CA12644, 58348, CA46368, CA16385, CA37981, CA58416, CA63845, CA58882, CA45450, CA35262, CA86780, CA35281, CA58666, CA22433, CA31946, CA71323, CA47559, CA41287, CA32291, CA04326, CA47577, CA35091, CA35113, CA11028, CA12449, CA16450, CA114558-02, CA04457, CA77651, CA77658, CA08025, CA02599, CA11789, CA60138, CA77440, CA03927 (to the Cancer and Leukemia Group B), CA33601 (to the Cancer and Leukemia Group B Statistical Center), CA27525, and CA21115 (to the Eastern Cooperative Oncology Group) from the National Cancer Institute (Bethesda, Maryland); by National Cancer Institute Contracts No. U01-CA70172-01 and N01-CM-17003 (to the Southwest Oncology Group); by the National Cancer Institute of Canada (Kingston, Ontario, Canada); by a Merit Review grant from the Department of Veterans Affairs; and by unrestricted grants from Novartis Pharmaceuticals.
Appendix
We thank the following groups of the GIST Meta-Analysis group (MetaGIST).
Steering Committee
Representatives of the European Organisation for Research and Treatment of Cancer (EORTC), Southwest Oncology Group (SWOG), Italian Sarcoma Group (ISG), Eastern Cooperative Oncology Group (ECOG), Cancer and Leukemia Group B (CALGB), Australasian Gastro-Intestinal Trials Group (AGITG), and National Cancer Institute of Canada Clinical Trials Group (NCIC CTG).
EORTC.
J. Verweij (Erasmus Medical Center, Rotterdam, NL), J-Y. Blay (Centre Leon Berard, Lyon, FR), M. Debiec-Rychter (University Hospital Gasthuisberg, Leuven, Belgium), and M. Van Glabbeke (EORTC Headquarters, Brussels, Belgium).
SWOG and CALGB.
G. Demetri (Dana Farber/Harvard Cancer Center, Boston, US), and M. Heinrich (Portland VA Medical Center and Oregon Health and Science, University Knight Cancer Institute, Portland, OR).
SWOG.
E. Borden (Cleveland Clinic Foundation, Cleveland, OH), B. Redman (University of Michigan Comprehensive Cancer Center, Ann Arbor, MI), C. Blanke (University of British Columbia and British Columbia Cancer Agency, Vancouver, Canada), C. Rankin (SWOG Statistical Center, Seattle, WA), and J. Crowley (SWOG Statistical Center, Seattle, WA).
ISG.
P. Casali (Istituto dei Tumori, Milano, Italy).
ECOG.
M. von Mehren (Fox Chase Cancer Center, Philadephia, PA).
CALGB.
C. Fletcher (Brigham and Women's Hospital, Boston, MA), J. Fletcher (Brigham and Women's Hospital, Boston, MA), and K. Owzar (CALGB Statistical Center, Durham, NC).
AGITG.
J. Zalcberg (Peter Mac Callum Cancer Institute, Melbourne, Australia) and J. Simes (NHMRC Clinical Trials Centre, Sydney, Australia).
NCIC CTG.
V. Bramwell (Tom Baker Cancer Institute, Calgary, Canada).
Principal Other Coinvestigators of > 2% of Total Accrual
R. Benjamin (M. D. Anderson Cancer Center, Houston, TX), A. Le Cesne (Institut Gustave Roussy, Villejuif, France), P. Reichard, P. Hohenberger (Charite, Universitaetsmedizin Berlin, Berlin, Germany), J-Y. Blay (Centre Léon Bérard, Lyon, France), R. Issels (Klinikum Grosshadern, Ludwig-Maximilians University, Muenchen, Germany), I. Judson (Royal Marsden Hospital, London, United Kingdom), R. Maki (Memorial Sloan-Kettering Cancer Center, New York, NY), M. Tanaka (University of California at Davis, Sacramento, CA), P. Rosen (University of California at Los Angeles, Los Angeles, CA), A. van Oosterom (UZ Gasthuisberg, Leuven, Belgium), and S. Leyvraz (Center Hospitalier Vaudois, Lausanne, Switzerland).
Writing Committee for This Work
Martine VanGlabbeke, Jaap Verweij, Jean-Yves Blay, Maria Debiec-Rychter, George Demetri, Michael Heinrich, Ernest Borden, Charles Blanke, John Crowley, Cathryn Rankin, Paolo Casali, Margaret Von Mehren, Christopher Fletcher, Jonathan Fletcher, Kouros Owzar, John Zalcberg, John Simes, and Vivien Bramwell.
Fig A1.
Hazard function compared between dosage arms.
,,
Table A1.
Baseline Demographic and Clinical Characteristics
| Characteristic | Total (N = 1,640) |
Study |
χ2P | ||||
|---|---|---|---|---|---|---|---|
| EU-AUS (n = 946) |
US-CDN (n = 694) |
||||||
| No. | % | No. | % | No. | % | ||
| Sex | |||||||
| Male | 949 | 57.9 | 573 | 60.6 | 376 | 54.2 | .0096 |
| Female | 691 | 42.1 | 373 | 39.4 | 318 | 45.8 | |
| WHO performance status | |||||||
| 0 | 701 | 42.7 | 436 | 46.1 | 265 | 38.2 | .0277 |
| 1 | 692 | 42.2 | 383 | 40.5 | 309 | 44.5 | |
| 2 | 177 | 10.8 | 92 | 9.7 | 85 | 12.2 | |
| 3 | 56 | 3.4 | 35 | 3.7 | 21 | 3.0 | |
| Missing | 14 | 0.9 | 0 | 0.0 | 14 | 2.0 | |
| Site of disease origin | |||||||
| Stomach | 569 | 34.7 | 316 | 33.4 | 253 | 36.5 | .06 |
| Small bowel | 539 | 32.9 | 327 | 34.6 | 212 | 30.5 | |
| Other | 486 | 29.6 | 303 | 32.0 | 183 | 26.4 | |
| Missing | 46 | 2.8 | 0 | 0.0 | 46 | 6.6 | |
| Prior surgery | |||||||
| No | 233 | 14.2 | 143 | 15.1 | 90 | 13.0 | .30 |
| Yes | 1,389 | 84.7 | 802 | 84.8 | 587 | 84.6 | |
| Missing | 18 | 1.1 | 1 | 0.1 | 17 | 2.4 | |
| Prior radiotherapy | |||||||
| No | 1,473 | 89.8 | 883 | 93.3 | 590 | 85.0 | .0036 |
| Yes | 134 | 8.2 | 63 | 6.7 | 71 | 10.2 | |
| Missing | 33 | 2.0 | 0 | 0.0 | 33 | 4.8 | |
| Prior chemotherapy | |||||||
| No | 1,138 | 69.4 | 635 | 67.1 | 503 | 72.5 | < .0001 |
| Yes | 462 | 28.2 | 311 | 32.9 | 151 | 21.8 | |
| Missing | 40 | 2.4 | 0 | 0.0 | 40 | 5.8 | |
| Age, years | |||||||
| Median | 60.2 | 59.9 | 61.0 | .0094 | |||
| Range | 17.0-94.0 | 18.2-91.5 | 17.0-94.0 | ||||
| < 40 | 144 | 8.8 | 72 | 7.6 | 72 | 10.4 | .1442 |
| 40-50 | 266 | 16.2 | 176 | 18.6 | 90 | 13.0 | |
| 50-60 | 403 | 24.6 | 228 | 24.1 | 175 | 25.2 | |
| 60-70 | 451 | 27.5 | 282 | 29.8 | 169 | 24.4 | |
| > 70 | 376 | 22.9 | 188 | 19.9 | 188 | 27.1 | |
| Time since initial diagnosis, months | |||||||
| Median | 13.9 | 11.1 | 17.2 | < .0001 | |||
| Range | 0.0-359.2 | 0.0-331.6 | 0.0-359.2 | ||||
| < 12 | 750 | 45.7 | 493 | 52.1 | 257 | 37.0 | < .0001 |
| > 12 | 866 | 52.8 | 453 | 47.9 | 413 | 59.5 | |
| Missing | 24 | 1.5 | 0 | 0.0 | 24 | 3.5 | |
| Largest diameter of lesions, mm | |||||||
| Median | 77.0 | 80.0 | 75.0 | .62 | |||
| Range | 0.0-800.0 | 0.0-800.0 | 8.0-560.0 | ||||
| < 40 | 348 | 21.2 | 201 | 21.2 | 147 | 21.2 | .64 |
| 40-80 | 510 | 31.1 | 296 | 31.3 | 214 | 30.8 | |
| 80-120 | 379 | 23.1 | 225 | 23.8 | 154 | 22.2 | |
| > 120 | 369 | 22.5 | 218 | 23.0 | 151 | 21.8 | |
| Missing | 34 | 2.1 | 6 | 0.6 | 28 | 4.0 | |
| Hemoglobin, g/dL | |||||||
| Median | 12.6 | 12.7 | 12.5 | .038 | |||
| Range | 7.4-25.1 | 7.6-25.1 | 7.4-22.7 | ||||
| < 11 | 370 | 22.6 | 212 | 22.4 | 158 | 22.8 | .068 |
| 11-12.5 | 427 | 26.0 | 230 | 24.3 | 197 | 28.4 | |
| 12.5-14 | 475 | 29.0 | 277 | 29.3 | 198 | 28.5 | |
| > 14 | 362 | 22.1 | 227 | 24.0 | 135 | 19.5 | |
| Missing | 6 | 0.4 | 0 | 0.0 | 6 | 0.9 | |
| Neutrophils, 109/L | |||||||
| Median | 4.7 | 4.8 | 4.5 | .0021 | |||
| Range | 0.3-35.3 | 1.5-30.6 | 0.3-35.3 | ||||
| < 4 | 587 | 35.8 | 317 | 33.5 | 270 | 38.9 | .0007 |
| 4-5 | 353 | 21.5 | 195 | 20.6 | 158 | 22.8 | |
| 5-6.5 | 313 | 19.1 | 195 | 20.6 | 118 | 17.0 | |
| > 6.5 | 375 | 22.9 | 239 | 25.3 | 136 | 19.6 | |
| Missing | 12 | 0.7 | 0 | 0.0 | 12 | 1.7 | |
| White blood cells, 109/L | |||||||
| Median | 7.3 | 7.6 | 7.1 | .0008 | |||
| Range | 2.6-53.0 | 2.7-32.5 | 2.6-53.0 | ||||
| < 6 | 436 | 26.6 | 231 | 24.4 | 205 | 29.5 | .0004 |
| 6-7.5 | 425 | 25.9 | 223 | 23.6 | 202 | 29.1 | |
| 7.5-9 | 299 | 18.2 | 181 | 19.1 | 118 | 17.0 | |
| > 9 | 444 | 27.1 | 280 | 29.6 | 164 | 23.6 | |
| Missing | 36 | 2.2 | 31 | 3.3 | 5 | 0.7 | |
| Platelets, 109/L | |||||||
| Median | 290.0 | 298.0 | 277.0 | .0083 | |||
| Range | 28.0-1,245.0 | 28.0-1,245.0 | 103.0-1,188.0 | ||||
| < 200 | 238 | 14.5 | 120 | 12.7 | 118 | 17.0 | .0067 |
| 200-300 | 646 | 39.4 | 365 | 38.6 | 280 | 40.5 | |
| 300-400 | 384 | 23.4 | 239 | 25.3 | 145 | 20.9 | |
| > 400 | 367 | 22.4 | 222 | 23.5 | 145 | 20.9 | |
| Missing | 5 | 0.3 | 0 | 0.0 | 5 | 0.7 | |
| Albumin, g/dL | |||||||
| Median | 3.9 | 3.9 | 3.8 | .0001 | |||
| Range | 0.4-8.3 | 0.4-7.0 | 0.7-8.3 | ||||
| < 3.5 | 383 | 23.4 | 191 | 20.2 | 192 | 27.7 | < .0001 |
| 3.5-3.9 | 377 | 23.0 | 182 | 19.2 | 195 | 28.1 | |
| 3.9-4.3 | 383 | 23.4 | 225 | 23.8 | 158 | 22.8 | |
| > 4.3 | 237 | 14.5 | 156 | 16.5 | 81 | 11.7 | |
| Missing | 260 | 15.9 | 192 | 20.3 | 68 | 9.8 | |
| Creatinine, mg/dL | |||||||
| Median | 0.9 | 0.9 | 0.9 | .14 | |||
| Range | 0.0-11.0 | 0.4-2.2 | 0.0-11.0 | ||||
| < 0.75 | 369 | 22.5 | 209 | 22.1 | 160 | 23.1 | .43 |
| 0.75-0.9 | 501 | 30.5 | 270 | 28.5 | 231 | 33.3 | |
| 0.9-1.1 | 475 | 29.0 | 313 | 33.1 | 162 | 23.3 | |
| > 1.1 | 289 | 17.6 | 154 | 16.3 | 135 | 19.5 | |
| Missing | 6 | 0.4 | 0 | 0.0 | 6 | 0.9 | |
| Bilirubin, mg/dL | |||||||
| Median | 0.5 | 0.6 | 0.5 | .0094 | |||
| Range | 0.0-8.0 | 0.1-8.0 | 0.0-8.0 | ||||
| < 0.4 | 525 | 32.0 | 245 | 25.9 | 280 | 40.3 | < .0001 |
| 0.4-0.55 | 292 | 17.8 | 185 | 19.6 | 107 | 15.4 | |
| 0.55-0.8 | 469 | 28.6 | 289 | 30.5 | 180 | 25.9 | |
| > 0.8 | 343 | 20.9 | 227 | 24.0 | 116 | 16.7 | |
| Missing | 11 | 0.7 | 0 | 0.0 | 11 | 1.6 | |
| No. of patients with mutation data | 772 | 378 | 394 | ||||
| Mutation site | |||||||
| Wild type | 117 | 15.2 | 50 | 13.2 | 67 | 17.0 | .0172 |
| KIT exon 9 | 91 | 11.8 | 59 | 15.6 | 32 | 8.1 | |
| KIT exon 11 | 530 | 68.7 | 249 | 65.9 | 281 | 71.3 | |
| KIT exon 13 | 11 | 1.4 | 6 | 1.6 | 5 | 1.3 | |
| KIT exon 17 | 6 | 0.8 | 3 | 0.8 | 3 | 0.8 | |
| KIT exon 8 | 1 | 0.1 | 0 | 0.0 | 1 | 0.3 | |
| All PDGFRα | 16 | 2.1 | 11 | 2.9 | 5 | 1.3 | |
Abbreviations: EU-AUS, joint studies in Europe and Australia; US-CDN, joint studies in United States and Canada.
Table A2.
Prognostic Factors by Survival Type
| Variable | Survival Data |
|||
|---|---|---|---|---|
| All Patients |
Patients With Mutation Data |
|||
| Hazard Ratio | P > χ2 | Hazard Ratio | P > χ2 | |
| Progression-free survival | ||||
| Female sex | 0.829 | .0021 | 0.740 | .0008 |
| WHO performance status | 1.173 | < .0001 | 1.234 | .0004 |
| Hemoglobin, g/dL | 0.938 | .0001 | 0.936 | .0062 |
| Neutrophils, 109/L | 1.054 | < .0001 | 1.054 | < .0001 |
| Site of origin by overall test | — | .0054 | — | .0477 |
| Other sites v stomach | 1.129 | .1017 | 0.856 | .1588 |
| Small bowel v stomach | 1.259 | .0012 | 1.119 | .2856 |
| Mutations | < .0001 | |||
| KIT exon 9 | 2.206 | < .0001 | ||
| Other | 2.659 | < .0001 | ||
| Wild type | 1.717 | < .0001 | ||
| Overall survival | ||||
| Age, years | 1.013 | < .0001 | 1.011 | .0150 |
| Female sex | 0.777 | .0026 | 0.677 | .0016 |
| WHO performance status | 1.289 | < .0001 | 1.519 | < .0001 |
| Tumor diameter, cm | 1.002 | .0018 | 1.003 | .0001 |
| Neutrophils, 109/L | 1.062 | < .0001 | 1.049 | .0060 |
| Albumin | 0.758 | .0002 | 0.717 | .0008 |
| Prior chemotherapy | 1.326 | .0014 | NS | |
| Mutations | — | < .0001 | ||
| KIT exon 9 | 2.187 | < .0001 | ||
| Other | 1.612 | .0692 | ||
| Wild type | 1.755 | .0010 | ||
Abbreviation: NS, not significant.
Table A3.
Predictive Value of KIT Exon 9 Mutations for Progression-Free Survival
| Treatment by Group | No. of Patients | No. of Events | Wald P | Log-Rank P | Wilcoxon P | Adjusted HR | 95% CI | Nonadjusted HR | 95% CI | Interaction P |
|---|---|---|---|---|---|---|---|---|---|---|
| All patients with mutation data | .012 | |||||||||
| Imatinib 400 mg | 383 | 287 | .7323 | .7961 | .3393 | .97 | 0.83 to 1.14 | 0.98 | 0.83 to 1.15 | |
| Imatinib 800 mg | 389 | 295 | ||||||||
| Patients with KIT exon 9 mutations | ||||||||||
| Imatinib 400 mg | 42 | 40 | .0171 | .0115 | .0030 | 0.58 | 0.38 to 0.91 | 0.57 | 0.37 to 0.89 | |
| Imatinib 800 mg | 49 | 42 | ||||||||
| Patients without KIT exon 9 mutations | ||||||||||
| Imatinib 400 mg | 341 | 247 | .8586 | .8155 | .8331 | 1.02 | 0.85 to 1.21 | 1.02 | 0.86 to 1.22 | |
| Imatinib 800 mg | 340 | 253 |
Abbreviation: HR, hazard ratio.
Footnotes
Presented in part at the Clinical Science Symposium on Practical Management of Gastrointestinal Stromal Tumors, 43rd Annual Meeting of the American Society of Clinical Oncology, June 1-5, 2007, Chicago, IL.
The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Disclosures of potential conflicts of interest and author contributions for the MetaGIST Group writing committee are found at the end of this article.
Clinical trial information can be found for the following: NCT00685828, NCT00009906.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Jaap Verweij, Novartis (C); Jean-Yves Blay, Novartis (C), Pfizer (C), GlaxoSmithKline (C); George Demetri, Novartis, Pfizer, ARIAD (C), Johnson & Johnson, Genentech, Infinity (C), ZIOPHARM Oncology, Alnylam, Idera Pharmaceuticals (C), Bayer Pharmaceuticals, EMD Serono, Amgen (C), Daiichi Sankyo, PamGene, Plexxikon (C), N-of-One, Kolltan (C); Michael Heinrich, Novartis (C), MolecularMD (C), Pfizer (C); Charles Blanke, Roche (C), Pfizer (C), Novartis (C), Genentech (C); Paolo Casali, Novartis (C); Margaret Von Mehren, Novartis, Pfizer (C); John Zalcberg, Novartis (C) Stock Ownership: George Demetri, Plexxikon, N-of-One, Kolltan; Michael Heinrich, Molecular MD Honoraria: Jaap Verweij, Novartis; Jean-Yves Blay, Novartis, Pfizer, GlaxoSmithKline; Maria Debiec-Rychter, Novartis; George Demetri, Novartis, Pfizer; Michael Heinrich, Novartis, Pfizer; Paolo Casali, Novartis; Jonathan Fletcher, Novartis; John Zalcberg, Novartis Research Funding: Martine VanGlabbeke, Novartis; Jean-Yves Blay, Novartis, Roche; George Demetri, Novartis, Pfizer, ARIAD, Johnson & Johnson, Bristol-Myers Squibb, Infinity; Michael Heinrich, Novartis, Pfizer; Charles Blanke, Novartis; Paolo Casali, Novartis; Margaret Von Mehren, Novartis, Pfizer; John Zalcberg, Novartis; John Simes, Novartis Expert Testimony: None Other Remuneration: Martine VanGlabbeke, Novartis; Paolo Casali, Novartis; John Zalcberg, Novartis
AUTHOR CONTRIBUTIONS
Conception and design: Martine VanGlabbeke, Jaap Verweij, Jean-Yves Blay, George Demetri, Michael Heinrich, Ernest Borden, John Crowley, Paolo Casali, Margaret Von Mehren, Jonathan Fletcher, John Zalcberg, Vivien Bramwell
Financial support: Michael Heinrich, Jonathan Fletcher
Administrative support: Martine VanGlabbeke, George Demetri, Michael Heinrich, Jonathan Fletcher
Provision of study materials or patients: Jaap Verweij, Jean-Yves Blay, George Demetri, Charles Blanke, Paolo Casali, Margaret Von Mehren, Christopher Fletcher, Jonathan Fletcher, John Zalcberg, John Simes, Vivien Bramwell
Collection and assembly of data: Martine VanGlabbeke, Jaap Verweij, Jean-Yves Blay, Maria Debiec-Rychter, George Demetri, Michael Heinrich, John Crowley, Cathryn Rankin, Paolo Casali, Christopher Fletcher, Jonathan Fletcher
Data analysis and interpretation: Martine VanGlabbeke, Jaap Verweij, Jean-Yves Blay, Maria Debiec-Rychter, George Demetri, Michael Heinrich, Charles Blanke, John Crowley, Paolo Casali, Margaret Von Mehren, Jonathan Fletcher, Kouros Owzar, John Zalcberg, John Simes
Manuscript writing: Martine VanGlabbeke, Jaap Verweij, Jean-Yves Blay, Maria Debiec-Rychter, George Demetri, Michael Heinrich, Charles Blanke, John Crowley, Paolo Casali, Jonathan Fletcher, Kouros Owzar, John Zalcberg, Vivien Bramwell
Final approval of manuscript: Martine VanGlabbeke, Jaap Verweij, Jean-Yves Blay, Maria Debiec-Rychter, George Demetri, Michael Heinrich, Ernest Borden, Charles Blanke, John Crowley, Cathryn Rankin, Paolo Casali, Margaret Von Mehren, Christopher Fletcher, Jonathan Fletcher, Kouros Owzar, John Zalcberg, John Simes, Vivien Bramwell
REFERENCES
- 1.Nilsson B, Bümming P, Meis-Kindblom J, et al. Gastrointestinal stomal tumors: The incidence, prevalence, clinical course and prognostication in the preimatinib era—A population based study in western Sweden. Cancer. 2005;103:821–829. doi: 10.1002/cncr.20862. [DOI] [PubMed] [Google Scholar]
- 2.Joensuu H, Fletcher C, Dimitrijevic S, et al. Management of malignant gastrointestinal stromal tumours. Lancet Oncol. 2002;3:655–664. doi: 10.1016/s1470-2045(02)00899-9. [DOI] [PubMed] [Google Scholar]
- 3.van Oosterom AT, Judson I, Verweij J, et al. For the European Organisation for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group: Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours—A phase I study. Lancet. 2001;358:1421–1423. doi: 10.1016/s0140-6736(01)06535-7. [DOI] [PubMed] [Google Scholar]
- 4.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 5.Verweij J, van Oosterom A, Blay JY, et al. Imatinib mesylate (STI-571 Glivec, Gleevec) is an active agent for gastrointestinal stromal tumours, but does not yield responses in other soft-tissue sarcomas that are unselected for a molecular target: Results from an EORTC Soft Tissue and Bone Sarcoma Group phase II study. Eur J Cancer. 2003;39:2006–2011. [PubMed] [Google Scholar]
- 6.Verweij J, Casali PG, Zalcberg J, et al. For the EORTC Soft Tissue and Bone Sarcoma Group, the Italian Sarcoma Group and the Australasian Gastrointestinal Trials Group: Progression-free survival in gastrointestinal stromal tumours with high-dose imatininb—Randomized trial. Lancet. 2004;364:1127–1134. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 7.Blanke CD, Rankin C, Demetri GD, et al. Phase III randomized, intergroup trial assessing imatinib mesylate at two dose levels in patients with unresectable or metastatic gastrointestinal stromal tumors expressing the KIT receptor tyrosine kinase: S0033. J Clin Oncol. 2008;26:626–632. doi: 10.1200/JCO.2007.13.4452. [DOI] [PubMed] [Google Scholar]
- 8.Casali PG, Verweij J, Kotasek D, et al. Imatinib mesylate in advanced Gastrointestinal Stromal Tumors (GIST): Survival analysis of the EORTC ISG AGITG randomized trial in 946 patients. European Journal of Cancer. 2005;(Supplements 3) abstract 711. [Google Scholar]
- 9.Rankin C, von Mehren M, Blanke C, et al. Dose effect of imatinib in patients with metastatic GIST: Phase III sarcoma group study S0033. J Clin Oncol. 2004;22(suppl):14S. abstr 9005. [Google Scholar]
- 10.Zalcberg JR, Verweij J, Casali PG, et al. For the EORTC Soft Tissue And Bone Sarcoma Group, the Italian Sarcoma Group, and the Australasian Gastrointestinal Trials Group: Outcome of patients with advanced gastro-intestinal stromal tumours crossing over to a daily imatinib dose of 800 mg after progression on 400 mg. Eur J Cancer. 2005;41:1751–1757. doi: 10.1016/j.ejca.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 11.Debiec-Rychter M, Sciot R, Le Cesne A, et al. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer. 2006;43:1093–1103. doi: 10.1016/j.ejca.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 12.Heinrich MC, Owzar K, Corless CL, et al. Correlation of kinase genotype and clinical outcome in the North American Intergroup phase III trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 study by Cancer and Leukemia Group B and Southwest Oncology Group. J Clin Oncol. 2008;26:5360–5367. doi: 10.1200/JCO.2008.17.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Glabbeke M, Verweij J, Casali PG, et al. Initial and late resistance to imatinib in advanced gastrointestinal stroma tumors are predicted by different prognostic factors: A European Organisation for Research and Treatment of Cancer—Italian Sarcoma Group, Australian Gastointestinal Trials Group study. J Clin Oncol. 2005;23:5795–5804. doi: 10.1200/JCO.2005.11.601. [DOI] [PubMed] [Google Scholar]
- 14.Van Glabbeke M, Verweij J, Casali PG, et al. Predicting toxicities for patients with advanced gastrointestinal stromal tumours treated with imatinib: A study of the European Organisation for Research and Treatment of Cancer, the Italian Sarcoma Group, and the Australasian Gastro-Intestinal Trials Group (EORTC-ISG-AGITG) Eur J Cancer. 2006;42:2277–2285. doi: 10.1016/j.ejca.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 15.Sauerbrei W, Schumacher M. A bootstrap resampling procedure for moel building: Application to the Cox regression model. Stat Med. 1992;11:2093–2109. doi: 10.1002/sim.4780111607. [DOI] [PubMed] [Google Scholar]
- 16.Harrell FE, Jr, Lee KL, Califf RM, et al. Regression modeling strategies for improved prognostic prediction. Stat Med. 1984;3:143–152. doi: 10.1002/sim.4780030207. [DOI] [PubMed] [Google Scholar]
- 17.Dileo P, Rankin CJ, Benjamin RS, et al. Incidence and reasons for dose modification of standard-dose vs. high-dose imatinib mesylate in the phase III Intergroup Study S0033 of patients with unresectable or metastatic gastrointestinal stromal tumor (GIST) J Clin Oncol. 2004;22(suppl):14S. abstr 9032. [Google Scholar]
- 18.Heinrich MC, Corless CL, Blanke CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006;24:4764–4774. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 19.Debiec-Rychter M, Cools J, Dumez H, et al. Mechanisms of resistance to imatinib mesylate in gastrointestinal stromal tumors and activity of the PKC412 inhibitor against imatinib-resistant mutants. Gastroenterology. 2005;128:270–279. doi: 10.1053/j.gastro.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 20.Corless CL, Fletcher AJ, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22:3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]