Abstract
The secondary metabolites VM55599 (4) and pre-paraherquamide (5) have been identified by LC-MSn analysis as natural metabolites in cultures of Penicillium fellutanum, whereas pre-paraherquamide has been identified only in cultures of Aspergillus japonicus. In accord with a previous proposal, the identification of both metabolites, which have a diastereomeric relationship, provides indirect support for a unified biosynthetic scheme.
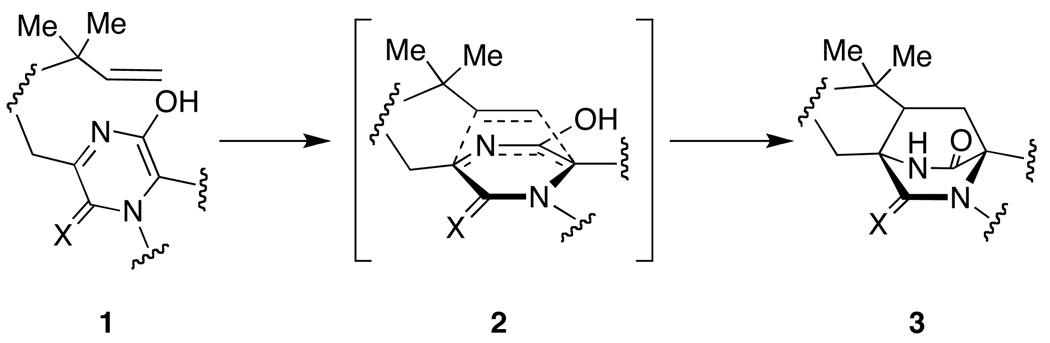
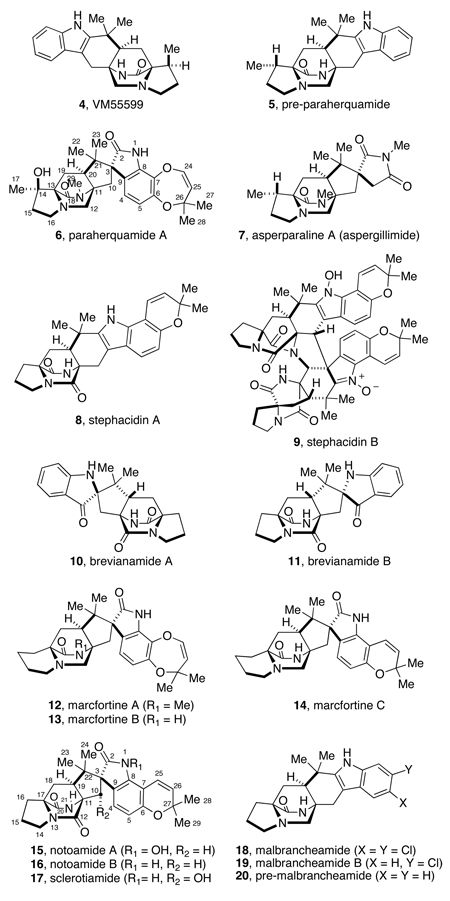
The paraherquamides (6),1 together with the asperparalines (7),2 stephacidins,3 brevianamides,4 marcfortines,5 notoamides,6 sclerotamide,7 and malbrancheamides8 are secondary metabolites derived from fungi that feature a common bicyclo[2.2.2]diazaoctane core. It has been postulated that this ring system is generated through an intramolecular Diels-Alder cycloaddition of the C5 moiety across the α-carbons of the amino acid subunits, as depicted in Scheme 1.9
Scheme 1.
Proposed intramolecular Diels-Alder construction of the bicyclo[2.2.2]diazaoctane core.
Members of this unique family of metabolites are all derived from tryptophan, isoprene units of mevalonate origin, and a cyclic amino acid residue consisting of either proline, β-methylproline (and derivatives), or pipecolic acid. In 1993, Everett and coworkers described the isolation of VM55599 (4), a minor metabolite from culture extracts of a Penicillium sp. (IMI332995) that also produces paraherquamide A (6), among other paraherquamides.10 Taking into account the structural similarities between these co-occurring metabolites, these authors proposed that 4 might indeed be a biosynthetic precursor of paraherquamide A.10 The relative stereochemistry of 4 was assigned by Everett and coworkers through extensive 1H NMR NOE experiments, but the small quantity of this compound isolated precluded the determination of its absolute configuration.10 Our laboratory has previously determined the absolute configuration of VM55599 produced by Penicillium sp. IMI332995 by an asymmetric, biomimetic total synthesis.11 We further demonstrated that 4 is not a biosynthetic precursor to paraherquamide A through the synthesis of double 13C-labeled, racemic VM55599 (4) for which the lack of incorporation into 6 in cultures of P. fellutanum cast doubt on the intermediacy of this species in paraherquamide biosynthesis.12
Asperparalines A (7) and C and several members of the paraherquamide family, including VM55599 have been identified as being biosynthesized from β-methylproline.13 Previous studies from our laboratories have revealed that (S)-isoleucine (L-Ile) serves as the biosynthetic precursor of the β-methyl-β-hydroxyproline residue in paraherquamide A as well as the β-methylproline residue of 7.13 This mandates that the L-Ile side chain stereochemistry is retained at C-14 in paraherquamide A and that hydroxylation occurs with net retention at C-14. Accordingly, these results brought into question the capacity of VM55599 (4) to serve as a biosynthetic precursor to the paraherquamides. If, as one could reasonably speculate, L-Ile is not only a biosynthetic precursor to the paraherquamides, but also to VM55599, the absolute stereochemistry of this compound must be that depicted in Scheme 2; we have rigorously confirmed the relative and absolute stereochemistry of 4 as mentioned above.11 Thus, the absolute configuration of the bicyclo[2.2.2]diazaoctane core of VM55599 (4) is enantiomorphic to that of virtually all of the other members of the paraherquamide family.14
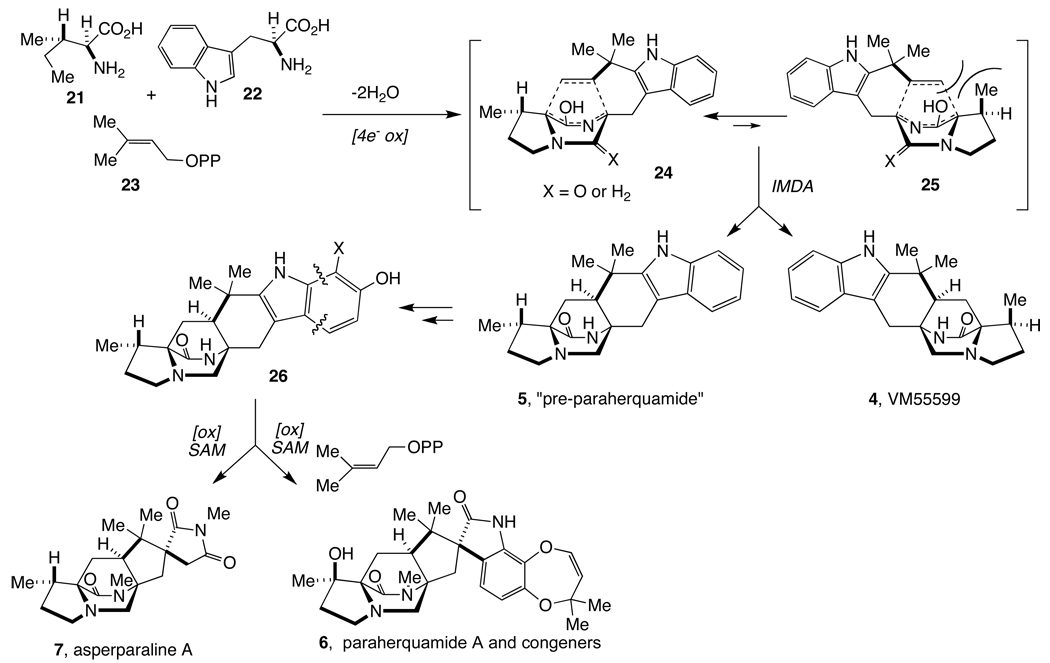
Scheme 2.
Proposed unified biogenesis of paraherquamides and asperparalines.
These experimental observations led us to propose a unified biosynthesis of the paraherquamides and VM55599 (4), as shown in Scheme 2. In this proposal, the biosynthetic precursors of the paraherquamides and that of 4 would arise as diastereomeric products of the putative intramolecular Diels-Alder cycloaddition of a common azadiene through two of four possible diastereomeric transition states (24 and 25). The major pathway (via 24), produces species 5 (named “pre-paraherquamide”), which is further processed in the respective organisms to produce the paraherquamides and asperparalines. In support of this hypothesis, we have synthesized double 13C-labeled species 5 and have demonstrated that this compound incorporates into paraherquamide A (6) in cultures of P. fellutanum.12 Despite the incorporation of 5 into the biosynthesis of paraherquamide A, this putative intermediate has heretofore not been detected as a secondary metabolite in either paraherquamide- or asperparaline-producing fungi. Herein, we demonstrate that: (i) paraherquamides A (6) and B are produced in both P. fellutanum and Aspergillus japonicus; (ii) both VM55599 (4) and pre-paraherquamide (5) are natural metabolites of the paraherquamide-producing organism P. fellutanum; and (iii) pre-paraherquamide (5) is also observed as a natural metabolite from A. japonicus cultures. The observation of VM55599 (4) and pre-paraherquamide (5) provides additional, indirect support for the unified biogenesis outlined in Scheme 2.
Results and Discussion
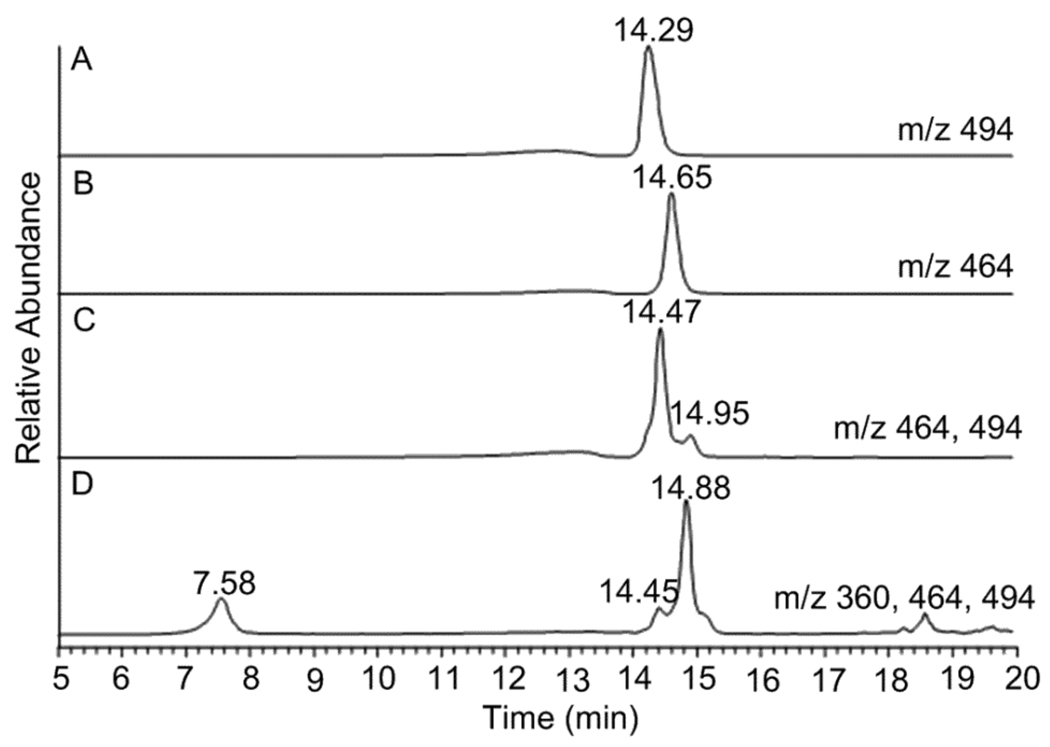
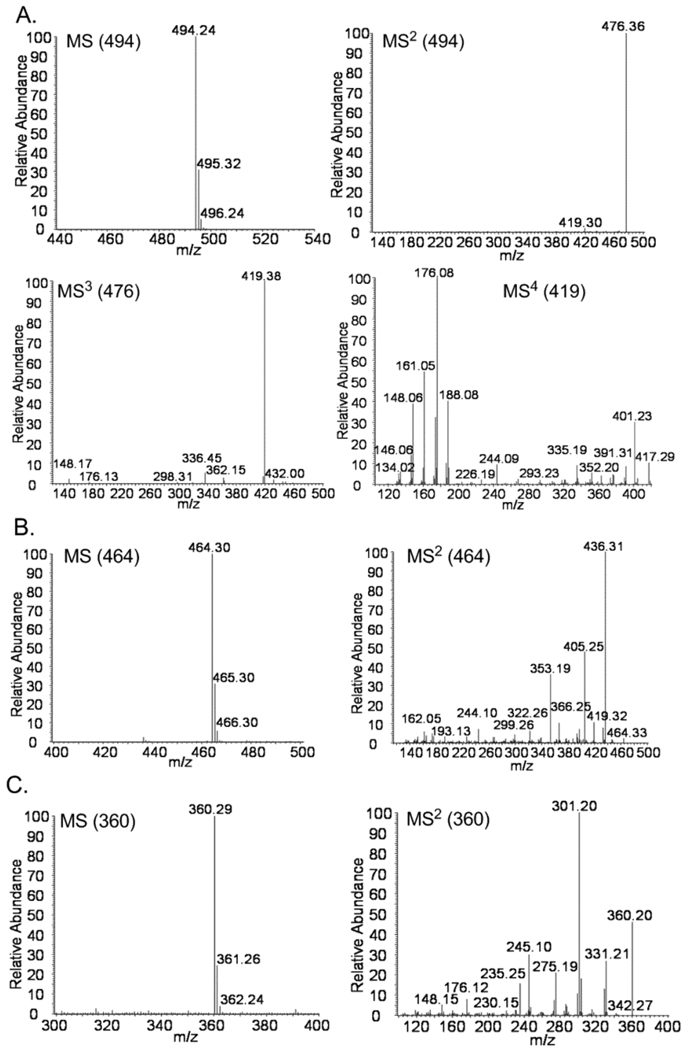
P. fellutanum and A. japonicus JV-23 were cultured and first examined for viable production of paraherquamide A (6), asperparaline A (7) and their derivatives, respectively. Authentic paraherquamide A was first analyzed by LC-MSn and its MS, MS2, MS3, and MS4 spectra were informative in identifying this alkaloid, establishing the successful application of LC-MSn analysis in this study (Figure S1, Supporting Information). Next, paraherquamide A (6) in the P. fellutanum isolation was eluted at 14.47 min in the selected-ion monitoring (SIM) chromatograph and exhibited an ion at m/z 494.36 (calculated [M+H]+: 494.26) by MS analysis (Figure 1C and Figure 2A). Further MS2, MS3, and MS4 analyses produced identical spectra to those of authentic paraherquamide A (Figure 2A and Figure S1). Along with paraherquamide A, paraherquamides B–G have previously been isolated from P. fellutanum (previously named P. charlesii).1 In this study, one compound at m/z 464.29 was detected in the fungal isolation and had a retention time of 14.95 min (Figure 1C and Figure 2B). This compound was identified as paraherquamide B (calcd [M+H]+: 464.25) by comparing its retention time and MS2 spectrum to those of an authentic specimen (Figure 1 and Figure S1).
Figure 1.
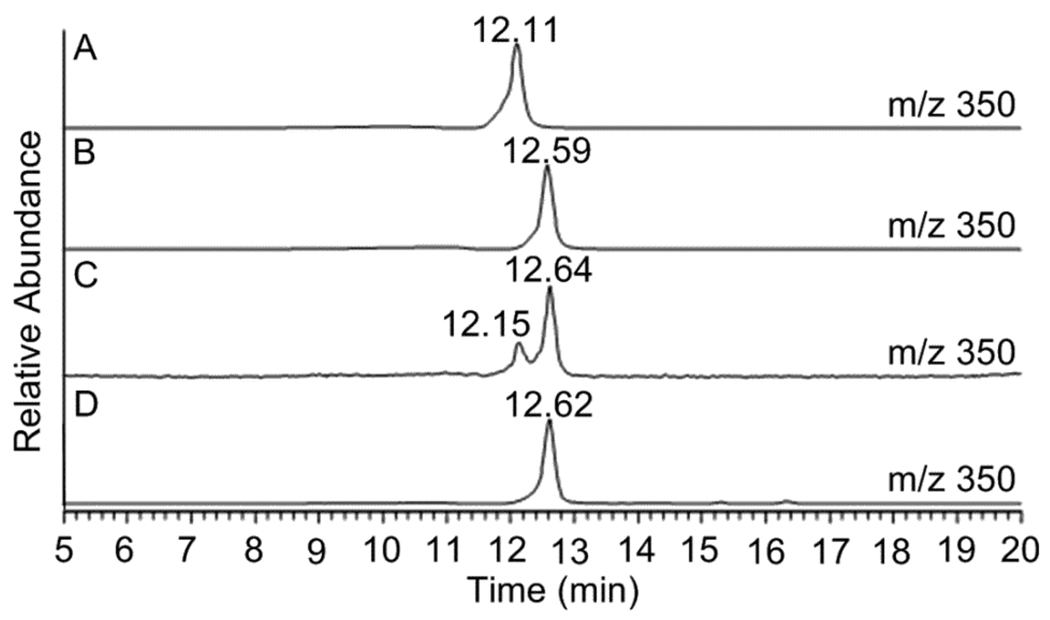
Selected-ion monitoring (SIM) chromatographs corresponding to authentic paraherquamide A (6) (m/z 494) (A), authentic paraherquamide B (m/z 464) (B), isolation from P. fellutanum culture (m/z 464 and 494) (C), and isolation from A. japonicus JV-23 (m/z 360, 464, and 494) (D).
Figure 2.
MSn spectra of paraherquamide A (6) (A) and paraherquamide B (B) from the isolation of P. fellutanum cultures, and asperparaline A (7) (C) from the extracts of A. japonicus JV-23. The integral m/z values of ions for each MSn analysis are included in corresponding graphs.
In extracts from A. japonicus JV-23 cultures, asperparaline A (7) had the retention time of 7.58 min and exhibited an ion at m/z 360.29 (calcd [M+H]+: 360.22) (Figure 1D and Figure 2C). This metabolite was further analyzed by MS2 analysis. In previous reports, Aspergillus species IMI 3376642 and A. sclerotiorum7 represented the first organisms outside of Penicillium sp. to produce paraherquamide congeners. In this study, we investigated paraherquamide production in A. japonicus JV-23. By comparing their retention times and MSn spectra to those of authentic compounds, both paraherquamide A (14.45 min) and paraherquamide B (14.88 min) were identified in this Aspergillus sp. isolation, further indicating that both Penicillium spp. and Aspergillus spp. are able to produce these anthelmintic alkaloid metabolites (Figure 1, and Figure S2). Moreover, this result strongly suggested one common biosynthetic pathway shared by both asperparalines and paraherquamides in this fungus.
The A. japonicus JV-23 strain produced more paraherquamide B than paraherquamide A (6) in PDB medium under the growth conditions given. A. japonicus JV-23 is the first reported Aspergillus sp. to produce paraherquamide A itself, to the best of our knowledge, although Everett and co-workers isolated the paraherquamide congeners VM54159, SB203105 and SB200437 from Aspergillus strain IMI 337664.2c In extracts from P. fellutanum, two metabolites with m/z of 350 were separated and identified by LC-MS/MS analysis (Figure 3). The first peak had a retention time of 12.15 min while the second metabolite was eluted at 12.64 min. These peaks were initially proposed to be VM55599 (4) and pre-paraherquamide (5), considering their theoretical molecular weights (C22H27N3O, 349.22) and the previous isolation of 4 as the minor metabolite from Penicillium sp. IMI337664.10
Figure 3.
Selected-ion monitoring (SIM) chromatographs corresponding to the LC-MSn analysis of authentic VM55599 (4) (m/z 350) (A), authentic pre-paraherquamide (5) (m/z 350) (B), isolation from P. fellutanum cultures (m/z 350) (C), and isolation from A. japonicus JV-23 (m/z 350) (D).
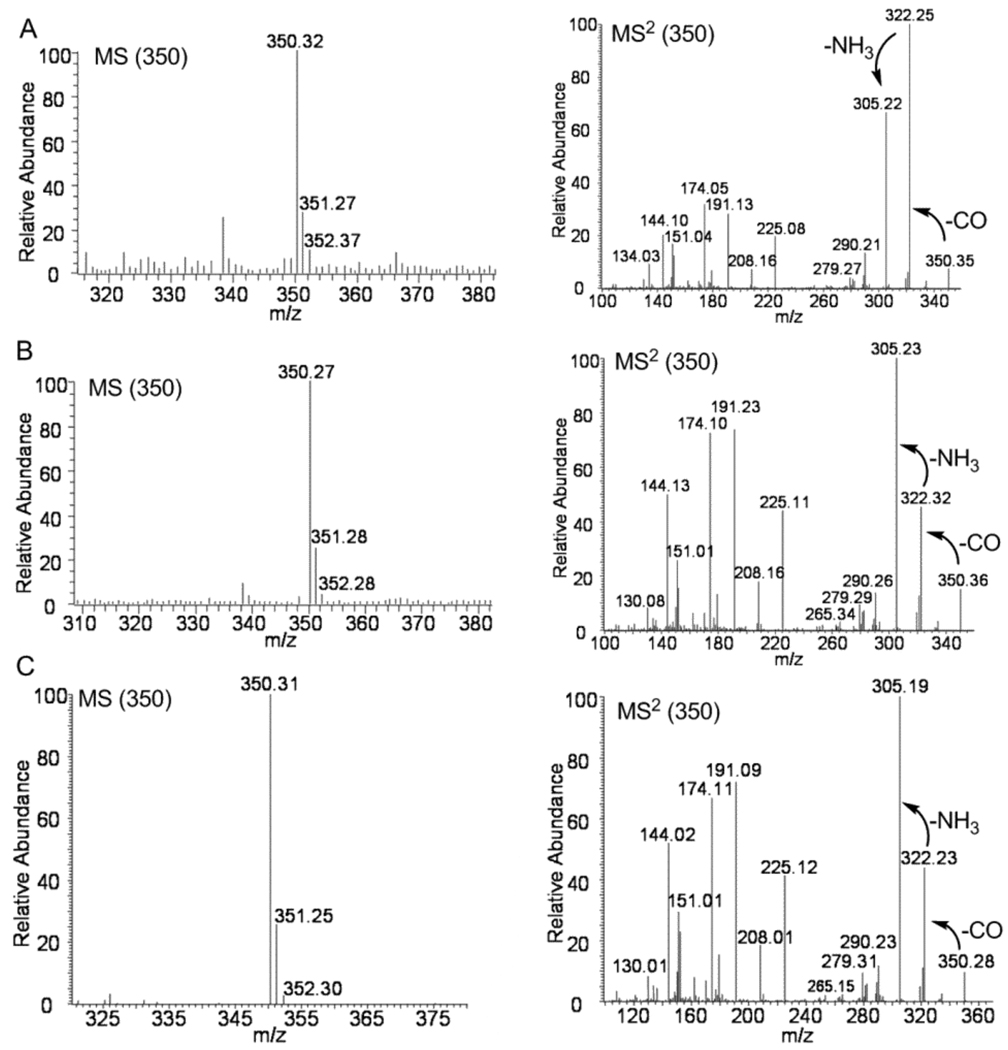
To further identify these metabolites, synthetic and authentic (racemic) samples of 4 and 5 were used to secure standard MS and MS2 fragmentation data (Figure S3). Interestingly, these two compounds, which are diastereomers, exhibited different fragmentation patterns in their MS2 spectra. The ratio of the peak at m/z 322.26 to the peak at m/z 305.25 in VM55599 MS2 spectrum was larger than 1.0 while this ratio was significantly smaller than 1.0 in the pre-paraherquamide (5) MS2 spectrum with the fragment at m/z 305.24 as the most intense peak, which likely serves as the distinctive feature of the MS2 spectra of these two compounds (Figure S3). CO was lost from the peptide bond of parent ions to produce the first fragment at m/z 322.26, which was further fragmented to give a signal at m/z 305.25 by losing NH3 (Figure 4 and Figure S3). The same fragmentation pathway was observed for 5. The fragmentation discrepancy observed in the MS2 spectra of 4 and 5 were apparently affected by the single relative stereochemical difference at C-14 (paraherquamide numbering) between these two compounds. Comparing their MS2 spectra with those of authentic compounds, VM55599 (4) (Rt = 12.15 min) and pre-paraherquamide (5) (Rt = 12.64 min) were identified in the extract from P. fellutanum cultures (Figure 3A–C and Figure 4A–B). Pre-paraherquamide was thus observed as one natural metabolite, further strongly validating the putative pathway in Scheme 2.
Figure 4.
MS and MS2 spectra of VM55599 (4) (A) and pre-paraherquamide (5) from the isolation from P. fellutanum culture (B), and pre-paraherquamide (5) from the extracts of A. japonicus JV-23 cultures (C). The integral m/z values of ions for each analysis are included in corresponding graphs.
In extracts from A. japonicus JV-23 cultures, only one metabolite at m/z 350 was identified by LC-MS/MS analysis (Figure 3D). When compared to the MS and MS2 spectra of authentic standards, the metabolite with the retention time of 12.62 min was validated as pre-paraherquamide (Figure 4C). This represents the first identification of this putative precursor in an asperparaline-producing organism. Pre-paraherquamide (5) has been proposed by this laboratory as the key, common biosynthetic precursor to both the paraherquamides and the asperparalines.9,13c The identification of this substance in the paraherquamide- and asperparaline-producing A. japonicus JV-23 strain further supports the unified biogenetic hypothesis detailed in Scheme 2. Curiously, VM55599 was not detected as a natural metabolite from the A. japonicus JV-23 cultures.
Numerous alkaloids that display a wide spectrum of biological activities have been isolated from various fungi. An important and growing family of prenylated indole alkaloids is constituted by a common bicyclo[2.2.2]diazaoctane core derived mainly from tryptophan, proline, substituted proline derivatives, and isoprene units. This family includes the paraherquamides, asperparalines,2 stephacidins,3 brevianamides,4 marcfortines,5 notoamides,6 sclerotiamide,7 and the malbrancheamides8. Among these alkaloids, malbrancheamide is further distinguished by the presence of two chlorine atoms in the indole aromatic nucleus. Pre-malbrancheamide, which lacks the C-14 methyl group found in pre-paraherquamide, was recently isolated and identified as the precursor in malbrancheamide B biosynthesis.15 By LC-MS/MS analysis, this natural metabolite exhibited a similar fragmentation pattern to VM55599 (4) and pre-paraherquamide (5). Similar to the results we obtained in precursor feeding experiments with 5, synthetic doubly 13C-labeled pre-malbrancheamide was also successfully incorporated into malbrancheamide B in Malbranchea aurantiaca.12,15 The identification of both pre-paraherquamide and pre-malbracheamide as natural, trace metabolites suggests that structurally related common precursors may be involved in the biosynthesis of other sub-groups of alkaloids within this family and that the structural diversity of these alkaloids are likely introduced by downstream tailoring enzymes following the construction of the bicyclo[2.2.2]diazaoctane core.
LC-MS/MS analysis has several important advantages with respect to sensitivity and selectivity in the detection of trace natural metabolites. In natural products identification, NMR techniques are widely used to directly provide structural information when sufficient amounts of purified substances are available. However, in the case of biosynthetic intermediates, there is often a paucity of material that is thus insufficient for NMR structural studies. This is a manifestation of biosynthetic intermediates being largely consumed by downstream tailoring enzymes, and thus these substrates do not accumulate. LC-MS/MS is an effective and powerful alternative in these cases and has been successfully used to identify many natural products in crude extracts by comparison with the respective reference compounds.16 Herein, we were able to successfully deploy LC-MS/MS analysis to identify the presence of paraherquamide A (6), paraherquamide B, asperparaline A (7), VM55599 (4), and pre-paraherquamide (5) in crude fungal extracts using this technique.
In conclusion, we have demonstrated for the first time that pre-paraherquamide (5) is a natural secondary metabolite of the paraherquamide-producing organism P. fellutanum and the paraherquamide- and asperparaline-producing organism A. japonicus JV-23. This report constitutes the first confirmatory evidence for the natural existence of pre-paraherquamide, albeit at low concentration levels. This provides additional support for the unified biogenesis we have proffered.9,11–13 VM55599 (4) is also produced by P. fellutanum and is consistent with the initial identification of this substance from the related paraherquamide-producing organism Penicillium sp. IMI332995 described by Everett and co-workers.10 As the identification of new paraherquamide-producing fungi are discovered in various environments around the world, we speculate here that VM55599 can be expected to be detected as a co-metabolite. It should be further noted that Miller and co-workers recently described the detection of VM55599 as a metabolite in several strains of Penicillium paneum on the basis of mass spectrometric data.17 It is entirely possible that these workers might have instead detected pre-paraherquamide (or both), which has the same mass as VM55599. As in the previously established case of P. fellutanum, we conclude that VM55599 is a shunt (dead-end) metabolite as it possesses the incorrect absolute (and relative) stereochemistry to be processed further to a paraherquamide-like structure. Likewise, we speculate that in A. japonicus, the major pathway metabolite 5 is largely consumed by the down-stream biosynthetic machinery responsible for the substantial oxidative elaboration of 5 into the asperparalines and paraherquamides. We are currently pursuing the synthesis and labeling of several plausible metabolites down stream of pre-paraherquamide to gain insight into the sequence of events following the construction of this early pathway metabolite in both paraherquamide- and asperparaline-producing fungi. Efforts to clone the biosynthetic gene clusters for the biosynthesis of these structurally unique and biologically important natural products are also currently under investigation in our laboratories.
Experimental Section
Chemicals and Strains
Ethyl acetate and methanol were HPLC grade from Sigma Aldrich (St. Louis, MO) while acetonitrile used in LC-MSn analysis was LC-MS grade from J. T. Baker (Phillipsburg, NJ). A MilliQ H2O purification system (Millipore Ltd., Bedford, MA) generated water for LC-MSn analysis. Trifluoroacetic acid (99%, reagent plus) and formic acid (>98%, ACS reagent) were also purchased from Sigma Aldrich. Authentic paraherquamide A (6),18 VM55599 (4),11,12 and pre-paraherquamide (5)12 were synthesized following previously published procedures. Paraherquamide B (as the unnatural enantiomer) was obtained by total synthesis.18b P. fellutanum (ATCC20841) was purchased from American Type Culture Collection (Manassas, VA) while A. japonicus JV-23 was provided by Dr. Hideo Hayashi of Osaka Prefecture University.
Cultures of P. fellutanum and A. japonicus JV-23
P. fellutanum and A. japonicus JV-23 were initially grown in solid medium (20 g malt extract, 20 g glucose, 1 g peptone, and 20 g agar in 1 L deionized water) and solid (20 g potato-dextrose-broth and 20 g agar in 1 L deionized water), respectively, at 25 °C in the dark for 2 weeks. Fungal mycelia and spores were then transferred into 300 mL sterile corn steep liquor medium (22 g corn steep liquor and 40 g glucose per liter deionized water) for P. fellutanum or 300 mL sterile potato-dextrose-broth (PDB) (24 g potato-dextrose-broth per liter deionized water) for A. japonicus JV-23, in 2 L Erlenmeyer flasks. Both fungal strains were then grown at 25 °C in the dark for 4 weeks.
Sample Extraction
The cultures with fungal mycelium were adjusted to pH 10–12 by 10 M KOH. The cultures were then extracted with an equal volume of ethyl acetate twice. The combined organic layer from each culture was washed with water, dried over anhydrous magnesium sulfate, and evaporated to dryness. The residues were re-dissolved in methanol prior to LC-MSn analysis.
LC-MSn Analysis
LC-MSn analysis was performed using a ThermoFinnigan LTQ linear ion-trap instrument equipped with electrospray source and Surveyor HPLC system at room temperature. Separations were carried out with a Waters XBridge™ C18 (3.5 µm, 2.1×150mm) column at a flow rate of 210 µL/min with solvent A (water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). Solvent B was kept at 15% in solvent A for 2.5 min and then was gradually increased to 40% over 12.5 min, 80% over 2 min, and then maintained at 80% for 6 min to elute fungal metabolites. The column was further re-equilibrated with 15% solvent B for 25 min. For mass spectrometry, the capillary temperature was set to 275 °C with the source voltage at 3.6 kV, the source current at 3.5 µA, the capillary voltage at 30 V, and the tube lens at 119 V. Sheath gas flow was set to 28 psi and auxiliary gas flow was 5 arbitrary units. The normalized collision energy for ion fragmentation was 30%. The injection volume was 5–10 µL and spectra were recorded in the positive-ion mode. Selected-ion monitoring (SIM) chromatographs were obtained at the selected m/z values.
Supplementary Material
Acknowledgment
We gratefully acknowledge financial support from the National Institutes of Health (CA70375) and an Eli Lilly & Company pre-doctoral fellowship (to Y.D.). We are indebted to Professor Hideo Hayashi for generously providing us with a culture of Aspergillus japonicus JV-23 and authentic specimens of asperparaline. Mass spectra (CSU) were obtained on instruments supported by the NIH Shared Instrumentation Grant GM49631.
Footnotes
Supporting Information Available. MSn spectra of authentic paraherquamide A (6) and authentic paraherquamide B; MSn spectra of two metabolites at 14.45 min and at 14.88 min from the extract from A. japonicus JV-23; MS and MS2 spectra of authentic VM55599 (4) and authentic pre-paraherquamide (5). This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.(a) Yamazaki M, Okuyama E, Kobayashi M, Inoue H. Tetrahedron Lett. 1981;22:135–136. [Google Scholar]; (b) Ondeyka JG, Goegelman RT, Schaeffer JM, Kelemen L, Zitano L. J. Antibiot. 1990;43:1375–1379. doi: 10.7164/antibiotics.43.1375. [DOI] [PubMed] [Google Scholar]; (c) Liesch JM, Wichmann CF. J. Antibiot. 1990;43:1380–1386. doi: 10.7164/antibiotics.43.1380. [DOI] [PubMed] [Google Scholar]; (d) Blanchflower SE, Banks RM, Everett JR, Manger BR, Reading C. J. Antibiot. 1991;44:492–497. doi: 10.7164/antibiotics.44.492. [DOI] [PubMed] [Google Scholar]
- 2.(a) Hayashi H, Nishimoto Y, Nozaki H. Tetrahedron Lett. 1997;38:5655–5658. [Google Scholar]; (b) Hayashi H, Nishimoto Y, Akiyama K, Nozaki H. Biosci. Biotechnol. Biochem. 2000;64:111–115. doi: 10.1271/bbb.64.111. [DOI] [PubMed] [Google Scholar]; (c) Banks RM, Blanchflower SE, Everett JR, Manger BR, Reading C. J. Antibiot. 1997;50:840–846. doi: 10.7164/antibiotics.50.840. [DOI] [PubMed] [Google Scholar]
- 3.(a) Qian-Cutrone J, Huang S, Shu Y-Z, Vyas D, Fairchild C, Menendez A, Krampitz K, Dalterio R, Klohr SE, Gao Q. J. Am. Chem. Soc. 2002;124:14556–14557. doi: 10.1021/ja028538n. [DOI] [PubMed] [Google Scholar]; (b) Qian-Cutrone J, Krampitz K, Shu Y-Z, Chang LP. U.S. Patent 6,291,461. 2001
- 4.(a) Birch AJ, Wright JJ. J. Chem. Soc. Chem. Commun. 1969:644–645. [Google Scholar]; (b) Birch AJ, Wright JJ. Tetrahedron. 1970;26:2329–2344. doi: 10.1016/s0040-4020(01)92812-1. [DOI] [PubMed] [Google Scholar]; (c) Birch AJ, Russell RA. Tetrahedron. 1972;28:2999–3008. [Google Scholar]; (d) Bird BA, Remaley AT, Campbell IM. Appl. Environ. Microbiol. 1981;42:521–525. doi: 10.1128/aem.42.3.521-525.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Bird BA, Campbell IM. Appl. Environ. Microbiol. 1982;43:345–348. doi: 10.1128/aem.43.2.345-348.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Robbers JE, Straus JW. Lloydia. 1975;38:355–356. [PubMed] [Google Scholar]; (g) Paterson RRM, Hawksworth DL. Trans. Br. Mycol. Soc. 1985;85:95–100. [Google Scholar]; (h) Wilson BJ, Yang DTC, Harris TM. Appl. Microbiol. 1973;26:633–635. doi: 10.1128/am.26.4.633-635.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Coetzer J. Acta Crystallogr. 1974;B30:2254–2256. [Google Scholar]
- 5.(a) Polonsky J, Merrien M-A, Prange T, Pascard C. J. Chem. Soc. Chem. Comm. 1980:601–602. [Google Scholar]; (b) Prange T, Buillion M-A, Vuilhorgne M, Pascard C, Polonsky J. Tetrahedron Lett. 1980;22:1977–1980. [Google Scholar]
- 6.Kato H, Yoshida T, Tokue T, Nojiri Y, Hirota H, Ohta T, Williams RM, Tsukamoto S. Angew. Chem. Int. Ed. 2007;46:2254–2256. doi: 10.1002/anie.200604381. [DOI] [PubMed] [Google Scholar]
- 7.Whyte AC, Gloer JB. J. Nat. Prod. 1996;59:1093–1095. doi: 10.1021/np960607m. [DOI] [PubMed] [Google Scholar]
- 8.(a) Martinez-Luis S, Rodriguez R, Acevedo L, Gonzalez MC, Lira-Rocha A, Mata R. Tetrahedron. 2006;62:1817–1822. [Google Scholar]; (b) Figueroa M, del Carmen González M, Mata R. Nat. Prod. Res. 2008:0000–0000. doi: 10.1080/14786410802012361. [DOI] [PubMed] [Google Scholar]; (c) Miller KA, Welch TR, Greshock TJ, Ding Y, Sherman DH, Williams RM. J. Org. Chem. 2008;73:3116–3119. doi: 10.1021/jo800116y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Stocking E, Williams RM. Angew. Chem. Int. Ed. 2003;42:3078–3115. doi: 10.1002/anie.200200534. [DOI] [PubMed] [Google Scholar]; (b) Cox RJ, Williams RM. Acc. Chem. Res. 2003;36:127–139. doi: 10.1021/ar020229e. [DOI] [PubMed] [Google Scholar]; (c) Williams RM. Chem. Pharm. Bull. 2002;50:711–740. doi: 10.1248/cpb.50.711. [DOI] [PubMed] [Google Scholar]; (d) Williams RM, Sanz-Cervera JF, Stocking E. In: In Topics in Current Chemistry, Volume on Biosynthesis-Terpenes and Alkaloids. Leeper F, Vederas JC, editors. vol 209. Berlin: Springer-Verlag; 2000. pp. 97–173. [Google Scholar]
- 10.Blanchflower SE, Banks RM, Everett JR, Reading C. J. Antibiot. 1993;46:1355–1363. doi: 10.7164/antibiotics.46.1355. [DOI] [PubMed] [Google Scholar]
- 11.Sanz-Cervera JF, Williams RM. J. Am. Chem. Soc. 2002;124:2556–2559. doi: 10.1021/ja017425l. [DOI] [PubMed] [Google Scholar]
- 12.(a) Stocking EM, Sanz-Cervera JF, Williams RM. J. Am. Chem. Soc. 2000;122:1675–1683. [Google Scholar]; (b) Stocking EM, Sanz-Cervera JF, Williams RM. Angew. Chem. Int. Ed. 2001;40:1296–1298. doi: 10.1002/1521-3773(20010401)40:7<1296::aid-anie1296>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 13.(a) Stocking E, Sanz-Cervera JF, Williams RM, Unkefer CJ. J. Am. Chem. Soc. 1996;118:7008–7009. [Google Scholar]; (b) Stocking EM, Martinez RA, Silks LA, Sanz-Cervera JF, Williams RM. J. Am. Chem. Soc. 2001;123:3391–3392. doi: 10.1021/ja005655e. [DOI] [PubMed] [Google Scholar]; (c) Gray C, Sanz-Cervera JF, Williams RM. J. Am. Chem. Soc. 2003;125:14692–14693. doi: 10.1021/ja037687i. [DOI] [PubMed] [Google Scholar]; (d) Stocking E, Sanz-Cervera JF, Unkefer CJ, Williams RM. Tetrahedron. 2001;57:5303–5320. [Google Scholar]
- 14.Both enantiomers of stephacidin A and notoamide B have recently been isolated from distinct Aspergillus spp., see: Greshock TJ, Grubbs AW, Jiao P, Wicklow DT, Gloer JB, Williams RM. Angew. Chem. Int. Ed. 2008;47:3573–3577. doi: 10.1002/anie.200800106.
- 15.Ding Y, Miller KA, Greshock TJ, Sherman DH, Williams RM. (unpublished results) [Google Scholar]
- 16.(a) Hayasaka Y, Wilkinson KL, Elsey GM, Raunkjaer M, Sefton MA. J. Agric. Food Chem. 2007;55:9195–9201. doi: 10.1021/jf072171u. [DOI] [PubMed] [Google Scholar]; (b) Koh HL, Wang H, Zhou S, Chan E, Woo SO. J. Pharm. Biomed. Anal. 2006;40:653–661. doi: 10.1016/j.jpba.2005.08.001. [DOI] [PubMed] [Google Scholar]; (c) Ding B, Zhou T, Fan G, Hong Z, Wu Y. J. Pharm. Biomed. Anal. 2007;45:219–226. doi: 10.1016/j.jpba.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen KF, Sumarah MW, Frisvad JC, Miller JD. J. Agric. Food Chem. 2006;54:3756–3763. doi: 10.1021/jf060114f. [DOI] [PubMed] [Google Scholar]
- 18.(a) Williams RM, Cao J, Tsujishima H, Cox RJ. J. Am. Chem. Soc. 2003;125:12172–12178. doi: 10.1021/ja036713+. [DOI] [PubMed] [Google Scholar]; (b) Cushing TD, Sanz-Cervera JF, Williams RM. J. Am. Chem. Soc. 1993;115:9323–9324. [Google Scholar]; (c) Domingo LR, Zaragozá RJ, Williams RM. J. Org. Chem. 2003;68:2895–2902. doi: 10.1021/jo020564g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.