Abstract
Common variants in the region of the UMOD gene, which encodes uromodulin (Tamm-Horsfall protein), associate with chronic kidney disease (CKD) and estimated GFR (eGFR). Whether uromodulin levels associate with UMOD variants or with the risk for developing CKD is unknown. We conducted an age- and gender-matched case-control study (n = 200) of incident CKD (eGFR <60 ml/min per 1.73 m2) within the Framingham Heart Study (FHS). Baseline urinary uromodulin concentrations were related to case-control status 9.9 yr later and to genotype at rs4293393. As a replication set, we tested the genotype association with uromodulin concentration in the Atherosclerosis Risk in Communities (ARIC) Study (n = 42). Geometric means of uromodulin concentrations were 51% higher in case than in control subjects (P = 0.016). The adjusted odds ratio of CKD per 1-SD higher concentration of uromodulin was 1.72 (95% confidence interval 1.07 to 2.77; P = 0.03) after accounting for CKD risk factors and baseline eGFR. We observed lower urinary uromodulin concentrations per each copy of the C allele at rs4293393 in both cohorts. In summary, elevated uromodulin concentrations precede the onset of CKD and associate with a common polymorphism in the UMOD region.
Chronic kidney disease (CKD) constitutes a serious public health burden worldwide.1 CKD prevalence increases with age and affects >10% of the US adult population2; estimates from the United Kingdom3 and other European and Asian study populations are similar.1 CKD can progress to ESRD that requires dialysis or transplantation and affects approximately 2 million individuals worldwide.1 In addition, CKD causes substantial morbidity and is an independent risk factor for cardiovascular disease4 and overall mortality.5
In addition to major known risk factors for CKD—hypertension and diabetes—multiple studies have provided evidence for a genetic component to kidney disease.6 Identification of genetic risk variants for CKD may provide novel insights into underlying biologic mechanisms and into CKD pathophysiology. We therefore recently conducted genome-wide association studies of CKD and estimated GFR (eGFR), a measure of kidney function, in 19,877 participants from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium. We identified common genetic variants in the UMOD gene, including rs4293393, of minor allele frequency 0.18, in association with CKD and eGFR.7
Rare mutations in UMOD have previously been described as a cause of monogenetic forms of kidney disease.8 The UMOD gene is exclusively transcribed in the kidney,9 and its protein product is the Tamm-Horsfall protein (THP),10 also known as uromodulin.11 Although the protein was isolated from urine almost 60 yr ago and is the most abundant urinary protein in healthy individuals, the physiologic role of the protein remains unclear,9 and its association with non-Mendelian, complex forms of CKD warrants further exploration.
Our study had two objectives: First, to address whether urinary uromodulin concentrations are associated with incident CKD in a community-based study, and, second, to determine whether a single-nucleotide polymorphism (SNP) in the UMOD region, rs4293393, is associated with urinary uromodulin concentrations. We asked these questions in a nested case-control study of incident CKD in the Framingham Heart Study (FHS). The association between genotype at rs4293393 and urinary uromodulin concentrations was examined in the FHS sample and confirmed in a subsample of Atherosclerosis Risk in Communities (ARIC) Study participants selected to represent equally the three genotypes at rs4293393.
Results
Baseline Study Characteristics
By design, the mean age of the FHS case and control subjects was similar (63.8 yr; Table 1), and 60% of both groups were women. Uromodulin concentrations ranged from 0.2 to 70.9 μg/ml in case patients and 0.2 to 49.9 μg/ml in control subjects. The overall geometric mean uromodulin concentration was 4.5 μg/ml; mean values were similar in women (5.1 μg/ml) and men (3.8 μg/ml; P = 0.11). Log uromodulin concentrations were not associated with CKD risk factors in FHS (Table 2), with the exception of a negative correlation with fasting glucose levels (P = 0.05) and a positive correlation with log urinary albumin-to-creatinine ratio (UACR; P = 0.008).
Table 1.
Baseline study sample characteristics by case-control status (FHS) and overall (ARIC)
| Characteristic | FHS |
ARIC (n = 42) | |||
|---|---|---|---|---|---|
| Control Subjects (n = 100) |
Case Patients (n = 100) |
||||
| Baseline | Follow-up | Baseline | Follow-up | ||
| Age (yr; mean ± SD) | 63.8 ± 7.3 | 74.0 ± 7.0 | 63.8 ± 7.2 | 73.4 ± 7.4 | 68.8 ± 3.1 |
| Women (%) | 60 | 60 | 60 | 60 | 0 |
| BMI (kg/m2; mean ± SD) | 27.2 ± 4.3 | 27.3 ± 5.1 | 27.9 ± 5.3 | 28.3 ± 5.4 | 29.0 ± 4.1 |
| SBP (mmHg; mean ± SD) | 127 ± 18 | 135 ± 17 | 135 ± 20 | 129 ± 18 | 134 ± 22 |
| HDL cholesterol (mg/dl; mean ± SD) | 55 ± 17 | 60 ± 18 | 51 ± 17 | 55 ± 19 | 43 ± 17 |
| Diabetes (%) | 5 | 15 | 11 | 22 | 19 |
| Smoking (%) | 10 | 7 | 11 | 3 | 7 |
| Hypertension treatment (%) | 23 | 49 | 50 | 77 | 55 |
| Hypertension (%) | 48 | 61 | 74 | 81 | 52 |
| eGFR baseline (ml/min per 1.73 m2; mean ± SD) | 98.0 ± 54.0 | 81.0 ± 12.5 | 80.0 ± 22.0 | 50.0 ± 6.5 | 79.0 ± 15.5 |
| UACR (median [25th, 75th percentiles]) | 4.8 (2.2, 11.2) | 8.1 (4.2, 16.0) | 4.7 (2.3, 8.7) | 8.2 (3.6, 16.8) | 3.3 (1.4, 6.3) |
BMI, body mass index; SBP, systolic BP.
Table 2.
Correlations between baseline log uromodulin concentrations and selected CKD risk factors in 200 FHS participants
| Characteristic | Pearson Correlation | P |
|---|---|---|
| Age | 0.09 | 0.20 |
| BMI | −0.05 | 0.47 |
| SBP | 0.02 | 0.81 |
| Fasting glucose | −0.14 | 0.05 |
| HDL cholesterol | 0.06 | 0.41 |
| ln(UACR) | 0.19 | 0.008 |
All correlations are age-adjusted except for age. BMI, body mass index; SBP, systolic BP.
Uromodulin Levels in Case Patients and Control Subjects
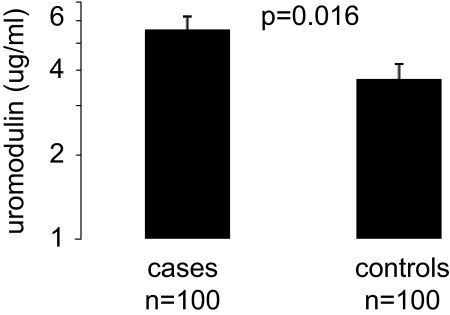
The median (quartile 1, quartile 3) of uromodulin concentrations was 6.12 μg/ml (3.40, 12.55) in case patients and 4.97 μg/ml (1.55, 9.12) in control subjects (Supplemental Figure 2). Baseline geometric mean concentrations of uromodulin were higher among case patients (5.58 μg/ml) than control subjects (3.71 μg/ml; paired t test P = 0.016; Figure 1). Per 1-SD increase in log uromodulin, the multivariable-adjusted odds ratio for incident CKD was 1.72 (95% confidence interval [CI] 1.07 to 2.77; P = 0.03; Table 3). Adding UACR to the model as an additional covariate did not materially change the results. When urinary uromodulin was indexed to urinary creatinine, similar results were observed (odds ratio 1.69; P = 0.04, fully adjusted model). Similar results were also observed in participants free of diabetes and in older and younger participants (Table 3).
Figure 1.
Geometric mean uromodulin concentrations in the case patients and control subjects at baseline are shown. Error bars represent SEs.
Table 3.
Association of uromodulin concentrations at baseline and incident CKD case/control status in the FHS
| Parameter | OR (95% CI) | P |
|---|---|---|
| MVa-adjusted model | ||
| uromodulin concentrations | 1.55 (1.06 to 2.26) | 0.02 |
| uromodulin-to-creatinine ratio | 1.69 (1.02 to 2.79) | 0.04 |
| MVa + baseline eGFR | 1.72 (1.07 to 2.77) | 0.03 |
| MVa + baseline eGFR + ln(UACR) | 1.72 (1.06 to 2.79) | 0.03 |
| Stratified analyses | ||
| free of diabetes (n = 184)a | 1.76 (1.07 to 2.89) | 0.03 |
| age below median (n = 88)a | 1.53 (0.69 to 3.36) | 0.29 |
| age above median (n = 112)a | 1.62 (0.75 to 3.48) | 0.22 |
Data were analyzed using conditional logistic regression and are presented per 1-SD increase of ln(uromodulin) levels. MV, multivariable; OR, odds ratio.
aAnalyses are adjusted for systolic BP, hypertension treatment, diabetes, HDL cholesterol, smoking, and body mass index with age and gender matched by design, with the exception of diabetes adjustment for the analyses in individuals who were free of diabetes.
SNP rs4293393 and Log Uromodulin Concentrations
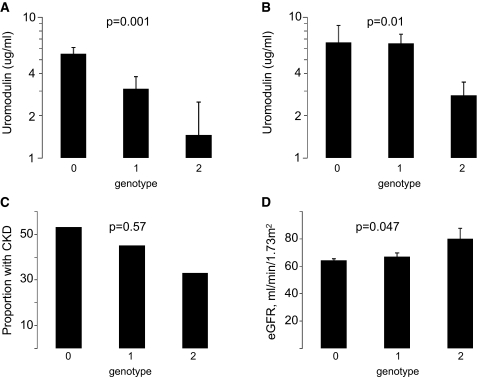
The study sample characteristics by genotype are shown in Supplemental Table 1. Genotype at rs4293393 was strongly associated with log uromodulin concentrations in FHS (P = 0.001; Figure 2): For each copy of the C allele at rs4293393, log uromodulin concentrations were significantly lower (geometric means 5.5, 3.1, and 1.5 μg/ml for zero, one, and two copies, respectively; P = 0.001), whereas eGFR was significantly higher, indicating better kidney function (64, 67, and 80 ml/min per 1.73 m2; P = 0.047; Figure 2).
Figure 2.
Distribution of uromodulin concentrations and kidney function measures in the FHS and ARIC samples by number of copies of the C allele at rs4293393 is shown. (A) Geometric mean uromodulin concentrations in FHS. (B) Geometric mean uromodulin concentrations in ARIC. (C) Proportion with CKD at FHS follow-up. (D) Mean eGFR at FHS follow-up. Genotype and phenotype information was available for 169 FHS participants: 121 had zero copies of the C allele at rs4293393, 42 had one copy, and six had two copies. Error bars represent SEs. P values in A, C, and D were obtained from regression models accounting for the matched design.
Replication in the ARIC Study
Characteristics of the ARIC sample are also listed in Table 1. Uromodulin concentrations measured by immunoassay ranged from 0.7 to 20.0 μg/ml, with an overall median (25th, 75th percentile) of 3.3 (1.4, 6.3) μg/ml. For zero, one, and two copies of the C allele at rs4293393, geometric mean uromodulin concentrations on the basis of the immunoassay were 6.6, 6.5, and 2.8 μg/ml (P = 0.01 for trend; Figure 2). A significant association between more copies of the C allele at rs4293393 and lower uromodulin concentrations in the ARIC samples were also observed on the basis of the gel-based methods to quantify uromodulin (P = 0.013 for trend; Supplemental Figure 3). The correlation between uromodulin concentrations in urine supernatant obtained by immunoassay and by gel electrophoresis was 0.79 (P < 0.0001). Gel-based quantification showed that the majority of uromodulin is present in urine supernatant, and the correlation of supernatant and total urine uromodulin was 0.96.
Discussion
In our nested case-control study of incident CKD, elevated urinary uromodulin concentrations preceded the development of CKD. Urinary uromodulin concentrations were associated with a CKD risk polymorphism in the UMOD gene region, rs4293393, indicating that a polymorphism in the UMOD gene may be one of the factors responsible for the association of altered uromodulin concentrations and incident CKD.
This study identified a novel disease marker through the follow-up of results from genome-wide association studies that were free of previous biologic hypotheses. Thus far, genome-wide association studies have mostly resulted in the establishment of statistical associations between genetic markers and disease rather than in translation of the findings into clinical settings. Despite the typically modest associations observed between common genetic risk variants and disease (in this case, the odds ratio of CKD was 0.76 per copy of minor protective C allele at rs4293393),7 we were able to identify a marker that was lower among individuals who remained free of CKD as compared with those who developed CKD after 10 yr of follow-up. The findings of this study highlight that even modest disease associations with SNPs can translate into novel insights about potential alternative phenotype measures and biomarkers. Such studies may also provide a basis for important and novel insights into underlying mechanisms of disease.
We consistently observed significant differences in urinary uromodulin concentrations by genotype at rs4293393 in two independent study samples, despite their modest sample sizes. Moreover, we observed these differences with two different laboratory methods to quantify uromodulin: Immunoassay and gel electrophoresis. It is therefore unlikely that the results from immunoassay quantification could be explained by differential antibody binding to potentially altered epitopes in carriers of the UMOD variant. In addition, the quantification by gel electrophoresis showed that both urine supernatant and urine pellet, separated by centrifugation, contain uromodulin with the majority present in supernatant. The high correlation of supernatant and total urine uromodulin suggests that its measurement using an immunoassay in supernatant is a valid approach to quantifying total urinary uromodulin.
The UMOD gene is exclusively transcribed in the cells lining the thick ascending limb of the loop of Henle and the early distal convoluted tubule.9 From here, uromodulin is cleaved into urine, where it can be found free or as an aggregate. Previous studies of individuals with rare monogenetic UMOD-related disease found substantially lower amounts of urinary uromodulin in patients compared with control subjects.12–14 This may result from the suppression of wild-type uromodulin excretion by the harmful intracellular effects of mutant uromodulin.14 In small, patient-oriented studies, lower concentrations of urinary uromodulin have also been observed in individuals with other forms of advanced kidney disease,15 and reduced urinary uromodulin concentrations have therefore been considered a marker of distal tubular cell damage.9
Conversely, in our study, we observed higher concentrations of urinary uromodulin preceding the development of CKD. Moreover, we observed an association of the protective C allele at SNP rs4293393 with lower uromodulin concentrations, highlighting the internal consistency of our findings. In our previous work, we reported an association between the rs4293393 C allele, along with seven other correlated SNPs in and upstream of UMOD, as associated with less CKD and higher eGFR.7 A possible explanation for the discrepancy of our findings compared with patients with monogenetic UMOD disease is that different mutations in the UMOD gene may have different effects on the gene product, resulting in differential changes in urinary uromodulin concentrations. Evidence from monogenetic disease suggests that these mutations cause protein-processing defects, which may result in a harmful effect of mutated uromodulin retained in the kidney.16–18 Higher amounts of urinary uromodulin, as we observed, could be due to increased transcription, faster protein maturation, less efficient binding of its anchor, or increased cleavage into urine. The association of higher urinary uromodulin concentrations with incident CKD may indicate that higher concentrations of uromodulin in urine are harmful or that wild-type uromodulin has a protective function inside the cell before its secretion. Alternatively, the genetic variant may result in altered glycosylation of uromodulin, which may affect some of the previously described functions of the protein, such as a role in innate immunity.19
The physiologic function of uromodulin and the biologic mechanism of the association we observed require further study. Regardless of the causal mechanism, however, individuals who developed CKD during 10 yr of follow-up had 50% higher geometric mean uromodulin concentrations in the urine at baseline. The most commonly used marker to estimate kidney function, serum creatinine, is elevated only in advanced stages of impaired renal function. Novel biomarkers of CKD for early identification of at-risk individuals are therefore needed. This study can serve as a proof-of-principle follow-up study from genome-wide association, but further studies are needed to determine whether altered uromodulin concentrations in urine could represent such a novel CKD biomarker. In particular, uromodulin should be measured in large population-based samples to determine whether it improves CKD risk prediction among individuals free of CKD at baseline, as well as its distribution and association with CKD in unselected samples. Moreover, further studies are needed to determine whether the uromodulin pathway represents a potential target for interventions aimed at reducing CKD and CKD progression.
Limitations of our study pertain to the small sample size of 200 and 42 individuals, respectively; however, our preliminary findings can serve as proof-of-principle of how genome-wide association studies can aid in the identification of markers for further evaluation as an alternative marker of the phenotype and also how the polymorphism we previously identified in the UMOD gene may contribute to the pathogenesis of CKD. Our participants were white of European descent and middle-aged to elderly; the generalizability of the data to younger individuals or other ethnicities/races is unknown, and further studies in larger, population-based cohorts are therefore necessary. We excluded individuals with microalbuminuria, further limiting the generalizability of our findings. We were not able to test formally the magnitude of the association between uromodulin concentrations and CKD after adjusting for rs4293393, because of the modest sample size in our study. In addition, the ARIC findings did not follow the additive pattern observed in our previous genome-wide association analysis7 and the FHS results in this article, likely because of the small sample size or the selected nature of the sample. Replication of our findings and additional investigation in large population-based as well as disease-specific studies is warranted. Future studies should also seek to address the potential impact of additional factors on urinary uromodulin concentrations, such as intraindividual variation,20 as well as a potential effect of pH and urine concentration.15,21,22
In summary, the presence of elevated uromodulin levels preceded the development of CKD in our study, and uromodulin levels were associated with a common SNP in the UMOD gene region in two independent study samples. Our findings identify a potentially useful target for diagnostic testing and therapeutic intervention.
Concise Methods
Study Samples
Framingham Heart Study.
Participants for this study were drawn from the Framingham Offspring Study, which began in 1971 with the enrollment of 5124 men and women. Offspring cohort members participated in examinations approximately every 4 yr; the design and method have been described elsewhere.23 This investigation includes Offspring cohort participants examined during the sixth examination cycle (1995 through 1998). This cycle was chosen because of the contemporaneous measurement of both serum creatinine and urinary albumin excretion and the 10-yr follow-up window. The study was approved by the institutional review boards of the Boston University Medical Center, and all participants provided written informed consent.
Among the 2966 participants with both serum creatinine measurements and available urine, we selected 200 individuals for a nested case-control study of incident CKD. All participants were free of CKD at baseline (defined as eGFR ≥60 ml/min per 1.73 m2 and UACR <25 mg/g in women and 17 mg/g in men, according to national guidelines).24 Case patients were selected as the 100 individuals with the lowest eGFR at the eighth examination cycle (2005 through 2008). Control subjects were required to have a follow-up eGFR ≥60 ml/min per 1.73 m2 and were matched to the case patients on age (±1 yr) and gender.
Atherosclerosis Risk in Communities.
The ARIC Study is a population-based, prospective, multicenter study of 15,792 men and women. Participants were aged 45 to 64 yr at enrollment from 1987 through 1989 and attended four study visits approximately every 3 yr. The study has been described in detail previously.25 Participants for this study were selected at study visit 4 (1996 through 1998), when urine samples were obtained. Written informed consent was obtained, and the institutional review boards of each study center approved the study protocols.
Individuals for urinary uromodulin measurements were selected among 1345 self-reported white men who were above the median age at visit 4 and did not have microalbuminuria, to reduce variability in urine composition as a result of differences in gender and age, and had available information on hypertension and diabetes status. From these, 15 individuals were randomly selected within each genotype category (among 893, 405, and 47 individuals with genotypes 0 [no minor C alleles (TT)], 1 [one minor C allele (CT)], and 2 [two minor C alleles (CC)]). The final study sample consisted of 42 individuals with sufficient amounts of stored urine (n = 13, 15, and 14 for genotypes 0, 1, and 2, respectively).
Kidney Function Measurements and Definition of CKD
Kidney function was evaluated as GFR (eGFR) estimated from the simplified Modification of Diet in Renal Disease (MDRD) Study Equation.26 CKD was defined as eGFR <60 ml/min per 1.73 m2 on the basis of the National Kidney Foundation Kidneys Disease Outcomes Quality Initiative (KDOQI) working group.24 Serum creatinine was measured using a modified Jaffe method; because variations in the quantification of serum creatinine can occur, we calibrated serum creatinine as described previously.27 Briefly, mean serum creatinine values were aligned with those of different gender and age groups (20 to 39, 40 to 59, 60 to 69, and ≥70 yr) among white participants of the nationally representative Third National Health and Nutrition Examination Survey (NHANES III). In the NHANES III, serum creatinine values had been calibrated to the Cleveland Clinic Laboratory, where the MDRD Study equation was developed.28
In FHS, spot urine samples collected at the sixth examination were stored at −20°C. Using a modified Jaffe method, urinary creatinine concentration was measured. The urine albumin concentration was measured using immunoturbimetry (Tina-quant Albumin assay; Roche Diagnostics, Indianapolis, IN). Microalbuminuria was defined as UACR of at least 25 mg/g in women and 17 mg/g in men. In ARIC, urine samples were collected at the fourth study visit, frozen within 12 h, and stored at −70°C. Urinary albumin levels were measured using a nephelometric method on either the Dade Behring BN100 or the Beckman Image Nephelometer, and urinary creatinine levels were measured using the Jaffe method.
Uromodulin Measurement
Urinary uromodulin concentrations in both FHS and ARIC were measured via immunoassay using a bead Luminex platform (Rules Based Medicine, Austin, TX) after centrifugation at 4000 rpm for 5 min. For this assay, the interassay coefficient of variation is 11.4% at a mean concentration of 9.4 μg/ml and 5.0% at a mean concentration of 37 μg/ml. Seven participants in FHS and one in ARIC had uromodulin concentrations below the limits of detection; their values were set to the lowest value in the data set in both studies. In ARIC, all samples were measured in duplicate. The intra-assay coefficient of variation was 7.1%, and the pair-wise correlation between the two measurements was 0.996 (P < 0.0001).
In ARIC, urinary uromodulin concentrations were also quantified using gel electrophoresis, and uromodulin was confirmed by a THP-specific antibody. First, 1 ml of urine was centrifuged at 3000 rpm for 10 min to separate supernatant and pellet. The pellet was resuspended in 0.5% Triton X-100 and 20 mM EDTA (pH 7.5; 500 μl) followed by a 1:1 (vol:vol) dilution with 2× concentrated SDS sample buffer (Invitrogen). Proteins were resolved by SDS-PAGE (10% or 4 to 12% NuPage Bis-Tris gel; Invitrogen; 1.0 mm × 8 cm × 8 cm) including a standard curve of chromatographically purified human THP standard (Biotechmedical Technologies, Stoughton MA). Samples were placed in random order. Gels were stained with silver and scanned, and the gel bands were quantified using Progenesis (Nonlinear Dynamics, Durham, NC) as described previously.29 Uromodulin concentrations were obtained from these raw values by fitting a centered quadratic model of the logged raw uromodulin values: Y = β0 + β1(log(x) − 2) + β2(log(x) − 2)^2, where Y was divided by 1,000,000. In addition, gels were transferred to nitrocellulose, probed with anti-THP polyclonal antibody (Biotechmedical Technologies; 1:5000), and detected using horseradish peroxidase goat anti-rabbit from Santa Cruz Biotechnology (Santa Cruz, CA) and ECL Western Blotting Detection Systems (GE Healthcare, Pittsburgh, PA).30 An example of uromodulin quantification using gel-based methods is shown in Supplemental Figure 1. Western blots showed that there were few or no degradation products.
CKD Covariate Assessment
In both FHS and ARIC, participants underwent blood testing and were assessed for CKD risk factors. Type 2 diabetes was defined as fasting blood glucose ≥126 mg/dl (7.0 mmol/L) at the examination or the use of insulin and/or oral hypoglycemic treatment. Hypertension was defined as a systolic BP ≥140 mmHg or a diastolic BP ≥90 mmHg on the basis of the mean of two readings obtained by an examining physician or receiving medication for treatment of hypertension. HDL cholesterol was measured from fasting morning samples. Current smoking status was defined as smoking at least one cigarette per day during the year before the examination. Body mass index was defined as the weight (in kilograms) divided by the height (in meters squared).
Genotyping
In FHS, genotyping of rs4293393 was performed in 3294 participants using an ABI PRISM 7900HT Sequence Detection System and TaqMan SNP Genotyping Assays developed by ABI. The instrument is regularly maintained by Applied Biosystems service engineers. A total of 148 samples were run in duplicate, with a 100% concordance rate. The call rate was 94.4%, and Hardy-Weinberg equilibrium was maintained (P = 0.89). Of the 200 participants with measured uromodulin levels, 169 had genotype information available. Individuals with available genotypes were slightly older than those without (64 versus 62 yr; P = 0.04), but there was no difference in ln(uromodulin) concentrations (P = 0.17), eGFR (P = 0.11), or CKD (P = 0.84).
In ARIC, participants were genotyped for whole-genome association studies using the Affymetrix 6.0 array. Genotype at rs4293393 was imputed on the basis of information from 602,642 high-quality genotyped SNPs as described previously.7 Briefly, imputation methods combine genotype data from each sample with a reference sample (phased HapMap CEU haplotypes, release 21) and then infer genotypes at untyped polymorphisms probabilistically. The average of the observed (i.e., imputed) to expected variance ratio of the true underlying genotypes at rs4293393 was 0.996, indicating excellent quality of imputation. The SNP rs4293393 was a perfect proxy (r2 = 1 in HapMap CEU) of the previously highlighted SNP rs12917707, and its association with CKD in our recently published genome-wide association study of CKD was of comparable magnitude and significance.7
Statistical Analyses
In both FHS and ARIC, the distribution of basic demographic variables among case patients and control subjects was assessed as mean (SD) for continuous and % (n) for categorical variables. Uromodulin concentrations were skewed and therefore natural log-transformed to improve normality. The geometric mean (exponentiated mean of the log-transformed values) was used to present results on the original scale (μg/ml). In FHS, a paired t test was used on the basis of the age- and gender-matched pairs to assess whether there was a difference in log uromodulin concentrations in case patients as compared with control subjects at baseline. Conditional logistic regression was used to perform multivariable regression and to adjust further for systolic BP, hypertension treatment, diabetes, HDL, smoking, body mass index, and baseline eGFR. Covariates were selected as those predictive of incident CKD on the basis of previous work.27 Furthermore, PROC GENMOD was used to assess whether the SNP rs4293393 was associated with log uromodulin concentrations and eGFR; an additive genetic model was used. Conditional logistic regression was used to assess whether SNP rs4293393 was associated with CKD status at follow-up. In ARIC, the association of genotype at rs4293393 and log uromodulin concentrations was assessed using linear regression and an additive genetic model. A type I error threshold of 0.05 was used to indicate statistical significance. All statistical analyses were performed using SAS 9.1 (SAS Institute, Cary, NC) in FHS and Stata 10.1 (Stat Corp, College Station, TX) and R 2.7.1. in ARIC.
Disclosures
None.
Supplementary Material
Acknowledgments
This work was supported by the National Heart, Lung, and Blood Institute's Framingham Heart Study (N01-HC-25195), the Center for Population Studies, National Heart, Lung, and Blood Institute intramural program, N01-HC-55020, R01-DK076770-01, R01-HL087652, 5UL1 RR025005-3, 1U54RR023561-01A1, NIH N01-HV-28180, AG028321, and HL092577-01A1.
We acknowledge the expert technical assistance of Yaping Wang. We thank the staff and participants of the Framingham Heart Study and the ARIC Study for important contributions.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Uromodulin and Translational Medicine: Will the SNPs Bring Zip to Clinical Practice?” on pages 204–206.
Supplemental information for this article is available online at http://www.jasn.org/.
References
- 1.Zhang QL, Rothenbacher D: Prevalence of chronic kidney disease in population-based studies: systematic review. BMC Public Health 8: 117, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, Eggers P, Van Lente F, Levey AS: Prevalence of chronic kidney disease in the United States. JAMA 298: 2038–2047, 2007 [DOI] [PubMed] [Google Scholar]
- 3.de Lusignan S, Chan T, Stevens P, O'Donoghue D, Hague N, Dzregah B, Van Vlymen J, Walker M, Hilton S: Identifying patients with chronic kidney disease from general practice computer records. Fam Pract 22: 234–241, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Sarnak MJ, Levey AS, Schoolwerth AC, Coresh J, Culleton B, Hamm LL, McCullough PA, Kasiske BL, Kelepouris E, Klag MJ, Parfrey P, Pfeffer M, Raij L, Spinosa DJ, Wilson PW; American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention: Kidney disease as a risk factor for development of cardiovascular disease: A statement from the American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention. Hypertension 42: 1050–1065, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Weiner DE, Tighiouart H, Amin MG, Stark PC, MacLeod B, Griffith JL, Salem DN, Levey AS, Sarnak MJ: Chronic kidney disease as a risk factor for cardiovascular disease and all-cause mortality: A pooled analysis of community-based studies. J Am Soc Nephrol 15: 1307–1315, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Satko SG, Sedor JR, Iyengar SK, Freedman BI: Familial clustering of chronic kidney disease. Semin Dial 20: 229–236, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Köttgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, Smith AV, Arking DE, Astor BC, Boerwinkle E, Ehret GB, Ruczinski I, Scharpf RB, Ida Chen YD, de Boer IH, Haritunians T, Lumley T, Sarnak M, Siscovick D, Benjamin EJ, Levy D, Upadhyay A, Aulchenko YS, Hofman A, Rivadeneira F, Uitterlinden AG, van Duijn CM, Chasman DI, Paré G, Ridker PM, Kao WH, Witteman JC, Coresh J, Shlipak MG, Fox CS: Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 41: 712–717, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart TC, Gorry MC, Hart PS, Woodard AS, Shihabi Z, Sandhu J, Shirts B, Xu L, Zhu H, Barmada MM, Bleyer AJ: Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J Med Genet 39: 882–892, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serafini-Cessi F, Malagolini N, Cavallone D: Tamm-Horsfall glycoprotein: Biology and clinical relevance. Am J Kidney Dis 42: 658–676, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Tamm I, Horsfall FL, Jr: Characterization and separation of an inhibitor of viral hemagglutination present in urine. Proc Soc Exp Biol Med 74: 106–108, 1950 [PubMed] [Google Scholar]
- 11.Muchmore AV, Decker JM: Uromodulin: A unique 85-kilodalton immunosuppressive glycoprotein isolated from urine of pregnant women. Science 229: 479–481, 1985 [DOI] [PubMed] [Google Scholar]
- 12.Rampoldi L, Caridi G, Santon D, Boaretto F, Bernascone I, Lamorte G, Tardanico R, Dagnino M, Colussi G, Scolari F, Ghiggeri GM, Amoroso A, Casari G: Allelism of MCKD, FJHN and GCKD caused by impairment of uromodulin export dynamics. Hum Mol Genet 12: 3369–3384, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Dahan K, Devuyst O, Smaers M, Vertommen D, Loute G, Poux JM, Viron B, Jacquot C, Gagnadoux MF, Chauveau D, Büchler M, Cochat P, Cosyns JP, Mougenot B, Rider MH, Antignac C, Verellen-Dumoulin C, Pirson Y: A cluster of mutations in the UMOD gene causes familial juvenile hyperuricemic nephropathy with abnormal expression of uromodulin. J Am Soc Nephrol 14: 2883–2893, 2003 [DOI] [PubMed] [Google Scholar]
- 14.Bleyer AJ, Hart TC, Shihabi Z, Robins V, Hoyer JR: Mutations in the uromodulin gene decrease urinary excretion of Tamm-Horsfall protein. Kidney Int 66: 974–977, 2004 [DOI] [PubMed] [Google Scholar]
- 15.Thornley C, Dawnay A, Cattell WR: Human Tamm-Horsfall glycoprotein: Urinary and plasma levels in normal subjects and patients with renal disease determined by a fully validated radioimmunoassay. Clin Sci (Lond) 68: 529–535, 1985 [DOI] [PubMed] [Google Scholar]
- 16.Bernascone I, Vavassori S, Di Pentima A, Santambrogio S, Lamorte G, Amoroso A, Scolari F, Ghiggeri GM, Casari G, Polishchuk R, Rampoldi L: Defective intracellular trafficking of uromodulin mutant isoforms. Traffic 7: 1567–1579, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Vylet'al P, Kublová M, Kalbácová M, Hodanová K, Baresová V, Stibůrková B, Sikora J, Hůlková H, Zivný J, Majewski J, Simmonds A, Fryns JP, Venkat-Raman G, Elleder M, Kmoch S: Alterations of uromodulin biology: A common denominator of the genetically heterogeneous FJHN/MCKD syndrome. Kidney Int 70: 1155–1169, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Williams SE, Reed AA, Galvanovskis J, Antignac C, Goodship T, Karet FE, Kotanko P, Lhotta K, Morinière V, Williams P, Wong W, Rorsman P, Thakker RV: Uromodulin mutations causing familial juvenile hyperuricaemic nephropathy lead to protein maturation defects and retention in the endoplasmic reticulum. Hum Mol Genet 18: 2963–2974, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su SJ, Chang KL, Lin TM, Huang YH, Yeh TM: Uromodulin and Tamm-Horsfall protein induce human monocytes to secrete TNF and express tissue factor. J Immunol 158: 3449–3456, 1997 [PubMed] [Google Scholar]
- 20.Kobayashi K, Fukuoka S: Conditions for solubilization of Tamm-Horsfall protein/uromodulin in human urine and establishment of a sensitive and accurate enzyme-linked immunosorbent assay (ELISA) method. Arch Biochem Biophys 388: 113–120, 2001 [DOI] [PubMed] [Google Scholar]
- 21.Lynn KL, Shenkin A, Marshall RD: Factors affecting excretion of human urinary Tamm-Horsfall glycoprotein. Clin Sci (Lond) 62: 21–26, 1982 [DOI] [PubMed] [Google Scholar]
- 22.Dawnay AB, Thornley C, Cattell WR: An improved radioimmunoassay for urinary Tamm-Horsfall glycoprotein: Investigation and resolution of factors affecting its quantification. Biochem J 206: 461–465, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP: An investigation of coronary heart disease in families. The Framingham Offspring Study. Am J Epidemiol 110: 281–290, 1979 [DOI] [PubMed] [Google Scholar]
- 24.National Kidney Foundation: K/DOQI clinical practice guidelines for chronic kidney disease: Evaluation, classification, and stratification. Am J Kidney Dis 39: S1–S266, 2002 [PubMed] [Google Scholar]
- 25.The Atherosclerosis Risk in Communities (ARIC) Study: Design and objectives. The ARIC investigators. Am J Epidemiol 129: 687–702, 1989 [PubMed] [Google Scholar]
- 26.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D: A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med 130: 461–470, 1999 [DOI] [PubMed] [Google Scholar]
- 27.Fox CS, Larson MG, Leip EP, Culleton B, Wilson PW, Levy D: Predictors of new-onset kidney disease in a community-based population. JAMA 291: 844–850, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Coresh J, Astor BC, McQuillan G, Kusek J, Greene T, Van Lente F, Levey AS: Calibration and random variation of the serum creatinine assay as critical elements of using equations to estimate glomerular filtration rate. Am J Kidney Dis 39: 920–929, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Fu Q, Garnham CP, Elliott ST, Bovenkamp DE, Van Eyk JE: A robust, streamlined, and reproducible method for proteomic analysis of serum by delipidation, albumin and IgG depletion, and two-dimensional gel electrophoresis. Proteomics 5: 2656–2664, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Matt P, Fu Z, Carrel T, Huso DL, Dirnhofer S, Lefkovits I, Zerkowski HR, Van Eyk JE: Proteomic alterations in heat shock protein 27 and identification of phosphoproteins in ascending aortic aneurysm associated with bicuspid and tricuspid aortic valve. J Mol Cell Cardiol 43: 792–801, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.