Abstract
Protein phosphorylation is an important component of vasopressin signaling in the renal collecting duct, but the database of known phosphoproteins is incomplete. We used tandem mass spectrometry to identify vasopressin-regulated phosphorylation events in isolated rat inner medullary collecting duct (IMCD) suspensions. Using multiple search algorithms to identify the phosphopeptides from spectral data, we expanded the size of the existing collecting duct phosphoproteome database from 367 to 1187 entries. Label-free quantification in vasopressin- and vehicle-treated samples detected a significant change in the phosphorylation of 29 of 530 quantified phosphopeptides. The targets include important structural, regulatory, and transporter proteins. The vasopressin-regulated sites included two known sites (Ser-486 and Ser-499) present in the urea channel UT-A1 and one previously unknown site (Ser-84) on vasopressin-sensitive urea channels UT-A1 and UT-A3. In vitro assays using synthetic peptides showed that purified protein kinase A (PKA) could phosphorylate all three sites, and immunoblotting confirmed the PKA dependence of Ser-84 and Ser-486 phosphorylation. These results expand the known list of collecting duct phosphoproteins and highlight the utility of targeted phosphoproteomic approaches.
Vasopressin plays a central role in collecting duct physiology. Signaling through the V2 receptor results in an increase of cAMP levels and causes activation of protein kinase A (PKA).1,2 In addition, over 200 other serine/threonine protein kinases are expressed in native collecting duct cells,3 and some of these have been shown to play important roles in the response to vasopressin.4–10 Vasopressin signaling is important not only for regulation of water transport through aquaporins11 but also for regulation of urea12 and sodium transport.13,14 Vasopressin also regulates long-term gene expression of collecting duct proteins, such as aquaporins.15,16 Because protein phosphorylation plays a central role in vasopressin signaling, the identification and quantification of phosphorylated proteins in response to vasopressin are essential to understanding the mechanism of action of this hormone in collecting duct.
In a previous study,17 we used tandem mass spectrometry (LC-MS/MS)-based quantitative phosphoproteomics to partially annotate the phosphoproteome of rat inner medullary collecting duct (IMCD). We subsequently quantified the differential phosphorylation of four serine residues (Ser-256, Ser-261, Ser-264, and Ser-269) in the C-terminal tail of rat aquaporin-2 (AQP2) in response to short-term exposure to the vasopressin analog dDAVP.17,18 We also found a number of phosphorylation sites on the vasopressin-sensitive urea channel, UT-A.17 However, because of the limited sensitivity of the experimental approach, we were unable to quantify changes in phosphorylation at these sites in UT-A, despite evidence for such sites on the basis of previous studies.19,20
One of the primary aims of this study was to increase the sensitivity of our MS-based workflow to annotate a larger portion of the IMCD phosphoproteome. This was accomplished in three ways: (1) by implementing an effective, chromatography-based stratification technique for our peptide samples, (2) by using a higher resolution mass spectrometer, and (3) by using multiple proteomic search algorithms to process the MS data. Using these combined approaches, we increased coverage of the IMCD phosphoproteome by approximately 3-fold compared with the previous study.17 In addition, we present large-scale phosphoproteomic data quantifying the effect of vasopressin on phosphorylation of IMCD proteins. Last, we identify and quantify six phosphorylation sites on the vasopressin-sensitive urea channel (isoforms A1 and A3) and demonstrate that three of these sites undergo large increases in phosphorylation in response to short-term dDAVP treatment.
Results
Phosphoproteomic Profiling of IMCD
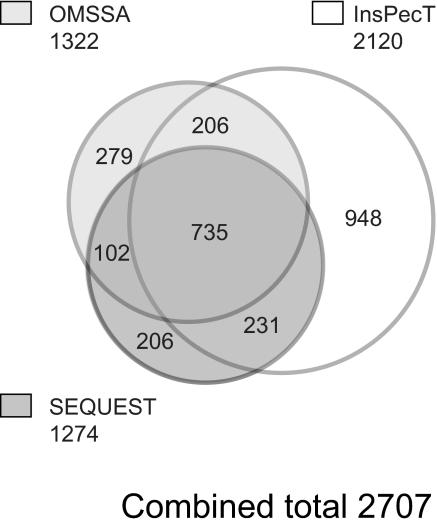
Our previous study using an LC-MS/MS “shotgun” approach identified 223 unique phosphoproteins in native IMCD cells.17 One of the aims of this study was to expand the size of the identified collecting duct phosphoproteome by adding methods that would increase overall sensitivity; viz., using sample fractionation via strong cation exchange (SCX) and employing multiple MS search algorithms. SCX was performed on a single dDAVP-treated IMCD sample (Figure 1A) as described in “Concise Methods,” followed by phosphopeptide enrichment via immobilized metal affinity chromatography (IMAC). Samples representing 24 SCX fractions were analyzed on a Thermo LTQ mass spectrometer, and the resulting spectra were searched using three different search algorithms: SEQUEST, InsPecT, and OMSSA. Figure 2 shows a Venn diagram of unique phosphopeptides identified from each of the three search algorithms. All datasets were filtered for a <2% false-discovery rate on the basis of target-decoy analysis.21 Overlapping regions consist of phosphopeptides that were identified by more than one search algorithm and indicate a match not only of the phosphopeptide primary amino acid sequence but also for the exact site(s) of phosphorylation. A total of 735 phosphopeptides were identified by all three search algorithms (as shown by the central overlap region). InsPecT identified the largest number of unique phosphopeptides (2120), with the highest number of identifications that were not shared with either of the other two programs (948). Annotated phosphopeptide data from all three searches are accessible online at the Collecting Duct Phosphoprotein Database (http://dir.nhlbi.nih.gov/papers/lkem/mpkccdprot/). This study increases the size of the Collecting Duct Phosphoprotein Database from 367 unique phosphoproteins to 1187 unique phosphoproteins.
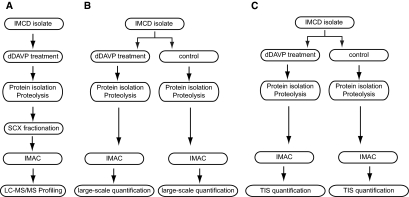
Figure 1.
The three different LC-MS/MS experiments performed are as follows: (A) The initial phosphoproteomic profiling experiment consisted of a single dDAVP-treated sample that was processed using SCX fractionation. (B) The second experiment was performed for large-scale quantification using a nonselective “profiling” mode. (C) The third experiment was performed for targeted quantification by selection of precursor ion m/z ratios, so-called targeted ion selection (TIS) mode.
Figure 2.
Venn diagram shows the number of unique phosphopeptide identifications that resulted from each of three search algorithms (SEQUEST, InsPecT, and OMSSA). A false-discovery rate stringency of <2% was used for each of the searches.
Verification of IMCD Response to dDAVP
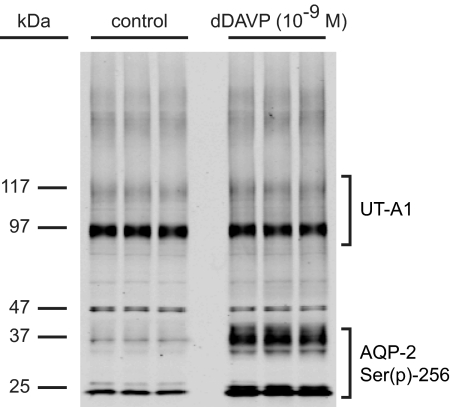
After profiling the dDAVP-treated IMCD phosphoproteome, we performed large-scale profiling experiments on paired vehicle and dDAVP-treated IMCD samples to determine which phosphoproteins were regulated by vasopressin. Phosphorylation on Ser-256 of AQP2 occurred in response to short-term vasopressin exposure,17 and immunoblotting with a phosphospecific antibody was used to verify the expected increase in Ser(p)-256 abundance in vasopressin-treated samples. Figure 3 shows that dDAVP increased the amount of Ser(p)-256-AQP2 compared with control samples (bands at 29 and 37 kDa). Total protein abundance of AQP2 does not change with dDAVP treatment, as shown by Hoffert et al.18
Figure 3.
Immunoblot showing that dDAVP increased phosphorylation of Ser-256 of AQP2. An immunoblot of IMCD protein isolate shows five pairs of control and dDAVP-treated samples. Bands at 29 and 37 kDa indicate Ser(p)-256 bands (nonglycosylated and glycosylated forms, respectively).
Large-Scale Quantitative Phosphoproteomics Reveals Potential Downstream Targets of Vasopressin Signaling
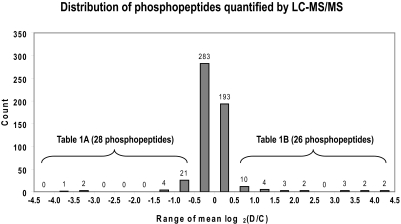
We isolated IMCD suspensions and prepared five paired control and dDAVP-treated samples all from different rats. Samples were digested with trypsin; phosphopeptide enrichment was performed using IMAC; and the samples were analyzed on the LTQ-Orbitrap MS platform using collision-induced dissociation (CID) fragmentation (Figure 1B). All 10 samples were searched with SEQUEST (false-discovery rate <2%), and relative quantification was performed using QUOIL.22 A total of 530 unique phosphopeptides from 278 proteins were quantified (Figure 4). For statistical analysis, values were converted to log2 of the ratio (D/C), where D and C refer to the normalized areas of the extracted peptide ion chromatograms under dDAVP-treated and control conditions, respectively. Brackets in Figure 4 indicate the subset of 54 phosphopeptides with log2(D/C) values either greater than 0.58 or less than −0.58. Of these 54 phosphopeptides, 29 represented phosphopeptides with statistically significant (P < 0.05) changes in abundance between the five paired control and dDAVP-treated IMCD tubules. These phosphopeptides are shown in Table 1 and are marked with a superscript letter a. Phosphopeptides whose abundance did not change significantly are marked with a superscript letter c and were included as an internal negative control showing that the changes in phosphopeptide abundances found are most likely not due to changes in the amount of protein analyzed. Supplementary Table 1 is a complete list of phosphopeptides with mean log2(D/C) values between −0.58 and 0.58.
Figure 4.
Histogram showing distribution of the mean log2(D/C) for all peptides quantified using LC-MS/MS. The vast majority of peptides did not change with dDAVP treatment (476 (90.0%) of 530), having mean log2(D/C) values between −0.58 and 0.58.
Table 1.
List of phosphopeptides that show significant change in abundance or no significant change with dDAVP treatment compared with control samples
| No. | Protein Name | Accession No. | Sequence (Charge) | Mean D/C | Log2 (D/C) | Sites |
|---|---|---|---|---|---|---|
| Part A | ||||||
| 1 | aquaporin 2 (collecting duct) | NP_037041 | R.QSVELHS*PQSLPR.G(+2) | 0.06 | −3.95 ± 0.24a | S261 |
| 2 | aquaporin 2 (collecting duct) | NP_037041 | R.RQSVELHS*PQSLPR.G(+3) | 0.10 | −3.39 ± 0.13a | S261 |
| 3 | aquaporin 2 (collecting duct) | NP_037041 | R.QSVELHS*PQSLPR.G(+3) | 0.11 | −3.20 ± 0.15a | S261 |
| 4 | ArfGAP with FG repeats 1 | NP_001129068 | K.SLLGESAPALHLNKGT*PT*QSPVVGR.S(+3) | 0.34 | −1.55 ± 0.27a | T177,b T179b |
| 5 | septin 9 isoform 2 | NP_789826 | R.LVDTLSQRS*PKPSLR.R(+3) | 0.41 | −1.30 ± 0.08a | S67b |
| 6 | ArfGAP with FG repeats 1 | NP_001129068 | K.SLLGESAPALHLNKGT*PT-QS*PVVGR.S(+3) | 0.44 | −1.19 ± 0.21a | T177, S181 |
| 7 | ysv oncogene homolog isoform A | NP_110484 | R.TIYVRDPT*S*NKQQRPVP-ES*QLLPGQR.F(+3) | 0.50 | −1.01 ± 0.37c | T37,b S38,b S48b |
| 8 | (ABC1), member 1 | NP_835196 | K.HVKAEMEQMALDVGLP-PS*KLK.S(+3) | 0.50 | −1.00 ± 0.49c | S964 |
| 9 | tensin like SH2 domain containing 1 | XP_341257 | R.WDSY*ENMSADGEVLHT-QGPVDGSLYAK.V(+3) | 0.51 | −0.98 ± 1.12c | Y410b |
| 10 | hsp 90, alpha (cytosolic), class A member 1 | NP_786937 | R.DKEVS*DDEAEEKEEK.E(+3) | 0.52 | −0.93 ± 0.82c | S231 |
| 11 | calpastatin isoform a | NP_445747 | K.NEAITGPLPDS*PKPMGID-HAIDALSSDFTCS*SPTGK.Q(+3) | 0.55 | −0.86 ± 1.15c | S260,b S280b |
| 12 | calpastatin isoform a | NP_445747 | K.NEAITGPLPDS*PKPMGID-HAIDALSSDFTCSS*PTGK.Q(+3) | 0.55 | −0.86 ± 1.15c | S260,b S281b |
| 13 | calpastatin isoform a | NP_445747 | K.NEAITGPLPDS*PKPMGID-HAIDALSSDFTCSSPT*GK.Q(+3) | 0.56 | −0.84 ± 1.13c | S260,b T283b |
| 14 | tensin | XP_237286 | R.HPVGSHQVPGLHSGVVTT-PGS*PSLGR.H(+4) | 0.60 | −0.73 ± 0.43c | S1523b |
| 15 | tx elongation factor A (SII) 1 isoform 2 | XP_001059640 | K.KKEPAISSQNS*PEAR.E(+3) | 0.62 | −0.70 ± 0.77c | S100b |
| 16 | tight junction protein 2 | NP_446225 | R.SQEES*PVPQPR.T(+2) | 0.62 | −0.69 ± 0.09a | S463 |
| 17 | nuclear RNA export factor 3 | XP_001053711 | K.S*SNINS*ILELFPK.L(+2) | 0.62 | −0.69 ± 0.23a | S287,b S292b |
| 18 | PDLIM1 interacting kinase 1 like | NP_001101454 | R.LDTS*DLEPT*LKVADFGLS-KVCS*ASGQNPEEPVSVNK.C(+3) | 0.62 | −0.68 ± 0.86c | S180,b T185,b S198b |
| 19 | (urea transporter), member 2 isoform 2 | NP_808877 | K.LYESELSSPT*WPSSSQDTH-PALPLLEMPEEK.D(+3) | 0.63 | −0.67 ± 0.93c | T37b |
| 20 | epsin 3 | NP_001019962 | R.TPVLPSGPPITDPWAPSSPT*PK.L(+2) | 0.63 | −0.66 ± 0.57c | T361b |
| 21 | (urea transporter), member 2 isoform 2 | NP_808877 | K.DLRS*S*DEDSHIVK.I(+3) | 0.63 | −0.66 ± 0.59c | S62, S63 |
| 22 | tensin | XP_237286 | K.VSS*SPVANGMAS*PSGSST-VSFSHTLPDFSK.Y(+3) | 0.63 | −0.66 ± 0.43c | S1667,b S1676b |
| 23 | calnexin | NP_742005 | K.SDAEEDGGTGS*QDEEDSK-PK.A(+3) | 0.64 | −0.65 ± 0.59c | S563b |
| 24 | AHNAK nucleoprotein isoform 1 | XP_001078032 | K.LEGEIKVPDVDISS*PGVNVE-APDIHVK.A(+4) | 0.64 | −0.65 ± 0.19c | S1807b |
| 25 | prostaglandin E synthase 3 (cytosolic) | NP_001124461 | K.DWEDDS*DEDMSNFDR.F(+2) | 0.64 | −0.64 ± 1.10c | S113b |
| 26 | beta-2-syntrophin | XP_001071043 | R.GLGPPS*PPAPPR.G(+2) | 0.64 | −0.64 ± 0.37c | S120 |
| 27 | PC4 and SFRS1 interacting protein 1 | NP_786941 | K.NLAKPGVT*STSDS*EEDDD-QEGEK.K(+3) | 0.66 | −0.60 ± 1.07c | T269,b S274b |
| 28 | tight junction protein 2 | NP_446225 | R.KVQVAPLQGS*PPLSHDDR.G(+3) | 0.66 | −0.59 ± 0.24c | S107b |
| Part B | ||||||
| 1 | AHNAK nucleoprotein isoform 1 | XP_001078032 | R.NRS*NS*FSDEREFSAPST-PTGTLEFAGGEGK.G(+3) | 1.51 | 0.59 ± 0.25c | S2454,b S2456b |
| 2 | AHNAK nucleoprotein isoform 1 | XP_001078032 | R.NRSNS*FS*DEREFSAPSTP-TGTLEFAGGEGK.G(+3) | 1.51 | 0.59 ± 0.25c | S2456,b S2458b |
| 3 | leucine-rich repeat (in FLII) interacting protein 2 | NP_001019932 | R.RGSGDTSS*LIDPDTSLSE-LR.E(+3) | 1.55 | 0.63 ± 0.29c | S138b |
| 4 | aquaporin 2 (collecting duct) | NP_037041 | R.RRQS*VELHS*PQSLPR.G(+3) | 1.55 | 0.63 ± 0.06a | S256, S261 |
| 5 | tankyrase 1-binding protein of 182 kDa | XP_215763 | K.RAS*VSTNQNTDENDQELR.M(+3) | 1.67 | 0.74 ± 0.18a | S883b |
| 6 | aquaporin 2 (collecting duct) | NP_037041 | R.RQS*VELHS*PQSLPR.G(+2) | 1.73 | 0.79 ± 0.17a | S256, S261 |
| 7 | PCTAIRE protein kinase 3 | NP_001093976 | R.RFS*MEDLNKR.L(+3) | 1.77 | 0.82 ± 0.05a | S66b |
| 8 | AHNAK nucleoprotein isoform 1 | XP_001078032 | R.NRS*NS*FSDEREFSAPSTPT-GTLEFAGGEGK.G(+4) | 1.91 | 0.93 ± 0.41c | S2454,b S2456b |
| 9 | aquaporin 2 (collecting duct) | NP_037041 | R.RRQS*VELHS*PQSLPR.G(+2) | 1.93 | 0.95 ± 0.21a | S256, S261 |
| 10 | calcium regulated heat stable protein 1 | NP_690003 | R.DRS*PS*PLRGNVVPSPLPTR.R(+3) | 2.28 | 1.19 ± 0.27a | S30, S32 |
| 11 | desmoplakin isoform I isoform 2 | XP_225259 | R.SMS*FQGIR.Q(+2) | 2.28 | 1.19 ± 0.77c | S2216b |
| 12 | PDZ and LIM domain 5 | NP_445778 | R.RGS*QGDIKQQNGPPR.K(+3) | 2.64 | 1.40 ± 0.42a | S228 |
| 13 | beta-catenin | NP_445809 | R.RTS*MGGTQQQFVEGVR.M(+3) | 2.69 | 1.43 ± 0.04a | S552b |
| 14 | beta-catenin | NP_445809 | R.RTSMGGT*QQQFVEGVR.M(+3) | 2.69 | 1.43 ± 0.04a | T556b |
| 15 | PCTAIRE protein kinase 3 | NP_001093976 | R.RFS*MEDLNK.R(+2) | 3.05 | 1.61 ± 0.21a | S66 |
| 16 | beta-catenin | NP_445809 | R.RTS*MGGTQQQFVEGVR.M(+2) | 3.29 | 1.72 ± 0.19a | S552b |
| 17 | aquaporin 2 (collecting duct) | NP_037041 | R.RQS*VELHS*PQS*LPR.G(+3) | 3.92 | 1.97 ± 0.07a | S256, S261, S264 |
| 18 | aquaporin 2 (collecting duct) | NP_037041 | R.RRQS*VELHSPQS*LPR.G(+3) | 4.14 | 2.05 ± 0.13a | S256,b S264b |
| 19 | plexin domain containing 2 | NP_001101892 | R.RGS*GHPAYAEVEPVGEK-EGFIVSEQC.-(+3) | 4.38 | 2.13 ± 0.24a | S507 |
| 20 | aquaporin 2 (collecting duct) | NP_037041 | R.RRQS*VELHSPQSLPR.G(+3) | 9.00 | 3.17 ± 0.20a | S256 |
| 21 | aquaporin 2 (collecting duct) | NP_037041 | R.RRQS*VELHS*PQS*LPR.G(+2) | 9.38 | 3.23 ± 0.39a | S256, S261, S264 |
| 22 | kinesin 13B | NP_998791 | R.RRS*S*GLQPQGAPEAR.R(+3) | 10.93 | 3.45 ± 0.20a | S1718, S1719 |
| 23 | aquaporin 2 (collecting duct) | NP_037041 | R.RQS*VELHS*PQS*LPR.G(+2) | 12.30 | 3.62 ± 0.41a | S256, S261, S264 |
| 24 | aquaporin 2 (collecting duct) | NP_037041 | R.RQS*VELHSPQSLPR.G(+2) | 14.93 | 3.90 ± 0.52a | S256 |
| 25 | (urea transporter), member 2 isoform 1 | NP_062220 | R.RKS*VFHIEWSSIR.R(+4) | 17.51 | 4.13 ± 0.22a | S486 |
| 26 | (urea transporter), member 2 isoform 1 | NP_062220 | R.RKS*VFHIEWSSIR.R(+3) | 19.84 | 4.31 ± 0.31a | S486 |
Mean and SE values are calculated from log2(D/C), where D and C are peak areas for dDAVP-treated and control samples, respectively. Phosphorylation sites are indicated with an asterisk.
aP < 0.05; significant change in abundance compared with control samples.
bAmbiguity in phosphorylation site assignment by the Ascore algorithm, despite certainty in phosphopeptide identification.
cNo significant change in abundance compared with control samples.
Regulation of β-Catenin Phosphorylation by dDAVP
One of the phosphorylation sites that increased in response to dDAVP was Ser-552 of β-catenin (Table 1, part B). Because β-catenin has been proposed to play a role in regulation of aquaporin-2,23 we investigated this response further using a phosphospecific antibody. Figure 5A shows an immunoblot performed on protein isolates from the same five pairs of animals used in our large-scale quantitation experiment. The blot was probed with an antibody recognizing Ser(p)-552-β-catenin. As shown in Figure 5A, Ser(p)-552-β-catenin was increased 3-fold in response to dDAVP (band densities: vehicle, 4.8 ± 0.2; dDAVP, 14.8 ± 0.3; P < 0.05). An immunoblot for total β-catenin was also performed and shows that dDAVP-treatment does not (P > 0.40) increase total protein abundance (Figure 5A, bottom). Thus, we were able to show convergent results for increased β-catenin phosphorylation on Ser-552 in response to dDAVP treatment using both LC-MS/MS and immunoblotting.
Figure 5.
dDAVP increases β-catenin phosphorylation in rat IMCD. (A) Top panel: Immunoblot of IMCD suspensions showing five pairs of control and dDAVP-treated samples. Band at 97 kDa is Ser(p)-552-β-catenin. dDAVP causes increased phosphorylation of Ser-552 of β-catenin. Bottom panel: Immunoblot of IMCD suspensions showing the same five pairs of control and dDAVP-treated samples using antibody recognizing total β-catenin. (B) Top panel: Immunoblot of whole inner medulla from Brattleboro rats showing increased Ser(p)-552-β-catenin in response to dDAVP given intramuscularly. Bottom panel: Immunoblot of whole inner medulla from Brattleboro rats showing that there is no increase in total β-catenin abundance in response to dDAVP given intramuscularly.
To address whether a similar change in β-catenin phosphorylation occurs in the inner medulla of intact kidney, Brattleboro rats were injected intramuscularly with dDAVP or vehicle, and in 45 minutes, the whole inner medulla was isolated for immunoblot analysis. As shown in Figure 5B, Ser(p)-552-β-catenin was increased nearly 2-fold in response to dDAVP (band densities: vehicle, 10.9 ± 1.1; dDAVP, 20.1 ± 2.6; P < 0.05).
dDAVP Increases Phosphorylation of the Vasopressin-Sensitive Urea Channel
Before examining the effect of short-term dDAVP treatment on UT-A phosphorylation, it was necessary to establish that total UT-A protein abundance does not change as a result of short-term dDAVP treatment, and thus was not contributing to changes in abundance of the UT-A phosphopeptides. The immunoblot in Figure 6 was probed with an antibody recognizing total UT-A1.24 The 97- and 117-kD bands represent UT-A1. There was no significant difference between control and treated samples (P = 0.11). The same immunoblot was also probed with a phosphospecific antibody recognizing Ser(p)-256 on AQP2 to verify the responsiveness of IMCD tubules to dDAVP stimulation.
Figure 6.
Immunoblot shows that dDAVP treatment of IMCD does not result in increased total protein abundance of UT-A1. Immunoblot shows three pairs of control and dDAVP-treated samples. Proteins at 29 and 37 kDa are Ser(p)-256-AQP2 (nonglycosylated and glycosylated forms, respectively). Protein at 97 and 117 kDa is UT-A1.
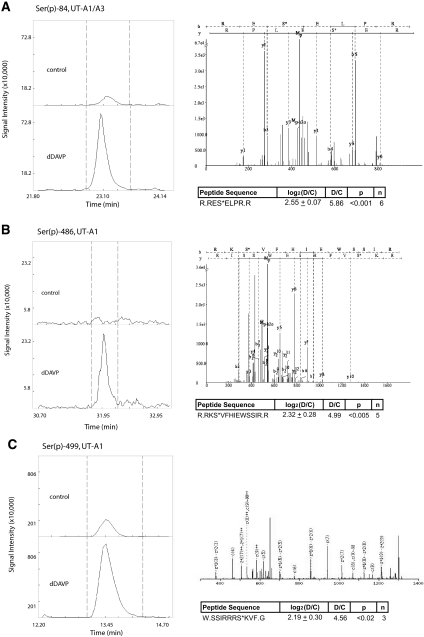
From our SCX profiling data, we identified phosphorylation of the vasopressin-sensitive urea channel (isoforms A1 and A3) on Ser-10, Ser-62, Ser-63, Ser-84, and Ser-486 (http://dir.nhlbi.nih.gov/papers/lkem/mpkccdprot/). Of these five sites, Ser-10 and Ser-84 have not been previously reported. Ser-486 was identified by Hoffert et al.17 using a similar approach and was subsequently found to be functionally important for urea transport activity by Blount et al.20 The same study also demonstrated the importance of phosphorylation at Ser-499 for the transport function of UT-A1.20 For this study, a trypsin digest could not be used to identify the peptide containing Ser-499, because the resulting peptide was too short for LC-MS/MS detection. Alternatively, sample digestion with chymotrypsin was predicted to produce a peptide of suitable length. However, attempts to identify the Ser-499-containing chymotryptic peptide were precluded by poor fragmentation of the parent ion using CID as the fragmentation technique. Therefore, we used an alternative fragmentation technique, electron transfer dissociation (ETD), to successfully identify the chymotryptic peptide containing Ser-499 (Figure 7C), and CID was used for the remaining sites (Ser-10, Ser-62/63, Ser-84, and Ser-486).
Figure 7.
MS quantification of UT-A phosphorylation is illustrated by reconstructed peptide ion chromatograms from QUOIL software. The data show an increase in peak areas with dDAVP treatment compared with control for (A) Ser(p)-84 on both UT-A1 and UT-A3, (B) Ser(p)-486 on UT-A1, and (C) Ser(p)-499 on UT-A1. Also shown in each subfigure are mean and SE values for all quantifications along with a representative MS2 spectrum for the identified peptide.
On the basis of phosphorylation sites identified from our profiling data and evidence showing that Ser-499 is functionally important for urea transport,20 we used targeted selection of precursor ion masses, so-called targeted ion selection (TIS) (Figure 1C), to quantify changes in phosphorylation in response to dDAVP for each of the sites. dDAVP treatment resulted in no change in phosphorylation at Ser-10 (Supplementary Figure 1) or phosphorylation at both Ser-62 and Ser-63 (Supplementary Figure 2). However, phosphorylation at Ser-84, Ser-486, and Ser-499 increased significantly (P < 0.02) in response to dDAVP. Figure 7 shows representative reconstructed ion chromatograms for Ser-84, Ser-486, and Ser-499 phosphopeptides analyzed by LC-MS/MS. Each of the three subfigures also contains a representative MS2 fragmentation spectrum and a table summarizing all measurements for the listed peptide. The ratios of the integrated peak areas from reconstructed ion chromatograms were used to calculate changes in abundances of phosphorylated peptides in response to dDAVP. Of these sites, only Ser-84 is shared by UT-A1 and UT-A3.
Protein Kinase A Can Directly Phosphorylate Ser-84, Ser-486, and Ser-499 of UT-A in Vitro
UT-A1 contains several putative PKA phosphorylation motifs,25–27 including Ser-84, Ser-486, and Ser-499. To determine whether PKA can, in fact, phosphorylate these three sites, we performed in vitro assays using purified PKA catalytic subunit and synthetic UT-A peptides as substrates. Figure 8A shows phosphorylation on the C-terminal tail of aquaporin-2 on Ser-256, which served as a positive control. In addition, PKA also was able to phosphorylate peptides containing Ser-486, Ser-84, and Ser-499 sites on UT-A1 (Figure 8, B, C, and D, respectively).
Figure 8.
PKA can phosphorylate Ser-84 (UT-A1 and UT-A3), Ser-486-UT-A1, and Ser-499-UT-A1 in vitro. In each of the windows, the top half shows the reconstructed ion chromatogram for the phosphopeptide when PKA was not present in the reaction mixture; the bottom half shows the corresponding profiles for the phosphopeptide when PKA was added to the reaction mixture. (A) The C-terminal tail of AQP-2, which served as a positive control, is phosphorylated by PKA on Ser-256. Phosphorylation occurred only in the presence of kinase for (B) Ser-486-UT-A1, (C) Ser-84 (UT-A1 and UT-A3), and (D) Ser-499-UT-A1.
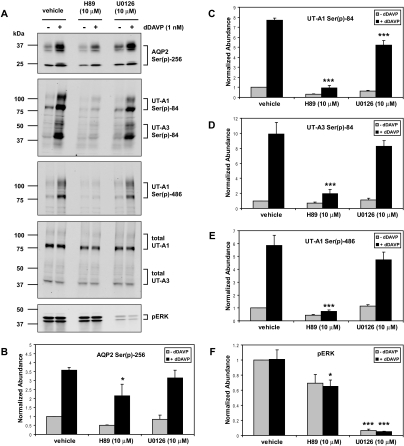
Phosphorylation of Ser-84 in UT-A1/3 and Ser-486 in UT-A1 Is Increased by dDAVP and Is Blocked by Inhibition of PKA
To test whether PKA regulates phosphorylation of UT-A1 and UT-A3 in vivo, isolated rat IMCD samples were incubated with dDAVP in the presence or absence of 10 μM PKA inhibitor H89, followed by immunoblotting with newly generated antibodies that recognize phosphorylated UT-A1 and UT-A3 at Ser-84, and phosphorylated UT-A1 at Ser-486 (Figure 9A). Phosphorylation at both Ser-84 and Ser-486 was significantly reduced in the presence of H89, suggesting that PKA may directly or indirectly regulate phosphorylation at these sites in response to short-term dDAVP treatment (Figure 9, A, C, D, and E). Inhibition of Ser(p)-256-AQP2 was used as a positive control to demonstrate that the H89 drug was working (Figure 9, A and B).
Figure 9.
Phosphorylation of both UT-A1 and UT-A3 is blocked by an inhibitor of PKA. (A) Rat IMCD samples were treated with either vehicle, the PKA inhibitor H89, or the MEK inhibitor U0126 for 10 minutes, followed by incubation with dDAVP for 20 minutes, followed by immunoblotting with antibodies to AQP2 Ser(p)-256, UT-A1 and UT-A3 Ser(p)-84, UT-A1 Ser(p)-486, total UT-A1 and UT-A3, and pERK. (B to F) Quantification of band densities from these immunoblots as well as other trials (n = 3; *P < 0.05 versus vehicle; ***P < 0.001 versus vehicle).
A recent study has also implicated the MEK-ERK pathway in regulating UT-A phosphorylation in response to forskolin in rat IMCD.28 To test whether extracellular signal-regulated kinase (ERK) mitogen-activated protein (MAP) kinase is involved in regulating UT-A phosphorylation, isolated rat IMCD samples were incubated with dDAVP in the presence or absence of 10 μM MEK1/2 (map kinase kinase 1 and 2) inhibitor U0126. Immunoblotting for phosphorylated ERK (pERK) demonstrated that dDAVP treatment did not increase ERK phosphorylation and that U0126 significantly blocked ERK phosphorylation in both control and dDAVP-treated samples (Figure 9, A and F). U0126 also produced a slight but significant reduction in UT-A1 phosphorylated at Ser-84 in dDAVP-treated samples (Figure 9C). However, inhibition of the ERK pathway failed to significantly affect the level of AQP2 phosphorylated at Ser-256, UT-A3 phosphorylated at Ser-84, and UT-A1 phosphorylated at Ser-486 (Figure 9, B, D, and E, respectively).
Discussion
The new advances in proteomic methodologies used here allow an unbiased, large-scale, discovery-based approach for the study of cell signaling. Here, we have applied these methodologies to vasopressin signaling in the renal collecting duct. Because water and urea transport are known to be vasopressin-regulated, we decided to emphasize our data focusing on phosphorylation of aquaporins and urea channels. However, a large-scale LC-MS/MS approach also allows unanticipated observations to be made regarding other parts of the signaling network. Here, we discuss examples of both types of observations: (1) regulated phosphorylation of UT-A urea channel isoforms and (2) regulation of β-catenin phosphorylation by vasopressin.
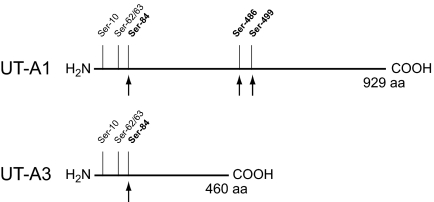
Our data identify and quantify multiple vasopressin-regulated phosphorylation sites on the urea channel (UT-A1 and UT-A3). A schematic of these sites and their relative locations on UT-A1 and UT-A3 is summarized in Figure 10. A previous study17 identified four phosphorylation sites (Ser-35, Ser-62, Ser-63, and Ser-486). In this study, we confirm previously identified sites and also report two previously unknown phosphorylation sites (Ser-10 and Ser-84). Of the sites reported in this study, we demonstrate that three (Ser-84, Ser-486, and Ser-499) are strongly regulated by vasopressin both by LC-MS/MS (Figure 6) and immunoblot (Figure 9) analysis. In a previous study using transfected LLC-PK1 cells, Blount et al.20 showed that S486A and S499A mutants each had reduced 32P incorporation into UT-A1 as well as attenuated urea flux in response to forskolin treatment. The importance of these two sites was highlighted by the fact that the S486A/S499A double mutants did not show any significant urea flux increase in response to forskolin. Because of the lack of site specificity of the 32P labeling technique employed in the study, quantitative conclusions concerning phosphorylation of these sites could not be made. In our study, we show that phosphorylation at each of these two sites increased approximately 4-fold with dDAVP exposure for 30 minutes. Because surrounding amino acid sequences from both sites are compatible with PKA as the kinase involved,27 we performed in vitro assays that showed PKA can phosphorylate both Ser-486 and Ser-499 (Figure 8). However, several other kinases have specificities similar to PKA,29 and consequently the kinase responsible for in vivo phosphorylation of these sites cannot be specified with certainty.
Figure 10.
Schematic of the two urea channels (UT-A1 and UT-A3) expressed in IMCD with the relative position of phosphorylation sites identified and quantified by LC-MS/MS. Sites marked with an arrow were found to be phosphorylated in response to vasopressin.
Our study also demonstrated a previously unknown site (Ser-84) on the N-terminal tail of the urea channel that is phosphorylated in response to dDAVP. Ser-84 is contained within a putative PKA motif,27 and an in vitro assay showed that PKA can phosphorylate this site (Figure 8). This site is present in both UT-A1 and UT-A3, and it is the only regulated site shared by both isoforms. The use of newly developed phosphospecific UT-A antibodies definitively showed that phosphorylation of this site is indeed increased by dDAVP treatment in both isoforms.
Further evidence supporting a role for PKA in regulating phosphorylation of collecting duct urea channels was found by using the inhibitor H89, which nearly completely blocked phosphorylation at both Ser-84 and Ser-486. Results from U0126 inhibitor studies suggested that the MEK-ERK pathway may also play a role in regulating phosphorylation of UT-A1 at Ser-84 but not at Ser-486. However, U0126 did not block the ability of dDAVP to increase phosphorylation of UT-A1 at either site or UT-A3 at Ser-84. Furthermore, neither Ser-84 nor Ser-486 is part of a proline-directed consensus motif, which would be compatible with direct phosphorylation by a MAP kinase.27
Regarding the evolutionary conservation of these sites, Ser-499 is highly conserved across rat, mouse, horse, cow, and human UT-A1. Ser-486 was also conserved in all of these species except for cow, where it is an asparagine residue. Ser-84 was less conserved, being replaced by aspartic acid in human and arginine in horse. The aspartic acid in human has the same charge (−1) as a phosphorylated serine and could conceivably represent a constitutively activated form.
Other UT-A sites that were quantified but were shown to not be regulated by vasopressin were Ser-10, Ser-62, and Ser-63. None of these sites have neighboring amino acids suggestive of a PKA or any other basophilic motif. However, these sites may still be important for other regulatory pathways.
In addition to UT-A, our study successfully quantified changes in site-specific phosphorylation of multiple other proteins, including Ser-552 on β-catenin (Table 1 and Figure 5). Ser-552 on β-catenin was found to undergo a large increase in phosphorylation in response to vasopressin, both in vitro (Figure 5A) and in vivo (Figure 5B). β-Catenin is a well-studied cytoplasmic protein that has two general roles in cells: (1) a structural role in cadherin-mediated cell adhesion and (2) a signaling role as a transcriptional coregulator.30 It is involved in the highly conserved canonical Wnt signaling pathway. Although involvement of β-catenin in the embryonic development of the collecting duct has been well studied,31–33 there is only indirect evidence regarding its relationship to vasopressin signaling in the mature collecting duct.23 The amino acid sequence surrounding Ser-552 of β-catenin (…QRRTS552MGGT…) is consistent with a PKA motif.27 Through 32P labeling and site-directed mutagenesis, it has been shown previously that PKA can phosphorylate Ser-552.34 Subsequent studies demonstrated that phosphorylation of Ser-552 by PKA can facilitate the interaction between β-catenin and T cell factor, a transcription factor that up-regulates gene expression related to cell growth.35 Given the fact that we have shown vasopressin causes increased phosphorylation of Ser-552 on β-catenin in the collecting duct, an important future direction will be to elucidate whether similar interactions among vasopressin, β-catenin, and expression of growth-promoting genes are also relevant for the collecting duct.
Gene Ontology analysis and Conserved Domain analysis of proteins identified in Table 1 did not point to any single category of proteins that are regulated by dDAVP (Supplementary Data). However, several proteins whose phosphorylation is increased have potential roles in regulation of aquaporin-2 and/or UT-A isoforms. For example, the PCTAIRE-3 protein kinase was strongly phosphorylated at Ser-66, a putative PKA site, in response to dDAVP. This kinase is differentially expressed in IMCD versus non-collecting duct cells3 and is thought to be involved in membrane trafficking.36 In addition, plexin domain containing-2 is also strongly phosphorylated at a putative PKA site (Ser-507). This protein has been shown to bind cortactin, which is an important regulator of actin polymerization at cell adhesion sites.37 In contrast, several phosphorylation sites were found to be strongly down-regulated in proteins that have relevance to aquaporin-2 and/or UT-A regulation. For example, septin-9 phosphorylation was strongly down-regulated at Ser-67, a putative MAP kinase site. Septins are polymeric GTP-binding proteins that associate with cell membranes as well as the actin and microtubule cytoskeletons.38 Furthermore, a protein called ArfGAP with FG repeats 1 also undergoes a marked decrease in phosphorylation in response to dDAVP at several proline-directed sites. This protein is believed to be involved in vesicle docking and fusion in acrosome formation in spermatozoa39 and could play a role in regulation of membrane trafficking in the collecting duct. Finally, epsin-3 underwent a marked decrease in phosphorylation at Thr-361 a putative MAP kinase site. This protein functions as a ubiquitin-dependent clathrin adaptor and may be involved in regulation of membrane trafficking in the collecting duct.40
Concise Methods
IMCD Isolation
Sprague-Dawley rats were obtained between 1 and 3 months of age (Animal Care and Use Committee protocol number H-0110). Furosemide was injected into the rats (intraperitoneally, 0.5 mg) 30 minutes before euthanization. Both kidneys were removed, and the inner medullas were dissected out. IMCD isolation was performed using a modified version of previously outlined methods.41,42 Inner medullas were minced with a razorblade and digested for approximately 75 minutes at 37°C in an enzyme solution containing 3 mg/ml collagenase B and 3 mg/ml hyaluronidase dissolved in sucrose buffer (250 mM sucrose and 10 mM triethanolamine, pH 7.6). After enzyme digestion, a low-speed spin (70 × g, 20 seconds) was used to pellet IMCD cells. Cells were rinsed twice with sucrose buffer and once with bicarbonate buffer (118 mM NaCl, 25 mM NaHCO3, 5.5 mM glucose, 5 mM KCl, 4 mM Na2HPO4, 2 mM CaCl2, and 1.2 mM MgSO4, pH 7.4). Cells were resuspended in bicarbonate buffer, and samples were treated with either 1 nM dDAVP or vehicle for 30 minutes at 37°C in a pH- and temperature-controlled chamber. For studies using H89 and U0126, the drug was added for a 10-minute preincubation period followed by addition of 1 nM dDAVP for 20 minutes. Data in the phosphoproteome profiling experiment were derived only from a dDAVP-treated sample without a vehicle-treated control (Figure 1A).
Reduction, Alkylation, and In-Solution Protease Digestion
After dDAVP treatment, IMCD tubules were resuspended in lysis buffer (8 M urea, 75 mM NaCl, 50 mM Tris-HCl, with protease and phosphatase inhibitors). Cells were sonicated with 0.5-second pulses for 30 cycles followed by centrifugation at 10,000 × g for 10 minutes. Protein concentration in the supernatants was determined using the BCA protein assay (Pierce, Rockford, IL). Approximately 1 mg of protein was used for large-scale phosphoproteome profiling, whereas 500 μg was used for quantitative studies. Samples were reduced for 1 hour with 10 mM dithiothreitol, followed by alkylation with 40 mM iodoacetamide for 1 hour in the dark. 40 mM dithiothreitol was added to quench unreacted iodoacetamide. Samples were diluted to a urea concentration of <1 M and digested with trypsin (Promega, Madison, WI) overnight at 37°C in 50 mM ammonium bicarbonate. For samples digested with chymotrypsin (Roche, Penzberg, Germany), the reaction was performed for 16 hours at 25°C in 100 mM Tris-HCl and 5 mM CaCl2.
Desalting, Phosphopeptide Enrichment, and Sample Fractionation
After protease digestion, peptide samples were desalted using a 1-cc HLB cartridge (Waters, Milford, MA) and dried by SpeedVac. The dried peptides were resuspended in 300 μl of solvent A (5 mM KH2PO4, 25% acetonitrile (ACN), pH 2.7) and injected onto a PolySulfoethyl A SCX column (4.6-mm inner diameter × 20-cm length, 5-μm particle size, 300-Å pore size). SCX chromatography was carried out on an Agilent HP1100 system at 1 ml/min flow rate using the following gradient: 0% solvent B for 2 minutes; 0 to 20% solvent B for 40 minutes; 20% to 100% solvent B for 5 minutes; and 100% solvent B held for 5 minutes (solvent B: 5 mM KH2PO4, 25% ACN, and 350 mM KCl, pH 2.7). UV absorbance at 214 nm was monitored while fractions were collected at 1.5-ml intervals and dried down by SpeedVac. All 24 fractions were desalted on 1-cc HLB cartridges (Waters) before phosphopeptide enrichment. SCX was not used in other analysis methods, such as TIS or label-free LC-MS/MS quantitation. Phosphopeptide enrichment was performed as described17 using immobilized metal affinity chromatography (IMAC) with a Ga3+ matrix (Phosphopeptide Isolation Kit; Pierce). Before analysis by MS/MS, samples were desalted using C18 Ziptips (Millipore) and resuspended in 0.1% formic acid.
LC-MS/MS Analysis on Linear Trap Quadrupole
An Agilent 1100 nanoflow LC system (Palo Alto, CA) connected to a Finnigan linear trap quadrupole (LTQ) mass spectrometer (Thermo Electron, San Jose, CA) was used for LC-MS/MS analysis. Peptide trapping was performed at a flow rate of 2 μl/min. Peptides were eluted onto a reversed-phase PicoFrit column (New Objective, Woburn, MA), and separation was performed using a linear ACN gradient (0 to 60%) in 0.1% formic acid. Peptides were introduced into the LTQ mass spectrometer using a nanospray ion source. The spray voltage was set to 2 kV and collision energy to 35%. Data-dependent acquisition mode (dynamic exclusion enabled) was used for profiling experiments. Survey MS1 scans were followed by five MS2 scans. TIS was performed with predetermined masses entered into an inclusion list (dynamic exclusion disabled).
LC-MS/MS Analysis on LTQ-Orbitrap
All dried fractions were analyzed on an LTQ-Orbitrap (Thermo-Fisher Scientific LLC) interfaced with an Eksigent nano-LC 1D plus system (Eksigent Technologies LLC, Dublin, CA) using CID fragmentation. Briefly, samples were loaded onto an Agilent Zorbax 300SB-C18 trap column (0.3-mm inner diameter × 5-mm length, 5-μm particle size) at a flow rate of 5 μl/min for 10 minutes. Reversed-phase C18 chromatographic separation of peptides was carried out on a prepacked PicoFrit column (75-μm inner diameter × 10-cm length; New Objective) at 300 nl/min using the following gradient: 2% to 5% solvent B for 5 minutes; 5% to 45% solvent B for 60 minutes; 45% to 50% solvent B for 5 minutes; and 50% to 95% solvent B for 5 minutes (solvent A: 0.1% formic acid in 98% water, 2% ACN; solvent B: 0.1% formic acid in 100% ACN). LTQ-Orbitrap settings were as follows: spray voltage 1.5 kV; full MS mass range: m/z 300 to 2000; and MS/MS mass range: m/z 100 to 2000. The LTQ-Orbitrap was operated in a data-dependent mode; i.e., one MS1 high resolution scan for precursor ions followed by three data-dependent MS2 scans for precursor ions above a threshold ion count of 2000 with collision energy of 35%. In some cases, parent ion masses were entered into an inclusion list for TIS experiments. MS3 scans were automatically triggered on the basis of the observation of neutral loss peaks of phosphoric acids from precursor ions in MS2 scans that met a threshold criterion of 500 counts.
ETD and Decision Tree-Driven Tandem MS/MS
ETD parameters were tuned using bovine growth hormone-releasing receptor (Sigma G0644). The reaction time was optimized at 85 milliseconds (with an AGC target for fluoranthene radical anion of 300,000). Online mass spectrometric experiments were performed by MS1 analysis in the Orbitrap at 30,000 resolution followed by five data-dependent MS/MS events with product ion analysis performed in the LTQ. The precursor ions selected for MS/MS were determined by intensity within MS1 (≥1000 counts) followed by dynamic exclusion (30 seconds or a peak list of 500) and charge state inclusion (ions with two or more charges). The form of MS/MS activation was either ETD-only, CID-only, or decision tree-based selection.43 When using the decision tree-based selection, the dissociation method for every MS/MS event was determined in real time in an automated fashion on the basis of the precursor z and m/z. This feature was fully incorporated into the LTQ Tune 2.5.5 instrument control software (default setting).
Data Searching, Scoring, Quantification, and Bioinformatics
Searches were performed using the latest version of the rat RefSeq database (National Center for Biotechnology Information), with concatenated forward and reversed sequences to allow for target-decoy analysis. The database also contained sequences for common MS contaminants, such as human keratin and porcine trypsin. Data from the phosphoproteome profiling experiment were searched with SEQUEST,44 InsPecT,45,46 and OMSSA.47 InsPecT and OMSSA searches used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). All of the other data were searched using SEQUEST only. Filtering and determination of false-discovery rate were done using methods described previously.48,49 All datasets were filtered for a false-discovery rate of <2%. Phosphorylation site localization was performed using Ascore50 and PhosphoScore.51 Label-free phosphopeptide quantification was performed using QUOIL.22 Gene Ontology and Conserved Domain Database analyses were performed using in-house software.
Brattleboro Rats
Brattleboro rats (Animal Care and Use Committee protocol number H-0110) were injected with dDAVP (2 nmol dissolved in saline) or vehicle intramuscularly and then euthanized after 45 minutes. Inner medullas were dissected, homogenized, and solubilized in Laemmli buffer as described previously.9
Antibodies
Affinity-purified rabbit polyclonal antibodies against rat Ser(p)-256-AQP252 and total UT-A1 (L448)24 have been previously described. Phosphospecific antibodies were generated against sequences surrounding residues Ser-84 and Ser-486 of rat UT-A1 (PhosphoSolutions, Aurora, CO). Specificity of these antibodies was established by dotblot (data not shown) and immunoblot. A phosphospecific antibody recognizing Ser(p)-552-β-catenin was obtained from Cell Signaling Technology (product no. 9566; Danvers, MA).
Immunoblot Analysis
Immunoblotting was used to assess the response of AQP2 and UT-A1 to dDAVP treatment and was carried out using previously described methods.17,48
In vitro Phosphorylation Assay
We performed an in vitro assay to determine whether particular serine residues contained within in silico-predicted PKA phosphorylation consensus motifs in UT-A1 could be phosphorylated by PKA. We obtained synthetic peptides corresponding to sequences surrounding the S84, S486, and S499 residues of UT-A1 (AnaSpec, San Jose, CA). We used a peptide with the amino acid sequence of the C-terminal tail of AQP218 as a positive control. We incubated a total of 5 μg of each peptide with 1× kinase buffer and 200 μM ATP (Cell Signaling Technology, Beverly, MA) in the presence or absence of PKA C-α kinase (Cell Signaling Technology, Beverly, MA) for 1 hour at 30°C. After incubation, the reaction mixtures were digested overnight with either trypsin at 37°C or chymotrypsin (for Ser-84 peptide) at 25°C. Samples were then desalted, resuspended in 0.1% formic acid, and analyzed on an LTQ mass spectrometer (Thermo Electron).
Disclosures
None.
Supplementary Material
Acknowledgments
This work was funded by the intramural budget of the National Heart, Lung, and Blood Institute (Z01-HL-01285-KE). A.D.B. is supported by the Howard Hughes Medical Institute-National Institutes of Health Research Scholars Program and the New York University School of Medicine. We would like to thank Guozhong Ma, Center for Biomedical Informatics, National Heart, Lung, and Blood Institute, National Institutes of Health, for his assistance with the Web publishing of the Collecting Duct Phosphoproteome Database.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Nielsen S, Frokiaer J, Marples D, Kwon TH, Agre P, Knepper MA: Aquaporins in the kidney: From molecules to medicine. Physiol Rev 82: 205–244, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Brown D: The ins and outs of aquaporin-2 trafficking. Am J Physiol Renal Physiol 284: F893–F901, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Uawithya P, Pisitkun T, Ruttenberg BE, Knepper MA: Transcriptional profiling of native inner medullary collecting duct cells from rat kidney. Physiol Genomics 32: 229–253, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yano Y, Rodrigues AC, Jr., de Braganca AC, Andrade LC, Magaldi AJ: PKC stimulated by glucagon decreases UT-A1 urea transporter expression in rat IMCD. Pflugers Arch 456: 1229–1237, 2008 [DOI] [PubMed] [Google Scholar]
- 5.O'Connor PM, Cowley AW, Jr.: Vasopressin-induced nitric oxide production in rat inner medullary collecting duct is dependent on V2 receptor activation of the phosphoinositide pathway. Am J Physiol Renal Physiol 293: F526–F532, 2007 [DOI] [PubMed] [Google Scholar]
- 6.Umenishi F, Narikiyo T, Vandewalle A, Schrier RW: cAMP regulates vasopressin-induced AQP2 expression via protein kinase A-independent pathway. Biochim Biophys Acta 1758: 1100–1105, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Mordasini D, Bustamante M, Rousselot M, Martin PY, Hasler U, Feraille E: Stimulation of Na+ transport by AVP is independent of PKA phosphorylation of the Na-K-ATPase in collecting duct principal cells. Am J Physiol Renal Physiol 289: F1031–F1039, 2005 [DOI] [PubMed] [Google Scholar]
- 8.Tamma G, Klussmann E, Maric K, Aktories K, Svelto M, Rosenthal W, Valenti G: Rho inhibits cAMP-induced translocation of aquaporin-2 into the apical membrane of renal cells. Am J Physiol Renal Physiol 281: F1092–F1101, 2001 [DOI] [PubMed] [Google Scholar]
- 9.Pisitkun T, Jacob V, Schleicher SM, Chou CL, Yu MJ, Knepper MA: Akt and ERK1/2 pathways are components of the vasopressin signaling network in rat native IMCD. Am J Physiol Renal Physiol 295: F1030–F1043, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou CL, Christensen BM, Frische S, Vorum H, Desai RA, Hoffert JD, de Lanerolle P, Nielsen S, Knepper MA: Non-muscle myosin II and myosin light chain kinase are downstream targets for vasopressin signaling in the renal collecting duct. J Biol Chem 279: 49026–49035, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Nielsen S, Chou CL, Marples D, Christensen EI, Kishore BK, Knepper MA: Vasopressin increases water permeability of kidney collecting duct by inducing translocation of aquaporin-CD water channels to plasma membrane. Proc Natl Acad Sci U S A 92: 1013–1017, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sands JM, Nonoguchi H, Knepper MA: Vasopressin effects on urea and H2O transport in inner medullary collecting duct subsegments. Am J Physiol 253: F823–F832, 1987 [DOI] [PubMed] [Google Scholar]
- 13.Snyder PM: Minireview: Regulation of epithelial Na+ channel trafficking. Endocrinology 146: 5079–5085, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Tomita K, Pisano JJ, Knepper MA: Control of sodium and potassium transport in the cortical collecting duct of the rat. Effects of bradykinin, vasopressin, and deoxycorticosterone. J Clin Invest 76: 132–136, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiGiovanni SR, Nielsen S, Christensen EI, Knepper MA: Regulation of collecting duct water channel expression by vasopressin in Brattleboro rat. Proc Natl Acad Sci U S A 91: 8984–8988, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terris J, Ecelbarger CA, Nielsen S, Knepper MA: Long-term regulation of four renal aquaporins in rats. Am J Physiol 271: F414–F422, 1996 [DOI] [PubMed] [Google Scholar]
- 17.Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA: Quantitative phosphoproteomics of vasopressin-sensitive renal cells: Regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci U S A 103: 7159–7164, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffert JD, Fenton RA, Moeller HB, Simons B, Tchapyjnikov D, McDill BW, Yu MJ, Pistikun T, Chen F, Knepper MA: Vasopressin-stimulated increase in phosphorylation at ser-269 potentiates plasma membrane retention of aquaporin-2. J Biol Chem 283: 24617–24627, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blount MA, Klein JD, Martin CF, Tchapyjnikov D, Sands JM: Forskolin stimulates phosphorylation and membrane accumulation of UT-A3. Am J Physiol Renal Physiol 293: F1308–F1313, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Blount MA, Mistry AC, Frohlich O, Price SR, Chen G, Sands JM, Klein JD: Phosphorylation of UT-A1 urea transporter at serines 486 and 499 is important for vasopressin-regulated activity and membrane accumulation. Am J Physiol Renal Physiol 295: F295–F299, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP: Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: The yeast proteome. J Proteome Res 2: 43–50, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Wang G, Wu WW, Zeng W, Chou CL, Shen RF: Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: Reproducibility, linearity, and application with complex proteomes. J Proteome Res 5: 1214–1223, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Nielsen J, Hoffert JD, Knepper MA, Agre P, Nielsen S, Fenton RA: Proteomic analysis of lithium-induced nephrogenic diabetes insipidus: Mechanisms for aquaporin 2 down-regulation and cellular proliferation. Proc Natl Acad Sci U S A 105: 3634–3639, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford AD, Terris JM, Ecelbarger CA, Klein JD, Sands JM, Chou CL, Knepper MA: 97- and 117-kDa forms of collecting duct urea transporter UT-A1 are due to different states of glycosylation. Am J Physiol Renal Physiol 281: F133–F143, 2001 [DOI] [PubMed] [Google Scholar]
- 25.Karakashian A, Timmer RT, Klein JD, Gunn RB, Sands JM, Bagnasco SM: Cloning and characterization of two new isoforms of the rat kidney urea transporter: UT-A3 and UT-A4. J Am Soc Nephrol 10: 230–237, 1999 [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, Sands JM, Klein JD: Vasopressin rapidly increases phosphorylation of UT-A1 urea transporter in rat IMCDs through PKA. Am J Physiol Renal Physiol 282: F85–F90, 2002 [DOI] [PubMed] [Google Scholar]
- 27.Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, Olhovsky M, Pasculescu A, Alexander J, Knapp S, Blom N, Bork P, Li S, Cesareni G, Pawson T, Turk BE, Yaffe MB, Brunak S, Linding R: Linear motif atlas for phosphorylation-dependent signaling. Sci Signal 1: ra2, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Klein JD, Blount MA, Martin CF, Kent KJ, Pech V, Wall SM, Sands JM: Epac regulates UT-A1 to increase urea transport in inner medullary collecting ducts. J Am Soc Nephrol 20: 2018–2024, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown D, Hasler U, Nunes P, Bouley R, Lu HA: Phosphorylation events and the modulation of aquaporin 2 cell surface expression. Curr Opin Nephrol Hypertens 17: 491–498, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clevers H: Wnt/beta-catenin signaling in development and disease. Cell 127: 469–480, 2006 [DOI] [PubMed] [Google Scholar]
- 31.Schmidt-Ott KM, Barasch J: WNT/beta-catenin signaling in nephron progenitors and their epithelial progeny. Kidney Int 74: 1004–1008, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, Carroll TJ, Rajagopal J, Kobayashi A, Ren Q, McMahon AP: A Wnt7b-dependent pathway regulates the orientation of epithelial cell division and establishes the cortico-medullary axis of the mammalian kidney. Development 136: 161–171, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyons JP, Miller RK, Zhou X, Weidinger G, Deroo T, Denayer T, Park JI, Ji H, Hong JY, Li A, Moon RT, Jones EA, Vleminckx K, Vize PD, McCrea PD: Requirement of Wnt/beta-catenin signaling in pronephric kidney development. Mech Dev 126: 142–159, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taurin S, Sandbo N, Qin Y, Browning D, Dulin NO: Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J Biol Chem 281: 9971–9976, 2006 [DOI] [PubMed] [Google Scholar]
- 35.Taurin S, Sandbo N, Yau DM, Sethakorn N, Dulin NO: Phosphorylation of beta-catenin by PKA promotes ATP-induced proliferation of vascular smooth muscle cells. Am J Physiol Cell Physiol 294: C1169–C1174, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer KJ, Konkel JE, Stephens DJ: PCTAIRE protein kinases interact directly with the COPII complex and modulate secretory cargo transport. J Cell Sci 118: 3839–3847, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Nanda A, Buckhaults P, Seaman S, Agrawal N, Boutin P, Shankara S, Nacht M, Teicher B, Stampfl J, Singh S, Vogelstein B, Kinzler KW, St Croix B: Identification of a binding partner for the endothelial cell surface proteins TEM7 and TEM7R. Cancer Res 64: 8507–8511, 2004 [DOI] [PubMed] [Google Scholar]
- 38.Spiliotis ET, Nelson WJ: Here come the septins: Novel polymers that coordinate intracellular functions and organization. J Cell Sci 119: 4–10, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang-Decker N, Mantchev GT, Juneja SC, McNiven MA, van Deursen JM: Lack of acrosome formation in Hrb-deficient mice. Science 294: 1531–1533, 2001 [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Traub LM, Weixel KM, Hawryluk MJ, Shah N, Edinger RS, Perry CJ, Kester L, Butterworth MB, Peters KW, Kleyman TR, Frizzell RA, Johnson JP: Clathrin-mediated endocytosis of the epithelial sodium channel. Role of epsin. J Biol Chem 281: 14129–14135, 2006 [DOI] [PubMed] [Google Scholar]
- 41.Chou CL, DiGiovanni SR, Luther A, Lolait SJ, Knepper MA: Oxytocin as an antidiuretic hormone. II. Role of V2 vasopressin receptor. Am J Physiol 269: F78–F85, 1995 [DOI] [PubMed] [Google Scholar]
- 42.Stokes JB, Grupp C, Kinne RK: Purification of rat papillary collecting duct cells: functional and metabolic assessment. Am J Physiol 253: F251–F262, 1987 [DOI] [PubMed] [Google Scholar]
- 43.Swaney DL, McAlister GC, Coon JJ: Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods 5: 959–964, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D: Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem 67: 1426–1436, 1995 [DOI] [PubMed] [Google Scholar]
- 45.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V: InsPecT: Identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem 77: 4626–4639, 2005 [DOI] [PubMed] [Google Scholar]
- 46.Payne SH, Yau M, Smolka MB, Tanner S, Zhou H, Bafna V: Phosphorylation-specific MS/MS scoring for rapid and accurate phosphoproteome analysis. J Proteome Res 7: 3373–3381, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH: Open mass spectrometry search algorithm. J Proteome Res 3: 958–964, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Yu MJ, Pisitkun T, Wang G, Aranda JF, Gonzales PA, Tchapyjnikov D, Shen RF, Alonso MA, Knepper MA: Large-scale quantitative LC-MS/MS analysis of detergent-resistant membrane proteins from rat renal collecting duct. Am J Physiol Cell Physiol 295: C661–C678, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffert JD, Wang G, Pisitkun T, Shen RF, Knepper MA: An automated platform for analysis of phosphoproteomic datasets: Application to kidney collecting duct phosphoproteins. J Proteome Res 6: 3501–3508, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP: A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 24: 1285–1292, 2006 [DOI] [PubMed] [Google Scholar]
- 51.Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD: PhosphoScore: An open-source phosphorylation site assignment tool for MSn data. J Proteome Res 7: 3054–3059, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fenton RA, Moeller HB, Hoffert JD, Yu MJ, Nielsen S, Knepper MA: Acute regulation of aquaporin-2 phosphorylation at Ser-264 by vasopressin. Proc Natl Acad Sci U S A 105: 3134–3139, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.