Abstract
Efficient recognition of odorous objects universally shapes animal behavior and is crucial for survival. To distinguish kin from non-kin, mate from non-mate, food from non-food, organisms must be able to create meaningful perceptual representations of odor qualities and categories. It is currently unknown where, and in what form, the brain encodes information about odor quality. By combining functional magnetic resonance imaging (fMRI) with multivariate (pattern-based) techniques, we show that spatially distributed ensemble activity in human posterior piriform cortex (PPC) coincides with perceptual ratings of odor quality, such that odorants with more (or less) similar fMRI patterns were perceived as more (or less) alike. Critically, these effects were not observed in anterior piriform cortex, amygdala, or orbitofrontal cortex, demonstrating that ensemble coding of odor categorical perception is regionally specific for PPC. These findings substantiate theoretical models emphasizing the importance of distributed piriform templates for the perceptual reconstruction of odor object quality.
A key property of the brain is to create coherent, meaningful perceptual constructs from the complexity of the outside world. These internalized representations of the external environment provide a neural basis for object recognition, identification, and categorization, enabling organisms to focus cognitive resources, optimize behavioral responses, and generalize past experiences to novel events1,2.
Research on object processing has traditionally focused on visual object processing2,3, which tends to overshadow the critical ecological role of “odor objects” – here defined as the perceptual quality or character of a smell emitted from an odorous substance. Efficient recognition of odor objects universally shapes animal behavior and is crucial for survival. Indeed, the ubiquity of olfactory-guided adaptive behaviors across vertebrate and invertebrate species, including maternal bonding4, kinship recognition5, mate selection6, and territorial defense7, makes it clear that olfactory systems face important challenges to record and classify odor objects.
Where, and in what form, does the brain encode the perceptual quality of an odor object? Several elegant studies have demonstrated that odor-evoked spatial activity in the rodent olfactory bulb correlates with behavioral measures of odor similarity8–11, as indexed via paradigms of habituation or reinforcement learning, leading to the suggestion that neural representations of odor quality are reflected in ensemble bulbar activity. However, the obligatory use of indirect perceptual assays in animal models hampers efforts to establish an explicit link between neural response patterns and odor quality perception. Such limitations accentuate the unique advantages of studying human subjects, whose ability to verbalize their percepts12 and use perceptual rating scales offers an ideal research alternative for clarifying the neuroscientific underpinnings of odor quality perception.
One plausible location for the encoding and classification of odor quality information is primary olfactory (piriform) cortex, which receives direct input from the olfactory bulb and has extensive interconnections with intrinsic and extrinsic fiber systems, including amygdala, hypothalamus, entorhinal cortex, and orbitofrontal cortex13,14. The privileged access to sensory, affective, physiological, and motivational features of an olfactory stimulus perfectly endows piriform cortex with the capacity for weaving together odor representations with direct relevance for perception and behavior. This hypothesis receives support from both anatomical15,16 and computational14,17,18 models, which predict that odor percepts are embodied in spatially distributed patterns of piriform activity, providing a robust neural substrate for odor coding, memory, and recall. However, direct neurobiological evidence for this idea is currently not available, such that the functional architecture of odor object qualities in the olfactory brain remains unknown.
Here we combined high-resolution functional magnetic resonance imaging (fMRI) with olfactory multivariate analysis techniques19 (Fig. 1) to investigate spatial ensemble coding of odor qualities and categories in human posterior piriform cortex (PPC). Multivariate fMRI methods20–22 differ from conventional (univariate) fMRI analyses, in which data are averaged over space (voxels), time (scans), and subjects, obscuring potentially important information that may be contained at the level of individual voxels, scans, and subjects. These pattern-based approaches provide a robust new method for characterizing how (rather than just where) perceptual information is represented in the human brain23.
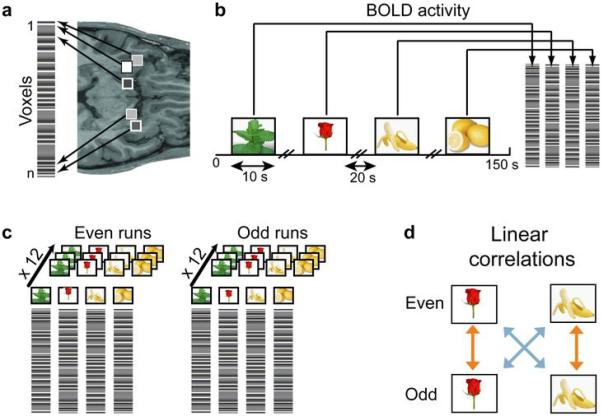
Fig. 1.
Schematic diagram of the correlation analysis. (a) The condition-specific spatial patterns of voxel activity in the PPC were transformed into linear vectors of voxel activity (pattern vectors). Voxels are represented by shaded squares on an axial slice of an anatomical MRI scan. The level of grey-scale intensity represents the blood oxygen level-dependent (BOLD) signal intensity. (b) Pattern vectors were composed of the peak BOLD activity across the stimulus presentation, shown here in the context of Experiment 1, for one run (150 s). (c) The entire dataset of pattern vectors was split into halves, one containing data from the 12 even runs and one from the 12 odd runs, and then averaged across runs, producing one mean pattern vector per odorant in each half of the data. (d) Averaged pattern vectors were used to calculate within-odorant (orange arrow) and across-odorant (blue arrow) pairwise correlation coefficients.
The present study consists of two independent experiments centered on the hypothesis that odor qualities and categories are encoded as distributed spatial ensembles in human PPC. In Experiment 1 we combined multivariate and cortical flattening techniques to test the following predictions: (a) qualitatively distinct odorants are associated with unique multi-voxel fMRI patterns in PPC, in the absence of mean activation differences; (b) odor-evoked fMRI representations in PPC are distributed and overlapping, without evidence for topographical clustering; and (c) olfactory ensemble codes of odor quality are regionally specific for PPC. In Experiment 2 we extended these hypotheses to a wider set of odorants, which in combination with multidimensional scaling techniques enabled us to examine whether fMRI ensemble patterns in PPC constitute an olfactory code that aligns with categorical perception of odor quality.
RESULTS
Experiment 1: odor-specific ensemble codes in PPC and OFC
In this first experiment we set out to test the hypothesis that neural codes of odor quality take the form of ensemble fMRI activity in PPC. Our decision to target PPC as a primary region of interest was based on recent animal and human studies suggesting that neural representations of odor quality are stored in this brain region24,25. During fMRI scanning, six subjects sniffed four easily distinguishable odorants [R-(−)-carvone (mint), phenethyl alcohol (rose), amyl acetate (banana), and citronellol (lemon)] across a total of 24 imaging runs. Upon each odorant presentation, subjects were visually cued to make a sniff and tacitly identify the quality of the odor.
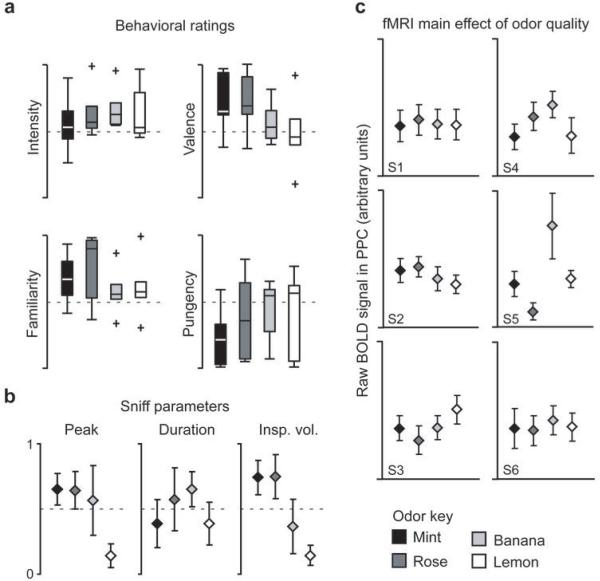
Behavioral ratings of the four odorants, acquired prior to scanning, did not significantly differ in odor intensity (F3,20 = 0.39, P = 0.76), pleasantness (F3,20 = 1.49, P = 0.25), pungency (F3,20 = 0.73, P = 0.55), or familiarity (F3,20 = 1.00, P = 0.41) (Fig. 2a). Moreover, pairwise ratings of odor quality similarity (total of six pairwise comparisons) indicated that the odorants were equally discriminable (F5, 35 = 1.73, P = 0.15). There were also no systematic differences in sniff peak amplitude (F3,20 = 2.78, P = 0.11), duration (F3,20 = 0.53, P = 0.67), or inspiratory volume (F3,20 = 2.04, P = 0.14) between odorants (Fig. 2b) that might have otherwise confounded the imaging analysis.
Fig. 2.
Behavioral data and univariate imaging analysis for Experiment 1. (a) Group-averaged behavioral ratings of odor intensity, pleasantness, pungency, and familiarity are depicted as boxplots indicating median (central line) and upper and lower quartiles (top and bottom of box, respectively) for each odorant. Whiskers denote extent of data between 10th and 90th percentiles. Outliers are indicated by crosses. Ratings did not significantly differ across any of these measures. (b) Mean normalized values (± between-subjects s.e.m.) for sniff peak amplitude, duration, and inspiratory volume (insp. vol.) did not significantly differ between the four odorants. (c) Plots of mean fMRI signal in PPC for each subject and odorant (± within-subject s.e.m.) revealed no significant difference except for subject 5 (S5).
We first considered whether wholesale differences in PPC activity between odorant conditions were detectable using a conventional (univariate) fMRI analysis. After all, if odor quality-specific information in PPC merely amounts to global changes in mean fMRI activity, then multivariate methods would not be necessary to confirm our original hypothesis. By averaging the fMRI signal in PPC across the set of voxels for each subject, we found no significant difference between odorants for 5/6 subjects (P's > 0.05; one-way ANOVAs; four levels [odorants]; one ANOVA per subject) (Fig. 2c). The mean fMRI signal significantly differed between odorants only in subject 5 (F3,92 = 3.71, P = 0.014), though follow-up pairwise t-tests indicated that not all odorants could be significantly discriminated from the others (e.g., mint vs. rose, T23 = 1.86, P = 0.076; mint vs. lemon, T23 = 0.28, P = 0.76; banana vs. lemon, T23 = 1.67, P = 0.11). These results indicate that if indeed a neural code for odor quality exists in PPC, it might be contained in a more fine-grained pattern of voxel activity that cannot be shown using standard imaging techniques.
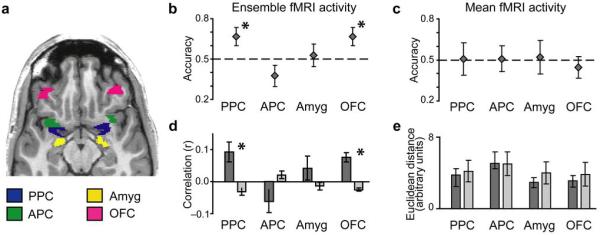
To test this hypothesis directly, we extracted odorant-specific voxel-wise patterns of fMRI ensemble activity within PPC, as determined by an independent odor localizer task. The 24-run set of spatial patterns, organized as linear vectors of voxel activity, was divided into 12 even and 12 odd runs20 and used to calculate linear correlations (Fig. 1) for all within-odor (e.g., rose/even vs. rose/odd) and all across-odor (e.g., rose/even vs. banana/odd) pairs. Odor identification accuracy, calculated as the proportion of within-odor correlations greater than across-odor correlations, significantly exceeded 50% chance in PPC across subjects (Fig. 3a,b) (T5 = 2.42, P = 0.030). Additionally, the average within-odor correlation was significantly greater than the average across-odor correlation across subjects in PPC (Fig. 3d) (T5 = 2.99, P = 0.030), an effect that was equally observed across the four odors (F3,20 = 2.10, P = 0.13, one-way ANOVA, four levels [difference between within- and across-odor correlations for each odorant]). These results demonstrate that odor-specific information is contained in multi-voxel PPC activity patterns, and that these effects were not driven by any one particular stimulus.
Fig. 3.
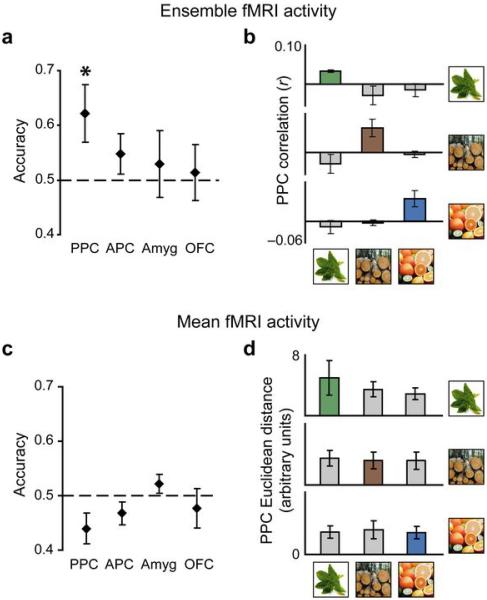
Pattern discrimination of odor quality in human PPC and OFC. (a) Axial slice of a T1-weighted structural scan showing anatomically defined regions of interest. Subsets of voxels from these brain regions (see Methods) were used in the pattern analyses. (b) Odor identification accuracy (mean ± between-subjects s.e.m; N = 6) calculated using fMRI patterns of ensemble activity exceeded chance (dashed line) across subjects in PPC and OFC, and the within-odor correlations (dark-grey bars) were greater than the across-odor correlations (light-grey bars) in both regions (d). In contrast, identification accuracy based on mean fMRI activity levels did not significantly differ from chance in any of the measured regions (c), nor were there significant differences between within-odor and across-odor Euclidean distances (e). *, P < 0.05.
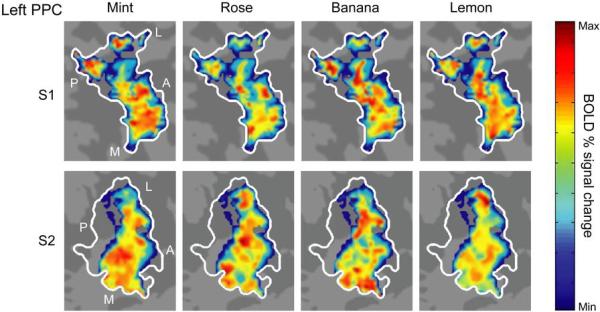
The above findings indicate that fMRI representations of odor quality are embedded in PPC ensemble activity. However, these analyses alone cannot reveal the specific topographical organization of brain activity underlying these putative odor codes (e.g., local vs. distributed; discrete vs. overlapping). In order to visualize the complex PPC anatomy in a single plane, we used cortical flattening (unfolding) techniques26 to generate odor “flat maps” in PPC. These maps were comprised of the flattened two-dimensional patterns of BOLD (blood oxygen level-dependent) signal in all odor-active piriform voxels, averaged across trials for each of the four odorants. Data from two representative subjects (Fig. 4) reveal that the topographical arrangement of left PPC activity was spatially distributed and unique for each odorant, in the absence of any obvious local clustering, and without topographical consistency between subjects. The implication is that olfactory percepts are represented in overlapping but distinct response profiles across subsets of voxels in PPC.
Fig. 4.
Odorant-specific spatial maps in PPC. The three-dimensional representations of odorant-evoked activity in PPC from two subjects were projected onto two-dimensional (flat) maps, allowing visualization of voxel-wise odor patterns within a single plane. Maps depict the odorant-evoked BOLD percent signal change in all odor-active voxels (liberally thresholded at P < 0.5), averaged across trials for each of the four odorants. The pseudocolor scale spans the full range of BOLD percent signal change within each map, from minimum (deep blue) to maximum (bright red). Each odorant elicited a distributed pattern of fMRI activity within left PPC (white outline) that overlapped with, but was distinct from, the other odorants. Unique distributed, overlapping profiles were also observed in right PPC (not shown). A, anterior; L, lateral; M; medial; P, posterior.
Is odor quality information encoded within PPC selectively, or are other cortical areas involved? To answer this question, we extended the analysis to three additional key olfactory brain regions. In anterior piriform cortex (APC) and amygdala, odor identification accuracy did not differ from chance (APC, T5 = 1.57, P = 0.089; amygdala, T5 = 0.34, P = 0.37), but interestingly, accuracy in orbitofrontal cortex (OFC) rivalled that in PPC and significantly exceeded chance (T5 = 2.48, P = 0.028) (Fig. 3a,b). A one-way ANOVA testing for an effect of region on the identification accuracy was significant (F3,20 = 3.45, P = 0.036), implying that the observed results are specific for PPC and OFC. Similarly, the within- vs. across-odor correlation difference was not significant in APC (T5 = 1.70, P = 0.15) or amygdala (T5 = 1.12, P = 0.31), but was highly significant in OFC (T5 = 5.40, P = 0.0029) (Fig. 3d). Once again a one-way ANOVA testing for region effects was significant (F3,20 = 4.91, P = 0.010), indicating that the correlation findings are specific to PPC and OFC. These additional results indicate that PPC and OFC both contain distributed odor-specific ensemble representations, in accord with prior animal and human data highlighting the role of OFC in olfactory coding27–30. That these patterns were only observed in PPC and OFC underscores the regional specificity of olfactory ensemble coding and validates the efficacy of multivariate techniques to delineate odor information processing in the human brain.
The above results indicate that PPC ensemble patterns can be used to discriminate odorant identity, but it remains possible that the mean fMRI activity in PPC could also contain discriminating information. Therefore, to directly compare classification performance based on fMRI multivariate and univariate data-sets, we performed a complementary analysis of the mean fMRI amplitudes, following the same classification methods used to analyze the fMRI patterns. Specifically, the global fMRI activity level was computed across all voxels in PPC, for each odorant and each run. The data were then split into even and odd runs, and pairwise Euclidean distances (as a proxy for linear correlations) were calculated between the means of within-odorant and across-odorant conditions. Identification accuracy (proportion of within-odor distances shorter than across-odor distances) did not differ from chance across the group in PPC, APC, amygdala, or OFC (Fig. 3c) (P's = 0.47, 0.48, 0.42, and 0.33, respectively). Furthermore, the within-odor distance was not significantly smaller than the across-odor distance in any of these regions (Fig. 3e) (P's = 0.73, 0.99, 0.47, and 0.67). These additional data suggest that mean fMRI amplitude does not contain sufficient information to distinguish between the odorants.
Finally, split-half classification methods were also applied to the trial-specific respiratory parameters (split into even and odd runs), to rule out potential contributions of sniffing to the observed PPC patterns. Odor identification accuracy was not significant across the group when classification was performed on sniff peak, duration, volume, or a linear combination of the three parameters (P's > 0.18; Supplementary Fig. 1a), demonstrating that any differences in sniff performance cannot account for the differential activity found in the PPC.
Experiment 2: categorical odor quality coding in PPC
The results from Experiment 1 are consistent with the idea that information about odor quality can be extracted from multi-voxel patterns of fMRI activity in human PPC (and OFC). However, to the extent that only one stimulus exemplar per odor category was used, the possibility remains that the observed fMRI effects may largely reflect odorant-specific differences, rather than more generalized differences in odor qualities and categories. Therefore, we conducted a second independent experiment using a more diverse set of odorants. This study, in conjunction with additional psychophysical measures and multidimensional scaling techniques, enabled us to definitively characterize ensemble coding of odor quality categories, as well as to quantify how well these fMRI codes coincide with perceived odor quality, on an odorant-by-odorant and subject-by-subject basis.
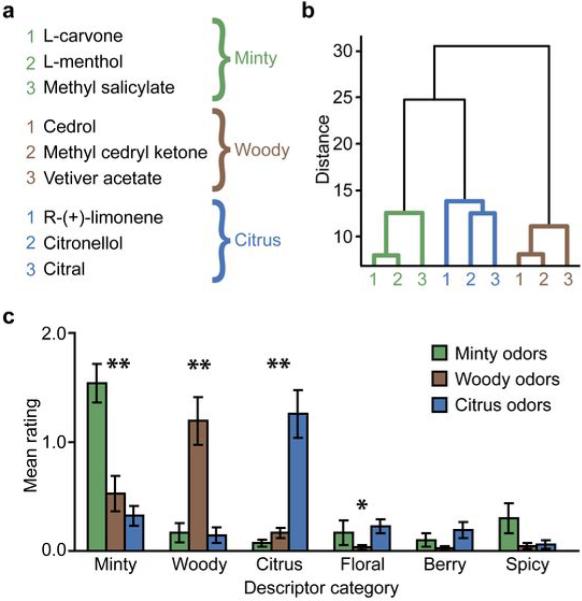
In this experiment, four additional participants smelled three exemplars per each of three odor quality categories: minty (R-(−)-carvone, L-menthol, methyl salicylate), woody (cedrol, methyl cedryl ketone, vetiver acetate), and citrus (citral, citronellol, (R)-(+)-limonene) (Fig. 5a). Pairwise similarity ratings of odor quality verified that subjects successfully grouped this nine-odorant set into three odor categories, as assessed using hierarchical cluster analysis (Fig. 5b). In addition, another ten independent participants, using a standardized questionnaire31 to rate the applicability of 146 odor descriptors to each stimulus (Methods), robustly classified the odorants into the relevant perceptual categories chosen a priori (minty category, χ2 = 16.63, P = 0.0002; woody, χ2 = 16.62, P = 0.0003; citrus, χ2 = 16.92, P = 0.0002; Friedman test) (Fig. 5c). A weaker category effect was also observed for the floral descriptors (χ2 = 6.74, P = 0.034), although the “floral” ratings were significantly lower than the “minty” ratings for minty odorants (Z = 3.67, P = 0.0002, Wilcoxon test), the “woody” ratings for woody odorants (Z = 3.87, P = 0.0001), and the “citrus” ratings for citrus odorants (Z = 2.88, P = 0.004). In fact, the “floral” ratings were no different than the “woody” (Z = 0.51, P = 0.61) or “citrus” (Z = 0.34, P = 0.73) ratings for the minty odorants, nor were they different from the “minty” (Z = 0.57, P = 0.57) or “woody” (Z = 0.99, P = 0.32) ratings for the citrus odorants. Together, these data show that the stimulus set closely conforms to the perceptual categories of odor quality proposed here. Finally, there were no significant category differences in behavioral ratings, sniffing, or mean PPC activity (Supplementary Fig. 2).
Fig. 5.
Odor stimuli and psychophysical ratings for Experiment 2. (a) The nine odorants included three stimuli per each of three quality categories (minty, woody, and citrus). (b) A dendrogram plot obtained from cluster analysis of the mean pairwise similarity ratings of odor quality revealed that the nine odorants sorted into three quality categories. Shorter distances indicate greater similarity. (c) Ratings of the applicability of 146 odor descriptors to the odor stimuli (descriptors pre-sorted into six different quality categories) indicated that subjects classifed the odorants into the appropriate categories (mean ± between-subjects s.e.m; N = 10). Non-parametric Friedman tests for related samples were separately conducted on each category (*, P < 0.05; **, P < 0.005).
During fMRI scanning the nine odorants were presented to the subjects 24 times each, spread over three days. Subjects tacitly identified the quality of the odor on each trial, and patterns of odor-evoked brain activity, extracted from the same four regions as in Experiment 1, were organized into vectors of voxel activity. The entire set of pattern vectors was divided into even and odd runs, and then linear correlations were calculated between every possible odorant pair (total of 72 unique correlations). Finally, classification accuracy was computed by testing the proportion of within-category correlations (e.g. mint-1/even vs. mint-2/odd) that were greater than across-category correlations (e.g. mint-1/even vs. woody-1/odd). Classification was significantly above chance across the group in PPC (T3 = 2.34, P = 0.050), but not in other regions (APC, T3 = 1.31, P = 0.14; amygdala, T3 = 0.48, P = 0.33; OFC, T3 = 0.27, P = 0.40) (Fig. 6a). Additionally, the average within-category correlation was greater than the across-category correlation only in PPC (T3 = 3.46, P = 0.041) (Fig. 6b), but not in APC (T3 = 0.78, P = 0.49), amygdala (T3 = 0.47, P = 0.67), or OFC (T3 = 0.47, P = 0.67). A one-way ANOVA testing for an effect of quality category on the within vs. across correlation differences in PPC was not significant (F2,9 = 0.30, P = 0.75), implying that the above results were not driven by one particular category.
Fig. 6.
fMRI pattern discrimination of odor categorical perception in PPC. (a, b) Classification performance calculated using fMRI patterns of ensemble activity. (a) Odor identification accuracy (mean ± between-subjects s.e.m; N = 4) was significantly greater than chance in PPC only. *, P < 0.05. (b) The within-category correlation was greater than the across-category correlation in PPC for all three odor quality categories, an effect that was separately observed for each category. (c, d) Classification performance calculated using fMRI mean activity. (c) Identification accuracy did not significantly differ from chance in any of the four regions, and there was no significant group difference between within-category and across-category Euclidean distances in PPC (d), or in APC, amygdala, or OFC (data not shown).
Following the same methods outlined in Experiment 1, we performed a split-halves classification analysis to determine whether mean fMRI activity contains reliable information about olfactory perceptual categories. Identification accuracy, calculated as the proportion of within-category distances shorter than across-category distances, was not significantly different from chance in any of the four regions of interest (Fig. 6c) (P's = 0.44, 0.39, 0.16, and 0.22, for PPC, APC, amygdala, and OFC, respectively). Furthermore, there were no significant differences in PPC between within-category and across-category distances (T3 = −1.23, P = 0.31; Fig. 6d). These data provide persuasive support for the idea that local (ensemble) fMRI activity in PPC, but not global (mean) fMRI activity, contains discriminating information about odor quality categories. At the same time, odor classification accuracy was not greater than chance when trial-specific respiratory parameters were entered into split-halves analyses (Supplementary Fig. 1b), indicating that sniff peak amplitude, duration, and volume (and their combination) are unable to account for the categorical effects in PPC.
In a subsequent analysis, we used multidimensional scaling (MDS) techniques32 to test the hypothesis that multi-voxel patterns of PPC activity coincide with perceptual ratings of odor quality, on an odorant-by-odorant basis. There were two related predictions: first, that odorants sharing a high (or low) degree of spatial overlap in PPC should be perceived as smelling more (or less) similar in quality; and second, that odorants exhibiting greater spatial overlap in PPC should be perceived as belonging to the same odor category. To implement this procedure, we assembled two “distance” matrices out of the nine-odorant data-set: a 9-by-9 imaging matrix composed of the multi-voxel fMRI correlations (averaged across subjects) in PPC for every odorant pair; and a 9-by-9 perceptual matrix composed of perceived differences in odor quality between every odorant pair. We then used classical MDS to project the distance matrices onto a common three-dimensional space, followed by a standard linear transformation algorithm (Procrustes alignment) to determine how well the imaging matrix aligned with the perceptual matrix.
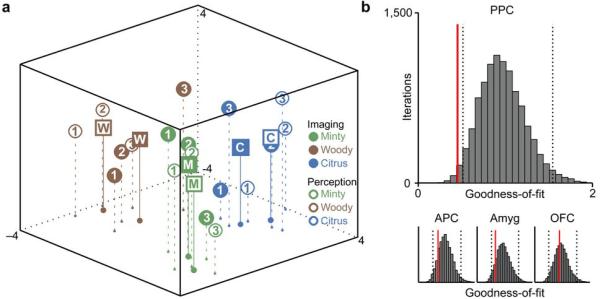
The MDS analysis demonstrated robust spatial correspondence between the projected PPC imaging map and the projected perceptual map (Fig. 7a): odorants belonging to the same category clustered together within each map, and clustering of odor quality categories (as well as of individual odorants) was closely aligned between the imaging and perceptual maps. Statistical significance of this latter effect was tested by calculating the “goodness-of-fit” (i.e., the sum of squared errors arising from alignment of the perceptual and imaging data-sets, where lower values indicate stronger fits). This parameter was plotted against a distribution of goodness-of-fits, generated by randomly shuffling the identities of each of the nine odors (10,000 iterations). The observed goodness-of-fit in PPC (0.53) was situated outside the lower bound of the 95% confidence interval of the randomly generated distribution (Fig. 7b), demonstrating a significant alignment between imaging and perception in this region. In contrast, when imaging matrices of APC, amygdala, and OFC were each aligned to the perceptual matrix, the goodness-of-fit (using the same methods) was not statistically significant (Fig. 7b). These findings reinforce the idea that ensemble coding of odor categorical perception is regionally specific for PPC.
Fig. 7.
Alignment of fMRI spatial patterns and perceived odor quality. (a) The group-averaged imaging and perceptual data-sets were each projected onto a common three-dimensional space using multidimensional scaling (MDS), revealing that the imaging maps of PPC linear correlations (filled circles) closely overlapped with the perceptual maps of odor quality similarity (empty circles), both for individual odorants and for odor quality categories. Squares labelled “M” (minty), “W” (woody), and “C” (citrus) represent centroids of each category for the imaging (solid squares) and perceptual (empty squares) data. (b) The observed goodness-of-fit in PPC (red line) fell outside the lower bound of the 95% confidence interval (dashed lines) of a randomly permuted distribution of goodness-of-fits, demonstrating a significant alignment between PPC activity and perception in this region. Alignment between imaging and perception was not significant in APC, amygdala, or OFC.
It is important to note that the three stimulus exemplars within each of the three odor categories are chemically and structurally diverse. Nonetheless, the possibility that unforeseen molecular similarities could explain some of the observed correlation effects still remains. To examine this issue, we obtained a set of 32 optimized molecular descriptors for each odorant, which have been shown to account well for variations in neural activity observed in the olfactory epithelium and olfactory bulb of different animal species33. Pairwise Euclidean distances between each odorant were calculated from the set of molecular descriptors and used to assemble a 9-by-9 molecular distance matrix, similar to the ones assembled for the imaging and perceptual data. We then implemented the permutation analysis as described above, but this time using the molecular distance matrix as the alignment target, in place of the perceptual matrix. The observed value of goodness-of-fit between imaging and molecular features fell inside of the 95% confidence interval of the randomly generated distribution in PPC, APC, amygdala, and OFC (Supplementary Fig. 3), indicating that relationships between odorants in molecular space cannot easily account for relationships between odorants in imaging space, at least with respect to these particular olfactory brain regions.
Finally, we combined MDS with multiple linear regression analysis to evaluate whether there is a consistent predictive relationship between PPC ensemble activity and odor quality perception across subjects. This question was tested by considering the three-dimensional MDS imaging projections of the PPC correlations as perceptual estimates of odor quality, closely following methods for predicting odor perception from olfactory bulb (or olfactory mucosal) activity in rats11,34. Importantly, a “leave-one-subject-out” approach was used to maintain data independence, such that the average PPC imaging correlation matrix of three subjects was used to estimate the perceptual matrix of a fourth (test) subject, with four-fold cross-validation, preventing the test set from influencing its own prediction. Multiple linear regression analysis showed that there was a highly significant predictive relationship between estimated odor quality and actual odor quality (R = 0.44; F1,24 = 18.84; P = 0.001), in the absence of significant across-subject variability (R = 0.16; F3,24 = 0.61; P = 0.615), suggesting that the capacity of PPC to encode information about odor quality generalizes across subjects.
Discussion
In this study we have integrated high-resolution olfactory fMRI, cortical flattening techniques, sensory psychophysical assays, and multivariate analyses to provide measurements of odor-evoked piriform spatial activity patterns and odor quality perception within the same set of subjects. Up until now pattern-based techniques had not been successfully implemented in the context of human olfactory imaging (though see Ref. 19), in spite of their widespread use to delineate odorant spatial mapping in animal imaging studies of the olfactory bulb35–37. The methods outlined here have enabled us to consider a new set of research questions not previously testable, centering around the idea that ensemble fMRI activity patterns may represent a viable signature of sensory perception that can be used to infer olfactory perceptual experience. That odor classification could be reliably attained from multi-voxel fMRI patterns, but not from mean fMRI responses (cf. Figs. 3 and 6), indicates that multivariate measures of fMRI ensemble activity are particularly suited to extract discriminating information about odor qualities and categories from the human olfactory brain.
These results show that the spatial arrangement of fMRI activity in PPC is distributed and overlapping for each odorant, without any obvious local clustering (Fig. 4), in keeping with the known anatomical organization of this region15,16. Odorants belonging to the same perceptual category exhibited similar pattern topography (Fig. 7), suggesting that perceptual information about odor quality may be reflected in distributed ensemble activity within PPC. Overall, the present findings suggest that olfactory codes of odor object categories are arranged much in the same way that visual object categories (houses, cows, chairs) are organized in inferotemporal cortex20, highlighting the critical sensory-associative nature of PPC. It is reasonable to speculate that the degree of correlation between odor-evoked PPC spatial patterns and pre-existing odor “templates”38 would provide a convenient metric by which the brain could infer similarities among odorants and classify odor objects into discrete, meaningful categories.
As a point of clarification, references here to “spatially distributed” codes and patterns are not meant to imply that the brain is abstracting and codifying information about the spatial location of odors in the external world. Such definitions are highly relevant to neural coding of visual and auditory stimuli, but are probably less pertinent to the olfactory modality (for further discussion of this topic see Ref. 39). Rather, as used in the present context, a “spatially distributed” olfactory code refers to a spatial ensemble representation of odor-evoked activity dispersed across a set of fMRI voxels. Ultimately, what matters is the formation of a unique spatial (or spatiotemporal) pattern of cortical activity that reliably encodes the perceptual identity of an odorant, in keeping with “content addressable memory” models that are predicated on unique distributed neuron ensembles in piriform cortex14,18,40,41.
Interestingly, odor-specific fMRI ensemble patterns were also reliably observed in OFC, but this finding was restricted to the first experiment (Fig. 3b,c). Considering that Experiment 1 included only one stimulus exemplar per odor category, the multivariate data across both experiments suggest that PPC and OFC each contain fMRI ensemble representations of individual odorants (odorant “identity”), but that PPC alone contains ensemble representations of odor perceptual category (odor “quality”). That we observed effects across diverse odor classes (minty, woody, citrus) substantiates the idea that PPC is a key substrate where categorical perceptions of odor objects are maintained. On the other hand, it remains unclear whether OFC may encode some discriminating perceptual feature other than quality, but the idea that the neural fidelity of individual odorants is preserved in OFC nicely accords with single-unit recordings in monkeys30 showing that the specificity of odor tuning also happens to be highest in this region.
Data from Experiment 2 demonstrated a statistically significant relationship between PPC spatial patterns and subjective similarity ratings of odor qualities and categories. Indeed, to the extent that PPC ensemble maps correlate with subjective perceptions of odor quality, these results suggest that it might be possible to read out olfactory percepts from spatially distributed fMRI activity in PPC. The pattern regression analysis further extended these findings, by showing that perceptual estimates of odor quality can be inferred from group-based three-dimensional MDS projections of PPC imaging correlations. That the perception of odor object categories can be estimated from group-averaged PPC correlations would fulfill an important criterion42 for establishing an olfactory code with direct relevance for odor quality perception. The ability of multivariate techniques to draw out these predictive relationships highlights a novel and robust method for characterizing the direct links between olfactory codes and odor percepts, and perhaps for assessing (or predicting) individual human variability in how an odorant's quality is perceived.
Several influential models of olfactory perception14,17,18,38,40 posit that a spatially (or spatiotemporally) distributed pattern of brain activity satisfies the requirements of an odor detection system faced with the challenge of extracting perceptual constancy from environmental inconstancy. Ecological variations in odor background, wind direction, and sniff sampling phase, as well as physical alterations in the odor source itself, can all degrade stimulus fidelity of the original input. The prevailing idea that “pattern completion” mechanisms and “content-addressable memory” systems14,18 can resolve corrupted odor inputs hinges on the presence of ensemble activity patterns in olfactory cortical structures, but these theories are principally derived from anatomical data and network simulations, in the absence of confirmatory functional data. Our findings are among the first to verify the presence of pattern-based perceptual representations of odor qualities and categories in human PPC, which would be indispensable for the reconstruction of odor objects, ensuring perceptual invariance in the wake of fragmentary inputs.
Methods
Subjects
Twenty subjects (age range: 22–35 years) gave informed consent to participate in this study, which was approved by the Northwestern University Institutional Review Board. Six subjects (4 women) participated in Experiment 1, four subjects (3 women) participated in Experiment 2, and ten subjects (8 women) completed odor questionnaires for Experiment 2.
Behavioral ratings
Prior to the first scanning day, subjects rated odor intensity, valence, pungency, and familiarity using visual analog scales43,44. Ratings were analyzed in Matlab (Mathworks) using one-way ANOVAs. Subjects also rated odor quality similarity between all possible odorant pairs on a visual analog scale (anchors “not alike at all” and “identical”). In Experiment 2 the 36 pairwise similarity ratings, averaged across subjects, were transformed into a hierarchical cluster tree using a single-linkage algorithm in Matlab and plotted as a dendrogram, with linkage distances (reflecting degree of quality similarity between odorants) indicated on the y-axis.
Ten independent participants completed a 146-item Odor Quality Evaluation questionnaire31 for each odorant. Ratings of the applicability of odor descriptors on the questionnaire range from 0 (“Absent”) to 5 (“Extremely”). Before acquiring ratings, and following prior techniques24,45, we identified the descriptors best fitting the three perceptual categories (minty, woody, citrus), plus three control categories (floral, berry, spicy) (Supplementary Table 1). For each descriptor category, odorant-specific ratings were averaged across descriptors belonging to that category, and then were averaged across the three odorants pre-assigned to each perceptual group. For example, all of the “minty” descriptor ratings were averaged together for each odorant, and then averaged across the three putative minty odorants, the three putative woody odorants, and the three putative citrus odorants, to form mean ratings for each odorant perceptual category. The applicability of each descriptor rating to each perceptual category was tested (Friedman tests for related samples and Wilcoxon sign-rank tests for paired samples).
Respiratory monitoring and analysis
Subjects were affixed with breathing belts to monitor respirations during scanning43. Sniff peak amplitude, duration, and inspiratory volume were computed for each trial, averaged across conditions and runs, normalized within subjects by subtracting the mean parameter value (across conditions) from each condition-specific value, and then entered into individual one-way ANOVAs for statistical analysis.
fMRI data acquisition
Gradient-echo T2-weighted echoplanar images (EPI) were acquired with a Siemens Trio 3T scanner using parallel imaging19, an eight-channel head-coil, and the following parameters: TR, 2 s; TE, 20 ms; matrix size, 128 × 120 voxels; field-of-view, 220 × 206 mm; in-plane resolution, 1.72 × 1.72 mm. slice thickness, 2 mm; gap, 1 mm; acquisition angle, 30° rostral to the intercommissural line. A 1-mm3 T1-weighted MRI scan was obtained to define anatomical regions of interest (ROIs).
Multivariate fMRI paradigm
In both experiments, there were eight fMRI runs on each of three consecutive days (total of 24 runs). In Experiment 1 each odorant was presented once per run for 10 s, with 20-s rest between odorants (Fig. 1). In Experiment 2 each odorant was presented once per run for 6 s with 12-s rest. Odorants were presented using an MRI-compatible olfactometer (airflow, 2.5 L/min)24. Upon odorant presentation, subjects were visually cued to sniff and to identify the odor quality covertly. Each run was separated by approximately 180 s to minimize olfactory fatigue. Odorant presentation was counterbalanced across runs such that each possible stimulus order was used once in Experiment 1, and each odorant appeared only once per run and a different stimulus order was used for each run in Experiment 2.
Odor localizer scan
We used an independent “odor localizer” fMRI scan (using the same imaging parameters as the main experiment) as an unbiased way to identify odor-active voxels44. In Experiment 1, on each scanning day, subjects underwent an odor detection task consisting of 12 odor and 12 no-odor (air only) trials, using four odorants (butanol, anisole, heptanol, and α-ionone). In Experiment 2, two odor localizer runs were performed on a fourth day of scanning. Odor and no-odor trials were presented for 2 s (stimulus-onset asynchrony, 12 s), followed by a button press to indicate whether an odor was present or absent. Images were spatially realigned, smoothed (6-mm full-width-half-maximum), and then analyzed using the general linear model (GLM) in SPM2 (www.fil.ion.ucl.ac.uk/spm/). After model estimation, we contrasted odor vs. no-odor conditions, collapsed across runs, to generate subject-specific statistical parametric maps of odor-active cortex.
Voxel selection procedure
Voxels included in the multivariate analyses were selected from anatomically defined ROIs, manually drawn on each subject's structural MRI scan using MRIcroN software (www.mricro.com) (Supplementary Fig. 4). Anatomical definition of PPC, APC, and amygdala was guided by a human brain atlas46. Delineation of OFC was based on an olfactory fMRI meta-analysis27. Subsequently, the voxels within each subject's ROI were functionally ranked, according to their T-values (obtained from the independent odor localizer scans), and specifically without reference to the pattern-based fMRI data or to odor classification performance. For each ROI, we considered the N most odor-active voxels for which anatomical data were maximally available from all subjects, such that N was set based on the subject with the smallest anatomical ROI. Thus, if the PPC of Subjects 1, 2, and 3 contained 200, 150, and 300 voxels, respectively, then N = 150 voxels. In this manner, the size of each ROI was different across regions, but the same across subjects for a given ROI. The respective voxel numbers used for analysis of PPC, APC, amygdala, and OFC in Experiment 1 were 186, 138, 312, and 195, and in Experiment 2 were 228, 155, 339, and 239.
Multivariate fMRI data pre-processing
After discarding the first six “dummy” volumes of each run, all functional images were spatially realigned to the first volume of the first run using SPM2. Subsequent steps, including (a) extraction of fMRI signal intensity from each voxel within an ROI for each run, (b) temporal detrending with a second-order polynomial47, and (c) assembly into linear vectors of odor-specific voxel activity, were done outside of SPM2, using customized scripts in Matlab. To preserve the native spatial fidelity of the fMRI signal, image normalization and smoothing were not performed. Control analyses indicated that spatial realignment across the three scanning days was robust (Supplementary Fig. 5, Supplementary Data).
Multivariate fMRI analysis
Ninety-six pattern vectors (Fig. 1) were extracted for Experiment 1 (one vector for each of the four odorants across 24 runs), and 216 pattern vectors were extracted for Experiment 2 (nine odorants, 24 runs). In both experiments, pattern vectors were split into even and odd halves, and the mean activity across all conditions was subtracted from each vector in that half20. These mean-corrected vectors were averaged across runs within each half, producing one pattern vector per condition per half, which were then used to calculate pairwise correlation coefficients within and between odorants (Experiment 1) and odor quality categories (Experiment 2).
Within-odor and across-odor correlations were compared directly, and were also used to estimate identification accuracy20. In Experiment 1, identification accuracy was calculated such that, for odor A, if (Aeven vs. Aodd) was greater than (Aeven vs. Bodd), it was counted as a correct identification. There were 6 such comparisons for each odor (given four unique odor types), resulting in 24 comparisons. In Experiment 2, identification accuracy was calculated such that if a within-category correlation (e.g. Mintyeven vs. Mintyodd) was greater than an across-category correlation (e.g. Mintyeven vs. Woodyodd), it was counted as a correct identification. Thus, for each of the three odor quality categories there were three within-category correlations and 18 across-category correlations, resulting in 54 comparisons per quality category. Note, the pairwise correlations (total of 72 correlations, excluding on-diagonal correlations to prevent estimation bias of the within-category correlations) were calculated before mean within- and across-category correlation coefficients were computed. Chance identification accuracy was 50%.
Piriform flat maps
In the first step, we specified a GLM using SPM2 software that included odor onset times from Experiment 1 as regressors of interest (collapsed across runs). Percent signal change was then estimated for each condition. Flattened cortical maps were made using mrVista software (http://white.stanford.edu/software/). In this process, each subject's T1-weighted anatomical image was segmented to isolate grey matter in PPC, followed by spatial flattening into a two-dimensional map. The functional volumes containing voxel-wise percent signal change information from the GLM were then coregistered to the T1 volume and flattened to two dimensions using the same transformation parameters.
Multidimensional scaling
Multidimensional scaling (MDS) is commonly employed to measure similarities among complex, high-dimensional datasets containing non-independent elements32. Here it was used to facilitate comparisons and statistical analysis between fMRI and perceptual data acquired in Experiment 2. First, a 9-by-9 imaging dissimilarity (distance) matrix for each ROI was created for each subject by subtracting the pairwise linear correlation coefficients from a value of 1, and scaling from 0 to 10. Second, a 9-by-9 perceptual dissimilarity (distance) matrix was created using each subject's similarity ratings of perceived odor quality for every odorant pair. These distance matrices were each averaged across subjects and entered into classical MDS analyses (Matlab). This generated three-dimensional maps (one for the imaging data, one for the perceptual data), with each odorant represented by a unique coordinate in x-y-z space. Finally, the three-dimensional map of imaging coordinates was aligned to the perceptual map using Procrustes analysis, which provides a quantitative measure of similarity (“goodness-of-fit”) between two sets of coordinates (ranging between 0 and 1, where lower values indicate better alignment). Selection of a three-dimensional MDS space was based on a “scree plot” of the perceptual data-set indicating that a three-dimensional projection best captured the variance in the nine-dimensional matrix.
To estimate the significance of goodness-of-fit, a random permutation (Monte Carlo) procedure was implemented, whereby the actual (measured) fit was compared to a distribution of fits generated by randomizing the odor condition assignments prior to MDS analysis and Procrustes alignment (10,000 iterations). This procedure scrambles the assignment of odorant condition (and category) in advance of MDS, preserving the values of all pairwise distances and minimizing overfitting of the data. Randomization statistics have been applied to studies comparing multivariate response profiles between different brain regions or between neural and perceptual data-sets48–50. An observed fit between imaging and perception was considered significant at P < 0.05, meaning that the observed fit was smaller (better fit) than 95% of randomly generated fits.
For presentation the goodness-of-fits, d, were transformed to normal values using Fisher z-transformation:
Odor classification: mean fMRI activity
To test whether odor-specific information is reliably contained in the mean fMRI signal amplitude, we performed a “split-halves” classification analysis that closely paralleled the methods employed to analyze fMRI ensemble activity. First, the raw BOLD signal elicited by each odorant presentation was averaged across all voxels from each ROI for each trial, resulting in 24 values per condition (corresponding to the 24 runs of data acquisition). Mean signal across conditions within each run was subtracted from the condition-specific values within that run for normalization. Normalized signals were split into halves, one containing values from even runs and one from odd runs. Data within each condition and half were averaged across runs, and pairwise Euclidean distances were calculated within and between conditions.
Odor classification: sniff parameters
To test whether trial-specific sniff parameters could be used to successfully classify the odors, a “split-halves” analysis was performed on sniff peak, duration, and inspiratory volume. Additionally, the three sniff parameters were combined into one three-component vector (analogous to vectors of voxel activity used in the pattern-based fMRI analyses) to test whether the combination of these parameters might contain odor-specific information. Data were normalized by subtracting the mean parameter value across conditions within each run from the condition-specific values. Data were then split into halves according to even and odd runs and averaged across runs in each half. Pairwise Euclidean distances were calculated within and across conditions.
Statistical analyses
Results are shown as means ± s.e.m. for participants or conditions. Where not otherwise indicated, statistical significance was determined using one-tailed t-tests (when comparing identification accuracy to chance), two-tailed t-tests (when comparing two conditions), or ANOVAs (when comparing more than two conditions). Results were considered significant at P < 0.05.
Supplementary Material
Acknowledgments
The authors would like to thank T. Egner, W. Li, J.-P. Magué, and T. Parrish for helpful suggestions. This work was supported by Grant # K08-DC007653 from the National Institute on Deafness and Other Communication Disorders to J.A.G.
References
- 1.Rosch EH. Principles of categorization. In: Rosch EH, Lloyd B, editors. Cognition and Categorization. Erlbaum Associates; Hillsdale: 1978. pp. 27–48. [Google Scholar]
- 2.Miller EK, Nieder A, Freedman DJ, Wallis JD. Neural correlates of categories and concepts. Curr. Opin. Neurobiol. 2003;13:198–203. doi: 10.1016/s0959-4388(03)00037-0. [DOI] [PubMed] [Google Scholar]
- 3.Reddy L, Kanwisher N. Coding of visual objects in the ventral stream. Curr. Opin. Neurobiol. 2006;16:408–414. doi: 10.1016/j.conb.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Kendrick KM, et al. Neural control of maternal behaviour and olfactory recognition of offspring. Brain Res. Bull. 1997;44:383–395. doi: 10.1016/s0361-9230(97)00218-9. [DOI] [PubMed] [Google Scholar]
- 5.Todrank J, Heth G, Johnston RE. Kin recognition in golden hamsters: evidence for kinship odours. Anim. Behav. 1998;55:377–386. doi: 10.1006/anbe.1997.0611. [DOI] [PubMed] [Google Scholar]
- 6.Jacob S, McClintock MK, Zelano B, Ober C. Paternally inherited HLA alleles are associated with women's choice of male odor. Nat. Genet. 2002;30:175–179. doi: 10.1038/ng830. [DOI] [PubMed] [Google Scholar]
- 7.Guerrieri FJ, d'Ettorre P. The mandible opening response: quantifying aggression elicited by chemical cues in ants. J. Exp. Biol. 2008;211:1109–1113. doi: 10.1242/jeb.008508. [DOI] [PubMed] [Google Scholar]
- 8.Cleland TA, Morse A, Yue EL, Linster C. Behavioral models of odor similarity. Behav. Neurosci. 2002;116:222–231. doi: 10.1037//0735-7044.116.2.222. [DOI] [PubMed] [Google Scholar]
- 9.Linster C, Johnson BA, Morse A, Yue E, Leon M. Spontaneous versus reinforced olfactory discriminations. J. Neurosci. 2002;22:6842–6845. doi: 10.1523/JNEUROSCI.22-16-06842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linster C, et al. Perceptual correlates of neural representations evoked by odorant enantiomers. J. Neurosci. 2001;21:9837–9843. doi: 10.1523/JNEUROSCI.21-24-09837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Youngentob SL, Johnson BA, Leon M, Sheehe PR, Kent PF. Predicting odorant quality perceptions from multidimensional scaling of olfactory bulb glomerular activity patterns. Behav. Neurosci. 2006;120:1337–1345. doi: 10.1037/0735-7044.120.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zelano C, Sobel N. Humans as an animal model for systems-level organization of olfaction. Neuron. 2005;48:431–454. doi: 10.1016/j.neuron.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Carmichael ST, Clugnet MC, Price JL. Central olfactory connections in the macaque monkey. J. Comp. Neurol. 1994;346:403–434. doi: 10.1002/cne.903460306. [DOI] [PubMed] [Google Scholar]
- 14.Haberly LB. Neuronal circuitry in olfactory cortex: anatomy and functional implications. Chem. Senses. 1985;10:219–238. [Google Scholar]
- 15.Illig KR, Haberly LB. Odor-evoked activity is spatially distributed in piriform cortex. J. Comp. Neurol. 2003;457:361–373. doi: 10.1002/cne.10557. [DOI] [PubMed] [Google Scholar]
- 16.Sharp FR, Kauer JS, Shepherd GM. Laminar analysis of 2-deoxyglucose uptake in olfactory bulb and olfactory cortex of rabbit and rat. J. Neurophysiol. 1977;40:800–813. doi: 10.1152/jn.1977.40.4.800. [DOI] [PubMed] [Google Scholar]
- 17.Barkai E, Bergman RE, Horwitz G, Hasselmo ME. Modulation of associative memory function in a biophysical simulation of rat piriform cortex. J. Neurophysiol. 1994;72:659–677. doi: 10.1152/jn.1994.72.2.659. [DOI] [PubMed] [Google Scholar]
- 18.Hasselmo ME, Wilson MA, Anderson BP, Bower JM. Associative memory function in piriform (olfactory) cortex: computational modeling and neuropharmacology. Cold Spring Harb. Symp. Quant. Biol. 1990;55:599–610. doi: 10.1101/sqb.1990.055.01.057. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Howard JD, Parrish TB, Gottfried JA. Aversive learning enhances perceptual and cortical discrimination of indiscriminable odor cues. Science. 2008;319:1842–1845. doi: 10.1126/science.1152837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haxby JV, et al. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science. 2001;293:2425–2430. doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- 21.Kamitani Y, Tong F. Decoding the visual and subjective contents of the human brain. Nat. Neurosci. 2005;8:679–685. doi: 10.1038/nn1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haynes JD, Rees G. Decoding mental states from brain activity in humans. Nat. Rev. Neurosci. 2006;7:523–534. doi: 10.1038/nrn1931. [DOI] [PubMed] [Google Scholar]
- 23.Kriegeskorte N, Bandettini P. Analyzing for information, not activation, to exploit high-resolution fMRI. Neuroimage. 2007;38:649–662. doi: 10.1016/j.neuroimage.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottfried JA, Winston JS, Dolan RJ. Dissociable codes of odor quality and odorant structure in human piriform cortex. Neuron. 2006;49:467–479. doi: 10.1016/j.neuron.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Kadohisa M, Wilson DA. Separate encoding of identity and similarity of complex familiar odors in piriform cortex. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15206–15211. doi: 10.1073/pnas.0604313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tootell RB, et al. Functional analysis of V3A and related areas in human visual cortex. J. Neurosci. 1997;17:7060–7078. doi: 10.1523/JNEUROSCI.17-18-07060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottfried JA, Zald DH. On the scent of human olfactory orbitofrontal cortex: meta-analysis and comparison to non-human primates. Brain Res. Brain Res. Rev. 2005;50:287–304. doi: 10.1016/j.brainresrev.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Rolls ET, Critchley HD, Treves A. Representation of olfactory information in the primate orbitofrontal cortex. J. Neurophysiol. 1996;75:1982–1996. doi: 10.1152/jn.1996.75.5.1982. [DOI] [PubMed] [Google Scholar]
- 29.Schoenbaum G, Eichenbaum H. Information coding in the rodent prefrontal cortex. I. Single-neuron activity in orbitofrontal cortex compared with that in pyriform cortex. J. Neurophysiol. 1995;74:733–750. doi: 10.1152/jn.1995.74.2.733. [DOI] [PubMed] [Google Scholar]
- 30.Tanabe T, Iino M, Takagi SF. Discrimination of odors in olfactory bulb, pyriform-amygdaloid areas, and orbitofrontal cortex of the monkey. J. Neurophysiol. 1975;38:1284–1296. doi: 10.1152/jn.1975.38.5.1284. [DOI] [PubMed] [Google Scholar]
- 31.Dravnieks A. Atlas of odor character profiles. ASTM; Philadelphia: 1985. [Google Scholar]
- 32.Borg I, Groenen PJF. Modern Multidimensional Scaling: Theory and Applications. Ed. 2 Springer; New York: 2005. [Google Scholar]
- 33.Haddad R, et al. A metric for odorant comparison. Nat. Methods. 2008 doi: 10.1038/nmeth.1197. [DOI] [PubMed] [Google Scholar]
- 34.Kent PF, Youngentob SL, Sheehe PR. Odorant-specific spatial patterns in mucosal activity predict perceptual differences among odorants. J. Neurophysiol. 1995;74:1777–1781. doi: 10.1152/jn.1995.74.4.1777. [DOI] [PubMed] [Google Scholar]
- 35.Johnson BA, Leon M. Chemotopic odorant coding in a mammalian olfactory system. J. Comp. Neurol. 2007;503:1–34. doi: 10.1002/cne.21396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori K, Takahashi YK, Igarashi KM, Yamaguchi M. Maps of odorant molecular features in the Mammalian olfactory bulb. Physiol. Rev. 2006;86:409–433. doi: 10.1152/physrev.00021.2005. [DOI] [PubMed] [Google Scholar]
- 37.Soucy ER, Albeanu DF, Fantana AL, Murthy VN, Meister M. Precision and diversity in an odor map on the olfactory bulb. Nat. Neurosci. 2009;12:210–220. doi: 10.1038/nn.2262. [DOI] [PubMed] [Google Scholar]
- 38.Freeman WJ. EEG analysis gives model of neuronal template-matching mechanism for sensory search with olfactory bulb. Biol. Cybern. 1979;35:221–234. doi: 10.1007/BF00344205. [DOI] [PubMed] [Google Scholar]
- 39.Stevenson RJ, Wilson DA. Odour perception: an object-recognition approach. Perception. 2007;36:1821–1833. doi: 10.1068/p5563. [DOI] [PubMed] [Google Scholar]
- 40.Haberly LB, Bower JM. Olfactory cortex: model circuit for study of associative memory? Trends Neurosci. 1989;12:258–264. doi: 10.1016/0166-2236(89)90025-8. [DOI] [PubMed] [Google Scholar]
- 41.Wilson M, Bower JM. Cortical oscillations and temporal interactions in a computer simulation of piriform cortex. J. Neurophysiol. 1992;67:981–995. doi: 10.1152/jn.1992.67.4.981. [DOI] [PubMed] [Google Scholar]
- 42.Laurent G. A systems perspective on early olfactory coding. Science. 1999;286:723–728. doi: 10.1126/science.286.5440.723. [DOI] [PubMed] [Google Scholar]
- 43.Li W, Luxenberg E, Parrish T, Gottfried JA. Learning to smell the roses: experience-dependent neural plasticity in human piriform and orbitofrontal cortices. Neuron. 2006;52:1097–1108. doi: 10.1016/j.neuron.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plailly J, Howard JD, Gitelman DR, Gottfried JA. Attention to odor modulates thalamocortical connectivity in the human brain. J. Neurosci. 2008;28:5257–5267. doi: 10.1523/JNEUROSCI.5607-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevenson RJ. Associative learning and odor quality perception: how sniffing an odor mixture can alter the smell of its parts. Learn. Motiv. 2001;32:154–177. [Google Scholar]
- 46.Mai JK, Assheuer J, Paxinos G. Atlas of the Human Brain. Elsevier Academic Press; San Diego: 2004. [Google Scholar]
- 47.Haynes JD, Rees G. Predicting the orientation of invisible stimuli from activity in human primary visual cortex. Nat. Neurosci. 2005;8:686–691. doi: 10.1038/nn1445. [DOI] [PubMed] [Google Scholar]
- 48.Kayaert G, Biederman I, Vogels R. Representation of regular and irregular shapes in macaque inferotemporal cortex. Cereb. Cortex. 2005;15:1308–1321. doi: 10.1093/cercor/bhi014. [DOI] [PubMed] [Google Scholar]
- 49.Op de Beeck H, Wagemans J, Vogels R. Inferotemporal neurons represent low-dimensional configurations of parameterized shapes. Nat. Neurosci. 2001;4:1244–1252. doi: 10.1038/nn767. [DOI] [PubMed] [Google Scholar]
- 50.Young MP, Yamane S. Sparse population coding of faces in the inferotemporal cortex. Science. 1992;256:1327–1331. doi: 10.1126/science.1598577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.