Lead-In
The interaction of mammalian cells with nanoscale topography has proven to be an important signaling modality in controlling cell function. Naturally occurring nanotopographic structures within the extracellular matrix present surrounding cells with mechanotransductive cues that influence local migration, cell polarization, and other functions. Synthetically nanofabricated topography can also influence cell morphology, alignment, adhesion, migration, proliferation, and cytoskeleton organization. Here we review the use of in vitro synthetic cell-nanotopography interactions to control cell behavior and influence complex cellular processes including stem cell differentiation and tissue organization. Future challenges and opportunities in cell-nanotopography engineering will also be discussed including the elucidation of mechanisms and applications in tissue engineering.
Keywords: nanotechnology, nanostructures, cell adhesion, tissue engineering
1. Introduction to Cell-Nanotopography Interactions
1.1. Native Cell-Nanotopography Interactions
Extracellular matrix (ECM) proteins exhibit abundant nanometer-scale structures that are hypothesized to contribute to cell-matrix signaling. The basement membranes of many tissues exhibit rich nanotopography that interact directly with adjacent cells.[1, 2] Nanotopography is also present in individual ECM molecules such as collagen, which are approximately 300 nm in length and 1.5 nm in width.[3] These molecules can form fibrils that extend for tens of microns in length and have diameters between 260 and 410 nm.[4] Cells interact with native topographical structures in many ways, often through a phenomenon known as contact guidance. Contact guidance is a leading example of a naturally occurring phenomenon that is characterized by the response of cells to structures on the micron and sub-micron scale. Contact guidance is an essential component in regulating cell migration, which is modulated by organized ECM proteins.[5] Migration can also be influenced by surrounding cells as in the case of fibroblast migration in vivo[6] and coordinated epithelial cell migration on a collagen substrate in vitro.[7] T cell migration is also known to be highly dependent upon cell-biomaterial interactions with native ECM proteins.[8] The role of contact guidance can also be important in the migration of individual cells, or groups of cells or tissue.[9] The migration of nematocytes in hydra is guided bidirectionally by a fibrous mat produced from epithelial muscle processes[10] despite the absence of any detectable chemotactic gradient. Contact guidance is also an important component in efficient organelle formation, such as axonal guidance and growth cone motility.[11]
1.2. Cell-Nanotopography Responses on Synthetic Substrates
Recent developments in advanced micro- and nanofabrication techniques have enabled the fabrication of substrates that are able to recapitulate the structure and length scale of native topography in two-dimensional substrates. Cells respond to two-dimensional synthetic topographic substrates in a wide array of responses, which depend upon many factors including cell type, feature size and geomertry,[12] or the physical properties of the bulk substrate material including substrate stiffness.[13] Of particular interest for this review is the effect of synthetic substrates with features of that exhibit long-range order and sizes with length scales between approximately 10 nm and 3 µm. Although these length scales include micro- and nanometer-sized structures, they will herein be categorized and referred to as nanotopography for simplicity. This selection criterion excludes a large body of work that has examined the collective effects of other systems that lack long-range order including nanofibers,[14] electrospun fibrous mats, and substrates with nanoroughness.[15–17] Three basic nanotopographic geometries that will be discussed are nanogratings, nanopost arrays, and nanopit arrays (Figure 1). Nanotopography affects basic cell function in almost all types of mammalian cells (Table 1 and Table 2). Cell-nanotopography interactions can induce different effects within a single cell type due to the coupled effect of nanotopography in combination with physicochemical properties of the substrate. Cell-nanotopography interactions also vary across cell type, feature size, and feature geometry as well. Nevertheless, there are some general trends that can be extricated from the rapidly growing body of literature.
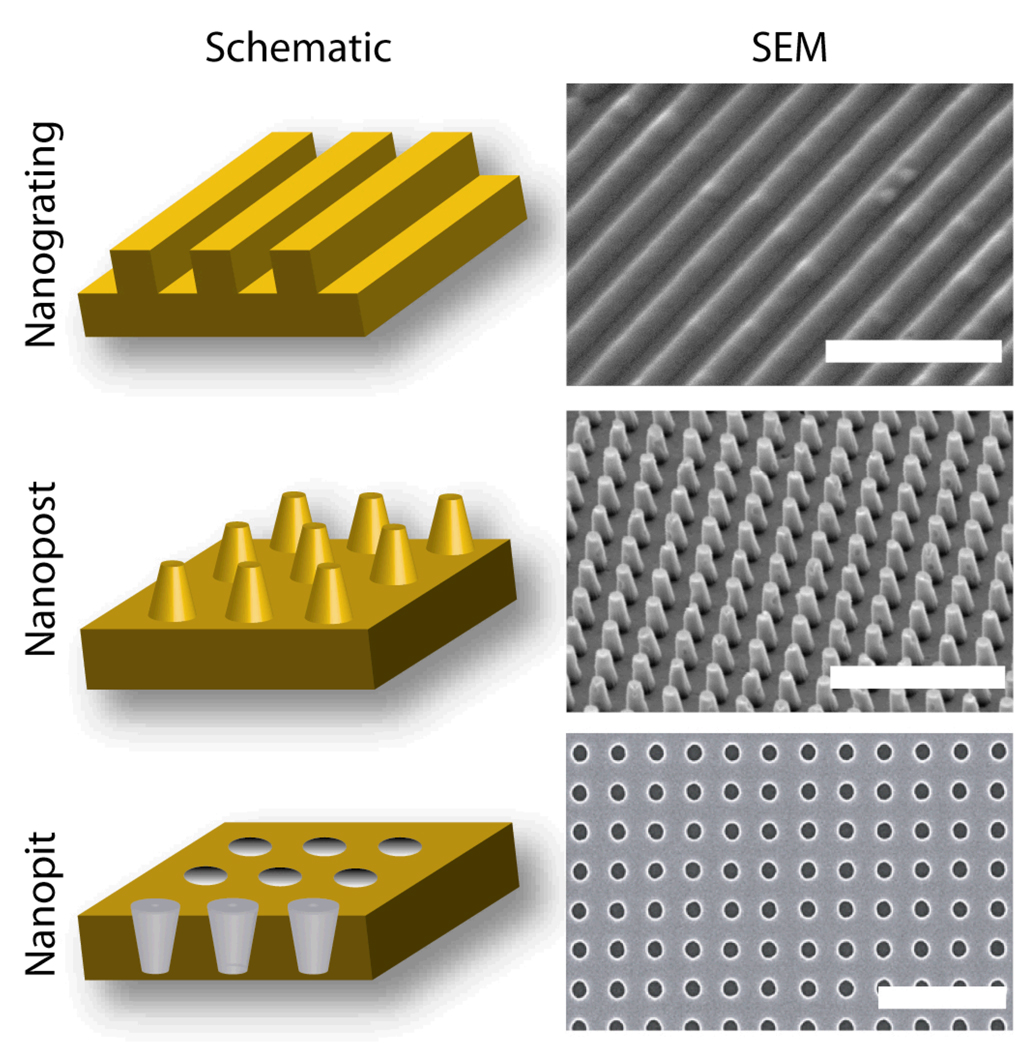
Figure 1.
Schematics and SEM Images of Representative Nanotopography Geometries. Three basic nanotopography geometries include nanograting (45° tilt, scale bar represents 5 µm), nanopost array (15° tilt, scale bar represents 5 µm) and nanopit array (0° tilt, scale bar represents 1 µm). Schematics not drawn to scale.
Table 1.
Generalized Cell Responses to Nanograting Topography.
| Cell Type | Feature Size |
Substrate Material |
Elongation, Alignment |
Cell Area | Attachment, Adhesion |
Proliferation | Biased Migration |
Other | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| hEndothelial Cells |
600 nm | PDMS | ++ | -- | -- | ++ | Organized into cellular superstructures |
[18] | |
| bEndothelial Cells |
2 µm | PGS | ++ | [19] | |||||
| rEndothelial Cells |
750 nm – 10 µm |
Ti | + | [20] | |||||
| hEmbryonic Stem Cells |
600 nm | PDMS | ++ | -- | -- | Cytoskeleton disrupting agents impact response |
[21] | ||
| hMesenchymal Stem Cells |
350 nm – 10 µm |
PDMS | ++ | -- | -- | Differentiation into neuronal lineage |
[22] | ||
| rC6 Glioma | 266 nm | PS | ++ | ++ | [23] | ||||
| hEKCs (HEK- 293) |
200–430 | PS | ++ | ++ | [24] | ||||
| bSmooth Muscle Cells |
350 nm | PDMS, PMMA |
++ | -- | Polarized MTOC | [25] | |||
| hFibroblasts | 50–600 nm | Si | ++ | - | - | [26] | |||
| Fibroblasts | 3–5 µm | Ti | ++ | 0 | Increase in fibronectin mRNA, incorporation |
[27] | |||
| hCorneal ECs | 70–2100 nm |
Si | ++ | -- | ++ | Biased lamellipodia extension |
[28, 29] |
||
| hCorneal ECs | 2–20 µm | PS | ++ | -- | ++ | [30] | |||
| hCorneal ECs | 1–4 µm | Quartz | ++ | Study of coupled topography, E-field, and soluble factors |
[31] | ||||
| PC12 | 70–1900 nm |
Si | ++ | Cooperative neurite extension |
[32] |
n/a: Data not available
++: Increase under all conditions
+: Increase under most conditions
0: No detectable change
-: Decrease under most conditions
--: Decrease under all conditions
Table 2.
Generalized Cell Responses to Nanopost and Nanopit Topography.
| Cell Type | Feature Size |
Substrate Material |
Spreading | Attachment, Adhesion |
Proliferation | Other | Ref. |
|---|---|---|---|---|---|---|---|
| hOsteoblasts | 300 nm (pit) |
PC | -- | Reduced area of adhesion complexes |
[33] | ||
| rCardiomyocytes | 150 nm | PEG | ++ | [34] | |||
| mP19EC Stem Cells |
300–500 nm |
PEG | ++ | [35] | |||
| hMesenchymal Stem Cells |
300 nm (pit) |
PMMA | Osteogenic Differentiation |
[36] | |||
| hBone Marrow Cells |
300 nm (pit) |
PC | -- | Constant filopodia formation |
[37] | ||
| hFibroblasts | 400–700 nm (post) |
PLA | ++ | -- | |||
| hFibroblasts | 50–600 (post) |
Si | - | 0 | [26] | ||
| hFibroblasts | 80 nm (pit) | Si | -- | Gene array analysis | [38] | ||
| hFibroblasts | 35–120 nm (pit) |
PCL | - | Increased filopodia | [39] | ||
| hFibroblasts | 35–120 nm (pit) |
PCL, PMMA |
-- | Biased orientation | [40] | ||
| mFibroblasts | 750–1500 nm (post) |
PDMS w Fn |
0 | Constant traction forces across geometry |
[41] | ||
| rFibroblasts | 60–150 nm (pit, post) |
PCL | - | Increased adhesion on random nanoposts |
[42] | ||
| HeLA | 160–1000 nm (post) |
PS | -- | 0 | [43] |
n/a: Data not available
++: Increase under all conditions
+: Increase under most conditions
0: No detectable change or trend in data set
-: Decrease under most conditions
--: Decrease under all conditions
1.2.1. Morphology
Perhaps the most palpable effect of nanotopography on cell function is the impact upon cell geometry. Many cell types typically respond to nanogratings by simultaneously aligning and elongating in the direction of the grating axis (Table 1). This response has been observed in various cell types across numerous species including fibroblasts, endothelial cells, stem cells, smooth muscle cells, epithelial cells (Figure 2), and Schwann cells.[44] Neurites extending from neuroblastoma cells (PC12) cultured on nanogratings have also been shown to exhibit enhanced alignment and extension when cultured in the presence of nerve growth factor (NGF). The morphological response is seen in cells cultured on substrates with features as small as 100 nm and depths as small as 75 nm.[45] Stronger responses have been observed across decreasing feature pitch and increased depth, the latter being the stronger effector in general. Other studies have demonstrated that some nanograting feature sizes induced alignment of cells both parallel and orthogonally to the nanograting axis.[46] There are several examples of cell types that do not respond to nanogratings including human-derived leukocytes, keratinocytes, and monocytes.[47] Hence, even widespread morphological effects of cell-nanotopography interactions are not universally observed across all cell types. Substrates with nanopost and nanopit features elicit a more subtle effect on cellular morphology. Many studies have demonstrated the reduction of spreading on nanoposts and nanopits, although the overall effect of these structures on cell area is unclear (Table 2). Other studies have observed either increased[39] or constant filopodia formation[37] and reduction in adhesion complex formation.[33]
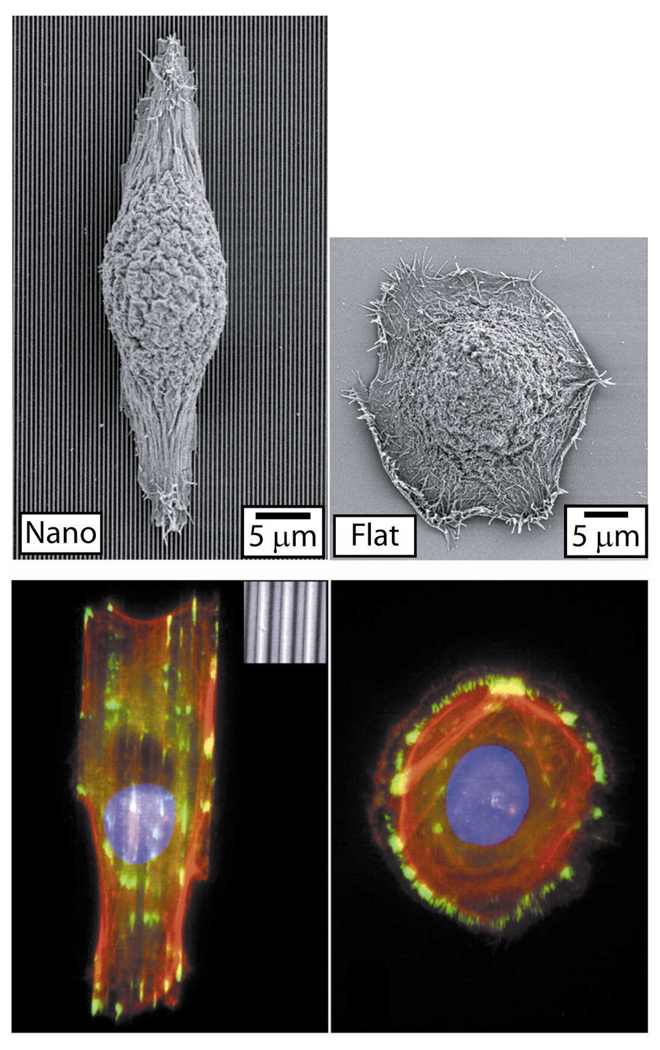
Figure 2.
Cell-Nanograting Response in Epithelial Cells. Epithelial cells respond to nanograting through alignment and elongation to the grating axis as evident through fluorescent and SEM micrographs. Other cell types exhibit similar morphological responses when cultured on nanograting substrates (Table 1). Reproduced with permission of the Company of Biologists.
1.2.2. Attachment and Adhesion
The length scale of synthetic nanotopography can be designed to mimic that of extracellular matrix proteins,[2] including collagen,[48] which may substantiate the hypothesis that nanotopography can enhance attachment and adhesion of mammalian cells. Nanogratings generally appear to enhance the adhesion in various cell-biomaterial-geometry combinations, nanoposts and nanopits generally reduce initial cell attachment (Table 1 and 2). Further studies must be aimed at elucidating the apparent dependence on feature size and geometry for differential adhesion.
1.2.3. Proliferation
Nanotopography has also been shown to affect the proliferation profiles of various cell types. In general, cells cultured on nanogratings exhibit reduced proliferation rates compared to cells cultured on planar substrates (Table 1). The effect of nanopost or nanopit substrates on proliferation is more ambiguous as some combinations of geometry, length scale, substrate material and cell types promote more rapid proliferation while others reduce proliferation rate (Table 2). There appear to be no obvious trends to predict the effect of nanopost or nanopit geometries on proliferation. Furthermore, there are currently no widely accepted hypotheses regarding the mechanism for the effect of cell-nanotopography interactions on cell proliferation.
1.2.4. Migration
The effect of nanotopography on migration is typically observed in cells cultured on nanogratings. Many cell types have exhibited biased migration direction in the direction of the grating axis and increased overall migration velocities including endothelial cells,[18] epithelial cells,[30, 31, 49] osteoblasts,[50] and C6 glioma cells[23] (Table 1). Nanotopography also biases markers for directional migration as shown in work by Yim et al. in which microtubule organization centers (MTOCs) were observed to be polarized as a direct consequence of the nanograting.[25] Furthermore, the polarization of MTOCs was observed to supersede directional migration cues from wound healing.[25] Enhanced migration is a response that is typically coupled with elongated morphology and alignment of the cell body with the nanograting axis.[18, 23, 50] There have been limited studies regarding cell migration on nanopit or nanopost arrays compared to nanogratings. One study by Tzvetkova-Chevolleau et al. studied the migration of normal (3T3) and malignant (SaI/N) mouse fibroblasts on poly(dimethylsiloxane) (PDMS) substrates with nanograting and nanopost arrrays.[51] Nanogratings biased the migration vector of both 3T3 and SaI/N cells along the grating axis. Rectangular arrays of nanoposts appeared to bias the direction of migration of 3T3 cells, but not SaI/N cells. Although there was no discernable effect of nanotopography on the migration velocities of 3T3 cells, there was a significant impact of nanoposts on SaI/N cells. SaI/N cells cultured on nanoposts exhibited a wide range of migration velocities including a high percentage of cells that attained high speeds. Additional studies must be conducted to further examine the potential impact of nanopost and nanopit geometries on migration profiles.
2. Cell-Nanotopography Interactions for Controlling Complex Function
2.1. Genotypic Alteration
Nanotopography is known to alter the gene expression profiles of various cell types. These genetic profiles have been analyzed first through analysis of individual genes followed by comprehensive gene analysis studies. A study by Chou et al. suggested that the mRNA levels and stability in human fibroblasts were influenced by nanogratings.[27] More specifically, fibroblasts cultured on titanium nanogratings expressed higher levels of fibronectin mRNA with increased stability. Furthermore, nanogratings induced higher levels of fibronectin incorporation into cell-matrix proteins. More recent work has utilized gene array techniques to probe the effects of nanotopography on genome-wide expression in fibroblasts[52] and mesenchymal stem cells.[22, 36] Fibroblasts cultured on nanopits exhibited widespread downregulation of many genes including those associated with apoptotic initiation, DNA repair, and transcription regulation.[38] Other genes are upregulated in this instance including TGF-βr2 and those involved in regulating G-protein signaling.
2.2. Differentiation
The concomitant impact of topography on both basic cell function and gene expression in many cells types suggests that nanotopography could potentially be utilized as a signaling modality for directing differentiation. There has been significant progress in this thrust despite the fact that coordinated work in this specific application of nanotopography has only recently been explored. Work by Yim et al suggests that human mesenchymal stem cells (hMSCs) cultured on nanogratings can be preferentially differentiated into neuronal lineages as determined by the presence of synaptophysin, tuj1, and nestin markers as well as the upregulation of MAP2.[22] The enhanced differentiation of hMSCs has also been explored using nanopit arrays. In work by Dalby et al., osteoprogenitor cells and hMSCs were cultured long-term on PMMA nanopit arrays of varying order.[36] The symmetry and order of the nanopits was found to significantly affect the expression of osteopontin and osteocalcin, two bone-specific ECM proteins, in both cell types (Figure 3). While hMSCs cultured on completely ordered or completely random nanopits did not lead to expression of these two proteins, hMSCs cultured on slightly irregular substrates did exhibit significant amounts of these proteins of interest. Increased bone nodule formation was also evident in hMSCs cultured on these substrates relative to substrates with either completely ordered or completely random features. Human MSCs were also cultured in three specific conditions: (1) nanopits, (2) planar substrates in the presence of dexamethasone (DEX), a soluble factor that can induce bone formation (positive control), and (3) planar substrates without DEX (negative control). Human MSCs cultured on nanopits expressed a similar level of many osteoblast-specific genes when compared to hMSCs cultured on flat substrates in the presence of DEX. Furthermore, some genes were specifically upregulated in hMSCs cultured on nanopits compared to hMSCs cultured with DEX alone. The results from these two studies demonstrate the potential of nanotopography to direct cell fate. Additionally, the complementary findings of hMSCs cultured on nanogratings and ordered-disordered nanopits suggest the potential for selective, controllable differentiation based solely on the geometry of the nanotopographic substrate.
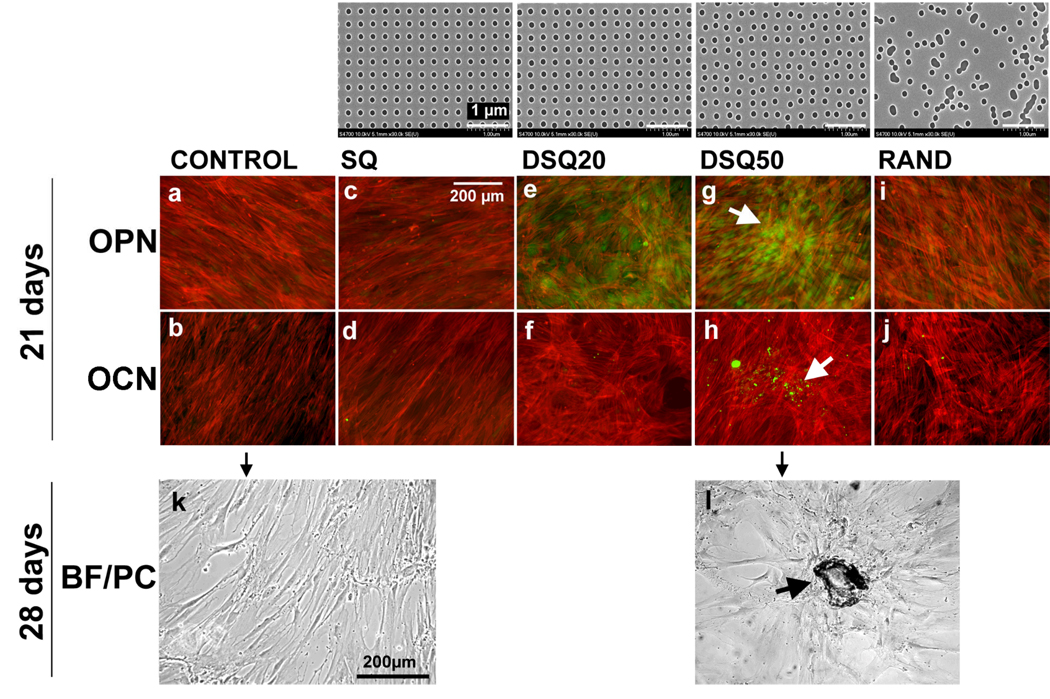
Figure 3.
Directed Differentiation of Human Mesenchymal Stem Cells to Osteoblast Lineage Using Nanopit Substrates. The top row shows images of nanopit arrays fabricated by electron beam lithography. All have 120-nm-diameter pits (100 nm deep, absolute or average 300 nm centre–centre spacing) with square (SQ), displaced square 20 (±20 nm from true centre, DSQ20), displaced square 50 (±50 nm from true centre, DSQ50) and random placements (RAND). Human MSCs cultured on a planar control substrate (a,f), note the fibroblastic appearance and no osteopontin (OPN) or osteocalcein (OCN) positive cells; on the SQ array (b,g), note the fibroblastic appearance and no OPN or OCN positive cells; on the DSQ20 array (c,h), note OPN positive cells; on the DSQ50 array (d,i), note OPN and OCN positive cells and nodule formation (arrows); on the RAND array (e,j), note the osteoblast morphology, but no OPN or OCN positive cells. (k,l) Phase-contrast/bright-field images showing that hMSCs cultured on the control (k) had a fibroblastic morphology after 28 d, whereas hMSCs cultured on DSQ50 arrays (l) exhibited mature bone nodules containing mineral. Reprinted by permission from Macmillan Publishers Ltd: Nature Materials (M Dalby, N Gadegaard, R Tare, A Andar, M O Riehle, P Herzyk, C D W Wilkinson, R O C Oreffo. Nat Mater 6. 997), copyright (2007).
2.3. Cell Superstructure
Modulating cell-nanotopography interactions also has the potential to influence cell-cell interactions with potential to generate complex multicellular structures. One such example of this is the culture of human endothelial progenitor cells (EPCs) on PDMS nanogratings.[18] EPCs responded to substrates though alterations in morphology with reduced proliferation and enhanced migration. The protein-level expression of endothelial markers in EPCs cultured on both nanograting and planar substrates was similar. However, EPCs cultured on nanogratings for up to 6 d formed segregated multicellular band structures (Figure 4) that were approximately 100 µm wide and spanned hundreds of microns in length. The morphology of EPCs organized into superstructures on nanogratings contrasted significantly with EPCs cultured on planar substrates, which formed confluent monolayers. The band structures found in EPCs cultured on nanogratings formed well-defined and organized capillary tubes in an in vitro matrigel assay. Confluent layers of EPCs formed during culture on planar substrates did not form distinct capillary tubes. This preferential formation found in EPCs cultured on nanotopography is hypothesized to occur for several reasons. First, alignment, elongation, and increased migration velocities biased morphology and cell-cell interactions. This biased contact produced band structures, which served as capillary tube precursors. Second, reduced proliferation of EPCs cultured on nanogratings prevented the formation of confluent monolayers. This reduced cell density could serve to enhance cell mobility during the induction of capillary tube formation with matrigel. Regardless of the possible mechanism, this study demonstrates the expanded potential for nanotopography to control the formation of multicellular structures.
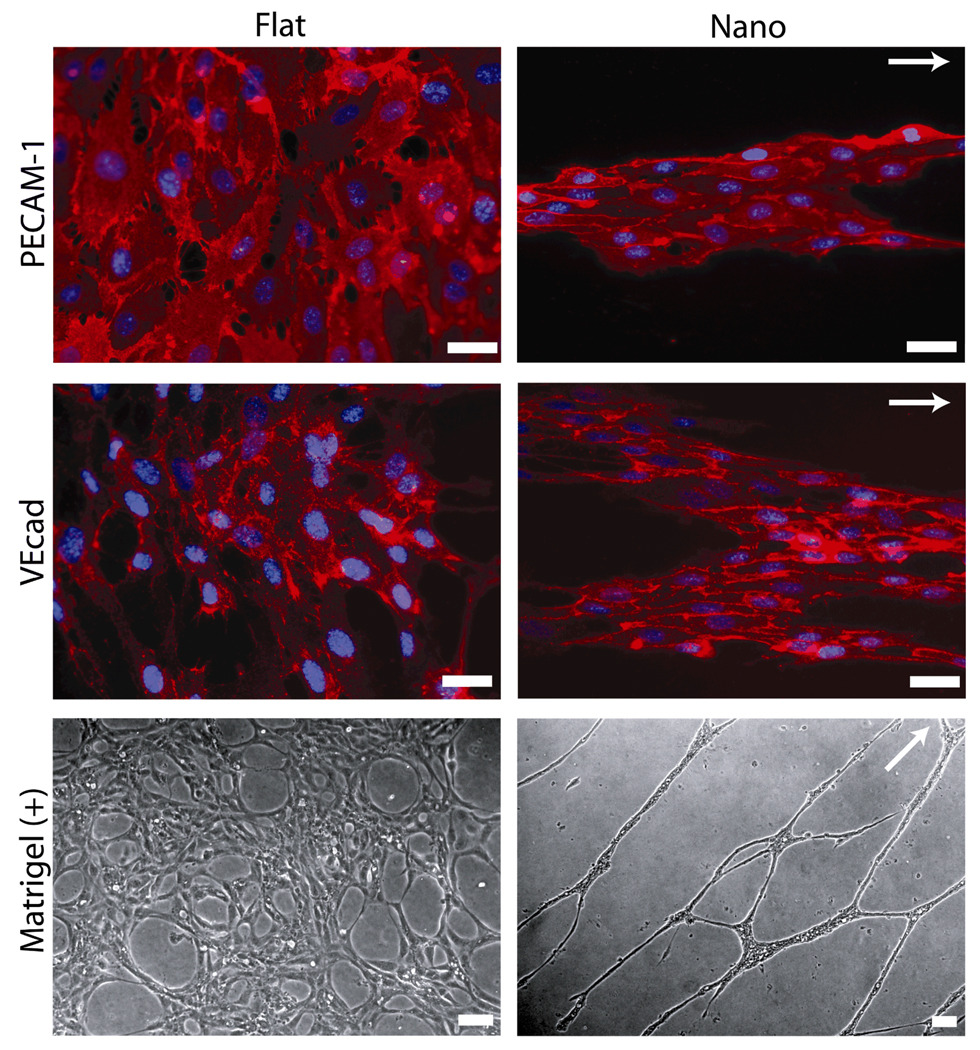
Figure 4.
Nanograting Substrates Promote Organized Multicellular Structures and Enhance In Vitro Capillary Tube Formation. Protein-level expression of platelet/endothelial cell adhesion molecule-1 (PECAM-1) and vascular endothelial cadherin (VEcad), two endothelial cell markers, was constant in endothelial cells cultured on both planar and nanograting substrates. However, endothelial cells cultured on planar substrates formed confluent monolayers while endothelial cells cultured on nanogratings were organized into multicellular band structures. These aligned band structures formed aligned capillary tubes in an in vitro matrigel assay (Matrigel (+)). The grating axis is indicated by the white arrows. Scale bars represent 50 µm. Adapted from C J Bettinger, Z Zhang, S Gerecht, J T Borenstein, R Langer: Enhancement of In Vitro Capillary Tube Formation by Substrate Nanotopography. Adv Mater. 2008. 20. 99–103. Copyright Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission.
3. Engineering Synthetic Nanotopographic Substrates for Tissue Engineering
3.1. Fabrication
There are a variety of advanced nanofabrication methods available for creating nanotopographic substrates with short and long-range order, which have been discussed in detail elsewhere.[53] The effect of order and symmetry of features has been demonstrated in numerous studies.[36, 40, 42] This suggests that the reaction of cells to substrates with feature roughness of a characteristic length scale should not be assumed to be equivalent to those cultured on ordered nanotopography of similar feature size. The nanofabrication of substrates with a long-range order across a wide range of feature sizes and geometries has been pursued using a variety of methods including traditional photolithography, e-beam photolithography, and interference lithography. Although suitable for creating ordered arrays of features, these processes are expensive, time consuming, and require access to intricate equipment. Alternative approaches have been pursued to fabricate polymeric substrates containing structures with long range order including the use of diblock copolymers[54] for nanograting lamellae[55] and nanosphere lithography[56] for nanopits.[57] Advanced fabrication techniques and compatible materials must eventually be synergized as a means to integrate cell-nanotopography interactions into advanced tissue engineering scaffolds.
There are several potential fabrication strategies for integrating nanotopographic cues into three-dimensional structures including advanced two-photon polymerization[58, 59] and micro-scale origami.[60, 61] The integration of nanotopographic cues into three-dimensional scaffold design and fabrication remains a challenging pursuit.
3.2. Utilization of Cell-Nanotopography Cues
Cell-nanotopography interactions have the ability to control stem cell differentiation and cellular superstructure, both of which have obvious implications for use in tissue engineering. However, the use of nanotopography as a cue to modulate basic cell function has potential use in scaffold design as well. For example, the influence of nanogratings on morphology can be used to form aligned populations of cells, which are important for the structure and function of smooth muscle cells and endothelial cells. Vascular tissue engineering scaffolds are of particular interest because of the correlation with alignment and cell function of multiple cell types within close proximity to one another. Recent work has led to the fabrication of a tubular scaffold with multiple nanograting surfaces.[61] The influence of nanotopography on adhesion could also serve as a method to create patterned arrays of cells without the need for direct patterning of proteins. Incorporating nanotopographic cues directly into a three-dimensional scaffold may therefore overcome the intrinsic planar limitations of microcontact printing[62–64] and related methods.[65, 66] Observed enhanced migration on nanogratings also has potential implications for the design of guidance channels for peripheral nerve regeneration. For example, tubular conduits modified with nanogratings could enhance the migration of Schwann cells into the injury cite to promote axonal regeneration.[67] These structures could also potentially promote the rapid migration of neurites across the nerve gap.
3.3. Material Selection
There has been substantial progress in the design and fabrication of nanotopography in a wide spectrum of materials systems. These techniques are typically designed to be used in combination with materials that are directly adapted or closely related to the semiconductor industry. Bulk materials processing and nanofabrication strategies for many nanotopographic surfaces are typically fine-tuned for silicon, silicon oxide, polycrystalline silicon, and other inorganic material systems such as titanium. These substrates can be used directly or serve as masters for replica-molding of organic polymers such as PDMS, polystyrene (PS), poly(methyl methacrylate) (PMMA), polycarbonate (PC), and poly(ethylene glycol) (PEG) for in vitro applications or biodegradable polymers such as poly(ε-caprolactone) (PCL), poly(L-lactic acid) (PLA), poly(glycolic acid) (PGA), and poly(L-lactic-co-glycolic acid) (PLGA) for potential use in vivo. Although the aforementioned biodegradable material candidates are ubiquitous in biomedical applications, there are significant drawbacks including bulk degradation and rigid mechanical properties in the case of PLA, PGA, and PLGA. Non-compliant materials can result in localized inflammation within the dynamic in vivo mechanical environment.[68–70] Novel material selection is of critical importance as cell-nanotopography interactions continue to be utilized in tissue engineering applications. Synthetic and natural materials must not only be selected on the basis of cell-biomaterial interactions, but also on the compatibility with nanofabrication processes. Natural proteins exhibit many advantages including favorable cell-biomaterial interactions. However difficulty in processing and the potential for immune response[71, 72] may limit widespread adoption. Synthetic biodegradable elastomers[68, 69, 73, 74] offer advantages such as ease of processing, variety of physical and mechanical properties, favorable tissue response and biodegradation kinetics,[75, 76] and compatibility with nanofabrication techniques.[19, 77, 78]
4. Mechanisms for Nanotopographic Sensing and Response
Studies demonstrating significant influence of nanoscale topographic features have yet to elucidate well-defined mechanisms of cell-nanotopography interaction. In particular, several characteristics of this field of study confound the pursuit of precise interaction mechanisms. There are virtually an infinite number of potential combinations of cell types, biomaterial composition, and topographic feature arrangements. Cell-nanotopography interactions are also transient, which confounds the difficulty in extricating a mechanistic view of the contact guidance response.[52] For example, cells in long-term culture can secrete additional extracellular matrix proteins, which can lead to convoluted topographic signaling.[79] The large potential set of experiments and cell-specific outputs has resulted in a primarily phenomenological approach to studying cell-nanotopography interactions. Despite this large body of work, little is known about the origin or underlying mechanism of the effect of topographical cues on cell function. Various theories have been proposed to explain such phenomenon as the alignment and elongation along the grating axis in nanograting substrates. Recent work has begun to interrogate cell-topography interactions with a focus on elucidating mechanism including identifying relevant signal transduction pathways and the role of organelles including the cytoskeleton. Nevertheless, there is a significant opportunity to further explore cell-nanotopography interactions, which could lead to refinement and more comprehensive predictive models of cell-nanotopography interactions.[80]
4.1. Current Theories
We suggest that the morphological response serves as both an indicator of relevant cell-nanotopography interactions and a basis for second order effects. The elongation and alignment of the nucleus is presumably another source for alteration of the gene profile as cells response to substrate nanotopography.[52, 81] The generalized consensus regarding the mechanism for the morphological response is anisotropic stress generation. However, the precise origin and specific role of the anisotropic stresses is still under debate. Theories for the basis of cell-nanotopography interactions will be discussed in the context of nanogratings. Contact guidance kinetics of fibroblasts to titanium nanogratings suggests that microtubules align within 20 min after attachment and preceded alignment of the overall cell.[82] This cluster of events is followed by the alignment of microfilament bundles at 40–60 min and focal adhesion contacts after 3 h. From this study, it is clear that there are numerous organelles that are responsible for initializing and transmitting the effect of surface topography throughout the cell to influence overall cell functions such as stress fiber formation, lamellipodia, and filopodia. One critical organelle that is thought to play an instrumental role in the contact guidance response is filopodia,[39] which could be modulated through Cdc42 activation.[28] While this can explain the mechanism of detection and transmission of cell-nanotopography interactions, several theories have been proposed to explain the origin of this response.
4.1.1. Intrinsic Protein Patterning Via Substrate Discontinuities
This idea suggests that discontinuities in features lead to preferential protein absorption, and subsequent protein patterning. Patterned protein deposition due to topography could induce preferential alignment just as micropatterned protein substrates can lead to preferential confinement of cells.[83] Micropatterned proteins of various feature sizes can induce dramatic changes in cell behavior including morphology, proliferation, differentiation, and apoptosis.[84–86] However, this theory is unlikely in light of more recent studies including observed contact guidance phenomenon in smooth, continuous features that are much larger than the length scale of proteins.[19, 87]
4.1.2. Spatial Biasing of Focal Adhesion Formation
Nanotopography can induce the overall alignment and elongation of cells by first inducing the alignment of focal adhesions. The initial alignment of focal adhesions could result from asymmetric probability of focal adhesion formation due to feature geometry, or geometrically restricted focal adhesion morphology. The alignment of focal adhesions could then lead to an overall response in the cell morphology through the aforementioned intimate signaling connection between focal adhesions and cytoskeleton proteins. Although this theory may explain the connection between aligned focal adhesions and the aligned, elongated gross morphology, it does not sufficiently address the initial alignment of focal adhesions.
4.1.3. Preferential Actin Polymerization
Actin polymerization dynamics involved in cytoskeleton rearrangement are essential for cell attachment[88] and serve as a driving force for directional migration and morphological alterations.[89–91] Filopodia are highly motile organelles involved in many cellular processes including migration[92] and sensing local topography.[93] Filopodia formation perpendicular to the ridge-groove features is hypothesized to occur less frequently due to unfavorable stress formation. Conversely, the formation of filopodia parallel to the ridge-groove features is more frequent, which leads to biased propagation, cytoskeleton rearrangement, polarization of the cell body, and ultimately a gross morphological effect of alignment and elongation. Highly dynamic filopodia serve as topographical sensors, which are able to detect the immediate surrounding environment. This theory is most consistent with the corpus of work that has since been conducted regarding this topic.[19]
4.2. Potential Role of Small GTPases
The Rho family of GTPases has been shown to control the formation and organization of filaments that compose the actin cytoskeleton.[94] This activation of Rho, Rac, and Cdc42 GTPases controls a wide spectrum of cell functions including cytoskeleton formation and remodeling, alterations in gene expression, cell cycle progression, cell morphogenesis, and cell migration in many cell types.[95, 96] Hence, we argue that these molecule switches likely play a vital role in the concerted response of cells to substrate nanotopography. Recent studies have only begun to explore the role of these signaling pathways in the context of cell-nanotopography interactions.[21, 97] One key function that could directly connect nanotopographic signaling to cell responses is spatially biased focal adhesion formation[98] through Rho activation, which could have dramatic downstream effects on cell migration and signaling.[99] Focal influence cell morphogenesis[80, 100] and have been shown to be sensitive to mechanical forces.[101] Future work must be conducted to further elucidate the dynamics between nanotopographic signaling and modulation of cell function.
4.3. Advanced Biological Techniques
The impact of nanotopography on gene expression is a somewhat intuitive extension of the effect on basic cell function. Recent quantification of the precise impact of nanotopography on genome-wide expression has provided an enormous body of data, but has yet to elucidate any clear mechanisms. Studies that investigate individual signaling pathways that are likely to be implicated in cell-nanotopography interactions may provide greater utility in understanding the origin of these responses. These studies can be conducted by examining cell-topography interactions in the presence of other signaling modalities; a trend that is evident in more recent studies that investigate the coupled effect of soluble factors and substrate nanotopography on cell function.[21, 31, 97] Future studies should ideally span multiple cell types and substrates to confirm the generality of subsequent findings. Genetic manipulation[102] and gene knockdown using siRNA[103] are other techniques which would serve to identify and investigate the role of specific organelles and signaling pathways implicated in cell-nanotopography interactions.
5. Summary and Outlook
5.1. Cell-Nanotopography Signaling
Cell-nanotopography interactions could serve as an alternative signaling mechanism to precisely control cell function. Cells respond to numerous chemical, physical, mechanical, and electrical stimuli, which can be engineered to control cell function. Substrate engineering encompasses several of these factors in an attempt to utilize cell-biomaterial interactions to control and tune such functions as cell fate,[104] differentiation,[105] and genetic manipulation.[106] Nanotopographic cues are a subset of substrate engineering, which can be used to control many aspects of cell behavior. Nanotopographic cues can be incorporated into large areas with relative ease, which allow for large scale cell culture. Synthetic nanotopography could act synergistically with soluble factors to enhance the efficiency of differentiation protocols. For example, synergistic cues from soluble factors and cell-nanotopography interactions could have the potential to dramatically increase the specificity and efficiency of lineage-specific stem cell differentiation.[22] Nanotopography could also be superimposed upon other modes of substrate engineering because they can be incorporated into many engineering and biomedical materials without significantly impacting the physicochemical properties of the bulk material. Therefore nanotopography could be utilized in addition to other types of cell-biomaterial surface conditions including microcontact printed chemistries[107] and bulk mechanical properties of the substrate, two additional examples of cues to guide cell function.[13, 104,105]
5.2. Nanotopography in Basic Science and Engineering
Engineering substrates to induce desired cell phenotype and genotype has the potential to become an important component of scaffold design for tissue engineering applications. Substrate nanotopography may also be utilized as a tool to study complex cell functions such as adhesion, migration, cytoskeleton reorganization, and cell polarization. For example, nanograting substrates can be used to study contact guidance and migration in vitro. Nanopost or nanopit substrates can be used to study the role of filopodia dynamics, focal adhesion formation, and other cytoskeleton functions in a controlled manner. As nanotopography aids in unveiling new discoveries in basic cell function, engineers could used these new discoveries as a basis for the design and fabrication of next-generation synthetic nanotopographic substrates. These advances could then be used to iteratively improve the impact of cell-nanotopography interactions for use in tissue engineering applications.[108]
Acknowledgements
The authors would first like to acknowledge the work of authors that could not be featured in this review article due to content limitations. The authors are grateful to Sarah Tao and Joseph Charest for their comments and assistance in writing this manuscript. CJB and JTB would like to thank the Charles Stark Draper Laboratory for funding (DL-H-550154). CJB, RL, and JTB would like to thank the National Institutes of Health for funding (R01-DE-013023-06, P41 EB002520-01A1 and 1R01HL076485-01A2). CJB was funded through a Charles Stark Draper Fellowship. The content of this paper does not necessarily reflect the position or the policy of the government, and no official endorsement should be inferred.
Biographies

Christopher J. Bettinger is a currently a postdoctoral fellow working in the field of organic electronics under the supervision of Prof. Zhenan Bao in the Department of Chemical Engineering at Stanford University. He studied at the Massachusetts Institute of Technology where he received an S.B. in Chemical Engineering in 2003, an M.Eng. in Biomedical Engineering in 2004, and a Ph.D. in Materials Science and Engineering in 2008. He completed his doctoral thesis under the supervision of Robert Langer where he worked to develop micro and nanoscale biomaterial systems for use in tissue engineering.

Robert Langer is currently an Institute Professor at the Massachusetts Institute of Technology working in the Department of Chemical Engineering. Dr. Langer is the most cited engineer in history with over 1,000 articles and more than 600 issued or pending patents worldwide. His current research focuses on advanced biomaterials and drug delivery systems for gene therapy, cancer treatment, and tissue engineering and regenerative medicine. Dr. Langer received a B.S. in 1970 from Cornell University and an Sc.D. in 1974 from MIT, both in Chemical Engineering.

Jeffrey Borenstein is a Distinguished Member of the Technical Staff at the Charles Stark Draper Laboratory in Cambridge, Massachusetts, and Program Leader for Tissue Engineering for the Center for Integration of Medicine and Innovative Technology. Dr. Borenstein has a Ph.D. in Physics and 25 years of experience in MEMS, nanotechnology and biomedical devices. His research is focused on microfabrication for tissue engineering and drug delivery, with support from the NIH and NSF. He has thirteen issued patents, as well as twenty published patent applications and over eighty peer-reviewed journal articles and conference proceedings.
References
- 1.Goodman SL, Sims PA, Albrecht RM. Biomaterials. 1996;17:2087. doi: 10.1016/0142-9612(96)00016-6. [DOI] [PubMed] [Google Scholar]
- 2.Abrams GA, Goodman SL, Nealey PF, Franco M, Murphy CJ. Cell Tissue Res. 2000;299:39. doi: 10.1007/s004419900074. [DOI] [PubMed] [Google Scholar]
- 3.Pamula E, De Cupere V, Dufrene YF, Rouxhet PG. J Colloid Interf Sci. 2004;271:80. doi: 10.1016/j.jcis.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Bozec L, van der Heijden G, Horton M. Biophys J. 2007;92:70. doi: 10.1529/biophysj.106.085704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf K, Muller R, Borgmann S, Brocker EB, Friedl P. Blood. 2003;102:3262. doi: 10.1182/blood-2002-12-3791. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland J, Denyer M, Britland S. J Anat. 2005;206:581. doi: 10.1111/j.1469-7580.2005.00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haga H, Irahara C, Kobayashi R, Nakagaki T, Kawabata K. Biophys J. 2005;88:2250. doi: 10.1529/biophysj.104.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedl P, Bröcker EB. Dev Immunol. 2000;7:249. doi: 10.1155/2000/56473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedl P. Curr Opin Cell Biol. 2004;16:14. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Campbell RD, Marcum BA. J Cell Sci. 1980;41:33. doi: 10.1242/jcs.41.1.33. [DOI] [PubMed] [Google Scholar]
- 11.Dent EW, Gertler FB. Neuron. 2003;40:209. doi: 10.1016/s0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- 12.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Biomaterials. 1999;20:573. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 13.Discher DE, Janmey P, Wang Y-l. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 14.Pham QP, Sharma U, Mikos AG. 2006;Vol. 12:1197. doi: 10.1089/ten.2006.12.1197. [DOI] [PubMed] [Google Scholar]
- 15.Variola F, Yi J-H, Richert L, Wuest JD, Rosei F, Nanci A. Biomaterials. 2008;29:1285. doi: 10.1016/j.biomaterials.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 16.Lim JY, Hansen JC, Siedlecki CA, Hengstebeck RW, Cheng J, Winograd N, Donahue HJ. Biomacromolecules. 2005;6:3319. doi: 10.1021/bm0503423. [DOI] [PubMed] [Google Scholar]
- 17.Dalby MJ, Riehle MO, Johnstone H, Affrossman S, Curtis ASG. Biomaterials. 2002;23:2945. doi: 10.1016/s0142-9612(01)00424-0. [DOI] [PubMed] [Google Scholar]
- 18.Bettinger CJ, Zhang Z, Gerecht S, Borenstein JT, Langer R. Adv Mater. 2008;20:99. doi: 10.1002/adma.200702487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bettinger CJ, Orrick B, Misra A, Langer R, Borenstein JT. Biomaterials. 2006;27:2558. doi: 10.1016/j.biomaterials.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 20.Lu J, Rao MP, MacDonald NC, Khang D, Webster TJ. Acta Biomater. 2008;4:192. doi: 10.1016/j.actbio.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Gerecht S, Bettinger CJ, Zhang Z, Borenstein J, Vunjak-Novakovic G, Langera R. Biomaterials. 2007;28:4068. doi: 10.1016/j.biomaterials.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yim EKF, Pang SW, Leong KW. Exp Cell Res. 2007;313:1820. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Ohlin CA, Lu Q, Hu J. Biomaterials. 2008;29:2049. doi: 10.1016/j.biomaterials.2007.12.047. [DOI] [PubMed] [Google Scholar]
- 24.Rebollar E, Frischauf I, Olbrich M, Peterbauer T, Hering S, Preiner J, Hinterdorfer P, Romanin C, Heitz J. Biomaterials. 2008;29:1796. doi: 10.1016/j.biomaterials.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 25.Yim EKF, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW. Biomaterials. 2005;26:5405. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi C-H, Hagvall SH, Wu BM, Dunn JCY, Beygui RE, Cj" Kim C-J. Biomaterials. 2007;28:1672. doi: 10.1016/j.biomaterials.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 27.Chou L, Firth JD, Uitto VJ, Brunette DM. J Cell Sci. 1995;108:1563. doi: 10.1242/jcs.108.4.1563. [DOI] [PubMed] [Google Scholar]
- 28.Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. J Cell Sci. 2003;116:1881. doi: 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karuri NW, Liliensiek S, Teixeira AI, Abrams G, Campbell S, Nealey PF, Murphy CJ. J Cell Sci. 2004;117:3153. doi: 10.1242/jcs.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalton BA, Walboomers F, Dziegielewski M, Evans MDM, Taylor S, Jansen JA, Steele JG. J Biomed Mater Res. 2001;56:195. doi: 10.1002/1097-4636(200108)56:2<195::aid-jbm1084>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Rajnicek AM, Foubister LE, McCaig CD. Dev Biol. 2007;312:448. doi: 10.1016/j.ydbio.2007.09.051. [DOI] [PubMed] [Google Scholar]
- 32.Foley JD, Grunwald EW, Nealey PF, Murphy CJ. Biomaterials. 2005;26:3639. doi: 10.1016/j.biomaterials.2004.09.048. [DOI] [PubMed] [Google Scholar]
- 33.Biggs M, Richards R, Gadegaard N, Wilkinson C, Dalby M. J Mater Sci-Mater M. 2007;18:399. doi: 10.1007/s10856-006-0705-6. [DOI] [PubMed] [Google Scholar]
- 34.Kim D-H, Kim P, Suh KY, Seung Kyu Choi A, Sang Ho Lee A, Byungkyu Kim A. In: IEEE-EMBS. Pilnam K, editor. 2005. p. 4091. [DOI] [PubMed] [Google Scholar]
- 35.Kim P, Kim DH, Kim B, Choi SK, Lee SH, Khademhosseini A, Langer R, Suh KY. Nanotechnology. 2005;16:2420. doi: 10.1088/0957-4484/16/10/072. [DOI] [PubMed] [Google Scholar]
- 36.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. Nat Mater. 2007;6:997. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 37.Hart A, Gadegaard N, Wilkinson C, Oreffo R, Dalby M. J Mater Sci-Mater M. 2007;18:1211. doi: 10.1007/s10856-007-0157-7. [DOI] [PubMed] [Google Scholar]
- 38.Dalby MJ, Gadegaard N, Wilkinson CDW. J Biomed Mater Res. 2008;84A:973. doi: 10.1002/jbm.a.31409. [DOI] [PubMed] [Google Scholar]
- 39.Dalby MJ, Gadegaard N, Riehle MO, Wilkinson CDW, Curtis ASG. Int J Biochem Cell B. 2004;36:2005. doi: 10.1016/j.biocel.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Curtis ASG, Gadegaard N, Dalby MJ, Riehle MO, Wilkinson CDW, Aitchison GA. IEEE Trans Nanobiosci. 2004;3:61. doi: 10.1109/tnb.2004.824276. [DOI] [PubMed] [Google Scholar]
- 41.Yang MT, Sniadecki NJ, Chen CS. Adv Mater. 2007;19:3119. [Google Scholar]
- 42.Curtis ASG, Casey B, Gallagher JO, Pasqui D, Wood MA, Wilkinson CDW. Biophysical Chemistry. 2001;94:275. doi: 10.1016/s0301-4622(01)00247-2. [DOI] [PubMed] [Google Scholar]
- 43.Nomura S, Kojima H, Ohyabu Y, Kuwabara K, Miyauchi A, Uemura T. J Artif Organs. 2006;9:90. doi: 10.1007/s10047-006-0329-0. [DOI] [PubMed] [Google Scholar]
- 44.Hsu S-h, Chen C-Y, Lu PS, Lai C-S, Chen C-J. Biotechnol Bioeng. 2005;92:579. doi: 10.1002/bit.20634. [DOI] [PubMed] [Google Scholar]
- 45.Loesberg WA, te Riet J, van Delft FCMJM, Schön P, Figdor CG, Speller S, van Loon JJWA, Walboomers XF, Jansen JA. Biomaterials. 2007;28:3944. doi: 10.1016/j.biomaterials.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 46.Teixeira AI, McKie GA, Foley JD, Bertics PJ, Nealey PF, Murphy CJ. Biomaterials. 2006;27:3945. doi: 10.1016/j.biomaterials.2006.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyle J, Gültig K, Nisch W. J Biomed Maters Res. 1995;29:81. doi: 10.1002/jbm.820290112. [DOI] [PubMed] [Google Scholar]
- 48.Bozec L, Horton M. Biophys J. 2005;88:4223. doi: 10.1529/biophysj.104.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diehl KA, Foley JD, Nealey PF, Murphy CJ. J Biomed Mater Res. 2005;75A:603. doi: 10.1002/jbm.a.30467. [DOI] [PubMed] [Google Scholar]
- 50.Lenhert S, Meier M-B, Meyer U, Chi L, Wiesmann HP. Biomaterials. 2005;26:563. doi: 10.1016/j.biomaterials.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 51.Tzvetkova-Chevolleau T, Stéphanou A, Fuard D, Ohayon J, Schiavone P, Tracqui P. Biomaterials. 2008;29:1541. doi: 10.1016/j.biomaterials.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 52.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CDW, Curtis ASG. Exp Cell Res. 2003;284:274. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 53.Norman JJ, Desai TA. Ann Biomed Eng. 2006;34:89. doi: 10.1007/s10439-005-9005-4. [DOI] [PubMed] [Google Scholar]
- 54.Cheng JY, Mayes AM, Ross CA. Nat Mater. 2004;3:823. doi: 10.1038/nmat1211. [DOI] [PubMed] [Google Scholar]
- 55.Ruiz R, Sandstrom RL, Black CT. Adv Mater. 2007;19:587. [Google Scholar]
- 56.Haynes CL, Duyne RPV. J. Phys. Chem. B. 2001;105:5599. [Google Scholar]
- 57.Kosiorek A, Kandulski W, Glaczynska H, Giersig M. Small. 2005;1:439. doi: 10.1002/smll.200400099. [DOI] [PubMed] [Google Scholar]
- 58.LaFratta CN, Li L, Fourkas JT. Proc Nat Acad Sci USA. 2006;103:8589. doi: 10.1073/pnas.0603247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pikulin A, Bityurin N. Phys Rev B. 2007;75:195430. [Google Scholar]
- 60.In HJ, Kumar S, Shao-Horn Y, Barbastathis G. Appl Phys Lett. 2006;88:083104. [Google Scholar]
- 61.Seunarine K, Meredith DO, Riehle MO, Wilkinson CDW, Gadegaard N. Microelec Eng. 2008 In Press, Corrected Proof. [Google Scholar]
- 62.Xia Y, Whitesides GM. Angew Chem Int Edit. 1998;37:550. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 63.Bernard A, Renault JP, Michel B, Bosshard HR, Delamarche E. Adv Mater. 2000;12:1067. [Google Scholar]
- 64.Lin CC, Co CC, Ho CC. Biomaterials. 2005;26:3655. doi: 10.1016/j.biomaterials.2004.09.051. [DOI] [PubMed] [Google Scholar]
- 65.Kim E, Xia Y, Zhao XM, Whitesides GM. Adv Mater. 1997;9:651. [Google Scholar]
- 66.Chiu DT, Jeon NL, Huang S, Kane RS, Wargo CJ, Choi IS, Ingber DE, Whitesides GM. Proc Nat Acad Sci USA. 2000;12:2408. doi: 10.1073/pnas.040562297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Torigoe K, Tanaka H-F, Takahashi A, Awaya A, Hashimoto K. Exp Neurol. 1996;137:301. doi: 10.1006/exnr.1996.0030. [DOI] [PubMed] [Google Scholar]
- 68.Nijst CLE, Bruggeman JP, Karp JM, Ferreira L, Zumbuehl A, Bettinger CJ, Langer R. Biomacromolecules. 2007;8:3067. doi: 10.1021/bm070423u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bettinger CJ, Bruggeman JP, Borenstein JT, Langer RS. Biomaterials. 2008;29:2315. doi: 10.1016/j.biomaterials.2008.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bruggeman JP, Bettinger CJ, Nijst CLE, Kohane DS, Langer R. Adv Mater. 2008;20:1922. [Google Scholar]
- 71.Ellingsworth LR, DeLustro F, Brennan JE, Sawamura S, McPherson J. J Immunol. 1986;136:877. [PubMed] [Google Scholar]
- 72.García-Domingo MI, Alijotas-Reig J, Cisteró-Bahima A, Tresserra F, Enrique E. J Investig Allergol Clin Immunol. 2000;10:107. [PubMed] [Google Scholar]
- 73.Wang Y, Ameer GA, Sheppard BJ, Langer R. Nat Biotechnol. 2002;20:602. doi: 10.1038/nbt0602-602. [DOI] [PubMed] [Google Scholar]
- 74.Bruggeman JP, Bettinger CJ, Nijst CLE, Kohane DS, Langer R. 2008;Vol. 9999:NA. [Google Scholar]
- 75.Wang Y, Kim YM, Langer R. J Biomed Mater Res A. 2003;66:192. doi: 10.1002/jbm.a.10534. [DOI] [PubMed] [Google Scholar]
- 76.Bettinger CJ, Bruggeman JP, Borenstein JT, Langer R. J Biomed Mater Res. 2008 doi: 10.1002/jbm.a.32306. (in press) [DOI] [PubMed] [Google Scholar]
- 77.Mahdavi A, Ferreira L, Sundback C, Nichol JW, Chan EP, Carter DJD, Bettinger CJ, Patanavanich S, Chignozha L, Ben-Joseph E, Galakatos A, Pryor H, Pomerantseva I, Masiakos PT, Faquin W, Zumbuehl A, Hong S, Borenstein J, Vacanti J, Langer R, Karp JM. Proc Nat Acad Sci USA. 2008;105:2307. doi: 10.1073/pnas.0712117105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bettinger CJ, Kulig KM, Vacanti JP, Langer R, Borenstein JT. Tissue Eng. 2008 doi: 10.1089/ten.tea.2008.0134. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamilton D, Wong K, Brunette D. Calcified Tissue Int. 2006;78:314. doi: 10.1007/s00223-005-0238-x. [DOI] [PubMed] [Google Scholar]
- 80.Kemkemer R, Jungbauer S, Kaufmann D, Gruler H. Biophys J. 2006;90:4701. doi: 10.1529/biophysj.105.067967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis ASG. Eur J Cell Biol. 2004;83:159. doi: 10.1078/0171-9335-00369. [DOI] [PubMed] [Google Scholar]
- 82.Oakley C, Brunette DM. J Cell Sci. 1993;106:343. doi: 10.1242/jcs.106.1.343. [DOI] [PubMed] [Google Scholar]
- 83.Tan JL, Liu W, Nelson CM, Raghavan S, Chen CS. Tissue Eng. 2004;10 doi: 10.1089/1076327041348365. [DOI] [PubMed] [Google Scholar]
- 84.Ito Y. Biomaterials. 2000;20:2333. doi: 10.1016/s0142-9612(99)00162-3. [DOI] [PubMed] [Google Scholar]
- 85.Lim JY, Donahue HJ. Tissue Eng. 2007;13:1879. doi: 10.1089/ten.2006.0154. [DOI] [PubMed] [Google Scholar]
- 86.Song HK, Toste B, Ahmann K, Hoffman-Kim D, Palmore GTR. Biomaterials. 2006;27:473. doi: 10.1016/j.biomaterials.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 87.Andersson A-S, Olsson P, Lidberg U, Sutherland D. Exp Cell Res. 2003;288:177. doi: 10.1016/s0014-4827(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 88.Pierres A, Benoliel A-M, Touchard D, Bongrand P. Biophys J. 2008;94:4114. doi: 10.1529/biophysj.107.125278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wojciak-Stothard B, Curtis AS, Monaghan W, McGrath M, Sommer I, Wilkinson CD. Cell Motil Cytoskel. 1995;31:147. doi: 10.1002/cm.970310207. [DOI] [PubMed] [Google Scholar]
- 90.Wojciak-Stothard B, Curtis A, Monaghan W, McGrath M, Sommer I, Wilkinson C. Cell Biol Int. 1995;19:1861. [Google Scholar]
- 91.Walboomers XF, Monaghan W, Curtis A, Jansen JA. J Biomed Mater Res. 1999;46:212. doi: 10.1002/(sici)1097-4636(199908)46:2<212::aid-jbm10>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 92.Nemethova M, Auinger S, Small JV. J Cell Biol. 2008;180:1233. doi: 10.1083/jcb.200709134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galbraith CG, Yamada KM, Galbraith JA. Science. 2007;315:992. doi: 10.1126/science.1137904. [DOI] [PubMed] [Google Scholar]
- 94.Hall A. Science. 1998;279:509. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 95.Jaffe AB, Hall A. Ann Rev Cell Dev Biol. 2005;21:247. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 96.Tzima E. Circ Res. 2006;98:176. doi: 10.1161/01.RES.0000200162.94463.d7. [DOI] [PubMed] [Google Scholar]
- 97.Dalby MJ, Hart A, Yarwood SJ. Biomaterials. 2008;29:282. doi: 10.1016/j.biomaterials.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 98.Uttayarat P, Toworfe GK, Dietrich F, Lelkes PI, Composto RJ. J Biomed Mater Res. 2005;75A:668. doi: 10.1002/jbm.a.30478. [DOI] [PubMed] [Google Scholar]
- 99.Wang R, Clark RAF, Mosher DF, Ren X-D. J Biol Chem. 2005;280:28803. doi: 10.1074/jbc.M501421200. [DOI] [PubMed] [Google Scholar]
- 100.Jungbauer S, Gao H, Spatz JP, Kemkemer R. Biophys J. 2008;95:3470. doi: 10.1529/biophysj.107.128611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bershadsky A, Kozlov M, Geiger B. Current Opinion in Cell Biology. 2006;18:472. doi: 10.1016/j.ceb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 102.Putnam D. Nat Mater. 2006;5:439. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 103.Mullard A. Nat Rev Mol Cell Biol. 2007;8:513. [Google Scholar]
- 104.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 105.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 106.Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Nat Mater. 2005;4:460. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 107.Charest JL, Eliason MT, Garcia AJ, King WP. Biomaterials. 2006;27:2487. doi: 10.1016/j.biomaterials.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 108.Khademhosseini A, Bettinger C, Karp JM, Yeh J, Ling Y, Borenstein J, Fukuda J, Langer R. J Biomater Sci Polym Ed. 2006;17:1221. doi: 10.1163/156856206778667488. [DOI] [PubMed] [Google Scholar]