Abstract
Background
MicroRNAs (miRNAs) are small non-coding RNAs that post-transcriptionally regulate gene expression in a variety of organisms, including insects, vertebrates, and plants. miRNAs play important roles in cell development and differentiation as well as in the cellular response to stress and infection. To date, there are limited reports of miRNA identification in mosquitoes, insects that act as essential vectors for the transmission of many human pathogens, including flaviviruses. West Nile virus (WNV) and dengue virus, members of the Flaviviridae family, are primarily transmitted by Aedes and Culex mosquitoes. Using high-throughput deep sequencing, we examined the miRNA repertoire in Ae. albopictus cells and Cx. quinquefasciatus mosquitoes.
Results
We identified a total of 65 miRNAs in the Ae. albopictus C7/10 cell line and 77 miRNAs in Cx. quinquefasciatus mosquitoes, the majority of which are conserved in other insects such as Drosophila melanogaster and Anopheles gambiae. The most highly expressed miRNA in both mosquito species was miR-184, a miRNA conserved from insects to vertebrates. Several previously reported Anopheles miRNAs, including miR-1890 and miR-1891, were also found in Culex and Aedes, and appear to be restricted to mosquitoes. We identified seven novel miRNAs, arising from nine different precursors, in C7/10 cells and Cx. quinquefasciatus mosquitoes, two of which have predicted orthologs in An. gambiae. Several of these novel miRNAs reside within a ~350 nt long cluster present in both Aedes and Culex. miRNA expression was confirmed by primer extension analysis. To determine whether flavivirus infection affects miRNA expression, we infected female Culex mosquitoes with WNV. Two miRNAs, miR-92 and miR-989, showed significant changes in expression levels following WNV infection.
Conclusions
Aedes and Culex mosquitoes are important flavivirus vectors. Recent advances in both mosquito genomics and high-throughput sequencing technologies enabled us to interrogate the miRNA profile in these two species. Here, we provide evidence for over 60 conserved and seven novel mosquito miRNAs, expanding upon our current understanding of insect miRNAs. Undoubtedly, some of the miRNAs identified will have roles not only in mosquito development, but also in mediating viral infection in the mosquito host.
Background
Culex and Aedes mosquitoes are members of the Culicinae subfamily that vector positive-sense RNA viruses from the family Flaviviridae. Many flaviviruses, such as West Nile virus (WNV), dengue virus (DENV), and yellow fever virus (YFV), are highly pathogenic in humans and pose an important health problem worldwide [1]. Each year, an estimated 50 million human cases of dengue fever occur due to infection with DENV. Since the introduction of WNV to the United States in 1999, over 28,000 cases have been reported to the CDC, with approximately 3,000 cases annually http://CDC.gov. Culex mosquitoes are primarily responsible for the transmission of WNV to humans (reviewed in [2]), although WNV has also been isolated from Aedes albopictus in the eastern United States (reviewed in [3]). Virus transmission from Cx. quinquefasciatus occurs as early as five days following an infectious blood meal [4], and virus can persist as long as four weeks in the midguts and salivary glands of infected mosquitoes [5,6].
Both Culex and Aedes mosquitoes are prevalent in tropical and subtropical regions around the world. Recently, Ae. albopictus has emerged as a major vector for Chikungunya virus, an alphavirus, in regions bordering the western Indian Ocean [7,8]. Ae. albopictus is also considered a secondary vector for dengue virus serotypes 1-4 (DENV1-4) and YFV, which are predominantly transmitted to humans by a mosquito from the same genus, Ae. aegypti. Ae. albopictus can potentially vector at least 22 known arboviruses (reviewed in [3]).
Of the over 3,000 mosquito species worldwide, microRNAs (miRNAs) have so far only been described in two species of African malaria mosquitoes, Anopheles gambiae and Anopheles stephensi, using direct cloning and computational methods. Over 55 miRNAs have been described for Anopheles mosquitoes, at least 49 of which have orthologs in Drosophila melanogaster and other insects [9-12]. The functions of these miRNAs in mosquitoes, and the identities of their mRNA targets, are not yet known.
miRNAs are a class of small, non-coding RNAs, from 19-24 nt in length, that post-transcriptionally regulate gene expression by binding to complementary regions in, primarily, the 3' untranslated region (3' UTR) of target messenger RNAs. First identified in Caenorhabditis elegans, miRNAs have now been found in a wide variety of organisms including insects, vertebrates, and plants [13-15]. Over 10,800 miRNAs are currently annotated in miRBase, many of which are conserved from worms to flies to humans [9]. In humans, miRNAs are predicted to regulate as much as one-third of all mRNAs [16], and thus, represent an important component in managing biological processes.
Much of what we understand about insect miRNAs comes from studies in the fruit fly D. melanogaster. D. melanogaster miRNAs were originally identified via direct cloning of small RNA molecules and many of these miRNAs exhibited significant sequence conservation with miRNAs expressed in C. elegans [17]. At present, 147 different miRNAs have been annotated for D. melanogaster, the majority of which have orthologous sequences in other winged insects [9]. With the identification of new miRNAs in a number of organisms, evolutionary sequence conservation has become a hallmark of miRNA biology [12,15,18,19].
Differential miRNA expression throughout the various stages of the Drosophila life cycle has revealed a role for miRNAs in important cellular processes such as apoptosis, cell division, and differentiation [20-22]. Additionally, miRNA expression profiles change in response to stress, inflammation, and infection [11,19]. For example, in Anopheles mosquitoes, expression levels of four miRNAs are altered during the response to Plasmodium infection [11].
The process of miRNA biogenesis is conserved, initiating with the cleavage of long, endogenous nuclear primary miRNA transcripts, ranging from hundreds to thousands of nucleotides in length, into pre-miRNAs [23,24]. Two proteins are required for this processing in insects, the RNAse III enzyme Drosha and its binding partner Pasha, which together excise the ~60 nt pre-miRNA hairpin from the pri-miRNA [25]. The pre-miRNA is then exported to the cytoplasm and processed by a second RNAse III enzyme called Dicer-1 to yield the ~22 bp miRNA:miRNA* duplex intermediate [13]. Mature miRNAs are selectively loaded into the multi-component RNA-induced silencing complex (RISC) which contains members of the Argonaute family (Ago). In Drosophila, strand selection has been shown to depend on the intrinsic structure of the miRNA:miRNA* duplex, which facilitates sorting into either Ago1- or Ago2-containing RISCs [26,27]. Recent comparative genomics studies have shown that the Anopheles, Aedes, and Culex mosquito genomes all encode orthologs of key proteins involved in the miRNA, as well as small interfering RNA (siRNA) and piwi RNA (piRNA), regulatory pathways [28]. Mature miRNAs are used as guide RNAs to direct RISC to complementary regions of mRNAs, resulting in the inhibition of translation and/or target mRNA degradation. Important for this targeting are nucleotides 2-8 from the 5' end of the mature miRNA, known as the "seed" [29,30]. Many studies have shown that miRNAs can target 3'UTRs of mRNAs [31,32]; however, recent studies have also revealed functional target sites within the ORFs of mRNAs [33,34].
Recent advances in mosquito genomics, such as the sequencing of the genomes of three mosquito species, Ae. aegypti, Cx. quinquefasciatus, and An. gambiae [35], together with technological advances in small RNA cloning methods, enabled us to interrogate the miRNA repertoire in two flavivirus mosquito vectors. In this study, we used deep sequencing to identify over 60 conserved and several novel miRNAs in Cx. quinquefasciatus mosquitoes and an Ae. albopictus cell line, C7/10, commonly used for in vitro flavivirus studies. We additionally investigated the effects of flavivirus infection on miRNA expression and found that miR-92 and miR-989 are significantly changed in response to WNV infection.
Results and Discussion
Deep sequencing of small RNAs
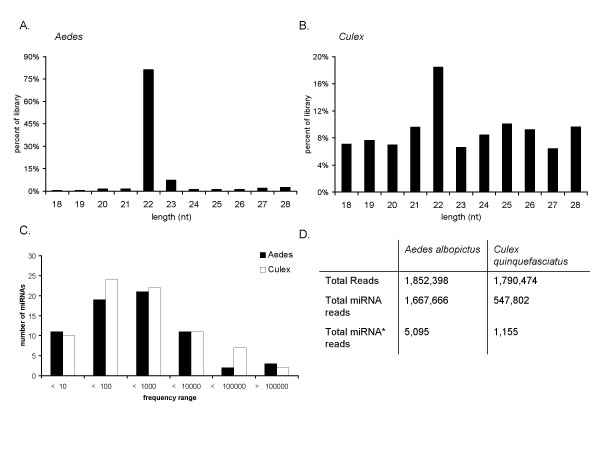
To identify miRNAs in Culex and Aedes mosquitoes, we isolated small RNAs (18-28 nt) from C7/10 Ae. albopictus cells and blood-fed, female Cx. quinquefasciatus mosquitoes. Small RNA libraries were subjected to Illumina-based high-throughput sequencing. After filtering for linker sequences, and removing ambiguous reads, a total of 1,852,398 reads for Ae. albopictus cells and 1,790,474 reads for Cx. quinquefasciatus mosquitoes, representing 41,056 and 281,918 non-redundant sequences, respectively, were obtained (Figure 1D). >90% of final reads for Ae. albopictus and >50% of reads for Cx. quinquefasciatus exhibited the predominantly ~22 nt size expected for insect miRNAs (Figure 1A, B).
Figure 1.
Deep sequencing of small RNA populations in Ae. albopictus C7/10 cells and Cx. quinquefasciatus mosquitoes. Size distributions of small RNA libraries (18-28 nt) from A) C7/10 cells (Aedes) and B) Cx. quinquefasciatus (Culex). C) Frequencies of read counts for individual, conserved miRNAs present in C7/10 cells and Culex mosquitoes. Expression levels, based on read counts, of individual miRNAs are separated into several ranges and span over 5 orders of magnitude. D) Breakdown of the total number of reads obtained for each library. The number of reads mapping to miRNA and miRNA* strands is reported.
Most mosquito miRNAs are orthologs of known insect miRNAs
We aligned sequencing reads to known miRNAs and miRNA* strands present in miRBase v14. 1,541,048 reads from the Ae. albopictus library corresponded to 53 distinct pre-miRNAs (61 miRNAs) (Table 1). For the Cx. quinquefasciatus library, 382,878 reads aligned to sequences present in miRBase, representing 69 distinct pre-miRNAs (74 miRNAs) (Table 2). For each miRNA, the sequence with the greatest number of reads was annotated and named according to the most similar match in miRBase [9]. In addition to mature miRNAs, we identified a number of miRNA* strands (Figure 1D, Tables 1, 2), which accounted for < 0.2% of the 20-24 nt population. 21 and 33 distinct miRNA* strands were identified in Ae. albopictus and Cx. quinquefasciatus respectively, and are orthologous to miRNA* strands in other winged insects (Tables 1, 2).
Table 1.
miRNAs identified in Ae. albopictus C7/10 cells and predicted in Ae. aegypti.
| C710 | # miRNA | # miRNA* | Sequence | Length | aga | ame | dme | Ae. aegypti | Start | End | Strand |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 184 | 1487481 | 0 | UGGACGGAGAACUGAUAAGGGC | 22 | Y | Y | Y | 1.496 | 143378 | 143399 | Minus |
| 275 | 23841 | 78 | UCAGGUACCUGAAGUAGCGC | 20 | Y | Y | Y | 1.24 | 486591 | 486610 | Plus |
| 277 | 4453 | 7 | UAAAUGCACUAUCUGGUACGAC | 22 | Y | Y | Y | 1.265 | 508860 | 508881 | Plus |
| 9 | 4085 | 602 | UCUUUGGUAUUCUAGCUGUAGA | 22 | Y | Y | Y | 1.785 | 186231 | 186252 | Plus |
| 8-3p | 3002 | - | UAAUACUGUCAGGUAAAGAUGUC | 23 | Y | Y | Y | 1.411 | 876091 | 876113 | Plus |
| 252.1 | 1608 | 13 | UAAGUACUAGUGCCGCAGGAG | 21 | Y | Y | Y | 1.56 | 1580060 | 1580080 | Minus |
| bantam-5p | 1384 | - | CCGGUUUUCAUUUUCGAUCUGAC | 23 | Y | Y | Y | 1.49 | 157893 | 157915 | Minus |
| 71 | 1246 | 17 | AGAAAGACAUGGGUAGUGAGAU | 22 | ? | Y | ? | 1.268 | 889428 | 889449 | Minus |
| 8-5p | 1244 | - | CAUCUUACCGGGCAGCAUUAGA | 22 | Y | Y | Y | 1.411 | 876052 | 876073 | Plus |
| 276-1 | 1209 | 4 | UAGGAACUUCAUACCGUGCUC | 21 | Y | Y | Y | 1.5 | 2769510 | 2769530 | Minus |
| 276-2 | - | - | UAGGAACUUCAUACCGUGCUC | 21 | Y | Y | Y | 1.134 | 39026 | 39046 | Plus |
| 317-1 | 1118 | 0 | UGAACACAGCUGGUGGUAUCU | 21 | Y | Y | Y | 1.265 | 429503 | 429523 | Plus |
| 317-2 | - | - | UGAACACAGCUGGUGGUAUCU | 21 | Y | Y | Y | 1.153 | 2154717 | 2154737 | Minus |
| 283 | 947 | 0 | CAAUAUCAGCUGGUAAUUCUGGGC | 24 | Y | Y | Y | 1.68 | 2729393 | 2729416 | Minus |
| 252.2 | 888 | - | CUAAGUACUAGUGCCGCAGGAG | 22 | Y | Y | Y | 1.56 | 1580060 | 1580081 | Minus |
| let-7 | 650 | 0 | UGAGGUAGUUGGUUGUAUAGU | 21 | Y | Y | Y | 1.43 | 1156331 | 1156351 | Plus |
| 2 | 708 | 1 | UAUCACAGCCAGCUUUGAAGAGC | 23 | Y | Y | Y | 1.268 | 888597 | 888619 | Minus |
| 998 | 561 | 0 | UAGCACCAUGAGAUUCAGC | 19 | ? | ? | Y | 1.744 | 322338 | 322356 | Plus |
| 92b | 530 | 0 | AAUUGCACUUGUCCCGGCCUG | 21 | Y | Y | Y | 1.116 | 1319201 | 1319221 | Plus |
| 1889-3p | 478 | - | CACGUUACAGAUUGGGGUUUCC | 22 | Y | ? | ? | 1.68 | 2720796 | 2720817 | Minus |
| bantam-3p | 475 | - | UGAGAUCAUUUUGAAAGCUGAU | 22 | Y | Y | Y | 1.49 | 1579555 | 1579576 | Minus |
| 306 | 454 | 81 | UCAGGUACUGAGUGACUCUCAG | 22 | Y | ? | Y | 1.785 | 213078 | 213099 | Plus |
| 281 | 398 | 1 | AAGAGAGCUAUCCGUCGAC | 19 | Y | Y | Y | 1.957 | 134462 | 134480 | Plus |
| 1889-5p | 378 | - | UAAUCUCAAAUUGUAACAGUGG | 22 | Y | ? | ? | 1.68 | 2720896 | 2720917 | Minus |
| 980 | 309 | 6 | UAGCUGCCUAGUGAAGGGC | 19 | ? | ? | Y | 1.23 | 1043069 | 1043087 | Plus |
| 278 | 286 | 53 | ACGGACGAUAGUCUUCAGCGGCC | 23 | Y | Y | Y | 1.16 | 3596269 | 3596291 | Plus |
| 989 | 242 | 0 | UGUGAUGUGACGUAGUGGUAC | 21 | Y | ? | Y | 1.115 | 804187 | 804207 | Minus |
| 14 | 247 | 0 | UCAGUCUUUUUCUCUCUCCUAU | 22 | Y | Y | Y | 1.249 | 1089019 | 1089040 | Minus |
| 11 | 222 | 31 | CAUCACAGUCUGAGUUCUUGCU | 22 | Y | ? | Y | 1.744 | 322062 | 322083 | Plus |
| 190 | 210 | 0 | AGAUAUGUUUGAUAUUCUUGGUUG | 24 | Y | Y | Y | 1.195 | 82254 | 82277 | Minus |
| 1 | 169 | 0 | UGGAAUGUAAAGAAGUAUGGAG | 22 | Y | Y | Y | 1.812 | 291373 | 291394 | Plus |
| 34 | 147 | 1 | UGGCAGUGUGGUUAGCUGGUUG | 22 | Y | ? | Y | 1.265 | 509536 | 509557 | Plus |
| 1890 | 123 | 0 | UGAAAUCUUUGAUUAGGUCUGG | 22 | Y | ? | ? | 1.204 | 1733356 | 1733377 | Plus |
| 988 | 118 | 24 | CCCCUUGUUGCAAACCUCACGC | 22 | Y | ? | Y | 1.442 | 623057 | 623078 | Minus |
| 957 | 99 | 0 | UGAAACCGUCCAAAACUGAGGC | 22 | Y | ? | Y | 1.7 | 464339 | 464360 | Plus |
| 305 | 96 | 4 | AUUGUACUUCAUCAGGUGCUCUGG | 24 | Y | Y | Y | 1.24 | 495601 | 495624 | Plus |
| 996 | 88 | 0 | UGACUAGAUUACAUGCUCGUC | 21 | Y | Y | Y | 1.437 | 567507 | 567527 | Minus |
| 87 | 79 | 0 | GUGAGCAAAUUUUCAGGUGUGU | 22 | Y | Y | Y | 1.36 | 1010846 | 1010867 | Plus |
| 12 | 68 | 3 | UGAGUAUUACAUCAGGUACUGGU | 23 | Y | Y | Y | 1.68 | 2720468 | 2720490 | Minus |
| 13 | 57 | 1 | UAUCACAGCCAUUUUGACGAGUU | 23 | Y | Y | Y | 1.268 | 888744 | 888766 | Minus |
| 92a | 44 | 0 | UAUUGCACUUGUCCCGGCC | 19 | Y | Y | Y | 1.116 | 1267254 | 1267272 | Plus |
| 33 | 40 | 0 | GUGCAUUGUAGUUGCAUUGCA | 21 | ? | Y | Y | 1.487 | 351656 | 351676 | Plus |
| 279 | 36 | 2 | UGACUAGAUCCACACUCAUUAA | 22 | Y | Y | Y | 1.437 | 572258 | 572279 | Minus |
| 79 | 33 | 6 | GCUUUGGCGCUUUAGCUGUAUGA | 23 | Y | Y | Y | 1.785 | 213277 | 213299 | Plus |
| 263 | 32 | 0 | AAUGGCACUGGAAGAAUUCACGG | 23 | Y | Y | Y | 1.981 | 164999 | 165021 | Minus |
| 7 | 32 | 0 | UGGAAGACUAGUGAUUUUGUUGU | 23 | Y | Y | Y | 1.1359 | 46041 | 46063 | Plus |
| **2945 | 32 | 0 | UGACUAGAGGCAGACUCGUUUA | 22 | Y | ? | ? | 1.43 | 481083 | 481105 | Plus |
| 100-5p | 30 | - | AACCCGUAGAUCCGAACUUGUG | 22 | Y | Y | Y | 1.43 | 1142184 | 1142205 | Plus |
| 100-3p | 25 | - | CAAGAACGGAUGUAUGGGAUUC | 22 | Y | Y | Y | 1.43 | 1142224 | 1142245 | Plus |
| 970 | 21 | 0 | UCAUAAGACACACGCGGCUAU | 21 | Y | ? | Y | 1.229 | 1045875 | 1045895 | Plus |
| 210.1 | 13 | 0 | CUUGUGCGUGUGACAACGG | 19 | Y | Y | Y | 1.512 | 515650 | 515668 | Plus |
| 999 | 14 | 0 | UGUUAACUGUAAGACUGUGUCU | 22 | ? | ? | Y | 1.100. | 2315145 | 2315166 | Plus |
| 308 | 16 | 4 | CGCGGUAUAUUCUUGUGGCUUG | 22 | Y | ? | Y | 1.107 | 508980 | 509001 | Plus |
| 125 | 7 | 3 | UCCCUGAGACCCUAACUUGUGA | 22 | Y | Y | Y | 1.43 | 1156615 | 1156636 | Plus |
| 210.2 | 6 | 0 | UUGUGCGUGUGACAACGGCUAU | 22 | Y | Y | Y | 1.512 | 515646 | 515667 | Plus |
| 307 | 6 | 0 | CACAACCUCCUUGAGUGAGCGA | 22 | Y | ? | Y | 1.16 | 1859430 | 1859451 | Minus |
| 1000-1 | 6 | 0 | AUAUUGUCCUGUCACAGCAGU | 21 | Y | Y | Y | 1.187 | 325478 | 325498 | Plus |
| 1000-2 | - | - | AUAUUGUCCUGUCACAGCAGU | 21 | Y | Y | Y | 1.3798 | 224 | 244 | Minus |
| 375 | 4 | 0 | UUUGUUCGUUUGGCUCGAGUUA | 22 | Y | Y | Y | 1.309 | 1318752 | 1318773 | Minus |
| 309-1 | 4 | - | UCACUGGGCAAAGUUUGUCGCA | 22 | Y | ? | Y | 1.15 | 907938 | 907959 | Plus |
| 309-2 | - | - | UCACUGGGCAAAGUUUGUCGCA | 22 | Y | ? | Y | 1.602 | 94088 | 94109 | Plus |
| 932 | 3 | - | UGCAAGCAAUGUGGAAGUGAAG | 22 | ? | Y | Y | 1.1064 | 154192 | 154213 | Minus |
| 315 | 2 | 0 | UUUUGAUUGUUGCUCAGAAAGC | 22 | Y | Y | Y | 1.612 | 104143 | 104164 | Plus |
| 927 | 1 | 0 | CAAAGCGUUUGGAUUCUGAAAC | 22 | Y | Y | Y | 1.26 | 2065461 | 2065482 | Plus |
| **2943 | 1 | 0 | UUAAGUAGGCACUUGCAGGC | 20 | Y | ? | ? | 1.348 | 212450 | 212470 | Plus |
| 1891-1 | predicted | UGAGGAGUUAAUUUGCGUGUUU | 22 | Y | ? | ? | 1.199 | 1109750 | 1109771 | Minus | |
| 1891-2 | predicted | UGAGGAGUUAAUUUGCGUGUUU | 22 | Y | ? | ? | 1.466 | 72802 | 72823 | Plus | |
| 1175 | predicted | AAGUGGAGUAGUGGUCUCAUCG | 22 | Y | ? | ? | 1.125 | 1648037 | 1648058 | Plus | |
| 1174 | predicted | UCAGAUCUACUUAAUACCCAU | 21 | Y | ? | ? | 1.125 | 1647921 | 1647941 | Plus | |
| 993 | predicted | UACCCUGUAGUUCCGGGCUUUU | 22 | Y | Y | Y | 1.056 | 256798 | 256819 | Plus | |
| 981 | predicted | UUCGUUGUCGACGAAACCUGCA | 22 | Y | Y | Y | 1.127 | 638380 | 638401 | Minus | |
| 965 | predicted | UAAGCGUAUAGCUUUUCCCAUU | 22 | Y | ? | Y | 1.51 | 2008701 | 2008722 | Plus | |
| 316 | predicted | UGUCUUUUUCCGCUUACUGCCG | 22 | ? | Y | Y | 1.289 | 891327 | 891348 | Minus | |
| 285 | predicted | UAGCACCAUUCGAAAUCAGUAC | 22 | ? | ? | Y | 1.26 | 3339153 | 3339174 | Minus | |
| 137-1 | predicted | UAUUGCUUGAGAAUACACGUAG | 22 | Y | Y | Y | 1.1191 | 97844 | 97865 | Plus | |
| 137-2 | predicted | UAUUGCUUGAGAAUACACGUAG | 22 | Y | Y | Y | 1.137 | 260684 | 260705 | Minus | |
| 133 | predicted | UUGGUCCCCUUCAACCAGCUGU | 22 | Y | Y | Y | 1.778 | 350306 | 350327 | Minus | |
| 124 | predicted | UAAGGCACGCGGUGAAUGC | 19 | Y | Y | Y | 1.6 | 1664623 | 1664641 | Minus | |
| 31 | predicted | UGGCAAGAUGUUGGCAUAGCUGA | 23 | ? | Y | Y | 1.636 | 483257 | 483279 | Minus | |
| 10 | predicted | ACCCUGUAGAUCCGAAUUUGUU | 22 | Y | Y | Y | 1.44 | 813557 | 813578 | Plus | |
| iab-4-1 | predicted | ACGUAUACUGAAUGUAUCCUGA | 22 | Y | Y | Y | 1.708 | 50265 | 50286 | Plus | |
| iab-4-2 | predicted | ACGUAUACUGAAUGUAUCCUGA | 22 | Y | Y | Y | 1.423 | 763526 | 763547 | Minus | |
| 2940 | 125253 | 4125 | UGGUUUAUCUUAUCUGUCGAGGC | 23 | ? | ? | ? | 1.222 | 643960 | 643982 | Plus |
| 2765 | 1162 | 0 | UGGUAACUCCACCACCGUUGGC | 22 | Y | ? | ? | 1.11 | 5248310 | 5248331 | Plus |
| 2951 | 1136 | 28 | AAGAGCUCAGUACGCAGGGG | 20 | ? | ? | ? | multiple | |||
| 2941-1 | 9 | 0 | UAGUACGGCUAGAACUCCACGG | 22 | ? | ? | ? | 1.385 | 413147 | 413168 | Minus |
| 2941-2 | - | - | UAGUACGGCUAGAACUCCACGG | 22 | ? | ? | ? | 1.385 | 413451 | 413472 | Minus |
aga = An. gambiae; ame = Ap. mellifera; dme = D. melanogaster.
Top group: miRNAs and miRNA* strands identified by deep sequencing
Middle group: predicted miRNAs identified in Cx. quinquefasciatus but absent from C7/10 cells were cross-referenced with the Ae. aegypti (AaegL1)
Bottom group: novel miRNAs identified in this study. miR-2765 (not present in miRBase v14) has recently been identified in Bombyx mori.
** novel miRNAs (not present in miRBase v14) recently identified in Ae. aegypti mosquitoes while this manuscript was under review
Table 2.
miRNAs identified in Cx. quinquefasciatus adult female mosquitoes.
| Culex | # miRNA | # miRNA* | Sequence | Length | aga | ame | dme | supercontig | Start | End | Strand |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 184 | 107190 | 0 | UGGACGGAGAACUGAUAAGGGC | 22 | Y | Y | Y | 3.567 | 240312 | 240333 | Minus |
| 317-1 | 71313 | 2 | UGAACACAGCUGGUGGUAUCU | 21 | Y | Y | Y | 3.36 | 1133209 | 1133229 | Plus |
| 317-2 | - | - | UGAACACAGCUGGUGGUAUCU | 21 | Y | Y | Y | 3.36 | 1134875 | 1134895 | Plus |
| 277 | 58628 | 0 | UAAAUGCACUAUCUGGUACGAC | 22 | Y | Y | Y | 3.36 | 1153785 | 1153806 | Plus |
| 1 | 36084 | 0 | UGGAAUGUAAAGAAGUAUGGAG | 22 | Y | Y | Y | 3.78 | 246250 | 246271 | Plus |
| 989 | 23667 | 0 | UGUGAUGUGACGUAGUGGUAC | 21 | Y | ? | Y | 3.315 | 321364 | 321384 | Plus |
| 275 | 13910 | 2 | UCAGGUACCUGAAGUAGCGC | 20 | Y | Y | Y | 3.291 | 329815 | 329834 | Plus |
| 957 | 11682 | 0 | UGAAACCGUCCAAAACUGAGGC | 22 | Y | ? | Y | 3.787 | 29593 | 29614 | Plus |
| 8-3p | 10950 | - | UAAUACUGUCAGGUAAAGAUGU | 22 | Y | Y | Y | 3.40 | 815865 | 815886 | Minus |
| 281 | 9322 | 95 | AAGAGAGCUAUCCGUCGACAGU | 22 | Y | Y | Y | 3.64 | 99744 | 99765 | Plus |
| Let-7 | 9266 | 5 | UGAGGUAGUUGGUUGUAUAGU | 21 | Y | Y | Y | 3.4 | 280610 | 280630 | Plus |
| 34 | 6301 | 3 | UGGCAGUGUGGUUAGCUGGUU | 21 | Y | Y | Y | 3.36 | 1154478 | 1154498 | Plus |
| 263 | 3749 | 2 | AAUGGCACUGGAAGAAUUCACGG | 23 | Y | Y | Y | 3.219 | 351848 | 351870 | Minus |
| 252-1 | 3157 | 2 | (C)UAAGUACUAGUGCCGCAGGAG | 21 | Y | Y | Y | 3.1787 | 6836 | 6856 | Minus |
| 252-2 | - | - | (C)UAAGUACUAGUGCCGCAGGAG | 21 | Y | Y | Y | 3.975 | 115594 | 115614 | Plus |
| 87 | 2364 | 0 | GUGAGCAAAUUUUCAGGUGUGU | 22 | Y | Y | Y | 3.431 | 379788 | 379809 | Plus |
| 71 | 2232 | 14 | AGAAAGACAUGGGUAGUGAGAU | 22 | ? | Y | ? | 3.366 | 117552 | 117573 | Minus |
| bantam-5p | 1459 | - | CCGGUUUUCAUUUUCGAUCUGAC | 21 | Y | Y | Y | 3.65 | 199737 | 199759 | Minus |
| 9 | 1138 | 440 | UCUUUGGUAUUCUAGCUGUAGA | 22 | Y | Y | Y | 3.83 | 64733 | 64754 | Plus |
| 11 | 888 | 5 | CAUCACAGUCUGAGUUCUUGCU | 22 | Y | ? | Y | 3.153 | 639669 | 639690 | Minus |
| 276-1 | 860 | 2 | UAGGAACUUCAUACCGUGCUCU | 22 | Y | Y | Y | 3.136 | 340911 | 340932 | Plus |
| 276-2 | - | - | UAGGAACUUCAUACCGUGCUCU | 22 | Y | Y | Y | 3.136 | 541192 | 541213 | Plus |
| 276-3 | - | - | UAGGAACUUCAUACCGUGCUCU | 22 | Y | Y | Y | 3.2457 | 930 | 951 | Plus |
| 210.1 | 720 | 5 | UUGUGCGUGUGACAACGGCUAU | 22 | Y | Y | Y | 3.549 | 157657 | 157678 | Minus |
| 927 | 703 | 21 | CAAAGCGUUUGGAUUCUGAAAC | 22 | Y | Y | Y | 3.11 | 560282 | 560302 | Plus |
| bantam-3p | 689 | - | UGAGAUCAUUUUGAAAGCUGA | 21 | Y | Y | Y | 3.65 | 199698 | 199718 | Minus |
| 8-5p | 594 | - | CAUCUUACCGGGCAGCAUUAGA | 22 | Y | Y | Y | 3.40 | 815904 | 815925 | Minus |
| 2 | 547 | 2 | UAUCACAGCCAGCUUUGAAGAGC | 23 | Y | Y | Y | 3.366 | 116861 | 116883 | Minus |
| 998 | 434 | 0 | UAGCACCAUGAGAUUCAGC | 19 | ? | ? | Y | 3.153 | 639527 | 639545 | Minus |
| 210.2 | 405 | - | CUUGUGCGUGUGACAACGGCUAU | 23 | Y | Y | Y | 3.549 | 157657 | 157679 | Minus |
| 14 | 358 | 0 | UCAGUCUUUUUCUCUCUCCUAU | 22 | Y | Y | Y | 3.676 | 52251 | 52272 | Minus |
| 285 | 324 | 5 | UAGCACCAUUCGAAAUCAGUAC | 22 | ? | ? | Y | 3.98 | 262290 | 262311 | Minus |
| 1890 | 287 | 0 | UGAAAUCUUUGAUUAGGUCUGG | 22 | Y | ? | ? | 3.64 | 982786 | 982807 | Minus |
| 190-1 | 231 | 0 | AGAUAUGUUUGAUAUUCUUGGUUG | 24 | Y | Y | Y | 3.181 | 347953 | 347976 | Minus |
| 190-2 | - | - | AGAUAUGUUUGAUAUUCUUGGUUG | 24 | Y | Y | Y | 3.351 | 105098 | 105121 | Minus |
| 283 | 224 | 0 | CAAUAUCAGCUGGUAAUUCUGGG | 23 | Y | Y | Y | 3.57 | 559462 | 559484 | Plus |
| 7 | 192 | 0 | UGGAAGACUAGUGAUUUUGUUGU | 23 | Y | Y | Y | 3.1 | 3357390 | 3357412 | Minus |
| 100 | 170 | 43 | AACCCGUAGAUCCGAACUUGUG | 22 | Y | Y | Y | 3.4 | 271414 | 271435 | Plus |
| 1891 | 167 | 1 | UGAGGAGUUAAUUUGCGUGUUU | 22 | Y | ? | ? | 3.829 | 180383 | 180404 | Minus |
| 999 | 165 | 0 | UGUUAACUGUAAGACUGUGUCU | 22 | ? | ? | Y | 3.14 | 96917 | 96938 | Plus |
| 309 | 33 | 1 | UCACUGGGCAUAGUUUGUCGCAU | 23 | Y | ? | Y | 3.145 | 66041 | 66063 | Minus |
| 375 | 144 | 0 | UUUGUUCGUUUGGCUCGAGUUAC | 23 | Y | Y | Y | 3.455 | 42584 | 42605 | Plus |
| 306 | 143 | 65 | UCAGGUACUGAGUGACUCUCAG | 22 | Y | ? | Y | 3.83 | 80436 | 80457 | Plus |
| 125 | 140 | 7 | UCCCUGAGACCCUAACUUGUGA | 22 | Y | Y | Y | 3.4 | 280975 | 280996 | Plus |
| 315 | 131 | 0 | UUUUGAUUGUUGCUCAGAAAGC | 22 | Y | Y | Y | 3.438 | 61926 | 61947 | Plus |
| 124 | 105 | 0 | UAAGGCACGCGGUGAAUGC | 19 | Y | Y | Y | 3.8 | 2074772 | 2074790 | Plus |
| 92b | 96 | 0 | AAUUGCACUUGUCCCGGCCUG | 21 | Y | Y | Y | 3.722 | 164913 | 164933 | Minus |
| 1889-5p | 89 | - | UAAUCUCAAAUUGUAACAGUGG | 22 | Y | ? | ? | 3.57 | 562555 | 562576 | Plus |
| 981-1 | 82 | 0 | UUCGUUGUCGACGAAACCUGCA | 22 | Y | Y | Y | 3.431 | 144482 | 144503 | Plus |
| 981-2 | - | - | UUCGUUGUCGACGAAACCUGCA | 22 | Y | Y | Y | 3.431 | 151371 | 151392 | Plus |
| 12 | 80 | 2 | UGAGUAUUACAUCAGGUACUGGU | 23 | Y | Y | Y | 3.57 | 563009 | 563031 | Plus |
| 31 | 76 | 2 | UGGCAAGAUGUUGGCAUAGCUGA | 23 | ? | Y | Y | 3.559 | 256577 | 256599 | Minus |
| 10 | 59 | 40 | CAAAUUCGGUUCUAGAGAGGUUU | 23 | Y | Y | Y | 3.12 | 96000 | 96022 | Minus |
| 1174 | 58 | 0 | CUGGGUAUUUUAGAUCAUCGGC | 22 | Y | ? | ? | 3.86 | 865901 | 865922 | Plus |
| **2945 | 52 | 0 | UGACUAGAGGCAGACUCGUUU | 20 | Y | ? | ? | 3.4 | 184461 | 184481 | Plus |
| 1000 | 49 | 0 | AUAUUGUCCUGUCACAGCAGU | 21 | Y | Y | Y | 3.153 | 102853 | 102873 | Minus |
| 13 | 37 | 3 | UAUCACAGCCAUUUUGACGAGU | 22 | Y | Y | Y | 3.366 | 116994 | 117015 | Minus |
| 996 | 36 | 2 | UGACUAGAUUACAUGCUCGU | 20 | Y | Y | Y | 3.19 | 1437010 | 1437029 | Minus |
| 137 | 33 | 0 | UAUUGCUUGAGAAUACACGUAG | 22 | Y | Y | Y | 3.1714 | 27566 | 27587 | Minus |
| 133 | 32 | 0 | UUGGUCCCCUUCAACCAGCUGU | 22 | Y | Y | Y | 3.1189 | 55748 | 55769 | Plus |
| 1175 | 35 | 7 | AAGUGGAGUAGUGGUCUCAUCG | 22 | Y | ? | ? | 3.86 | 866116 | 866137 | Plus |
| 279 | 26 | 21 | UGACUAGAUCCACACUCAUUAA | 22 | Y | Y | Y | 3.19 | 1441123 | 1441144 | Minus |
| 92a | 24 | - | UAUUGCACUUGUCCCGGCCUAU | 22 | Y | Y | Y | 3.722 | 174912 | 174933 | Minus |
| 932-3p | 22 | - | UGCAAGCAAUGUGGAAGUGA | 22 | ? | Y | Y | 3.261 | 301413 | 301432 | Minus |
| 970 | 20 | 0 | UCAUAAGACACACGCGGCUAU | 21 | Y | ? | Y | 3.495 | 35970 | 35990 | Plus |
| 316 | 18 | 0 | UGUCUUUUUCCGCUUACUGCCG | 22 | ? | Y | Y | 3.496 | 152508 | 152529 | Minus |
| 305 | 17 | 1 | AUUGUACUUCAUCAGGUGCUCU | 22 | Y | Y | Y | 3.291 | 339134 | 339155 | Plus |
| **2944a-1 | 13 | 1 | GAAGGAACUUCUGCUGUGAUC | 21 | Y | ? | ? | 3.66 | 328681 | 328701 | Minus |
| **2944a-2 | - | - | GAAGGAACUUCUGCUGUGAUC | 21 | Y | ? | ? | 3.145 | 66240 | 66260 | Minus |
| 988 | 11 | 5 | CCCUUGUUGCAAACCUCACGC | 21 | Y | ? | Y | 3.791 | 14331 | 14351 | Plus |
| 932-5p | 11 | - | UCAAUUCCGUAGUGCAUUGCAG | 22 | ? | Y | Y | 3.261 | 301450 | 301471 | Minus |
| 1889-3p | 7 | - | CACGUUACAGAUUGGGGUUUCC | 22 | Y | ? | ? | 3.57 | 562642 | 562663 | Plus |
| 993 | 4 | 1 | UACCCUGUAGUUCCGGGCUUUU | 22 | Y | Y | Y | 3.12 | 55487 | 55508 | Plus |
| 278 | 3 | 0 | UCGGUGGGACUUUCGUCCGUUU | 22 | Y | Y | Y | 3.16 | 1026212 | 1026233 | Plus |
| 965 | 2 | 0 | UAAGCGUAUAGCUUUUCCCAUU | 22 | Y | ? | Y | 3.48 | 484177 | 484198 | Plus |
| Iab-4 | 2 | 1 | ACGUAUACUGAAUGUAUCCUGA | 22 | Y | Y | Y | 3.12 | 681163 | 681184 | Plus |
| 980 | 2 | 0 | UAGCUGCCUAGUGAAGGGC | 19 | ? | ? | Y | 3.263 | 352922 | 352940 | Plus |
| 308 | 3 | 1 | CGCAGUAUAUUCUUGUGGCUUG | 22 | Y | ? | Y | 3.98 | 764133 | 764154 | Plus |
| 79 | 2 | 0 | GCUUUGGCGCUUUAGCUGUAUGA | 23 | Y | Y | Y | 3.83 | 80591 | 80613 | Plus |
| **2943 | 1 | 0 | UAAGUAGGCACUUGCAGGCAAAG | 23 | Y | ? | ? | 3.121 | 94164 | 94186 | Minus |
| **2944b-1 | 1 | 0 | GAAGGAACUCCCGGUGUGAUAU | 22 | Y | ? | ? | 3.66 | 328838 | 328859 | Minus |
| **2944b-2 | - | - | GAAGGAACUCCCGGUGUGAUAU | 22 | Y | ? | ? | 3.145 | 66389 | 66410 | Minus |
| 33 | predicted | GUGCAUUGUAGUUGCAUUGCA | 21 | ? | Y | Y | 3.1258 | 69381 | 69401 | Minus | |
| 2951 | 162309 | 342 | AAGAGCUCAGCACGCAGGGGUGGC | 24 | ? | ? | ? | multiple | |||
| 2952 | 2203 | - | UAGUACGGCCAUGACUGAGGGC | 22 | ? | ? | ? | 3.5 | 753922 | 753943 | Minus |
| 2941-1 | 1221 | 3 | UAGUACGGCUAGAACUCCACGG | 22 | ? | ? | ? | 3.5 | 753643 | 753664 | Minus |
| 2941-2 | - | 1 | UAGUACGGCUAGAACUCCACGG | 22 | ? | ? | ? | 3.5 | 753797 | 753818 | Minus |
aga = An. gambiae; ame = Ap. mellifera; dme = D. melanogaster.
Top group: miRNAs and miRNA* strands identified by deep sequencing
Middle group: predicted miR-33 was identified in C7/10 cells but absent from Cx. quinquefasciatus. miR-33 was cross-referenced with the Cx. quinquefasciatus genome.
Bottom group: novel miRNAs identified in this study.
** novel miRNAs (not present in miRBase v14) recently identified in Ae. aegypti mosquitoes while this manuscript was under review
miRNA expression levels, based on the number of reads obtained, varied greatly, spanning over five orders of magnitude for Cx. quinquefasciatus and six orders of magnitude for Ae. albopictus (Figure 1C, Tables 1, 2). For both species, the majority of miRNAs (>70%) were sequenced between 10 and 10,000 times (Figure 1C). miR-184 was the most highly expressed miRNA in both species, represented by 1,487,481 reads in the Ae. albopictus library and 107,190 reads in the Cx. quinquefasciatus library. In fact, miR-184 dominated the Ae. albopictus library, accounting for >95% of all miRNA reads. To date, miR-184 has been identified in over 39 organisms, but has no defined role in insects. Surprisingly, although small RNAs were prepared from blood-fed whole Cx. quinquefasciatus mosquitoes compared to Ae. albopictus C710 cells, these two species shared five out of the top ten most frequently occurring miRNAs: miR-184, miR-317, miR-277, miR-275, and miR-8 (Tables 1, 2). In Drosophila, miR-277 has predicted targets in metabolic pathways [20] while miR-8 plays a role in Wnt signaling [36]. miR-275 and miR-317 have no experimentally reported targets to date.
Mature miRNA species showed sequence lengths between 19 and 24 nt with a predominance of 22 nt and also exhibited strong bias for a 5' uracil (> 65% of all identified miRNAs) (Tables 1, 2). The presence of a 5' U is a characteristic of many miRNAs [37,38], at least in part, because strand selection of the miRNA from the miRNA:miRNA* duplex is based on the level of thermodynamic stability of the paired ends of the duplex [27,39,40].
Mosquito miRNAs are highly conserved
The Ae. albopictus genome is not yet sequenced. Since miRNA sequences are highly conserved between species, we mapped miRNAs cloned from the Ae. albopictus cell line to the Ae. aegypti genome. Interestingly, all Ae. albopictus miRNAs and miRNA* strands mapped with 100% identity to the Ae. aegypti genome, indicating evolutionary constraints on not only the mature miRNA sequences, but also the pre-miRNA hairpins. 72 of the 74 Culex miRNA sequences mapped with 100% identity to the Cx. quinquefasciatus genome [35]. The identified sequence for miR-309 (Table 2) differed by one nucleotide (nt 11) from the Cx. quinquefasciatus genomic sequence. miR-927, occurring 700 times in the Cx. quinquefasciatus library (Table 2), exhibited sequence differences at nucleotides 1 and 16 compared to the Cx. quinquefasciatus genome. When mapped to the Ae. aegypti genome, one nucleotide differed from the genomic sequence. These sequence variations could not be accounted for by miRNA editing.
Several miRNA sequences mapped to multiple locations in the Cx. quinquefasciatus and Ae. aegypti genomes. Six Cx. quinquefasciatus miRNAs, miR-317, miR-252, miR-276, miR-190, miR-981, and miR-2944, arise from at least two possible hairpin precursors (Table 2). In Aedes, four miRNAs, miR-276, miR-317, miR-1000, and miR-309 arise from two potential hairpin precursors (Table 1).
With the exception of miR-33, all Ae. albopictus miRNAs were also identified in Cx. quinquefasciatus mosquitoes. Additionally, 14 miRNAs present in Cx. quinquefasciatus mosquitoes, but absent from Ae. albopictus cells, mapped with 100% sequence identity to the Ae. aegypti genome, and are annotated as predicted (Tables 1 and 2). Of note, Cx. quinquefasciatus miR-1174 was not found in Ae. aegypti; however, the annotated mature miRNA sequence for An. gambiae miR-1174 aligns to the Ae. aegypti genome with 95% sequence identity. Table 1 contains the predicted miR-1174 sequence for Ae. aegypti. Interestingly, Cx. quinquefasciatus miR-1174 is orthologous not to the mature miR-1174 in An. gambiae, but to the predicted miR-1174* (19 out of 22 nt); only these 19 nucleotides are conserved between the Cx. quinquefasciatus and An. gambiae pre-miRNAs. In total, 75 Aedes and Cx. quinquefasciatus conserved miRNAs were identified, representing over 55 seed families (Tables 1, 2).
64 of the 75 miRNAs identified in Cx. quinquefasciatus and Ae. albopictus have orthologs in D. melanogaster. In addition to D. melanogaster, we examined orthologous miRNA sequences from two other winged insects, An. gambiae and Apis mellifera (Tables 1, 2). Five miRNAs, miR-1175, miR-1174, miR-1889, miR-1890, and miR-1891, have previously been identified in Anopheles mosquitoes but, to date, lack orthologs in D. melanogaster or A. mellifera. Interestingly, for miR-1890, only the miRNA sequenced is conserved between Anopheles, Culex, and Aedes, and extensive sequence variations occur in the remaining arm and loop of the precursor. While this manuscript was under review, eight additional novel mosquito-specific miRNAs were identified in Ae. aegypti mosquitoes [41]. miR-2944a/b is present at low levels in Cx. quinquefasciatus; miR-2943 and miR-2945 are present at low levels in both Cx. quinquefasciatus mosquitoes and C710 cells (Tables 1 and 2). While orthologs of these mosquito-specific miRNAs may be identified in other organisms in the future, this group of miRNAs appears to be restricted to mosquitoes and hence, may be of more recent evolutionary origin.
Sequence variation occurs predominantly at the 3' end of mature miRNAs
In each small RNA library, reads aligning to a given mature miRNA showed some degree of variability. Most variability occurred at the 3'ends of each mature miRNA, when compared to the 5' ends. Figure 2A depicts this variance for all conserved miRNAs present in the Culex library. Each canonical miRNA sequence is set at "0"; nucleotide truncations from either the 3' or 5' end are shown by negative numbers, whilst nucleotide additions are shown by positive numbers. 20.5% of miRNA reads exhibited 3' end variability compared to only 0.8% of reads for 5' variability. In accordance with other miRNA studies [18,42,43], we found that the majority of miRNAs, such as miR-1 (Figure 2B), followed this pattern of 5' sequence homogeneity and 3' heterogeneity. Precision at the mature miRNA 5' end has been reported for Drosophila miRNAs [44]. Such observations are congruent with the idea that the seed sequence, located within the 5' end of the miRNA, is evolutionarily constrained [15,29].
Figure 2.
Culex miRNA sequence variations. A) The total number of canonical miRNA sequence reads, annotated in Table 2, is set to "0" on the x-axis. Differences in the total numbers of canonical 3' versus 5' miRNA ends are due to greater diversity at the 3' end of a given miRNA. Negative numbers on the x-axis indicate 3' or 5' nucleotide truncations in a miRNA sequence, while positive numbers indicate 3' or 5' nucleotide additions. 58 Culex miRNAs (sequenced at least 10 times) are collectively represented. The numbers of reads with 3' or 5' nucleotide truncations or additions for individual miRNAs, B) miR-1, C) miR-210, and D) miR-252 are shown.
At least two miRNAs, however, did not match this trend. For both miR-210 and miR-252, two dominant miRNA species were identified (Figure 2C and 2D; Tables 1, 2). For miR-210, the most frequently occurring species was sequenced 301 times, while the second dominant species, one nucleotide longer with a cytosine at the 5' end, was sequenced 274 times. Due to variations in the 5' and 3' ends for the remaining 550 reads aligning to miR-210, the canonical 5' and 3' ends were actually represented by the second most frequently occurring sequence, which is annotated (Table 2). Interestingly, two dominant forms of miR-210, miR-210.1 and miR-210.2, one of which contains an extra 5' nucleotide, have been noted for D. melanogaster [18]. Furthermore, of the 19 reads aligning to miR-210 in the Ae. albopictus library, 13 (68%) contain an extra 5' cytosine. Only one copy of the miR-210 precursor is present in these insect genomes, therefore such differences cannot be attributed to processing from multiple pri-miRNAs. Mosquitoes and fruit flies diverged over 250 million years ago. Thus, it is striking that we see these same two forms of miR-210 expressed in mosquitoes. Our data provide strong evidence in support of the hypothesis that these two forms of miR-210 are evolutionarily conserved and are likely to function as at least partly distinct miRNAs in vivo.
miR-252, which maps to two loci within the Cx. quinquefasciatus genome, but only one locus in each of the Ae. aegypti and An. gambiae genomes, also exhibited similar variation at the 5' end (Figure 2D). The dominant, canonical miRNA species was sequenced 1,688 times, while the second dominant species, with a 5' cytosine addition, was sequenced 719 times. We also observed miR-252 variations in the Ae. albopictus library. 35% of the 2496 sequences aligning to Ae. albopictus miR-252 contained one extra 5' cytosine. The two 69 nt pri-miRNA stem-loops for Cx. quinquefasciatus miR-252 are 100% identical, and show 100% and 97% sequence identity with miR-252 pri-miRNA stem-loops present in the Ae. aegypti and An. gambiae genomes, respectively. Thus, these variations in the mature miRNA sequences, for both miR-252 and miR-210, do not appear to arise from differences in hairpin folding properties, and likely are a result of Drosha and/or Dicer processing.
The consequences of 5' variation can be severe, since an alteration to the 5' seed creates a new group of potential target mRNAs for a miRNA [29]. Depending on the length of the complementary seed match within a target mRNA, miRNAs arising from a single precursor, yet exhibiting 5' variation, could have both overlapping and distinct targets.
Whereas some miRNAs exhibited sequence differences at the 5' or 3' ends, we also noted differences in the ratios of miRNA:miRNA* reads when examining the Cx. quinquefasciatus and Ae. albopictus libraries. miRNA* strands for several miRNAs, including miR-8, miR-1889, and bantam, were sequenced a significant number of times, and thus are annotated with 5p or 3p (Tables 1, 2). In C7/10 cells, miR-1889-3p and miR-1889-5p were expressed at nearly identical levels, suggesting that both strands of the miRNA:miRNA* duplex are loaded equally into RISC as mature miRNAs. Interestingly, for miR-8 in the Ae. albopictus library, the ratio of 5p:3p miRNA reads was 1,244 miR-8-5p: 3,002 miR-8-3p (ratio of 0.41) (Table 1). In Cx. quinquefasciatus mosquitoes, however, the ratio was much different. miR-8-5p occurred only 594 times compared to miR-8-3p which occurred 10,950 times (ratio of 0.05) (Table 2). Of note, the dominant miRNA species for Ae. albopictus miR-8-3p contains one less 3' nucleotide compared to Cx. quinquefasciatus miR-8-3p.
We investigated the predicted miR-8 pre-miRNA structures in Ae. aegypti, Cx. quinquefasciatus, and An. gambiae. Ae.aegypti miR-8 pre-miRNA shares 98% and 92% sequence identity with the miR-8 pre-miRNA in Cx. quinquefasciatus and An. gambiae, respectively. Intriguingly, all nucleotide differences for miR-8 affect only the terminal loop of the pre-miRNA hairpin, which alters the pairing at the immediate base of the terminal loop. Thus, differences in the miRNA-5p:miRNA-3p ratios may reflect the RNA folding properties of the pre-miRNA within each species, which is known to influence strand selection. Furthermore, nucleotide diversity in the terminal loop for miR-8, a miRNA known to be involved in Wnt signaling in the fly [21,36], may help fine tune not only miRNA strand selection but also the miRNA sequence itself, thereby also fine tuning miRNA target regulation.
Whilst the total number of miRNA* strands accounted for a low percentage (<0.3%) of mapped reads in each small RNA library, some miRNA* strands were sequenced more frequently than individual miRNA species. For example, in total RNA from C7/10 cells, bantam-3p was sequenced 475 times, and therefore accounts for a greater percentage of the small RNA population than those mature miRNAs sequenced less than 400 times. Likewise, miR-281* in Cx. quinquefasciatus mosquitoes was sequenced 95 times, and thus accounts for a greater percentage of small RNAs than those occurring less than 95 times. Importantly, the biological relevance of the miRNA* population has been demonstrated in Drosophila; miRNA* strands can be loaded into Ago1-containing RISC and target complementary 3' UTRs of mRNAs [45].
Confirmation of mosquito miRNAs
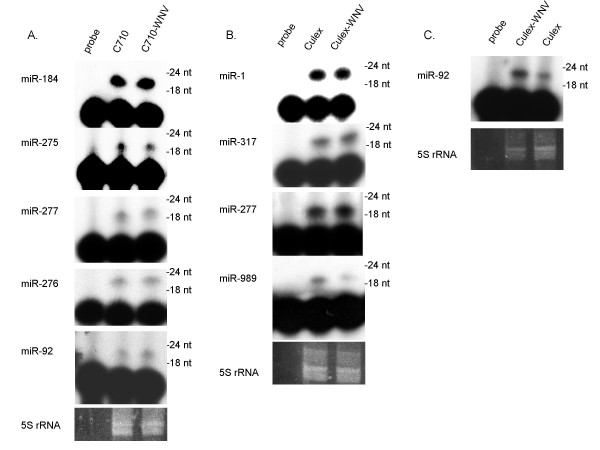
We used primer extension analysis to confirm the expression of several of the miRNAs represented in our sequencing data. Five miRNAs, miR-184, miR-275, miR-277, miR-276, and miR-92, were sequenced >500 times and were readily detectable in total RNA isolated from C7/10 cells (Figure 3A). Five miRNAs, miR-1, miR-317, miR-277, miR-989, and miR-92 were sequenced >120 times and were readily detectable in total RNA isolated from Cx. quinquefasciatus mosquitoes (Figure 3B). In general, the detection level of a given miRNA reflected the overall abundance of that miRNA in the sequenced library (Figure 3, Tables 1, 2). All miRNAs analyzed by this method exhibited the expected sizes.
Figure 3.
Primer extension analysis confirms miRNA expression. Total RNA was isolated from A) C7/10 cells or C7/10-WNV312 cells persistently infected with WNV replicons and B, C) Cx. quinquefasciatus mosquitoes fed a non-infectious blood meal (Culex) or a blood meal containing WNV-NY99 (Culex-WNV). 10 μg (A, C) or 4 μg (B) of RNA was used for primer extension to detect miRNAs. For each miRNA, free probe with no RNA is shown as a negative control. Ethidum bromide stained rRNA is shown as loading control.
Identification of novel mosquito miRNAs
To identify novel mosquito miRNAs, we used a combination of miRDeep [46] and MFold [47] to ask whether non-annotated sequences mapping to the mosquito genomes demonstrated folding properties of pre-miRNA hairpins. Each novel miRNA follows both expression and biogenesis criteria set forth for identifying new miRNAs, which include (i) a small RNA of appropriate and discrete length (19-24 nt), (ii) arising from one arm of a hairpin precursor, (iii) presence of the star strand, and (iv) evolutionary conservation [13,18,48].
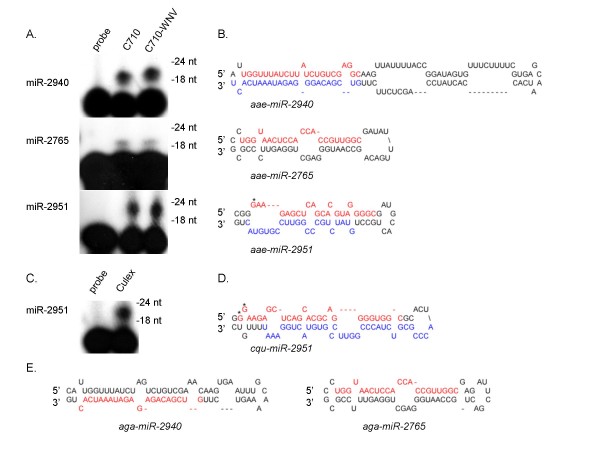
Four new Aedes miRNAs (five hairpins) and three new Cx. quinquefasciatus miRNAs (four hairpins) were identified (Tables 1, 2). Each miRNA arises from RNA structures which fold into canonical pre-miRNA hairpins (Figures 4 and 5). Four of the new miRNAs reside on the 5p arms of their respective precursors (Figure 4B and 4C), while the remaining three miRNAs reside on the 3p arms (Figure 5). Primer extension analysis confirmed the expression of five of these miRNAs (Figures 4 and 5).
Figure 4.
Identification of novel mosquito miRNAs. Primer extension analysis was used to confirm the expression of A) three novel Ae. albopictus miRNAs and C) one novel Cx. quinquefasciatus miRNAs. Total RNA was isolated from C7/10 cells or Cx. quinquefasciatus mosquitoes as described in Figure 3 and Methods. B) and D) Predicted pre-miRNA stem-loop structures for each novel miRNA. Ae. albopictus miRNAs were mapped to the Ae. aegypti genome, and therefore may not reflect the actual pre-miRNA structures. Mature miRNA sequences are shown in red, while corresponding miRNA* sequences identified in each library are shown in blue. For miR-2951, asterisks indicate additional 5' nucleotides present in a lower percentage of the reads mapping to each miRNA compared to the canonical sequence annotated in Tables 1 and 2. Ae. albopictus miR-2951 differs by one nucleotide from the Ae. aegypti genome. E) Pre-miRNA structures for two novel orthologous miRNAs mapping to the A. gambiae genome. The predicted mature miRNAs, based on sequence conservation, are shown in red.
Figure 5.
Novel mosquito miRNAs are clustered. A) Predicted pre-miRNA stem-loop structures for cqu-miR-2941-1, cqu-miR-2941-2, and cqu-miR-2952. B) Primer extension analysis confirms miR-2941 expression in total RNA isolated from Cx. quinquefasciatus adult female blood-fed mosquitoes (see Methods). C) Predicted pre-miRNA stem-loop structures for aae-miR-2941-1, and aae-miR-2941-2. As in Figure 4, mature miRNAs are shown in red, and miRNA* strands, when identified in a library, are shown in blue. D) Genomic location of the miR-2941 clusters in the Cx. quinquefasciatus and Ae. aegypti genomes. Numbers on the bottom indicate the nucleotide distance between pri-miRNA stem-loops for each miRNA.
miR-2940, which lacks seed homology to any known miRNA, was amongst the most frequently recovered miRNAs present in the Ae. albopictus library, sequenced 125,253 times; miR-2940* was sequenced 4,125 times (Table 1). Interestingly, miR-2940 and miR-2940* are separated by 60 nt of intervening sequence, resulting in a 104 nt pre-miRNA (Figure 4B). This pre-miRNA length is unusual for metazoan pre-miRNAs, which are normally ~60 nt in length [24]. Plant pre-miRNAs, however, can be as long as 200 nt [13], and several Drosophila miRNAs arise from long hairpins >100 nt. The D. melanogaster miR-989 precursor, for example, has 99 nt of intervening sequence between the miRNA and miRNA* [18].
Two novel Aedes miRNAs, miR-2765 and miR-2951, arise from pre-miRNAs with typical lengths of 59 nt and 57 nt, respectively (Figure 4B). Both miRNAs were sequenced ~1,100 times; however, primer extension analysis suggested that miR-2951 is expressed at higher levels than miR-2765 (Figure 4A). Like miR-2940, miR-2765 shows no seed sequence homology to any known miRNA present in miRBase v14. miR-2951 is 100% identical to cqu-miR-2951, expressed in Culex mosquitoes (Figure 4C).
Whilst the majority of new miRNAs exhibited discrete lengths, as determined from both sequencing data and primer extension analysis (Figure 4 and 5), we observed variations in the 5'ends of both Ae. albopictus miR-2951 and Cx. quinquefasciatus miR-2951, which affect the seed. 29% of Ae. albopictus miR-2951 reads contained an additional 5' G, while Cx. quinquefasciatus miR-2951 reads contained 5' GG (3.4%) or 5' G (30.2%) additions or single nucleotide 5' truncations (12%) compared to the canonical sequence (54.4% of reads). Furthermore, unlike Aedes miR-2951*, for which a distinct sequence was identified, over five equally abundant sequences for Cx. quinquefasciatus miR-2951* were observed, which affect the positioning of the star strand in the pre-miRNA hairpin (Table 1, 2). Only 15 nucleotides, excluding the potential 5' seed, are conserved between Aedes miR-2951*, and Cx. quinquefasciatus miR-2951*, contributing to differences in the predicted pre-miRNA hairpin structures. These differences are also due, in part, to nucleotide differences in the terminal loops (Figure 4B and 4D). These sequence variations might also be attributed to diversity in the flanking pri-miRNA sequences; miR-2951 maps to eight locations within each of the Ae. aegypti and Cx. quinquefasciatus genomes. Notably, within each genome, all pre-miRNA loci share 100% sequence identity.
We queried three mosquito genomes (Cx. quinquefasciatus, An. gambiae, Ae. aegypti) present in VectorBase for the presence of each new miRNA. Both miR-2940 and miR-2765 have orthologs in An. gambiae (Figure 4E). The predicted precursor for miR-2765 is 93% identical at the sequence level in An. gambiae, while the mature miRNA sequence is 100% conserved. Interestingly, for miR-2940, the orthologous sequence mapping to An. gambiae chromosome X with 95% sequence identity was actually miR-2940*. Given that miR-2940* was sequenced over 4,000 times, it is possible that both strands of the miR-2940:miR-2940* duplex are loaded into RISC and function as mature miRNAs. Notably, the predicted 5p arm for An. gambiae miR-2940 exhibits the same seed sequence as miR-2940-5p from Aedes, suggesting similar functions in mRNA targeting (Figure 4B and 4E). No orthologs for miR-2951 or miR-2952 were found in An. gambiae. In fact, miR-2952 appears to be specific to Cx. quinquefasciatus.
Two additional miRNAs, aae-miR-2941 and cqu-miR-2941, are also orthologs conserved in Aedes and Cx. quinquefasciatus. cqu-miR-2941 was readily detectable by primer extension analysis in Cx. quinquefasciatus (Figure 5B); however, miR-2941 was sequenced only nine times in the Ae. albopictus library, and thus was below the limit of detection. aae-miR-2941 and cqu-miR-2941 each arise from two different pre-miRNA hairpins that map to two loci (Figure 5A and 5C). cqu-miR-2941* strands from both of the Cx. quinquefasciatus pre-miRNAs were identified (Table 2), indicating that both hairpins are expressed and processed. The pre-miRNAs for both aae-miR-2941 and cqu-miR-2941 are clustered within a ~350 nt stretch which, for Cx. quinquefasciatus, also includes another novel miRNA, miR-2952 (Figure 5D). Notably, miR-2941 and miR-2952 share the first nine 5' nucleotides and thus, have the same seed (Table 2), suggesting these two miRNAs might regulate an overlapping set of target mRNAs.
Clusters of mosquito miRNA genes
The miR-2941 cluster represents a novel miRNA cluster present in both Cx. quinquefasciatus and Aedes mosquito genomes. To determine whether additional conserved miRNAs were clustered, we considered miRNAs which mapped to locations within 1 kb of each other. Nine mosquito miRNAs followed this pattern (Table 1, 2). The ordered distribution of each of the nine pre-miRNAs in the Ae. aegypti genome was similar to the distribution of pre-miRNAs in the Cx. quinquefasciatus genome, with two exceptions. miR-11 and miR-989 map to the plus strand in the Ae. aegypti genome, but map to the minus strand in Cx. quinquefasciatus. It is possible that this cluster is inverted in Cx. quinquefasciatus since (i) miR-11 and miR-989 are located on the plus strand in An. gambiae [35] and (ii) the order of miRNAs is still conserved. Based on sequencing reads, miRNAs within each cluster did not appear to be evenly expressed (Tables 1, 2).
Culex miR-989 and miR-92 expression levels are altered during flavivirus infection
miRNAs are known to be important regulators of development. Additionally, miRNA expression profiles can be altered in response to environmental changes such as stress or infection. Four An. gambiae miRNAs, miR-34, miR-1174, miR-1175, and miR-989, show changes in expression during Plasmodium infection [11]. Given that Cx. quinquefasciatus and Ae. albopictus are important flavivirus vectors, we asked whether any miRNAs were aberrantly expressed during infection with WNV.
We assayed miRNA expression in WNV-replicon C7/10 cells and WNV-NY99 infected Cx. quinquefasciatus using primer extension. Persistent infection of C7/10 cells with WNV-replicons had no significant effect on the expression levels of the miRNAs assayed (Figures 3A and 4A). A comparison of blood-fed, uninfected female Cx. quinquefasciatus mosquitoes to age- and sex-matched WNV-NY99 infected mosquitoes revealed that the majority of miRNAs were unaffected; however, we observed 2.8 fold downregulation of miR-989 following WNV-NY99 infection (Figure 3B; Additional file 1, Figure S1). In contrast, miR-92 expression was upregulated in WNV-infected Cx. quinquefasciatus (Figure 3C; Additional file 1, Figure S1). Notably, this pattern of miRNA expression for miR-989 and miR-92 is also found in deep sequencing reads of WNV-infected Cx. quinquefasciatus (Additional file 2, Table S1). We also observed changes in miR-957, miR-970, miR-980, and miR-33, among others (Additional file 2, Table S1).
The targets of miR-989 and miR-92 in mosquitoes are not yet known; however, several studies have examined expression of these miRNAs during development. In An. gambiae, An. stephensi, and Ae. aegypti, miR-989 expression is restricted to female mosquitoes and found predominantly in the ovaries [10,11]. While this manuscript was in review, Li et.al. reported 454 deep sequencing of miRNAs in Ae. aegypti mosquitoes; miR-989 is also present in the midgut while miR-92 is present in Ae. aegypti embryos [41]. In the silkworm Bombyx mori, miR-92 is associated with embryogenesis, a stage of high cellular proliferation and differentiation [49]. Furthermore, in vertebrates, miR-92 is a member of the conserved miR-17-92 cluster and is associated with oncogenesis and increased cellular proliferation. Given the dysregulation of miR-989 and miR-92 during WNV infection, it is interesting to speculate that the targets of these miRNAs may play roles in mediating flavivirus infection in the mosquito host.
Conclusions
This study provides experimental evidence for over 65 conserved and seven novel miRNAs present in Aedes and Cx. quinquefasciatus mosquitoes, and increases our current understanding of insect miRNAs. The majority of miRNAs identified here demonstrate conventional miRNA characteristics including evolutionary conservation, 5' end homogeneity, and an ~60 nt pre-miRNA. A small number of miRNAs were found that deviate from these standards. Cx. quinquefasciatus and Aedes miR-210, miR-252, and miR-2951 are examples of multiple, distinct miRNAs arising from one arm of a single hairpin (Figures 2 and 4). Aedes miR-2940, among others, arises from an unusually long pre-miRNA (Figure 4A). Additionally, the prevalence of the miRNA* strand for several miRNAs, such as miR-1889, miR-8, and bantam, expands the potential of miRNA regulation in an organism by adding to the number of possible miRNA seeds and thus adding new mRNA targets. Finally, of the novel miRNAs identified here, four currently lack orthologs in non-mosquito species, bringing the total mosquito-specific miRNAs to 16 [41].
Aedes and Culex mosquitoes are major arbovirus vectors, important in transmitting both alphaviruses and flaviviruses to humans. We found miR-989, a female-specific miRNA in Anopheles and Aedes mosquitoes, to be downregulated in WNV-infected Cx. quinquefasciatus while miR-92 is significantly upregulated. This is the first report of miRNA dysregulation following flavivirus infection of a natural mosquito host. Future research will elucidate the functions of these newly identified miRNAs in mosquito biology. Undoubtedly, some of the miRNAs identified here will have roles not only in mosquito development, like their Drosophila counterparts, but also in mediating viral infection in the mosquito host.
Methods
Mosquitoes and Cell Lines
Cx. quinquefasciatus mosquitoes (Sebring strain) were reared and maintained as previously described [50]. Female mosquitoes were fed a non-infectious blood meal containing 2 mL of Vero cells and media mixed with 2 mL of defibrinated sheep blood (Colorado Serum Company, Denver, CO) or an infectious blood meal containing 2 mL of WNV NY99 [51] infected Vero cells with media and 2 mL of sheep blood. The meals were presented separately to 200 female mosquitoes 3 to 5 day post-eclosion as previously described [50]. Mosquitoes were instantaneously killed in Eppendorf tubes by submersion in a dry ice/liquid nitrogen bath at 14 days post-blood meal and stored in RNAlater prior to RNA extraction. Ae. albopictus C7/10 cells were maintained at 28°C in Leibowitz L-15 media supplemented with 10% FCS, 10% tryptose phosphate broth, and antibiotics. C7/10-WNV replicon cells were generated by infecting C7/10 cells with GFP-expressing WNV replicon particles [52,53]. The cells were sorted for GFP expression 7 days post-infection, and monitored for GFP expression for one month prior to analysis to verify establishment of a persistent infection. Infection of both mosquitoes and C7/10 cells was confirmed by qRT-PCR [52].
RNA extraction and Primer Extensions
Total RNA was prepared from ~100 whole mosquitoes and two 80% confluent T75 flasks of Ae. albopictus cells using TRIzol (Invitrogen) according to the manufacturer's protocol. Primer extensions were performed with 4 μg (Cx. quinquefasciatus) or 10 μg (C7/10) of total RNA using the AMV PE kit according to manufacturer's protocol (Promega). Oligonucleotides used for probes are listed (Additional file 3, Table S2) and were end-labeled using gamma-[32P]-ATP and T4 polynucleotide kinase. To detect individual miRNAs, a master mix was prepared for each probe and divided equally amongst the reactions. Reverse transcription products were separated on 15% TBE-urea polyacrylamide gels, exposed to film, and subjected to analysis using NIH ImageJ (Additional file 1, Figure S1).
Small RNA cloning
Thirty micrograms of total RNA were size-fractionated on a 15% TBE-Urea polyacrylamide gel. Small RNA populations corresponding to 18-28 nt in size were extracted, eluted, and ligated to a 3' linker using T4 RNA ligase (Epicentre). 3' ligation reactions were loaded directly onto a 10% TBE-Urea polyacrylamide gel, and ligation products recovered by high-salt elution following electrophoresis. Next, a 5' linker was ligated, and products were used for SSII reverse transcription (Invitrogen). PCR reactions were carried out using the RT primer and 5' PCR primer. Linker and primer sequences are provided in Additional file 3, Table S2. Amplified cDNA products were gel-purified prior to submission for sequencing. High-throughput sequencing was performed by the Duke IGSP Sequencing Core Facility on an Illumina Genome Analyzer II.
Bioinformatics
Sequencing reads were parsed using in-house scripts according to the following criteria: a 5' and 3' linker match of at least 4 nt and an appropriate length (18-28 nt).
To find miRNA orthologs, sequences were mapped to known miRNAs, miRNA star strands, and hairpins present in miRBase v14.0 http://microrna.sanger.ac.uk using NCBI BLAST (word size = 17, p = 85, D = 2) allowing for a 2 nt mismatch, and parsed further using Perl scripts from the miRDeep pipeline [46]. Mosquito genomes (Cx. quinquefasciatus Johannesburg strain and Ae. aegypti Liverpool strain) were obtained from http://vectorbase.org and coordinates for miRNA sequences were extracted using BLAST. For new miRNA discovery, reads mapping to each mosquito genome were analyzed using the miRDeep pipeline [46]. To further confirm novel miRNAs, reads of 19-24 nt in length occurring at least 100 times in a library were mapped to mosquito genomes, and sequences of 200 nt in length surrounding the putative miRNA were extracted, and folded using MFold [47]. FASTA files containing all unique reads for the C7/10 and Culex libraries as well as miRNA precursor sequences are provided (Additional files 4567).
Authors' contributions
RLS prepared the small RNA libraries, analyzed the data, and drafted the manuscript. DLV and SH reared the mosquitoes and performed the WNV infections. FS provided the C7/10 cells and generated the C7/10-WNV replicon cells. BRC supervised the experiments and helped draft the manuscript. All authors read and commented on the final manuscript.
Supplementary Material
Figure S1, miRNA quantification in primer extension experiments shown in Figure 3. Primer extension experiments were quantified using NIH ImageJ. Signal ratios of (A) C7/10-WNV replicon cells: C7/10 cells and (B) WNV-infected Culex: uninfected Culex are graphed for individual miRNAs.
Table S1, miRNA reads in Cx. quinquefasciatus and WNV-infected Cx. quinquefasciatus. Table comparing mosquito miRNA counts from high-throughput sequencing of uninfected and WNV-infected Cx. quinquefasciatus. The WNV-infected Cx. quinquefasciatus library was prepared as described in Methods. Differences in miR-989 and miR-92 expression levels are highlighted. nd = not determined
Table S2, Oligonucleotides used in this study. Table of oligonucleotides used for primer extension and high-throughput sequencing.
Raw sequence data C710.fasta. FASTA file containing sequencing reads for C7/10 Ae. albopictus cells
Raw sequence data Culex.fasta. FASTA file containing sequencing reads for Cx. quinquefasciatus mosquitoes
miRNA precursor sequences Aedes_precursors.fasta. FASTA file containing miRNA, miRNA*, and precursor sequences for Ae. albopictus
miRNA precursor sequences Culex_precursors.fasta. FASTA file containing miRNA, miRNA*, and precursor sequences for Cx. quinquefasciatus
Contributor Information
Rebecca L Skalsky, Email: rebecca.skalsky@duke.edu.
Dana L Vanlandingham, Email: dlvanlan@utmb.edu.
Frank Scholle, Email: frank_scholle@ncsu.edu.
Stephen Higgs, Email: sthiggs@utmb.edu.
Bryan R Cullen, Email: culle002@mc.duke.edu.
Acknowledgements
This work was supported by NIH grant U54-AI-057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense to BRC, and by funds provided by the UTMB Department of Pathology to SH. DLV was supported by an NIH T32 grant (A107536). We thank Tonya Severson of the Duke Institute for Genome Sciences and Policy Core Sequencing Facility for assistance with high-throughput sequencing. We gratefully acknowledge the technical assistance of Jing H. Huang for the rearing of the Cx. p. quinquefasciatus mosquitoes.
References
- Gould EA, Solomon T. Pathogenic flaviviruses. Lancet. 2008;371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10:S98–109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Gratz NG. Critical review of the vector status of Aedes albopictus. Med Vet Entomol. 2004;18:215–227. doi: 10.1111/j.0269-283X.2004.00513.x. [DOI] [PubMed] [Google Scholar]
- Ebel GD, Rochlin I, Longacker J, Kramer LD. Culex restuans (Diptera: Culicidae) relative abundance and vector competence for West Nile Virus. J Med Entomol. 2005;42:838–843. doi: 10.1603/0022-2585(2005)042[0838:CRDCRA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Girard YA, Klingler KA, Higgs S. West Nile virus dissemination and tissue tropisms in orally infected Culex pipiens quinquefasciatus. Vector Borne Zoonotic Dis. 2004;4:109–122. doi: 10.1089/1530366041210729. [DOI] [PubMed] [Google Scholar]
- Girard YA, Popov V, Wen J, Han V, Higgs S. Ultrastructural study of West Nile virus pathogenesis in Culex pipiens quinquefasciatus (Diptera: Culicidae) J Med Entomol. 2005;42:429–444. doi: 10.1603/0022-2585(2005)042[0429:USOWNV]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Renault P, Solet JL, Sissoko D, Balleydier E, Larrieu S, Filleul L, Lassalle C, Thiria J, Rachou E, de Valk H. et al. A major epidemic of chikungunya virus infection on Reunion Island, France, 2005-2006. Am J Trop Med Hyg. 2007;77:727–731. [PubMed] [Google Scholar]
- Vazeille M, Moutailler S, Coudrier D, Rousseaux C, Khun H, Huerre M, Thiria J, Dehecq JS, Fontenille D, Schuffenecker I. et al. Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One. 2007;2:e1168. doi: 10.1371/journal.pone.0001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Mead EA, Tu Z. Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito, Anopheles stephensi. BMC Genomics. 2008;9:244. doi: 10.1186/1471-2164-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter F, Edaye S, Huttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35:6953–6962. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/S0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Black WCt, Hess AM, Foy BD. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics. 2008;9:425. doi: 10.1186/1471-2164-9-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E. et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc Natl Acad Sci USA. 2008;105:15417–15422. doi: 10.1073/pnas.0807763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/S0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Li S, Mead EA, Liang S, Tu Z. Direct sequencing and expression analysis of a large number of miRNAs in Aedes aegypti and a multi-species survey of novel mosquito miRNAs. BMC Genomics. 2009;10:581. doi: 10.1186/1471-2164-10-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5' precision of both MicroRNAs and their miRNA strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3' UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M. et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhang L, Li Q, Zhao P, Duan J, Cheng D, Xiang Z, Xia Q. MicroRNA expression profiling during the life cycle of the silkworm (Bombyx mori) BMC Genomics. 2009;10:455. doi: 10.1186/1471-2164-10-455. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Vanlandingham DL, Schneider BS, Klingler K, Fair J, Beasley D, Huang J, Hamilton P, Higgs S. Real-time reverse transcriptase-polymerase chain reaction quantification of West Nile virus transmitted by Culex pipiens quinquefasciatus. Am J Trop Med Hyg. 2004;71:120–123. [PubMed] [Google Scholar]
- Beasley DW, Whiteman MC, Zhang S, Huang CY, Schneider BS, Smith DR, Gromowski GD, Higgs S, Kinney RM, Barrett AD. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J Virol. 2005;79:8339–8347. doi: 10.1128/JVI.79.13.8339-8347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle F, Girard YA, Zhao Q, Higgs S, Mason PW. trans-Packaged West Nile virus-like particles: infectious properties in vitro and in infected mosquito vectors. J Virol. 2004;78:11605–11614. doi: 10.1128/JVI.78.21.11605-11614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi SL, Zhao Q, O'Donnell VK, Mason PW. Adaptation of West Nile virus replicons to cells in culture and use of replicon-bearing cells to probe antiviral action. Virology. 2005;331:457–470. doi: 10.1016/j.virol.2004.10.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, miRNA quantification in primer extension experiments shown in Figure 3. Primer extension experiments were quantified using NIH ImageJ. Signal ratios of (A) C7/10-WNV replicon cells: C7/10 cells and (B) WNV-infected Culex: uninfected Culex are graphed for individual miRNAs.
Table S1, miRNA reads in Cx. quinquefasciatus and WNV-infected Cx. quinquefasciatus. Table comparing mosquito miRNA counts from high-throughput sequencing of uninfected and WNV-infected Cx. quinquefasciatus. The WNV-infected Cx. quinquefasciatus library was prepared as described in Methods. Differences in miR-989 and miR-92 expression levels are highlighted. nd = not determined
Table S2, Oligonucleotides used in this study. Table of oligonucleotides used for primer extension and high-throughput sequencing.
Raw sequence data C710.fasta. FASTA file containing sequencing reads for C7/10 Ae. albopictus cells
Raw sequence data Culex.fasta. FASTA file containing sequencing reads for Cx. quinquefasciatus mosquitoes
miRNA precursor sequences Aedes_precursors.fasta. FASTA file containing miRNA, miRNA*, and precursor sequences for Ae. albopictus
miRNA precursor sequences Culex_precursors.fasta. FASTA file containing miRNA, miRNA*, and precursor sequences for Cx. quinquefasciatus