Summary
The ankyrin repeat (ANK) is the most common protein-protein interaction motif in nature and predominantly found in eukaryotic proteins. The genome sequencing of various pathogenic or symbiotic bacteria and eukaryotic viruses identified numerous genes encoding ANK-containing proteins that were proposed to have been acquired from eukaryotes by horizontal gene transfer. However, the recent discovery of additional ANK-containing proteins encoded in the genomes of archaea and free-living bacteria suggests either a more ancient origin of the ANK motif or multiple convergent evolution events. Many bacterial pathogens employ various types of secretion systems to deliver ANK-containing proteins into eukaryotic cells where they mimic or manipulate various host functions. Understanding the molecular and biochemical functions of this family of proteins will enhance our understanding of important host-microbe interactions.
Keywords: ankyrin repeat containing protein, secretion, evolution, prokaryotes, virus
The family of ankyrin-repeat containing proteins: definition and history
The ankyrin repeat (ANK) is a 33-residue motif that often occurs in tandem arrays that cooperatively fold into structures that mediate molecular recognition via protein-protein interactions. It is considered one of the most common protein-protein interaction motifs in nature [1]. ANK repeats are named after the human membrane-associated ankyrin protein, which is responsible for the attachment of the cytoskeleton to the plasma membrane [2–3]. The first characterized ANK-containing proteins were the yeast cell cycle regulator Swi6/Cdc10 and the Drosophila cell signaling Notch protein [4–6]. Until recently, it was thought that the members of this protein family were exclusively eukaryotic. They are involved in many cellular functions in eukaryotes, such as inhibition (Ink4, 53BP2) or development of tumors (Bcl3), transcriptional regulation (IκB, Mbp1, RFXANK), cell cycle, oncogenesis, signal transduction, and modulation of the inflammatory response mediated by NF-κB [7–8].
The availability of complete genome sequences has revealed the presence of ANK repeats in a vast number of deduced proteins from bacteria, archaea, eukaryotes and viruses. Homology based analyses of protein sequences in non-redundant databases such as Pfam [9], SMART [10] and INTERPRO [11] shows >64,000 ANK repeats in ~15,000 proteins. Most bacterial ANK-containing proteins are found in Proteobacteria, with ~1800 sequences believed to have been acquired through horizontal gene transfer (HGT); some of these proteins play important roles in microbial pathogenesis by mimicking or interfering with the host function [1, 12–13].
In this review, we discuss recent advances in our knowledge about ANK-containing proteins in bacteria and viruses, with emphasis on their structure, function and evolution. Despite the shared basic structure of the motif, microbial ANK-containing proteins appear to perform an impressive variety of functions.
Structure and function of the ANK-containing proteins
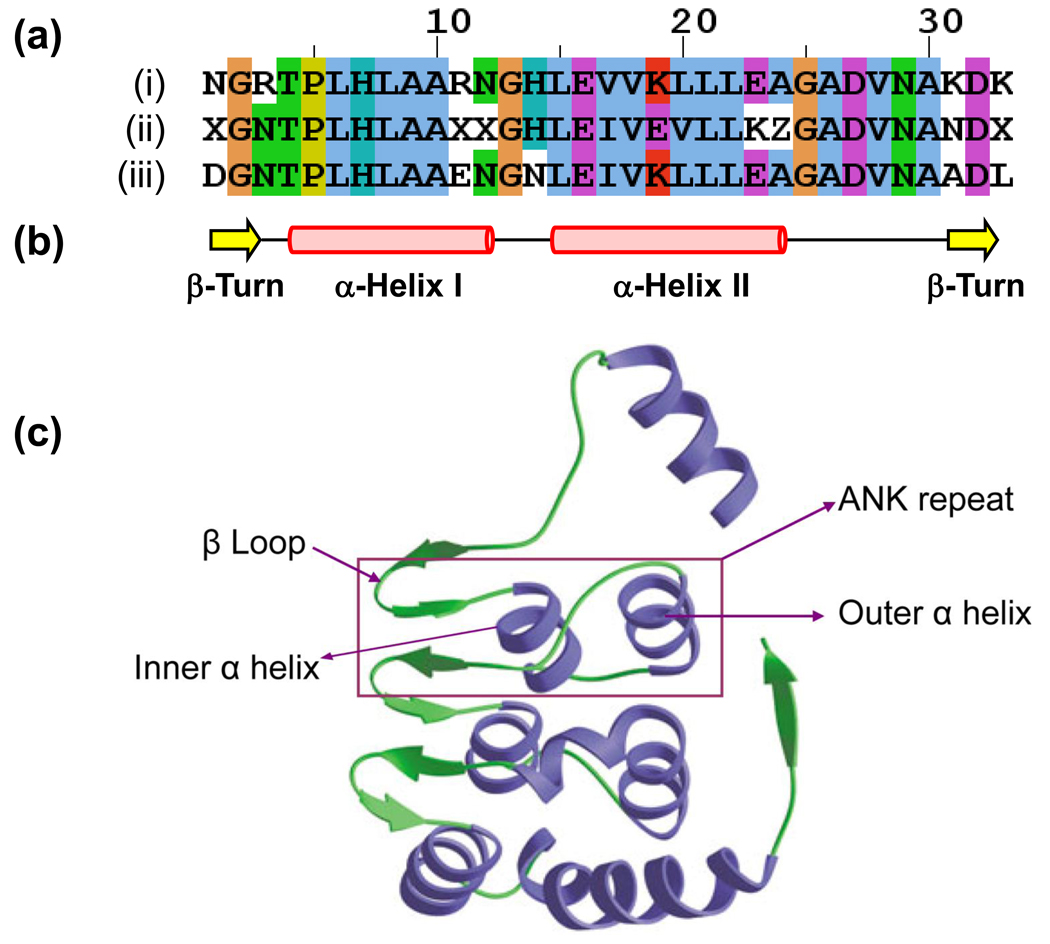
In spite of a strong degeneracy of the repeat sequences, multiple sequence alignments reveal a consistent pattern of key residues that preserve the structural integrity of the ANK motif [2, 14] (Figure 1). Certain residues in the prokaryotic ANK consensus sequence are more or less prevalent compared to the corresponding residues in eukaryotes. Overall, an ANK motif exhibits a canonical helix-turn-helix conformation. Crystallographic studies on eukaryotic ANK-containing proteins suggest that the two helices are arranged in an antiparallel fashion and the loop projects outward at an approximately 90° angle to facilitate the formation of hairpin-like β-sheets with neighboring loops [5, 15] (Figure 1b). Usually, the hairpin-like β-sheet structure consists of the last three residues of the preceding ANK motif and the first four residues of the next one. The ability of ANK to bind target proteins involves contact through the tips of the β loop, which are exposed to the solvent, and the surface of the helical bundle facing the concave part also termed the ANK groove [16]. Some residues are well conserved in both eukaryotic and bacterial ANK sequences (Figure 1a), which preserve integrity of the motif topology. Specifically, a highly prevalent TPLH tetrapeptide at positions 4–7 is involved in initiating the first α-helix and contributes to the stability of the motif [7]. The V/I-V-X-L/V-L-L motif (being X any hydrophilic amino acid) is central to the formation of the second α-helix and it contributes to formation of hydrophobic networks within and between ANK motifs to stabilize the protein [14]. The binding site for interacting proteins is constituted by a cluster of six non-conserved residues in an ANK repeat at the protein surface; these residues determine the specificity of the protein-protein interaction [17–18].
Figure 1.
Conserved structural features of the ANK motif. (a) Two ANK consensus sequences, derived mostly from eukaryotic proteins (i, ii) [2, 14] are compared to a consensus sequence from bacterial proteins (iii). The bacterial consensus sequence was produced using Jalview (http://www.jalview.org/) by aligning 3845 ANK sequences obtained from the SMART database; secondary structure was confirmed by using the JPRED3 server (http://www.compbio.dundee.ac.uk/). Residues are colored using the CLUSTALW color scheme implemented in Jalview. The Kohl et al. [14] sequence includes an X that denotes any amino acid except C, G, and P and Z that can be a H, N or Y. (b, c) An ANK repeat is composed of two α-helices arranged in an antiparallel fashion, and a β-loop that projects outward at an approximately 90° angle to facilitate the formation of hairpin-like β-sheets with neighboring loops. Panel (c) shows the crystal structure of ANK repeats for the cell cycle regulator Swi6 from yeast, reproduced from Ref. [4] with permission.
Crystallographic, spectroscopic and biochemical studies reveal that the core structure of the motif is sufficiently malleable to resist considerable sequence variation, while providing a stable scaffold for presenting surface contact residues [19]. The ANK repeats can be deleted or extended one at a time, without compromising the tertiary structure of the whole protein [20]. The tandem repeats exhibit tertiary structure-based elasticity and behave as a linear and fully reversible spring, as verified by atomic force microscopy [20]. In some biological systems, they might behave as multiple buffers linked in series to resist damaging effect by mechanical forces [21]. These physical characteristics of the ANK repeat along with its inherent ability of self repair and generation of mechanical forces during refolding might facilitate its use in many biotechnological and medical applications (Box 1).
Potential applications of ANK-containing proteins in biotechnology and medicine
The ability of unfolded ANK repeats to generate mechanical forces during their refolding mechanism — acting as springy nanostructures — could be important in the design of nanodevices with an inherent ability of self-repair [20]. Bio-nanomachines containing ANK repeats might be designed to target different cellular processes that use mechanical forces (e.g. replication, transcription, translation, organelle transport, cell adhesion, membrane fusion, cell crawling) [84]. Moreover, Plückthun and colleagues have developed ANK-containing proteins as an alternative to antibodies [85–86]. However, unlike most antibodies, these proteins are very stable, do not have disulfide bonds and, therefore, they are functional inside the cell [85–86]. They might be used as tumor-targeting reagents, or as intracellular inhibitors to study and influence signaling inside the cell.
The ANK repeats pile together to form an elongated structure ideally suitable for the presentation of multiple functional groups and/or recognition elements in a multivalent fashion [20]. The modular stacking of the ANK repeats allows incorporation of additional non-conserved residues that contribute to both structural stability of the domain and specificity of binding to the target molecule [14]. Furthermore, as in the cases of other repetitive structural motifs, ANK lends itself to sequence variation in overall domain size by simple sequence duplication or deletion. This allows the ANK fold to recognize and bind to target molecules that vary considerably in size and shape.
Analysis of the domain architecture of some ANK-containing proteins reveals that the ANK domain can co-exist with other functional modules, such as the F-box (a motif found in cyclin-F that interacts with the protein SKP1 [22]), domains for ion transport, protein kinase activity, etc. (Table 1). Eukaryotic F-box proteins are involved in protein ubiquitination, and normally harbor a substrate-binding domain such as WD40 (a protein sequence usually ending with Trp-Asp) or LRR (leucine rich repeat) [23]. ANK domains in non-canonical F-box proteins of microbial pathogens might act as substitutes for substrate-binding domains such as LRR or WD40 [23]. The most common domain partner in ANK-containing proteins is PRANC (Pox proteins repeats of ankyrin - C terminal), which is closely related to F-box and therefore might play a similar role. Proteins containing both ANK and PRANC domains are encoded in the genomes of poxviruses and Rickettsia (Table 1). In addition to those shown in Table 1, other domains found in ANK-containing proteins include: SOCS-box (suppressors of cytokine signaling), zf-DHHC domain (a Zn-finger domain first identified in the Drosophila putative transcription factor DNZ1), ion-transport domain, and protein kinase domain [9].Domains, like the ANK repeat, which participate in diverse domain architectures are called ‘promiscuous’ or mobile domains [24]. The promiscuity of the ANK repeat is much lower in bacteria than in eukaryotes (Table 1, Box 2).
Table 1.
Low promiscuity of the ANK repeat in bacterial and viral genomes, as compared to eukaryotic genomes.
| Domain partner a |
Seqs. b | Bacteria | Viruses | Eukaryotes |
|---|---|---|---|---|
| PRANC | 262 | Orientia tsutsugamushi, Wolbachia sp. | Poxviruses | None |
| Glutaminase | 49 | Francisella tularensis | None | 37 |
| F-box | 38 | Legionella pneumophila, Coxiella burnetti, Protochlamydia amoebophilus | Acanthamoeba polyphaga mimivirus | 34 |
| RHOD | 7 | Azoarcus sp., Polaromonas naphthalenivorans, Burkholderia vietnamiensis, Bradyrhizobium sp., Methlycoccus capsulatus, Xanthobacter autotrophicus | None | None |
| LRR | 47 | Rickettsia felis, Microscilla marina | None | 46 |
| SET | 53 | Legionella penumophila | None | 49 |
| ZnMc | 1 | Gleobacter violaceous | None | None |
| HTH_GNTR | 2 | Corynebacterium glutamicum | None | None |
| PQQ | 1 | Magnetospirillum magneticum | None | None |
| PDZ | 40 | Geobacter metallireducens | None | 39 |
| PBPb | 3 | Xanthobacter campestris, X. axonopodis | None | None |
| AAA ATPase | 21 | Thermoanaerobacter tengcongensis, Shigella dysenteriae, S. flexneri, S. sonne | None | 17 |
In the following explanations, PFAM (PF#) or SMART (SM#) accession numbers for each domain are listed in parentheses. Glutaminase (PF04960): core structural motif in glutaminases (which deaminate glutamine to glutamate). RHOD (SM00450): rhodanese domain, likely mediating protein interactions. LRR (SM00370): leucine-rich repeat, involved in cell signaling and protein interactions. SET (PF00856): probably involved in protein interactions, found in lysine methyltransferases. ZnMc (SM00235): zinc-dependent metalloprotease domain. HTH-GNTR (SM00345): transcriptional repressor domain. PQQ (PF01011): pyrrolo-quinoline quinone-dependent enzyme domain. PDZ (PF00595) anchors transmembrane proteins to the cytoskeleton and holds together signaling complexes. PBPb (SM00062): found in bacterial periplasmic substrate-binding proteins, involved in high-affinity binding of extracellular solutes. AAA ATPase (SM00382): found in certain ATPases. Functions for other domains are explained in the main text.
Total number of sequences (containing both ANK and the specified domain partner) found in PFAM or SMART databases.
Origin and evolution of the ANK repeat
ANK-containing proteins mediate diverse functions exclusively by facilitating protein-protein interactions. A key question is: how functional diversity is achieved while maintaining the structural integrity of the ANK repeat? Two key properties of the motif contribute to the functional diversity of ANK-containing proteins: (i) its ability to withstand considerable sequence variation (degeneracy), and (ii) its tendency to combine with additional structural and functional motifs (mobility or promiscuity) [87].
Functional diversity is further enhanced by domain promiscuity: the PFAM database includes 425 unique domain architectures among ANK-containing proteins. Intriguingly, the ANK motif appears to be significantly less promiscuous in bacteria than in eukaryotes: only 12 distinct domain architectures exist in known bacterial and viral genomes (Table 1) compared to several hundred in eukaryotes. Why is the ANK repeat more promiscuous in eukaryotes? Increasing phenotypic complexity correlates strongly with increase in domain promiscuity [88–90]. The reiterated use of any particular domain (e.g. ANK) in combination with several other modules seems central to eukaryotic evolution, in particular to the evolution of lineage-specific signaling networks [91]. Thus, the relative complexity of eukaryotic cellular function (relative to bacteria) might explain the observed differences in ANK promiscuity. Alternatively, the evolution of domain combinations in bacteria might have been limited because of a (hypothetical) recent origin of bacterial ANK-containing proteins — e.g. if they were acquired from eukaryotes by HGT [1].
The hypothesis that bacterial ANK-containing proteins are of eukaryotic origin was based on genomic analyses that indicated that some eukaryotic ANK-containing proteins were more similar to prokaryotic proteins than to other eukaryotic proteins. However, recent observations suggest that this hypothesis needs to be re-examined. First, bacterial genomes differ extensively in the number of resident ank genes (encoding ANK-containing proteins), which are more predominant in α-proteobacteria. This suggests the possibility of expansion or deletion of ank genes in bacterial genomes by recurrent duplication of genes or repeats (genes containing sequence repeats are especially susceptible to duplication and deletion) [92]. Second, the discovery of ank genes in archaeal and free-living bacterial genomes indicates a more ancient origin of prokaryotic ANK-containing proteins than that proposed by the HGT hypothesis. Thus, sequence similarity between eukaryotic and prokaryotic ANK-containing proteins might reflect convergent evolution, proposed earlier as a key mechanism for the evolution of nearly identical ANK-containing proteins in C. elegans, Drosophila and mice [93]. This idea is also supported by the fact that these proteins facilitate protein-protein interactions, which are universally used by all living organisms. As more archaeal and free-living bacterial genomes become available, deeper phylogenetic analyses should provide fundamental insights into the origin and evolution of prokaryotic ANK-containing proteins.
Common features of ANK-containing proteins of bacteria
The first bacterial gene coding for an ANK-containing protein was identifed in Serratia liquefaciens [25]. Subsequently, other members of the protein family have been found in various bacterial taxa (Table 1, Figure 2). Remarkably, interactions between pathogenic or symbiotic bacteria with their eukaryotic hosts are in part mediated by the secretion of ANK-containing proteins into the host cell [26]. At least seven different protein secretion systems have been described and are designated types I to VII [27–31]. Legionella pneumophila [13, 32], Coxiella. burnetii [13], Anaplasma spp. [33–34] and Rickettsia spp. [35] translocate their ANK-containing effectors via a type IV secretion system (T4SS). Another interesting case is an ANK-containing protein of Pseudomonas aeruginosa; this is an integral membrane protein in the inner membrane, with its N-terminal domain facing the cytoplasm and its C-terminal domain in the periplasm [36]. Below, we further discuss specific examples where the microbial ANK-containing proteins have been implicated in host-microbe interactions.
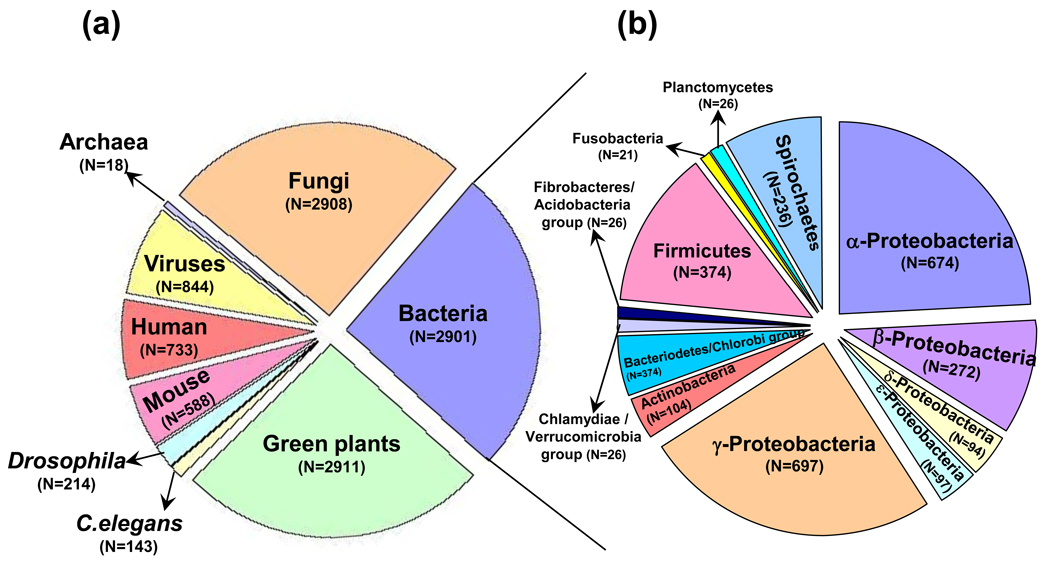
Figure 2.
Distribution of ANK-containing proteins among different organisms. (a) Genes encoding ANK-containing proteins are found in the genomes of eukaryotes (where they are ubiquitous), eukaryotic viruses, bacteria and archaea. The number (N) of these proteins seems to increase with increasing phenotypic complexity among eukaryotes (compare Caenorhabditis elegans, Drosophila, mouse and human). Differences in biological complexity among eukaryotes and bacteria might explain why ANK-24 containing proteins appear to be underrepresented among bacterial genomes (Box 2). (b) ANK-containing proteins in bacteria (only groups with N ≥ 20 are shown). These analyses were performed with Interpro ver 18.0 [11] and Pfam ver 23.0 [9].
The AnkA protein of Anaplasma phagocytophilum and Ehrlichia chaffeensis
Human granulocytic anaplasmosis is caused by the intracellular bacterium Anaplasma phagocytophilum [37], which infects, survives and propagates in neutrophils [34]. One of its proteins, AnkA, contains 11 ANK repeats in its N-terminal part [38], and is translocated by a T4SS to the host cell, where it localizes in the cytosol and the nucleus [34]. Interestingly, AnkA is phosphorylated by certain tyrosine kinases of the host (Abl-1 and Src) [34]. The tyrosine phsophorylation of AnkA is essential for its interaction with the host SHP-1 (Src homology phosphatase-1) during infection [34]. Once in the condensed chromatin areas, AnkA binds nuclear proteins and forms complexes with AT-rich DNA sequences, some of which are promoter sites, resulting in the modulation of host gene transcription [34, 39]. Recently, it has been shown that the rapid accumulation of AnkA in the nucleus of infected cells is accompanied with decreased transcription of the host CYBB locus where different transcriptional regulators bind [40].
Both Ehrlichia and Rickettsia harbor an ankA homologue [37]. In Erlichia chaffeensis, AnkA induces a delay of dissociation of the IκB/NF-κB complex, which in turn prevents the translocation of the transcription factor (NF-κB) into the nucleus and results in blocking cytokine synthesis [38, 41]. This might be due to the ability of E. chaffeensis AnkA to mimic or interfere with the seven ANK repeats present in IκB. The reduced cytokine production facilitates bacterial evasion of the immune system. In infected cell cultures, E. chaffeensis AnkA is located in the area of condensed chromatin in the host nucleus [38]. This protein might affect the expression of cell-cycle regulators like Bcl-3, leading to retaining cells at the border of the G1 and S phases [38]. Bcl-3 contains seven ANK repeats, which are most closely related to those found in IκB proteins [42]. It functions as a transcriptional co-activator through its association with NF-κB homodimers [43]. Therefore, by mimicking the Bcl-3 protein in the infected cells, AnkA might inhibit IκB dissociation and NF-κB translocation to the nucleus, resulting in the inhibition of cytokine secretion and down-regulation of the inflammatory response.
Moreover, both E. chaffeensis and Ehrlichia canis have a protein designated p200 that contains 21 ANK repeats [44]. Upon infection, E. chaffeensis p200 is translocated to the nuclei of infected monocytes [45]. Within the nucleus, p200 interacts with an adenine-rich motif of the Alu-Sx elements that are located in promoters and introns of various host cell genes [45]. Alu-Sx are the most abundant repetitive elements and are mainly related to transcription, apoptosis and ATPase activity [46]. This suggests that p200 might affect gene transcription globally in the host cell [45].
ANK-containing proteins from Legionella pneumophila
L. pneumophila is ubiquitous in natural aquatic environments and artificial water systems, where it replicates within protozoan hosts [47]. Once transmitted to humans, L. pneumophila enters the lung where it infects epithelial cells and alveolar macrophages developing pneumonia [48]. The bacterium resides in a vacuole that evades lysosomal fusion, a process mediated by effectors secreted by a T4SS known as Dot/Icm [49–50].
Only 11 ank genes (encoding ANK-containing proteins), out of 15, are shared between the four sequenced genomes of L. pneumophila strains [51–52]. With the exception of ankH, which is similar to a gene from C. burnetii [53], ank genes from L. pneumophila do not show clear sequence homologies to other genes in databases [51], indicating that they are novel. These genes might have been acquired by HGT [54], or they might have evolved through gene duplication and active site convergence, which has been described for many virulence proteins [55].
All the 11 shared ANK-containing proteins are translocated into the host cell by the Dot/Icm T4SS [32, 56] (Habyarimana et al, unpublished). One of these effectors, known as AnkX [13] or AnkN [53] blocks microtubule-dependent transport of vesicles from the ER to Golgi [13]. It is possible that AnkX might mimic certain host ANK-containing proteins that are known to facilitate the normal assembly of Golgi membranes [57]. Despite its clear role in blocking ER-to-Golgi vesicular traffic, AnkN/AnkX does not play any detectable role in the intracellular replication of the bacterium [13, 53].
The AnkB protein of L. pneumophila bears an F-box domain [32]. A mutation in ankB results in a severe defect in replication of the pathogen within human macrophages and protozoan hosts [32], and within the mouse model of Legionnaires’ disease [58]. The F-box domain of AnkB interacts with the host ubiquitination machinery and is essential for decoration of the Legionella-containing vacuole with polyubiquitinated proteins within macrophages and amoebae [58]. It is sugested that AnkB might target certain proteins for ubiquitination by acting as a linker between the protein substrates (which might bind to the ANK domains in AnkB) and the ubiquitination machinery (which appears to interact with the F-box of AnkB) [58]. Mutations in either ankH or ankJ result only in a moderate defect in intracellular replication [53]. None of the ankB, ankH or ankJ mutants showed defects in evasion from the endocytic vacuole [32, 53]. The host targets for AnkB, AnkH and AnkJ are still to be identified.
ANK-containing proteins of Coxiella burnetii
Coxiella burnetii is the agent of Q fever in humans and of coxiellosis in animals. It is an obligate intracellular bacterium that survives and replicates within large, acidified, phagolysosome-like vacuoles within macrophages [59]. Similar to L. pneumophila, C. burnetii utilizes a T4SS to deliver its effectors to the host cell [60], and harbors homologues of all the L. pneumophila dot/icm genes coding for the translocation apparatus and chaperons, with the exception of icmR [35]. C. burnetii dot/icm genes are expressed during infection, and four of them complement the corresponding L. pneumophila mutants, including the genes for the chaperones IcmS and IcmW [61]. Therefore, L. pneumophila has been used as a surrogate model to study delivery of C. burnetii T4SS substrates [61]. AnkA, AnkB, AnkF and AnkG from the Nine Mile (NM) isolate of C. burnetii are translocated via the L. pneumophila T4SS into the host cell [13]. An antibody generated against AnkF confirmed that this protein was secreted during C. burnetii infection [13]. The amount of secreted AnkF decreased over time after treating the cells with the chloramphenicol (an inhibitor of bacterial protein synthesis) indicating a possible degradation of the protein via the host proteasome [13]. Moreover, addition of a proteasome inhibitor prevents the degradation of AnkF in the chloramphenicol-treated cells [13]. BLAST searches show that C. burnetii possesses a gene (CBUD1380, ankK), similar to L. pneumophila ankH, suggesting that it might perform similar functions.
Comparison of the NM genome to those of other isolates (G, K and Dugway) revealed that many ank genes are disrupted in NM, while being intact in the other isolates, indicating a high hetereogenity of ANK-containing proteins in this species [62– 63]. Ten Dugway ANK-containing proteins and one ANK-containing protein specific to the G and K endocarditis isolates are translocated into the host cytosol by the L. pneumophila T4SS [62]. Their C-terminal region (encompassing 10 amino acids) appears to be an essential signal for translocation, and some of these effectors required IcmS for secretion [62]. The translocated ANK-containing effectors localized to a variety of subcellular compartments, which suggests that they might perform different functions [62].
ANK-containing proteins of Wolbachia spp.
The gram-negative obligate endosymbiont Wolbachia pipientis infects 20–75% of insect species [64], as well as some spiders, mites, terrestrial crustaceans and filarial nematodes [65–66]. Wolbachia is maternally transmitted and can rapidly invade insect populations through the reproductive distortions that it generates in the host [67]. These include cytoplasmic incompatibility (CI), parthenogenesis, male killing and reversal of genetic sex determination [67]. The CI is a type of embryonic lethality that in its simplest form results when Wolbachia-infected males mate with uninfected females. The CI provides a reproductive advantage to infected hosts and as a result enhances the transmission of Wolbachia in host populations.
Early sequencing and annotation of the genome of a Wolbachia strain (wMel) revealed ank genes [68]. Remarkably, 60 ank genes have been recently found in the genome of a different strain (wPip), making it the highest number reported in a prokaryote [12]. Thirteen of the wPip ank genes are contained in several chromosomally-integrated prophage regions, which are similar in sequence to the wMel WO-B prophage [12]. Some of the ANK-containing proteins contain predicted signal peptides and transmembrane domains, suggesting that they might be secreted into the mosquito cell cytoplasm or presented on the surface of the bacterium, and three of them show differences in expression depending on host sex [12]. Out of the 60 ank genes in the wPip genome, only 25 are homologous to wMel genes, and out of the 23 ank genes found in wMel, 16 show homology to wPip genes. The discrepancy in numbers is due to the fact that several of the wMel ank genes, mainly associated with prophage regions, are duplicated in the genome of wPip [69]. The expansion of ank genes in the wPip strain [69–70] and the degree of sequence variability and sex-specific expression in adult mosquitoes suggest an important biological role in endosymbiosis of Wolbachia [12].
ANK-containing proteins in eukaryotic viruses
Genomic analysis of viruses that infect eukaryotic cells has revealed a large number of ank genes, especially in the Poxviridae and Polydnaviridae families [71]. Recent studies have implicated viral ANK-containing proteins in host cell tropism and permissiveness [72], manipulation of host cell ubiquitination machinery [73] and interference with NF-κB activation through molecular mimicry of IκBα [74].
The role of viral ANK-containing proteins in host tropism and permissiveness has recently been explored. The M-T5 ANK-containing protein of myxoma virus (MV) enables replication in human cells [75] and interacts directly with two key host factors, Akt and Cullin-1 (Cul-1) [76–77]. M-T5 binds and activates Akt (a serine/threonine kinase) resulting in Akt phosphorylation, which allows MV to replicate in human cancer cells [75]. It is postulated that M-T5 mimics a host ANK-containing protein, PIKE-A, which also binds and activates Akt [75]. M-T5 also mediates host cell tropism and permissiveness by directly interacting with the cell-cycle progression factor, Cul-1 [76]. Cells infected with MV lacking M-T5 are halted at the G0/G1 checkpoint, and this is correlated with increased levels and reduced turnover of the cell cycle regulator p27/Kip, a process that is dependent on Cul-1 [76]. This overcomes the innate antiviral mechanisms that are triggered by G0/G1 cell-cycle arrest, and allows the virus to replicate effectively [76].
Non-canonical F-box proteins with ANK repeats have been identified in the Poxviridae family [73, 78–79]. The viral ANK-containing proteins manipulate the host ubiquitination machinery by mimicking the F-box protein component of the SCF1 ubiquitin ligase complex [73, 78]. Orf virus (a poxvirus) harbors five F-box containing proteins containing 7–9 ANK repeats each, and these proteins have been shown to interact directly with components of the host SCF1 ubiquitin ligase [73]. It seems likely that poxvirus F-box proteins, through molecular mimicry of the SCF1 ubiquitin ligase complex, target specific host proteins for ubiquitination and subsequent destruction via the ubiquitin/proteasome pathway, and this process enables effective viral pathogenesis.
Poxvirus and polydnavirus ANK-containing proteins have been shown to inhibit activation of the host NF-κB pathway that is crucial for stress, immune and proinflammatory responses [80]. In another poxvirus, Myxoma, protein MNF (containing nine ANK repeats) interferes with the NF-κB dependent pro-inflammatory response [81]. The eighth ANK repeat is highly similar to an ANK repeat in IκBα that mediates nuclear localization of this protein and, therefore, might act via molecular mimicry to interfere with NF-κB signaling [81].
Polydnaviruses are obligate symbionts transferred to lepidopteran larvae via endoparasitoids [82]. These viruses harbor proteins with up to four ANK repeats that share homology with the NF-κB inhibitor IκBα [83]. It has been postulated that these polydnavirus proteins stably bind to NF-κB, sequestering it in the cytoplasm and down-regulating the inflammatory response triggered by NF-κB [74].
Concluding remarks and future directions
The origin and evolution of microbial ANK-containing proteins is not fully understood (Box 3). Although acquisition by HGT from eukaryotes seems a likely mechanism for endosymbionts and pathogens that associate intimately with their eukaryotic hosts, it does not explain the origin of ANK-containing proteins among free-living bacteria and archaea. We propose two likely scenarios: (i) ANK-containing proteins had an ancient origin prior to the evolution of eukaryotes, or (ii) they evolved independently in different lineages by convergent evolution that selected the modular design of some of these proteins.
Questions for future research
What are the selective pressures for acquisition of ank genes (encoding ANK-containing proteins) by pathogenic bacteria and viruses?
How did the ank genes evolve in archaea and free-living non-pathogenic bacteria?
What are the biochemical functions and interacting partners of the bacterial ANK-containing proteins in the host cells?
How are the microbial ANK domains involved in molecular mimicry of host proteins to modulate cellular processes?
How similar are the biochemical functions performed by the eukaryotic and the prokaryotic ANK-containing proteins?
Can the novel binding specificities of microbial ANK-containing proteins be exploited for therapeutic, preventive or diagnostic purposes in medicine?
The ANK repeats can be deleted or extended one at a time without compromising the tertiary structure of the whole protein, indicating a strong tendency of the ANK repeats to refold. Some microbial ANK-containing proteins clearly play a role in host-pathogen interaction and the evolution of infectious diseases. However, it is likely that continued studies will identify new functions for bacterial and viral ANK-containing proteins. Their functions might involve molecular mimicry of host proteins to modulate key host cell processes to ensure proliferation of the microbe.
Finally, recombinant proteins containing modified ANK domains might be used as probes to study cellular signaling mechanisms, or they could target cancer cells for new antitumor therapies.
Acknowledgements
YAK is supported by Public Health Service Awards R01AI43965 and R01AI069321 from NIAID and by the commonwealth of Kentucky Research Challenge Trust Fund. AK was supported by a Research Incentive Grant (IRIG-2008) by the University of Louisville.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins. 1993;17(4):363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 2.Mosavi LK, Minor DL, Jr, Peng ZY. Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci U S A. 2002;99(25):16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitazawa M, et al. Intracellular cAMP controls a physical association of V-1 with CapZ in cultured mammalian endocrine cells. Biochem Biophys Res Commun. 2005;331(1):181–186. doi: 10.1016/j.bbrc.2005.03.127. [DOI] [PubMed] [Google Scholar]
- 4.Foord R, et al. X-ray structural analysis of the yeast cell cycle regulator Swi6 reveals variations of the ankyrin fold and has implications for Swi6 function. Nat Struct Biol. 1999;6(2):157–165. doi: 10.1038/5845. [DOI] [PubMed] [Google Scholar]
- 5.Lubman OY, et al. The crystal structure of a partial mouse Notch-1 ankyrin domain: repeats 4 through 7 preserve an ankyrin fold. Protein Sci. 2005;14(5):1274–1281. doi: 10.1110/ps.041184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breeden L, Nasmyth K. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature. 1987;329(6140):651–654. doi: 10.1038/329651a0. [DOI] [PubMed] [Google Scholar]
- 7.Mosavi LK, et al. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13(6):1435–1438. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voronin DA, Kiseleva EV. Functional role of proteins containing ankyrin repeats. Tsitologiia. 2007;49(12):989–999. [PubMed] [Google Scholar]
- 9.Finn RD, et al. The Pfam protein families database. Nucleic Acids Research. 2008;36 suppl_1:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Research. 2009;37 suppl_1:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter S, et al. InterPro: the integrative protein signature database. Nucleic Acids Research. 2009;37 suppl_1:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker T, et al. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biol. 2007;5:39. doi: 10.1186/1741-7007-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan X, et al. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320(5883):1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohl A, et al. Designed to be stable: crystal structure of a consensus ankyrin repeat protein. Proc Natl Acad Sci U S A. 2003;100(4):1700–1705. doi: 10.1073/pnas.0337680100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel F, et al. Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family. Embo J. 2001;20(22):6180–6190. doi: 10.1093/emboj/20.22.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becerra C, et al. Ankyrin repeat-containing proteins in Arabidopsis: characterization of a novel and abundant group of genes coding ankyrin-transmembrane proteins. Gene. 2004;340(1):111–121. doi: 10.1016/j.gene.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Mosavi LK, et al. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13(6):1435–1438. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magliery TJ, Regan L. Sequence variation in ligand binding sites in proteins. BMC Bioinformatics. 2005;6:240. doi: 10.1186/1471-2105-6-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Mahajan A, Tsai M-D. ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45(51):15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 20.Lee G, et al. Nanospring behaviour of ankyrin repeats. Nature. 2006;440(7081):246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 21.Oberhauser AF, Carrion-Vazquez M. Mechanical biochemistry of proteins one molecule at a time. J Biol Chem. 2008;283(11):6617–6621. doi: 10.1074/jbc.R700050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1(5):REVIEWS3002. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11(6):725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 24.Basu MK, et al. Evolution of protein domain promiscuity in eukaryotes. Genome Research. 2008;18(3):449–461. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett V. Ankyrins. Adaptors between diverse plasma membrane proteins and the cytoplasm. J Biol Chem. 1992;267(13):8703–8706. [PubMed] [Google Scholar]
- 26.Pallen MJ, Chaudhuri RR, Henderson IR. Genomic analysis of secretion systems. Curr Opin Microbiol. 2003;6(5):519–527. doi: 10.1016/j.mib.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Simeone R, Bottai D, Brosch R. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr Opin Microbiol. 2009;12(1):4–10. doi: 10.1016/j.mib.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7(10):703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enninga J, Rosenshine I. Imaging the assembly, structure and activity of type III secretion systems. Cell Microbiol. 2009;11(10):1462–1470. doi: 10.1111/j.1462-5822.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- 30.Galan JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5(6):571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tseng TT, Tyler BM, Setubal JC. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 2009;9 Suppl 1:S2. doi: 10.1186/1471-2180-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Khodor S, et al. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol Microbiol. 2008;70(4):908–923. doi: 10.1111/j.1365-2958.2008.06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin M, et al. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 2007;9(11):2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- 34.IJdo J, Carlson AC, Kennedy EL. Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell Microbiol. 2007;9(5):1284–1296. doi: 10.1111/j.1462-5822.2006.00871.x. [DOI] [PubMed] [Google Scholar]
- 35.Sexton JA, Vogel JP. Type IVB secretion by intracellular pathogens. Traffic. 2002;3(3):178–185. doi: 10.1034/j.1600-0854.2002.030303.x. [DOI] [PubMed] [Google Scholar]
- 36.Howell ML, et al. AnkB, a periplasmic ankyrin-like protein in Pseudomonas aeruginosa, is required for optimal catalase B (KatB) activity and resistance to hydrogen peroxide. J Bacteriol. 2000;182(16):4545–4556. doi: 10.1128/jb.182.16.4545-4556.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dumler JS. Anaplasma and Ehrlichia infection. Ann N Y Acad Sci. 2005;1063:361–373. doi: 10.1196/annals.1355.069. [DOI] [PubMed] [Google Scholar]
- 38.Caturegli P, et al. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect Immun. 2000;68(9):5277–5283. doi: 10.1128/iai.68.9.5277-5283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J, et al. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell Microbiol. 2004;6(8):743–751. doi: 10.1111/j.1462-5822.2004.00400.x. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Garcia JC, et al. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect Immun. 2009;77(6):2385–2391. doi: 10.1128/IAI.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang H, et al. Proteomic analysis of and immune responses to Ehrlichia chaffeensis lipoproteins. Infect Immun. 2008;76(8):3405–3414. doi: 10.1128/IAI.00056-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatada EN, et al. The ankyrin repeat domains of the NF-kappa B precursor p105 and the protooncogene bcl-3 act as specific inhibitors of NF-kappa B DNA binding. Proc Natl Acad Sci U S A. 1992;89(6):2489–2493. doi: 10.1073/pnas.89.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naumann M, Wulczyn FG, Scheidereit C. The NF-kappa B precursor p105 and the proto-oncogene product Bcl-3 are I kappa B molecules and control nuclear translocation of NF-kappa B. Embo J. 1993;12(1):213–222. doi: 10.1002/j.1460-2075.1993.tb05647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBride JW, Comer JE, Walker DH. Novel Immunoreactive glycoprotein orthologs of Ehrlichia spp. Ann N Y Acad Sci. 2003;990:678–684. doi: 10.1111/j.1749-6632.2003.tb07443.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhu B, et al. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect Immun. 2009;77(10):4243–4255. doi: 10.1128/IAI.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polak P, Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molmeret M, et al. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 2005;71:20–28. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao L-Y, Harb OS, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalianand protozoan cells. Infect.Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tilney LG, et al. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J Cell Sci. 2001;114(Pt 24):4637–4650. doi: 10.1242/jcs.114.24.4637. [DOI] [PubMed] [Google Scholar]
- 50.Roy CR, Tilney LG. The road less traveled: transport of Legionella to the endoplasmic reticulum. J Cell Biol. 2002;158(3):415–459. doi: 10.1083/jcb.200205011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cazalet C, Buchrieser C. What do we learn from the genome of Legionella pneumophila? Med Sci (Paris) 2005;21(5):455–457. doi: 10.1051/medsci/2005215455. [DOI] [PubMed] [Google Scholar]
- 52.Glockner G, et al. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int J Med Microbiol. 2008;298:411–428. doi: 10.1016/j.ijmm.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Habyarimana F, et al. Role for the ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ Microbiol. 2008;10:1460–1474. doi: 10.1111/j.1462-2920.2007.01560.x. [DOI] [PubMed] [Google Scholar]
- 54.De Felipe KS, et al. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187(22):7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sikora S, Strongin A, Godzik A. Convergent evolution as a mechanism for pathogenic adaptation. Trends Microbiol. 2005;13(11):522–527. doi: 10.1016/j.tim.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 56.de Felipe KS, et al. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 2008;4(8):e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beck KA, Buchanan JA, Nelson WJ. Golgi membrane skeleton: identification, localization and oligomerization of a 195 kDa ankyrin isoform associated with the Golgi complex. J Cell Sci. 1997;110(Pt 10):1239–1249. doi: 10.1242/jcs.110.10.1239. [DOI] [PubMed] [Google Scholar]
- 58.Price CT, et al. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 2009 doi: 10.1371/journal.ppat.1000704. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beron W, et al. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect Immun. 2002;70(10):5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Segal G, Feldman M, Zusman T. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol Rev. 2005;29(1):65–81. doi: 10.1016/j.femsre.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Zusman T, Yerushalmi G, Segal G. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect Immun. 2003;71(7):3714–3723. doi: 10.1128/IAI.71.7.3714-3723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voth DE, et al. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol. 2009;191(13):4232–4242. doi: 10.1128/JB.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beare PA, et al. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun. 2009;77(2):642–656. doi: 10.1128/IAI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeyaprakash A, Hoy MA. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol. 2000;9(4):393–405. doi: 10.1046/j.1365-2583.2000.00203.x. [DOI] [PubMed] [Google Scholar]
- 65.Breeuwer JA, Jacobs G. Wolbachia: intracellular manipulators of mite reproduction. Exp Appl Acarol. 1996;20(8):421–434. doi: 10.1007/BF00053306. [DOI] [PubMed] [Google Scholar]
- 66.Bouchon D, Rigaud T, Juchault P. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc Biol Sci. 1998;265(1401):1081–1090. doi: 10.1098/rspb.1998.0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stouthamer R, Breeuwer JA, Hurst GD. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- 68.Wu M, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2(3):E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klasson L, et al. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol. 2008;25(9):1877–1887. doi: 10.1093/molbev/msn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nikoh N, et al. Wolbachia genome integrated in an insect chromosome: evolution and fate of laterally transferred endosymbiont genes. Genome Res. 2008;18(2):272–280. doi: 10.1101/gr.7144908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cameron C, et al. The complete DNA sequence of myxoma virus. Virology. 1999;264(2):298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 71.Werden SJ, McFadden G. The role of cell signaling in poxvirus tropism: the case of the M-T5 host range protein of myxoma virus. Biochim Biophys Acta. 2008;1784(1):228–237. doi: 10.1016/j.bbapap.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 73.Sonnberg S, et al. Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc Natl Acad Sci U S A. 2008;105(31):10955–10960. doi: 10.1073/pnas.0802042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Falabella P, et al. Characterization of the IkappaB-like gene family in polydnaviruses associated with wasps belonging to different Braconid subfamilies. J Gen Virol. 2007;88(Pt 1):92–104. doi: 10.1099/vir.0.82306-0. [DOI] [PubMed] [Google Scholar]
- 75.Werden SJ, et al. M-T5, the ankyrin repeat, host range protein of myxoma virus, activates Akt and can be functionally replaced by cellular PIKE-A. J Virol. 2007;81(5):2340–2348. doi: 10.1128/JVI.01310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnston JB, et al. Myxoma virus M-T5 protects infected cells from the stress of cell cycle arrest through its interaction with host cell cullin-1. J Virol. 2005;79(16):10750–10763. doi: 10.1128/JVI.79.16.10750-10763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang G, et al. Infection of human cancer cells with myxoma virus requires Akt activation via interaction with a viral ankyrin-repeat host range factor. Proc Natl Acad Sci U S A. 2006;103(12):4640–4645. doi: 10.1073/pnas.0509341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sperling KM, et al. The highly conserved orthopoxvirus 68k ankyrin-like protein is part of a cellular SCF ubiquitin ligase complex. Virology. 2008;374(2):234–239. doi: 10.1016/j.virol.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 79.van Buuren N, et al. Ectromelia virus encodes a novel family of F-box proteins that interact with the SCF complex. J Virol. 2008;82(20):9917–9927. doi: 10.1128/JVI.00953-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karin M, et al. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2(4):301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 81.Camus-Bouclainville C, et al. A virulence factor of myxoma virus colocalizes with NF-kappaB in the nucleus and interferes with inflammation. J Virol. 2004;78(5):2510–2516. doi: 10.1128/JVI.78.5.2510-2516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turnbull M, Webb B. Perspectives on polydnavirus origins and evolution. Adv Virus Res. 2002;58:203–254. doi: 10.1016/s0065-3527(02)58006-4. [DOI] [PubMed] [Google Scholar]
- 83.Tian SP, Zhang JH, Wang CZ. Cloning and characterization of two Campoletis chlorideae ichnovirus vankyrin genes expressed in parasitized host Helicoverpa armigera. J Insect Physiol. 2007;53(7):699–707. doi: 10.1016/j.jinsphys.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 84.Bustamante C, et al. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 85.Interlandi G, et al. Characterization and further stabilization of designed ankyrin repeat proteins by combining molecular dynamics simulations and experiments. J Mol Biol. 2008;375(3):837–854. doi: 10.1016/j.jmb.2007.09.042. [DOI] [PubMed] [Google Scholar]
- 86.Milovnik P, et al. Selection and characterization of DARPins specific for the neurotensin receptor 1. Protein Eng Des Sel. 2009;22(6):357–366. doi: 10.1093/protein/gzp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 88.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420(6912):218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 89.Tordai H, et al. Modules, multidomain proteins and organismic complexity. FEBS J. 2005;272(19):5064–5078. doi: 10.1111/j.1742-4658.2005.04917.x. [DOI] [PubMed] [Google Scholar]
- 90.Koonin EV, Aravind L, Kondrashov AS. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101(6):573–576. doi: 10.1016/s0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- 91.Basu MK, et al. Evolution of protein domain promiscuity in eukaryotes. Genome Res. 2008;18(3):449–461. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 93.Michaely P, Bennett V. Mechanism for binding site diversity on ankyrin. Comparison of binding sites on ankyrin for neurofascin and the Cl-/HCO3- anion exchanger. J Biol Chem. 1995;270(52):31298–31302. doi: 10.1074/jbc.270.52.31298. [DOI] [PubMed] [Google Scholar]