Abstract
We have developed and validated a microporous poly(ethylene terephthalate) membrane-based indirect co-culture system for human pluripotent stem cell (hPSC) propagation, which allows real-time conditioning of the culture medium with human fibroblasts while maintaining the complete separation of the two cell types. The propagation and pluripotent characteristics of a human embryonic stem cell (hESC) line and a human induced pluripotent stem cell (hiPSC) line were studied in prolonged culture in this system. We report that hPSCs cultured on membranes by indirect co-culture with fibroblasts were indistinguishable by multiple criteria from hPSCs cultured directly on a fibroblast feeder layer. Thus this co-culture system is a significant advance in hPSC culture methods, providing a facile stem cell expansion system with continuous medium conditioning while preventing mixing of hPSCs and feeder cells. This membrane culture method will enable testing of novel feeder cells and differentiation studies using co-culture with other cell types, and will simplify stepwise changes in culture conditions for staged differentiation protocols.
Keywords: Human pluripotent stem cells, microporous membranes, indirect co-culture, cell separation
INTRODUCTION
Human pluripotent stem cells (hPSCs) have gained interest primarily because of their dual capabilities of self-renewal and differentiation into multiple cell types [1-4]. Traditional culturing of hPSCs involves direct contact with a feeder cell layer, such as mouse embryonic fibroblasts (MEFs), to support their undifferentiated growth [5]. These feeder cells not only produce extracellular matrix (ECM) components crucial for attachment of hPSCs, but also secrete factors that help maintain hPSCs in a pluripotent state. An important issue, however, is that direct contact of hPSCs with a feeder layer results in intermixing of cell types and may cause xenotypic contamination [6]. Recent improvements in culture techniques include using human rather than mouse fibroblasts as feeder layers [7-10] eliminating potential cross-species pathogen contamination; however separation of the feeder cells from the hPSCs, as is required for most differentiation protocols, still remains a technical challenge. For this reason, feeder-free systems have been developed, usually using purified substrates such as Matrigel™, fibronectin or laminin for attachment and replacing the other feeder cell-derived components with medium that is preconditioned by culture with feeder cells or defined medium containing high levels of synthetic growth factors [11].
We report here an alternative method for culturing hPSCs that does not require mixing the cells with feeder cells, using pre-conditioned medium, or adding high levels of growth factors. We evaluated a microporous poly(ethylene terephthalate) membrane-based indirect co-culture (MBIC) system that physically separates hPSCs from the feeder layer, while allowing for continuous conditioning of the medium by the feeder cells. In this study, we describe the first use of an MBIC system for hPSC propagation. We show that hPSCs cultured on membrane coated with human fibroblast-derived extracellular matrix and physically separated from human feeder cells (Supplementary Figure 1) are phenotypically indistinguishable from those cultured in contact with feeder cells, having the same colony morphology, expression of pluripotency markers, in-vitro differentiation and global gene expression profiles. Use of a MBIC system for hPSC expansion allows for an economical alternative to synthetic media-based feeder-free system and is amenable to scaling up production of pure populations of hPSCs and their derivatives. The complete separation of the cell types will simplify the testing of multiple culture conditions for optimal hPSC growth, and will enable rapid changes in conditions, such as testing differentiation factors and/or other types of cells for co-culture, without the requirement for dissociating and replating the hPSCs.
MATERIALS AND METHODS
Generation of inactivated human feeder layers and acellular substrates from human feeders
Human foreskin fibroblasts (HFFs) were maintained as previously described [12]. Mitotic inactivation of the cells was achieved by incubation in 10μg/ml of Mitomycin C (MMc), for 2 hours. The cells were then plated at a density of 300,000 cells/35mm dish. Acellular substrates were generated by allowing the HFF cultures to proliferate for 6-8 days past 100% confluency in 6-well tissue culture dishes (Sigma-Aldrich) on Millicell® 1.0μm polyethylene terephthalate (PET) inserts with hanging geometry (Millipore, Billerica, MA) (Supplementary Figure 1). The inserts were washed with sterile distilled water followed by a short exposure to 20mM NH3 solution to generate the acellular substrate on the filter. The filters were then thoroughly washed with phosphate buffered saline (PBS).
Propagation of hPSCs and hPSC-derived cells
Karyotypically normal diploid WA09 hESC [2] and hiPSC (WiCell Research Institute, Madison, WI) [4] were transferred from mouse feeder layers onto the acellular matrices within the inserts and maintained in growth medium as previously described [13]. hESC-derived fibroblasts were generated as described [14] for use in global gene expression comparisons. These fibroblasts were grown in DMEM+10% FBS at 37°C/5% CO2 and fed on alternate days and passaged weekly with 0.05% trypsin (Gibco).
Immunostaining of hPSCs
Routine staining for alkaline phosphatase was performed according to manufacturer instructions (Vector Labs) and as published previously[15]. Immunostaining of hPSCs on feeders and in the MBIC system was also based on previously published protocols [13, 16]. Fixed cells were incubated with primary antibodies: anti-POU5F1(OCT4)(Santa Cruz Biotechnology) and SSEA-4 (Millipore, Temecula, CA). Goat anti-mouse IgG conjugated to Alexa 488 (Molecular Probes, Eugene, OR) was used as secondary antibody. Fluorescent images were acquired using a Nikon Eclipse TE 2000-S inverted microscope with image analysis software. All image settings were controlled for uniform acquisition between samples.
In vitro differentiation of hPSCs and histology of hPSC-derived embryoid bodies
To generate embryoid bodies (EBs), hPSC colonies were divided into clumps of about 100-300 cells and resuspended in ultra-low attachment culture dishes, in growth medium without bFGF, and medium was changed every 3-4 days for 15 days. EBs were prepared for histological analysis by fixation in 3.7% PFA in 1.5ml microfuge tubes at approximately 15-25 EBs per tube. Once fixed overnight, EBs were rinsed with PBS to remove PFA, resuspended in 200μl melted 4% low melting point agarose (Sigma Aldrich) at 42°C and incubated for 2 hours to allow settling. Final pelleting and agarose solidification was performed with brief room temperature centrifugation at 500g. Agarose-embedded samples were processed for paraffin sectioning in a Leica TP1020 tissue processor. Hematoxylin and eosin (H&E) staining was performed on microscope slide-mounted 5μm sections in a Leica Autostainer XL workstation. Images were acquired using a Nikon TS-100 microscope using the default imaging parameters.
Scanning Electron Microscopy (SEM) analysis of membranes and hPSC colonies
To verify open membrane pores in the MBIC system, insert samples were fixed and dehydrated prior to SEM analysis, as previously described [16]. SEM analysis of hPSC colonies on inserts was performed by fixation in 4% gluteraldehyde followed by three washes in 0.1M Cacodylate Buffer pH 7.2 with 0.1M Sucrose. Cells were treated with 1% Osmium Tetroxide in 0.1M Cacodylate Buffer pH 7.2 followed by a 0.1M Cacodylate Buffer pH 7.2 rinse. Samples were dried by incubation in increasing percentage of ethanol (25 to 100%). SEM images were acquired by coating with ~ 150Å gold for contrast enhancement and electrical continuity. Representative images were collected in the FEI Quanta FEG ESEM 200 under high vacuum at 15 keV.
RNA isolation and real time reverse transcription polymerase chain reaction
RNA was isolated from hPSCs propagated for 15 passages under different conditions and from EBs after 15 days in suspension using Trizol (Invitrogen, Carlsbad, CA) and quantified using BioMate3 UV-VIS Spectrophotometer (Thermo Scientific, Waltham, MA). cDNA was synthesized from 1μg of RNA using cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). Expression of pluripotent and germ-layer specific genes (Supplementary Table 1) was analyzed using quantitative real time RT-PCR. PCR was performed in an ABI HT7900 system and the data were acquired using Sequence Detection System software (SDS v2.2.1, Applied Biosystems). SDS software was used to estimate relative fold change values, using ΔCt quantification methods. Endogenous 18S ribosomal RNA was used for normalization. Relative gene expression for hPSCs propagated in the MBIC system was assessed against hPSCs propagated on acellular HFFs in TCPS dishes. Relative gene expression of differentiated EBs obtained from different conditions was assessed against undifferentiated hPSCs propagated under the original condition. Expression Index (EI) was used to determine the relative differentiation state of cells [17], and was based on the average Ct values from triplicate measurements. An expression ratio of two or more genes was determined using a mathematical model based on the geometric average of assessed genes, previously described in detail [18], given by the following equation:
E is the PCR efficiency calculated from dilution series of purified PCR products, CT is the threshold cycle, and m and n are the numbers of genes that are up and down regulated upon differentiation respectively. KRS is the relative sensitivity constant and was not determined as it does not affect relative comparisons between samples.
Gene expression profiling, data collection and analysis
Whole-genome gene expression profiles were obtained in duplicate from hESCs propagated in (a) direct co-culture with HFF (b) Matrigel™ with HFF-conditioned medium and (c) indirect co-culture in the MBIC system with HFFs. In addition, we profiled three replicates of hESC-derived fibroblasts, and two replicates of HFFs. Total RNA was extracted using the Mirvana Total RNA extraction kit (Ambion) and mRNA labeling and amplification was performed using the Totalprep kit (Ambion). Whole-genome gene expression profiling was performed using Illumina human WGA-6 V2 gene expression arrays according to the manufacturer’s protocol. Data processing and normalization was performed in BeadStudio (Illumina). Clustering and statistical analyses were performed using MATISSE, based on previously developed methods [19].
RESULTS and DISCUSSION
Indirect co-culture allows expansion of hPSCs without feeder cell contact
We observed that hPSC lines did not attach to tissue culture-treated 1.0 μm PET inserts, and found that coating the inserts with human acellular substrates allowed attachment. We demonstrated that both hPSC lines could be successfully cultured for over 10 generations on PET inserts in co-culture with HFFs. In all of our studies, the feeder cells were attached to the bottom of the well, rather than the basolateral face of the microporous membrane (Supplementary Figure 1). hPSCs cultured in the MBIC system retained cell and colony morphology characteristic of undifferentiated cultures (Supplementary Figure 2). Expression of alkaline phosphatase [20] was monitored to show that the hPSCs retained this stem cell marker in the MBIC system (Figure 1).
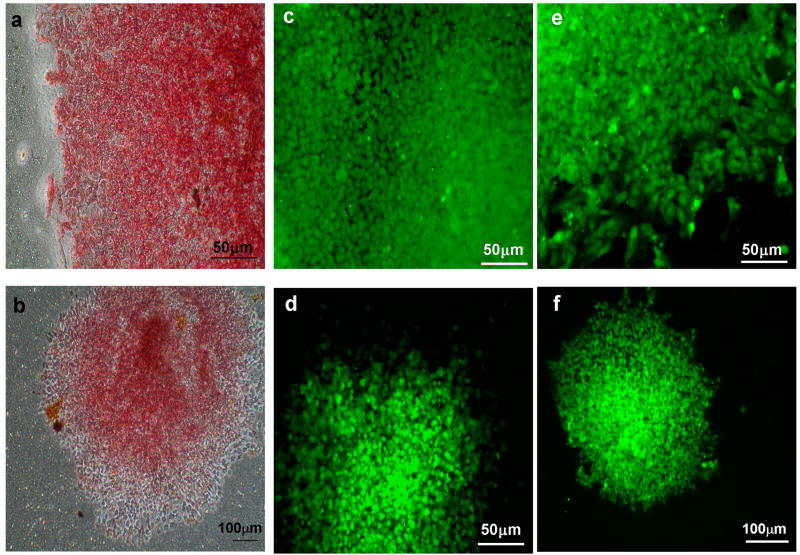
Figure 1.
hESCs and hiPSCs maintain alkaline phosphatase, Stage specific embryonic antigen (SSEA4) and POU5F1 (Oct4) expression after 10 passages in membrane-based culture. hESC (a,c,e) and hiPSC (b,d,f) are positive for alkaline phosphatase (a,b), SSEA4 (c,d) and POU5F1 (Oct4) (e,f).
Indirect co-culture maintains undifferentiated state of different hPSCs
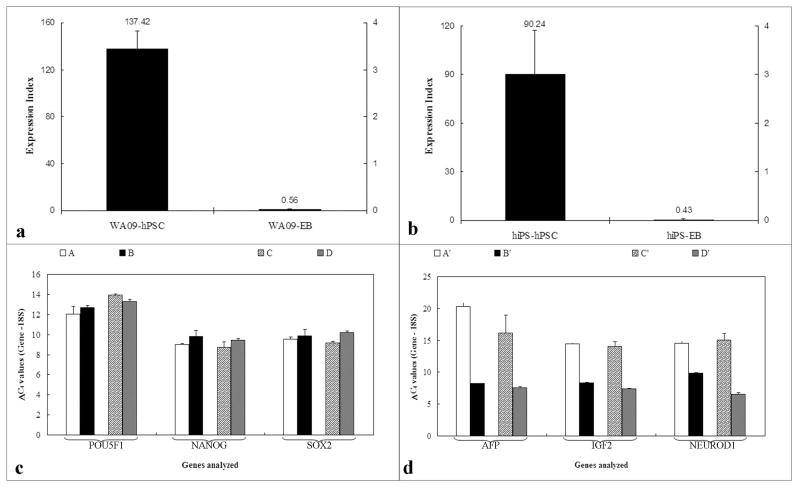
Immunocytochemical analysis for the pluripotency markers POU5F1(OCT4) and SSEA4 was used to validate the acellular substrates for hPSC propagation. The hPSCs grown on acellular substrates in the MBIC system continued to be positive for pluripotency markers over several passages (Figure 1). To evaluate the relative gene expression of key genes associated with pluripotency and differentiation, RNA from hESC and hiPSC maintained within the MBIC system for 15 passages was isolated and analyzed by quantitative RT-PCR. Samples were directly compared to hPSCs on substrate-coated TCPS. Expression of pluripotency markers (POU5F1(OCT4), SOX2 and NANOG) was measured as Ct values normalized against a housekeeping gene for each sample. As shown in Figure 2c, comparable ΔCt values across different culture conditions and cell lines indicated that the undifferentiated state was maintained in the MBIC system. Statistical analyses indicated no significant difference (p>0.05) between hPSCs propagated in the MBIC system and those propagated on substrate-coated TCPS. Further, the differentiation state of the hPSCs was quantified using an “expression index” as a metric to compare the undifferentiated hPSCs with differentiated cells (EBs). For the two cell lines (hESCs and hiPSCs) analyzed, the expression index of the undifferentiated samples at 15 passages was 90 and 137 respectively, while the expression index of the 15-day old EBs was 0.4 for the hESCs and 0.55 for the iPSCs (Figure 2 a &b).
Figure 2.
Quantitative real time polymerase chain reaction (QPCR) analysis of undifferentiated hPSCs and differentiated EBs derived from hPSCs. Differential expression index of a) hESCs and hESC-derived EBs and b) hiPSC and hiPSC-derived EBs, based on analysis of six genes (POU5F1(OCT4), NANOG, SOX2, AFP, IGF2 and NEUROD1). (c) Normalized gene expression of undifferentiated markers in hESC on acellular HFF (A); hESC in MBIC system (B); hiPSC on acellular HFF (C) and hiPSC in MBIC system (D). There is no significant difference (p>0.01) between the groups (n=3) indicating comparable pluripotent gene expression. (d) Normalized gene expression of differentiated markers in hESC in MBIC system (A’); EBs generated from hESC in MBIC system (B’); hiPSC in MBIC system (C’); and EBs generated from hiPSC in MBIC system (D’). Significant differential gene expression (p<0.01) in all three lineage specific markers was observed. It is important to note that ΔCt values (normalized against 18S) should be interpreted counter-intuitively; where a lower value indicated higher expression.
In-direct co-culture maintains in vitro differentiation potential
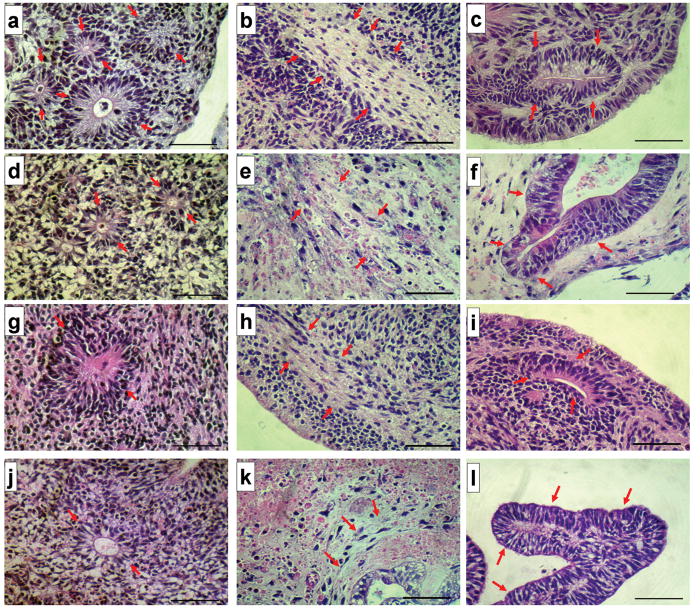
Functional pluripotency of the hPSCs cultured in the MBIC system was tested by in vitro differentiation into derivatives of the three germ layers. Statistical analyses indicated that the 15 day-old EBs had significantly high expression (p <0.01) of all germ layer specific markers tested (Figure 2d). In addition to the expression of markers indicative of germ layer formation in the EBs, histological studies were performed to assess the morphology of the differentiated tissue. As shown in Figure 3 (a-l), histologic evidence of tri-lineage maturation was present in the EB-sections from hPSCs propagated under both conditions, direct co-culture with feeder layers and in the MBIC system.
Figure 3.
Histological evidence of Tri-Lineage Differentiation in embryoid bodies generated from hPSCs. Shown are images of hematoxylin and eosin-stained histologic sections of EBs from hESC cells propagated on MEFs (top row, a-c), hiPSC propagated on MEFs (second row, d-f) as positive controls, hESC cells propagated in the MBIC system (third row, g-i) and hiPSC propagated in the MBIC system (bottom row, j-l). Tri-lineage potential is demonstrated as ectodermal (neuroepithelial) differentiation (a, d, g and j); mesodermal (fibrous connective) differentiation (b, e, h and k) and endodermal (intestinal) differentiation (c, f, i and l). Arrows point to the corresponding tissue in each figure. Magnification is 400x total (10x ocular, 40x objective). Each scale bar represents 50μm in length.
Global gene expression analysis validates indirect co-culture for hPSC propagation
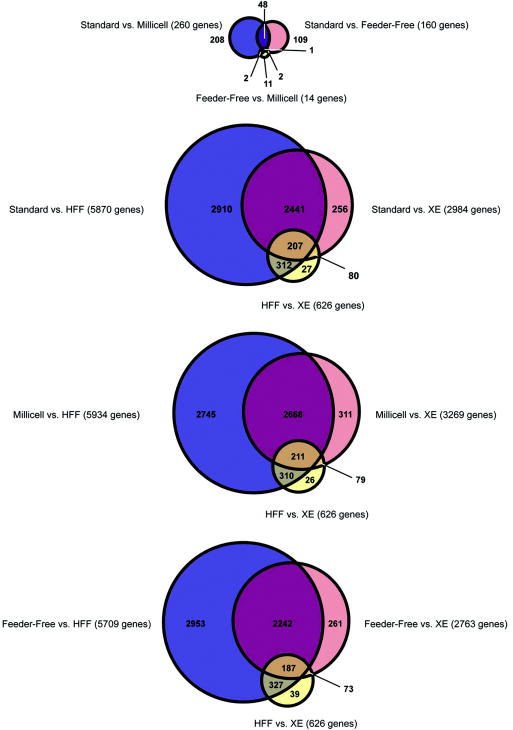
Genome-wide gene expression profiles of undifferentiated hESCs in direct co-culture with HFF feeder cells, in indirect co-culture (MBIC), and in feeder-free in HFF-conditioned media were very similar (Supplementary Figure 3). The correlation coefficients among the three culture conditions were very high (Supplementary Table 2), with only a small number of genes showing statistically significant differential expression (Figure 4). The whole genome expression profiles of hESC-derived fibroblasts resembled HFFs, as reported previously [21].
Figure 4.
Venn diagrams indicating the overlap between the sets of genes significantly differentially expressed (FDR<0.05) between pairs of samples. The top diagram shows results for the hESCs cultured in the three different conditions. The lower three diagrams show results among hESCs, HFFs and XE cells, with the data for each hESC culture conditions shown in a separate diagram. The sizes of the circles are proportional to the number of genes represented.
Although gene expression profiles of hESCs grown in all three conditions were remarkably similar, the hESCs grown under the MBIC and the feeder-free conditions were more similar to each other than hESCs grown in contact with feeder cells (Supplementary Figure 3). This is likely to be due to the fact that hESCs in both the feeder-free and MBIC conditions are exposed to HFF-generated soluble factors without direct contact with HFFs. Although only a few genes were differentially expressed in the non-contact (feeder-free and MBIC) conditions and the cells grown in contact with feeder cells, the identity of these genes may be significant. The list of differentially expressed genes was enriched for two GO categories (Development and Cell Communication; Supplementary Table 3) and for targets of ESC-related transcriptional regulators (14 transcriptional regulators examined, listed with references in Supplementary Table 4). The enrichment of targets of the pluripotency-associated transcription factors (POU5F1, NANOG, SOX2, TCF3) in the MBIC and feeder-free conditions suggests that these conditions may be more conducive to the maintenance of pluripotency than the standard co-culture methods.
Our results show for the first time that hPSCs can be successfully maintained in a pluripotent state in the MBIC system, with no contact between the stem cells and the feeder cells. An earlier study using porous membranes showed that hESCs maintained pluripotency when they were plated on the apical (top) face of the membrane and the feeder cells grown directly on the basolateral (bottom) face of the insert; however, in that system the hESCs could not be maintained if the feeder cells were instead cultured on the surface of the well containing the insert [22]. In our system, since the hPSCs and feeder cells are cultured in different compartments, the cells can be moved to other wells containing different cells or factors without having to dissociate the cultures. The ability to rapidly change the environment of the hPSCs will considerably reduce the effort involved in testing changes in culture conditions and may also reduce the stresses on the cells that have been shown to increase the frequency of selection for aneuploid variants[23].
CONCLUSIONS
We have demonstrated that hPSCs (both hESCs and hiPSCs) can be propagated and maintain pluripotency when cultured on a membrane separating them from a feeder layer. Our detailed gene expression analysis indicates that the hPSCs cultured under the MBIC conditions were virtually identical to cells cultured on feeder cells or in feeder cell-conditioned medium, and differentiation assays demonstrated that membrane-cultured cells remain fully pluripotent. The MBIC system could benefit researchers by reducing costs of feeder-free culture of hPSCs, and also aid in scale-up to larger culture systems composed of separated hPSC expansion and media conditioning compartments separated by a porous membrane partition. The system should also be useful for testing other feeder cell types and cell types that may induce specific differentiation, and for rapidly changing the cellular environment for testing for induction of specific signaling pathways and for determining the toxicity of pharmacological compounds.
Supplementary Material
Schematic of a Microporous membrane-based indirect co-culture system. HPSCs attach to extracellular matrix (ECM) coated microporous membranes of transwell inserts, which hang inside a 6-well tissue culture dish by plastic projections. HFF- Human foreskin fibroblast.
Scanning electron micrograph of WA09 hESCs co-cultured and passaged five times with human foreskin fibroblasts on tissue culture treated polystyrene (TCPS) (A), feeder free TCPS with conditioned media (B) or by indirect co-culture on 1mm PET membrane (C).
Agglomerative hierarchical clustering by average distance, using the Pearson correlation coefficient as the distance measure. The lengths of the branches on the dendrogram are proportional to the average distance in global expression between sample types, and the heat map showing expression displays row-normalized intensity values for the 7,373 genes significantly differentially expressed between at least two sample types. XE: WA09-derived fibroblasts, which are similar to extraembryonic endoderm. HFF: human foreskin fibroblasts. Undifferentiated hESCs were maintained with HFFs in indirect coculture (Millicell), on Matrigel™ with HFF-conditioned media (Feeder-Free), and on an HFF feeder layer (Standard).
Supplementary Table 1: List of primers used for determination of relative gene expression of pluripotency and differentiation genes as part of quantitative real time PCR analysis.
Supplementary Table 2: R2 correlation coefficients between expression profiles of different culture conditions (in pink boxes) and numbers of significantly differentially expressed genes (FDR < 0.05, minimum fold-change of 1.3) between pairs of culture conditions (in violet boxes).
Supplementary Table 3: TF-targets and GO categories enriched in sets of genes differentially expressed among the three hESC culture conditions. The numbers of genes in each category are listed in the second column (e.g. there are 36 genes expressed at significantly lower levels in the Standard culture condition compared to the Feeder-Free condition). Enrichments for genes in particular Gene Ontology (GO) categories are listed in column 3 (with the associated p-values in column 4), and enrichments for targets of transcription factors are indicated in the fifth column (with the associate p-values in column 6). p-values are Bonferroni corrected for multiple testing. NS = not significant.
Supplementary Table 4: 14 transcriptional regulators examined in the enrichment analysis in Supplementary Table 3. Lists of target genes were extracted from the referenced papers, which identified targets of transcriptional regulators from ChIP-Chip data.
Acknowledgments
Funding in part was provided by NIH 5K12HD000849-20 (LCL), CIRM RN2-00931 (LCL), Tel-Aviv University (IU), Israel Science Foundation 385/06 (RS), NIH T15 HL085028-01 (JFL), Alzheimer’s Association IIRG-05-14975 (JFL), CIRM RT1-01108 (JFL), NSF-CAREER 0744556 (RRR), NIH T15-HL074303 (RRR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reubinoff BE, Pera MF, Fong CY, et al. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nature biotechnology. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 2.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science (New York, N Y) 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (New York, N Y) 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Dravid G, Hammond H, Cheng L. Culture of human embryonic stem cells on human and mouse feeder cells. Methods in molecular biology (Clifton, N J) 2006;331:91–104. doi: 10.1385/1-59745-046-4:91. [DOI] [PubMed] [Google Scholar]
- 6.Martin MJ, Muotri A, Gage F, et al. Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nature medicine. 2005;11:228–232. doi: 10.1038/nm1181. [DOI] [PubMed] [Google Scholar]
- 7.Choo AB, Padmanabhan J, Chin AC, et al. Expansion of pluripotent human embryonic stem cells on human feeders. Biotechnology and bioengineering. 2004;88:321–331. doi: 10.1002/bit.20247. [DOI] [PubMed] [Google Scholar]
- 8.Amit M, Margulets V, Segev H, et al. Human feeder layers for human embryonic stem cells. Biology of reproduction. 2003;68:2150–2156. doi: 10.1095/biolreprod.102.012583. [DOI] [PubMed] [Google Scholar]
- 9.Hovatta O, Skottman H. Feeder-free derivation of human embryonic stem-cell lines. Lancet. 2005;365:1601–1603. doi: 10.1016/S0140-6736(05)66477-X. [DOI] [PubMed] [Google Scholar]
- 10.Amit M, Shariki C, Margulets V, et al. Feeder layer- and serum-free culture of human embryonic stem cells. Biology of reproduction. 2004;70:837–845. doi: 10.1095/biolreprod.103.021147. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig TE, Bergendahl V, Levenstein ME, et al. Feeder-independent culture of human embryonic stem cells. Nature methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 12.Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science (New York, N Y) 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 13.Calhoun JD, Rao RR, Warrenfeltz S, et al. Transcriptional profiling of initial differentiation events in human embryonic stem cells. Biochemical and biophysical research communications. 2004;323:453–464. doi: 10.1016/j.bbrc.2004.08.117. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez R, Loring J, Snyder E. Preparation of Autogenic Human Feeder Cells for Growth of Human Embryonic Stem Cells. John Wiley & Sons, Inc; 2008. [DOI] [PubMed] [Google Scholar]
- 15.Mitalipova M, Calhoun J, Shin S, et al. Human embryonic stem cell lines derived from discarded embryos. Stem cells (Dayton, Ohio) 2003;21:521–526. doi: 10.1634/stemcells.21-5-521. [DOI] [PubMed] [Google Scholar]
- 16.Sheridan SD, Gil S, Wilgo M, et al. Microporous membrane growth substrates for embryonic stem cell culture and differentiation. Methods Cell Biol. 2008;86:29–57. doi: 10.1016/S0091-679X(08)00003-4. [DOI] [PubMed] [Google Scholar]
- 17.Noaksson K, Zoric N, Zeng X, et al. Monitoring differentiation of human embryonic stem cells using real-time PCR. Stem cells (Dayton, Ohio) 2005;23:1460–1467. doi: 10.1634/stemcells.2005-0093. [DOI] [PubMed] [Google Scholar]
- 18.Stahlberg A, Aman P, Ridell B, et al. Quantitative real-time PCR method for detection of B-lymphocyte monoclonality by comparison of kappa and lambda immunoglobulin light chain expression. Clin Chem. 2003;49:51–59. doi: 10.1373/49.1.51. [DOI] [PubMed] [Google Scholar]
- 19.Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Syst Biol. 2007;1:8. doi: 10.1186/1752-0509-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connor MD, Kardel MD, Iosfina I, et al. Alkaline phosphatase-positive colony formation is a sensitive, specific, and quantitative indicator of undifferentiated human embryonic stem cells. Stem cells (Dayton, Ohio) 2008;26:1109–1116. doi: 10.1634/stemcells.2007-0801. [DOI] [PubMed] [Google Scholar]
- 21.Muller FJ, Laurent LC, Kostka D, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S, Ahn SE, Lee JH, et al. A novel culture technique for human embryonic stem cells using porous membranes. Stem cells (Dayton, Ohio) 2007;25:2601–2609. doi: 10.1634/stemcells.2006-0814. [DOI] [PubMed] [Google Scholar]
- 23.Mitalipova MM, Rao RR, Hoyer DM, et al. Preserving the genetic integrity of human embryonic stem cells. Nat Biotechnol. 2005;23:19–20. doi: 10.1038/nbt0105-19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic of a Microporous membrane-based indirect co-culture system. HPSCs attach to extracellular matrix (ECM) coated microporous membranes of transwell inserts, which hang inside a 6-well tissue culture dish by plastic projections. HFF- Human foreskin fibroblast.
Scanning electron micrograph of WA09 hESCs co-cultured and passaged five times with human foreskin fibroblasts on tissue culture treated polystyrene (TCPS) (A), feeder free TCPS with conditioned media (B) or by indirect co-culture on 1mm PET membrane (C).
Agglomerative hierarchical clustering by average distance, using the Pearson correlation coefficient as the distance measure. The lengths of the branches on the dendrogram are proportional to the average distance in global expression between sample types, and the heat map showing expression displays row-normalized intensity values for the 7,373 genes significantly differentially expressed between at least two sample types. XE: WA09-derived fibroblasts, which are similar to extraembryonic endoderm. HFF: human foreskin fibroblasts. Undifferentiated hESCs were maintained with HFFs in indirect coculture (Millicell), on Matrigel™ with HFF-conditioned media (Feeder-Free), and on an HFF feeder layer (Standard).
Supplementary Table 1: List of primers used for determination of relative gene expression of pluripotency and differentiation genes as part of quantitative real time PCR analysis.
Supplementary Table 2: R2 correlation coefficients between expression profiles of different culture conditions (in pink boxes) and numbers of significantly differentially expressed genes (FDR < 0.05, minimum fold-change of 1.3) between pairs of culture conditions (in violet boxes).
Supplementary Table 3: TF-targets and GO categories enriched in sets of genes differentially expressed among the three hESC culture conditions. The numbers of genes in each category are listed in the second column (e.g. there are 36 genes expressed at significantly lower levels in the Standard culture condition compared to the Feeder-Free condition). Enrichments for genes in particular Gene Ontology (GO) categories are listed in column 3 (with the associated p-values in column 4), and enrichments for targets of transcription factors are indicated in the fifth column (with the associate p-values in column 6). p-values are Bonferroni corrected for multiple testing. NS = not significant.
Supplementary Table 4: 14 transcriptional regulators examined in the enrichment analysis in Supplementary Table 3. Lists of target genes were extracted from the referenced papers, which identified targets of transcriptional regulators from ChIP-Chip data.