Abstract
Biofilm formation has been suggested to play an important role during Streptococcus pneumoniae nasopharyngeal colonization and may facilitate progression to pneumonia. To test whether the ability of S. pneumoniae to form biofilms was important for virulence we screened the ability of 30 invasive and 22 non-invasive clinical isolates of serotype 6A and 6B to form early biofilms on polystyrene microtiter plates and infect mice following intranasal and intratracheal challenge. We first determined that no correlation existed between the ability to form early biofilms and whether isolates were collected from healthy carriers or individuals with invasive disease. A disconnect between biofilm forming ability and the capacity to colonize the nasopharynx, cause pneumonia, and enter the bloodstream was also observed in mice. Importantly, S. pneumoniae mutants deficient in the established virulence determinants pneumolysin, CbpA, and hydrogen peroxide formed biofilms normally. Incidentally, we determined that robust biofilm production was dependent on the formation and coalescing of bacteria aggregates on a thin layer of bacteria attached to the plate surface. In summary, these studies suggest that the ability to form early biofilms in vitro does not reflect virulence potential. More complex studies are required to determine if biofilm formation is important for virulence.
Keywords: Streptococcus pneumoniae, adhesion, microtiter plate, biofilm, virulence
INTRODUCTION
Since the discovery that Pseudomonas aeruginosa forms biofilms in cystic fibrotic lungs, considerable attention has been placed on the role of bacteria biofilms during infectious diseases. To date bacteria in biofilms have been shown to have differences in metabolism, virulence gene expression, and protein production that contribute to surface adhesion and persistence of an infection. Because biofilm bacteria are enmeshed within an extracellular matrix, and in some instances metabolically inert, biofilm bacteria are more resistant to killing by leukocytes and antimicrobials, and serve as an recalcitrant source [of bacteria] during persistent infections [1, 2].
Considerable evidence suggests that biofilm formation is the underlying mechanism responsible for chronic otitis media. In regards to Streptococcus pneumoniae, the leading cause of otitis media, pneumococci have been detected on the surface of adenoid and mucosal epithelial cells biopsied from children with recurrent middle ear infections [3–5], occluded tympanostomy tubes isolated from the latter [6], and middle ear sections taken from challenged chinchillas [7, 8]. More recently, experimental evidence has been collected that suggests a role for pneumococcal biofilms during nasopharyngeal colonization and pneumonia. For example, Sanderson et al. found pneumococcal biofilms in biopsies of sinuses from individuals with chronic rhinosinusitis [9]. Following an in vitro screen of transposon mutants Munoz-Elias et al. identified 23 genes necessary for biofilm formation that were also required for nasopharyngeal colonization of mice [10]. Trappetti et al. found that treatment of mice with sialic acid, a condition that enhanced pneumococcal biofilm formation in vitro, increased bacterial counts in the nasopharynx of mice and instigated translocation of the bacteria into the lungs [11]. Finally, Oggioni and colleagues found that biofilm pneumococci had gene expression profiles similar to those of bacteria isolated from the lungs of mice; these profiles were distinct from planktonic bacteria isolated from either blood or culture media [12]. Thus, considerable evidence suggests that biofilms are an important aspect of pneumococcal pathogenesis.
To date many investigators have used the static polystyrene microtiter plate system to examine the molecular mechanisms underlying bacterial attachment to abiotic surfaces and early events during pneumococcal biofilm formation [10–16]. Advantages of this model system include that it is easy to establish, is applicable to high-throughput screens, and allows visualization of biofilm structures using an inverted microscope. Based on existing evidence supporting a role for biofilm formation during middle ear infection and nasopharyngeal colonization, we hypothesized that the ability to form biofilms might also contribute towards the ability to cause invasive pneumococcal disease (IPD). To test this hypothesis, we examined the ability of 30 invasive and 22 non-invasive low-passage clinical isolates of serotype 6A and 6B to attach to and form early biofilms (≤18 hours old) on untreated polystyrene 96-well microtiter plates and infect mice. In this manuscript we show that no correlation was found between biofilm production and the source of the clinical isolate, the ability of an isolate to colonize the nasopharynx, or cause invasive disease in mice. We conclude that the ability to form early biofilms in vitro had no correlation with virulence in mice and that extrapolations regarding in vitro biofilm formation with virulence are tenuous. These findings emphasize the importance of testing suspected pathogenic mechanisms using validated model systems along with a diverse panel of clinical isolates.
MATERIALS AND METHODS
Bacteria strains
Clinical isolates of S. pneumoniae were collected at The University of Texas Southwestern Medical Center in Dallas County, Texas, from February 1999 to January 2003. A total of 52 clinical isolates were examined, 23 serotype 6A isolates and 29 serotype 6B isolates. Non-invasive isolates were obtained from nasopharyngeal swabs of healthy carriers (strains 6A1–6A10 and 6B1–6B12). Invasive isolates were obtained from blood, cerebrospinal fluid, or aspirates of normally sterile sites from individuals with invasive disease. Phylogenetic relationships between the clinical isolates were extrapolated using comparative genomic hybridization data previously obtained for these strains [17]. TIGR4 is a virulent laboratory strain [18]. Isogenic mutants of TIGR4 deficient in pneumolysin (T4 Δpln), Choline binding protein A (T4 ΔcpbA), and hydrogen peroxide production (T4 ΔspxB) were made by insertion duplication mutagenesis with the suicide vector pJDC9 using previously described constructs [19]. Mutants were maintained with 1 µg/ml erythromycin.
In vitro screening for biofilm formation
We tested biofilm formation in the static polystyrene microtiter well system previously described by Allegrucci et al. [16]. Bacteria were streaked from blood agar plates (Remel, Lenexa, KS) into Todd Hewitt Broth (THB) (Difco, Detroit, MI) and grown at 37° C in 5% CO2. At mid-logarithmic phase growth (OD620=0.5) bacteria were diluted 1:20 in media containing 10% glycerol and frozen stocks were created. 96-well polystyrene microtiter plate (CELLSTAR, Greiner Bio-One, Monroe, NC) wells containing 270 µl of THB were inoculated with 30 µl of thawed stocks. Plates were incubated at 37° C, 10% CO2, overnight for 18 hours. The next day plates were washed with phosphate buffered saline (PBS), biofilms stained with 150 µl 0.5% crystal violet (CV) for 30 minutes, washed with PBS, and allowed to air dry. Biofilm formation was quantified by solubilizing the CV stain with 150 µl of 95% ethanol and measuring absorbance with a spectrophotometer at 540 nm (CV540). Images of the biofilms were captured using a Leica inverted microscope at 15X and 200X magnification prior to drying. Additional wells not inoculated with bacteria served as controls for inadvertent bacterial contamination of the media and served as background levels for CV staining. Experiments were done in triplicate with ≥3 replicate wells tested per strain in each experiment.
Animal infections
Female BALB/cJ mice (4–5 weeks old; The Jackson Laboratory) were maintained in animal biosafety level 2 facilities. For the initial virulence screen of all 52 isolates, a minimum of 2 replicate experiments were performed, each with 2–3 mice, for no less than 5 mice sampled for each clinical isolate tested. Briefly, mice were anesthetized with 2.5% inhaled isoflurane (Baxter Healthcare) and challenged intranasally with 107 colony forming units (CFU) suspended in 25 µL of PBS. Post-infection, blood was collected from the tail vein to test for bacteremia and nasal lavage performed to determine bacterial titers in the nasopharynx [20]. For long-term nasopharyngeal colonization studies, cohorts of 6–12 mice were challenged intranasally with 105 CFU in 10 µl PBS and examined over a 28-day period. For intratracheal challenge experiments cohorts of 6 mice were used. Mice were anesthetized, hung upright by their incisors, and 105 CFU in 100 µL of PBS placed in their throats. Aspiration was induced by gently pulling the tongue outward and covering the nostrils. Lung and blood samples were collected 2 days later. For blood and nasal lavage samples, serial dilution in PBS followed by plating and colony counting was used to determine the bacterial burden. For lung samples, the lungs were weighed and homogenized in 1 ml of PBS. Bacterial titers in the lungs were assessed per gram of homogenized tissue following serial dilution of the homogenate and colony counting. In all instances, the infectious dose administered was confirmed.
Statistical analyses
Differences between the highest and lowest biofilm producers in regards to biofilm production, blood, and nasopharyngeal bacterial titers were tested for significance using a two-tailed Student’s t-test. Comparisons regarding the incidence of positive blood cultures were done using a two-tailed Fisher’s exact test. Simple linear regression analysis was performed using SigmaStat 3.1 software (Systat Software Inc. Point Richmond, CA). For comparison of strains in the long-term nasopharyngeal colonization studies and the intratracheal challenge studies statistical analysis were performed using a One-Way ANOVA.
RESULTS
Early biofilm formation in vitro
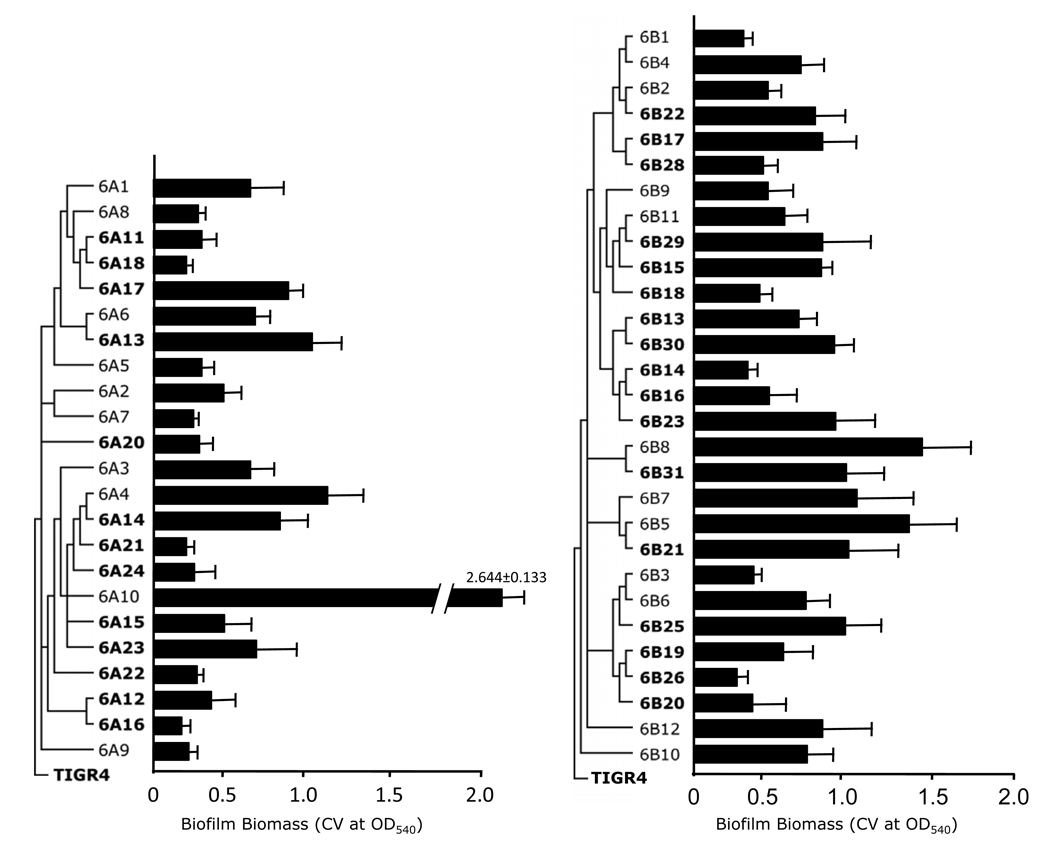
The static 96-well microtiter plate model tests for bacterial attachment to an abiotic surface and early biofilm formation. All clinical isolates tested were able to attach to the polystyrene surface; however, individual isolates had differential abilities to form early biofilms as measured by CV540, an indicator of biofilm biomass (Figure 1). For example, in serotype 6A the lowest early biofilm producer was 6A16 which had a CV540 = 0.241 ± 0.047, whereas the greatest biofilm producer was 6A10 which had a CV540= 2.64 ± 0.133 (6A16 versus 6A10; p<0.001). Likewise for 6B the lowest early biofilm producer was 6B26 which had a CV540 = 0.376 ± 0.060 and the highest was 6B8 with a CV540 = 1.457 ± 0.377 (6B26 versus 6B8; p=0.03). Due to the exceptionally high and divergent ability of 6A10 to produce early biofilms, no significant differences in early biofilm production were observed between serotype 6A and 6B when compared as a whole. For all 6A isolates the mean CV540 detected was 0.674 ± 0.099; whereas for all 6B isolates the mean CV540 was 0.768 ± 0.051 (p=0.375). Excluding 6A10, a statistically significant difference was observed between serotype 6A and 6B, with 6B isolates having 31% higher mean CV540 values (p=0.018).
Figure 1. Biofilm formation on 96-well polystyrene microtiter plates by individual isolates of S. pneumoniae.
Clinical isolates belonging to serotype 6A and 6B were collected from individuals with invasive disease (bold font) and healthy asymptomatic carriers (regular font). Media in the wells were inoculated with ~105 CFU and incubated overnight at 37° C at 10% CO2. Plates were washed, and biofilm formation was assessed by crystal violet staining (CV540) as described in the methods. Shown is the average of three independent experiments, each with >3 replicate wells for each clinical isolate tested. Error bars indicate the standard error of means. Phylogenetic trees on the left of the isolate name are based on comparative genomic analyses done using microarrays [17]. Branches indicate phylogenetic relationships between the clinical isolates in context of TIGR4, a serotype 4 isolate, from whose genomic DNA the microarray was designed.
As exemplified by 6A10, the ability to form biofilms was highly diverse between individual clinical isolates. Closely related strains, including those determined to be clonal (i.e. within the same phylogenetic clade) by previous comparative genomic hybridization studies [17], varied considerably in their attachment and early biofilm forming ability. For example, 6A17 and 6A18 differed by 2.4-fold (6A17 CV540 = 0.873 ± 0.100; 6A18 CV540 = 0.365 ± 0.044; p=0.009) despite belonging to the same phylogenetic clade. Importantly, no significant differences were observed for attachment/early biofilm formation when comparing clinical isolates from individuals with invasive disease versus those collected from healthy carriers (6A non-invasive CV540 = 0.825 ± 0.217; 6A invasive CV540 = 0.564 ± 0.066; p=0.199) (6B non-invasive CV540 = 0.809 ± 0.098; 6B invasive CV540 = 0.739 ± 0.058; p=0.513). Thus, we determined that initial attachment and early biofilm production was not correlated with the disease state of the individuals from whom the isolates were obtained, and that sufficient individual variation existed between clinical isolates (as much as 10-fold) that no statistically significant difference in early biofilm formation occurred between serotype 6A and 6B.
Early biofilm formation is not correlated with the ability to colonize the nasopharynx or cause invasive disease in mice
To determine if in vitro early biofilm formation was correlated with in vivo fitness, we first infected cohorts of Balb/cJ mice with 107 CFU of each strain and determined their ability to colonize the nasopharynx and cause bacteremia (Table 1). With exception of 6B10 that failed to colonize mice, we observed stable nasopharyngeal colonization for all isolates on day 7 that was comparable with levels observed in previous studies [20, 21]. Between strains, colonization levels were determined to be uniform with no statistical differences between the highest and lowest colonizers of each serotype (6A21, 6A23, respectively; p=0.111) (6B29, 6B2, respectively; p=0.194). Likewise no significant differences were observed between serotype 6A and 6B when compared as groups (average value of medians: 6A = 5.04 ± 0.944 × 105 CFU/ml nasal elute, 6B =3.11 ± 1.47 × 106 CFU/ml; p=0.111). Again, no differences where observed when comparing invasive disease isolates versus asymptomatic carrier isolates (average value of medians: 6A non-invasive = 3.33 ± 0.843 × 105 CFU/ml; 6A invasive = 5.82 ± 1.43 × 105 CFU/ml nasal elute; 6A invasive vs. noninvasive: p=0.192; 6B non-invasive = 9.68 ± 1.91 × 105 CFU/ml; 6B invasive = 4.65 ± 2.54 × 105 CFU/ml; 6B invasive vs. non-invasive: p=0.244). Thus excluding 6B10, clinical isolates from serotype 6A and 6B colonized mice stably and equally despite a differential ability in abiotic surface colonization.
Table 1.
Percentage of mice with positive blood cultures following intransal challenge and bacterial burden in the blood and nasopharynx*
| day 1 |
day 4 |
day 7 |
||||||
|---|---|---|---|---|---|---|---|---|
| n= | % bact. | blood (cfu/ml) | % bact. | blood (cfu/ml) | % bact. | blood (cfu/ml) | nasal lavage (cfu/ml) | |
| 6A1 | 5 | 0% | <103 | 0% | <103 | 20% | <103 | 8.00E+05 |
| 6A2 | 5 | 0% | <103 | 40% | <103 | 20% | <103 | 3.00E+05 |
| 6A3 | 5 | 60% | 7.00E+03 | 100% | 4.40E+06 | 80% | 1.97E+05 | 4.20E+04 |
| 6A4 | 5 | 80% | 3.00E+03 | 100% | 1.00E+06 | 60% | 5.00E+08 | 5.45E+04 |
| 6A5 | 5 | 0% | <103 | 20% | <103 | 20% | <103 | 6.00E+05 |
| 6A6 | 5 | 0% | <103 | 0% | <103 | 0% | <103 | 2.00E+05 |
| 6A7 | 5 | 0% | <103 | 20% | <103 | 60% | 8.70E+04 | 6.50E+05 |
| 6A8 | 5 | 0% | <103 | 40% | <103 | 0% | <103 | 2.00E+05 |
| 6A9 | 5 | 0% | <103 | 0% | <103 | 0% | <103 | 1.11E+05 |
| 6A10 | 5 | 60% | 4.00E+03 | 20% | <103 | 20% | <103 | 3.68E+05 |
| 6A11 | 5 | 60% | 2.00E+03 | 60% | 2.00E+03 | 20% | <103 | 7.00E+05 |
| 6A12 | 5 | 0% | <103 | 0% | <103 | 0% | <103 | 1.00E+06 |
| 6A13 | 5 | 20% | <103 | 0% | <103 | 0% | <103 | 2.00E+05 |
| 6A14 | 5 | 40% | <103 | 40% | <103 | 80% | 4.20E+06 | 1.00E+05 |
| 6A15 | 5 | 60% | 2.00E+03 | 40% | <103 | 60% | 1.68E+05 | 4.50E+05 |
| 6A16 | 5 | 0% | <103 | 0% | <103 | 0% | <103 | 6.00E+05 |
| 6A17 | 5 | 60% | 1.00E+03 | 20% | <103 | 0% | <103 | 2.00E+05 |
| 6A18 | 5 | 80% | 1.50E+04 | 80% | 5.70E+04 | 80% | 9.00E+03 | 8.50E+04 |
| 6A20 | 8 | 0% | <103 | 0% | <103 | 13% | <103 | 4.50E+05 |
| 6A21 | 5 | 40% | <103 | 20% | <103 | 60% | 1.00E+03 | 1.70E+06 |
| 6A22 | 6 | 50% | 3.00E+03 | 17% | <103 | 50% | 1.00E+03 | 1.55E+06 |
| 6A23 | 5 | 40% | <103 | 20% | <103 | 20% | <103 | 1.29E+05 |
| 6A24 | 6 | 67% | 9.34E+05 | 23% | <103 | 83% | 2.53E+08 | 4.00E+05 |
| 6B1 | 5 | 80% | 4.00E+03 | 40% | <103 | 20% | <103 | 1.20E+06 |
| 6B2 | 5 | 40% | <103 | 100% | 2.00E+03 | 20% | <103 | 2.00E+04 |
| 6B3 | 5 | 100% | 3.40E+04 | 80% | 1.00E+03 | 40% | <103 | 1.80E+06 |
| 6B4 | 5 | 60% | 1.20E+04 | 20% | <103 | 20% | <103 | 1.50E+06 |
| 6B5 | 5 | 60% | 1.00E+03 | 40% | <103 | 60% | 2.00E+03 | 6.00E+05 |
| 6B6 | 5 | 60% | 1.00E+03 | 100% | 1.50E+04 | 100% | 7.70E+04 | 1.10E+06 |
| 6B7 | 5 | 40% | <103 | 40% | <103 | 20% | <103 | 1.00E+06 |
| 6B8 | 5 | 60% | 1.10E+04 | 80% | 4.00E+03 | 20% | <103 | 1.34E+05 |
| 6B9 | 5 | 40% | <103 | 40% | <103 | 0% | <103 | 1.50E+06 |
| 6B10 | 5 | 0% | <103 | 0% | <103 | 0% | <103 | <103 |
| 6B11 | 5 | 20% | <103 | 60% | 7.00E+03 | 40% | <103 | 1.70E+06 |
| 6B12 | 5 | 40% | <103 | 60% | 3.00E+03 | 60% | 6.00E+03 | 9.50E+04 |
| 6B13 | 5 | 40% | <1O3 | 100% | 5.00E+03 | 80% | 1.00E+04 | 1.00E+06 |
| 6B14 | 5 | 60% | 1.00E+06 | 100% | 1.00E+04 | 40% | <103 | 4.90E+06 |
| 6B15 | 5 | 80% | 4.30E+04 | 100% | 2.50E+04 | 40% | <103 | 1.20E+06 |
| 6B16 | 5 | 80% | 1.00E+04 | 60% | 2.00E+03 | 80% | 4.00E+03 | 3.80E+06 |
| 6B17 | 5 | 40% | <103 | 60% | 9.00E+03 | 0% | <103 | 3.70E+06 |
| 6B18 | 6 | 50% | 4.50E+04 | 40% | <103 | 17% | <103 | 5.35E+06 |
| 6B19 | 5 | 80% | 1.00E+04 | 100% | 1.06E+05 | 80% | 1.00E+03 | 3.70E+06 |
| 6B20 | 7 | 29% | <103 | 100% | 3.10E+04 | 86% | 5.30E+04 | 3.80E+06 |
| 6B21 | 5 | 80% | 1.00E+03 | 100% | 1.10E+04 | 40% | <103 | 2.30E+06 |
| 6B22 | 5 | 40% | <103 | 80% | 2.00E+03 | 40% | <103 | 4.90E+06 |
| 6B23 | 5 | 20% | <103 | 100% | 1.87E+05 | 80% | 2.00E+03 | 3.10E+04 |
| 6B25 | 5 | 80% | 1.35E+04 | 60% | 5.00E+02 | 80% | 2.50E+03 | 2.49E+05 |
| 6B26 | 5 | 60% | 2.00E+03 | 100% | 3.00E+05 | 80% | 6.00E+03 | 2.50E+04 |
| 6B28 | 5 | 80% | 2.80E+04 | 80% | 5.00E+03 | 60% | 6.00E+03 | 2.05E+05 |
| 6B29 | 5 | 60% | 1.00E+03 | 40% | <103 | 0% | <103 | 4.34E+07 |
| 6B30 | 5 | 40% | <103 | 100% | 4.00E+03 | 0% | <103 | 2.17E+05 |
| 6B31 | 5 | 20% | <103 | 40% | <103 | 20% | <103 | 3.13E+05 |
Median values are shown
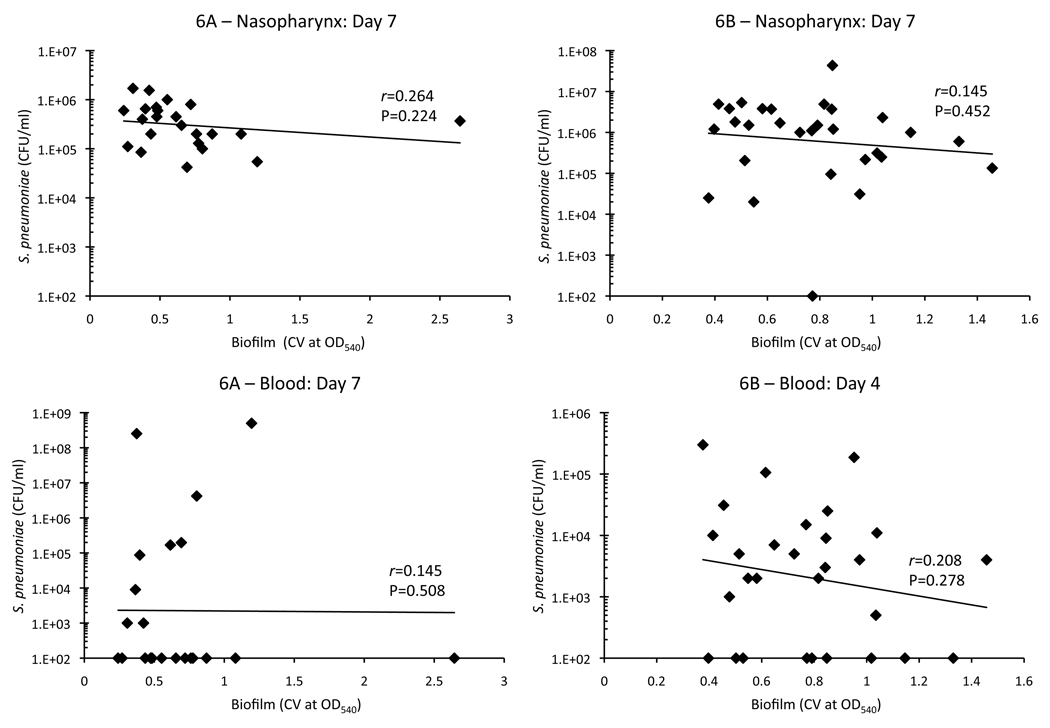
To assess invasiveness, the same mice were tail bled on days 1, 4, and 7 post-challenge and bacteria burden in the blood determined (Table 1). Most mice infected with 6A isolates were not bacteremic during the 7 days of observation (blood culture positive on day 1: 9/23, day 4: 4/23, day 7: 9/23); whereas in contrast the majority of 6B isolates were able to cause bacteremia (day 1: 16/29, day 4: 20/29, day 7 11/29). On day 4, the number of isolates able to cause bacteremia was determined to be statistically significant between the serotypes using a two-tailed Fisher’s Exact test (p<0.001). In general bacterial titers in the blood were low with little to no mortality due to infection. Of the 148 mice infected with serotype 6B only 9 died: 2 for 6B6, and 1 for 6B3, 6B14, 6B19, 6B20, 6B25, 6B28, and 6B31. Whereas for 6A, only 15 of the 120 mice infected died: 3 for 6A4 and 6A24, 2 for 6A3 and 6A14, and 1 for 6A7, 6A10 and 6A15. Importantly, the ability to cause bacteremia was not correlated with the source of the clinical isolate (Day 4, invasive vs. asymptomatic carrier: 6A p=1.0; 6B p=0.105), suggesting that a discrepancy also exists between the disease potential of S. pneumoniae in humans and mice. We next directly tested whether early biofilm formation on a microtiter plate was positively correlated with virulence in the mice. To do this, we performed linear regression analysis using CV540 biofilm production measures and nasal lavage and blood bacterial titers. For serotype 6A we used bacterial titers from day 7, as more mice were blood culture positive at this later time point. For serotype 6B we used day 4 bacterial titers for the same reason. Figure 2 shows that biofilm formation on microtiter plates in vitro was not correlated with the ability to colonize the nasopharynx or cause bacteremia for either serotype 6A or 6B.
Figure 2. Biofilm formation is not correlated with nasopharyngeal colonization or bacteremia.
Linear regression analysis was used to determine if biofilm formation was correlated with the ability to colonize the nasopharynx and cause invasive disease. For nasopharyngeal colonization, CV540 values were plotted against median bacterial titers in nasal lavage elutes from day 7. For 6A invasive disease, CV540 values was plotted against median bacteremia titers at day 7, the day the majority of mice had bacteria in the blood. For 6B invasive disease, CV540 values was plotted against median bacteremia titers at day 4 for the same reason. For both 6A and 6B no correlation was found between biofilm formation and invasive disease on days 1, 4 or 7 (data not shown).
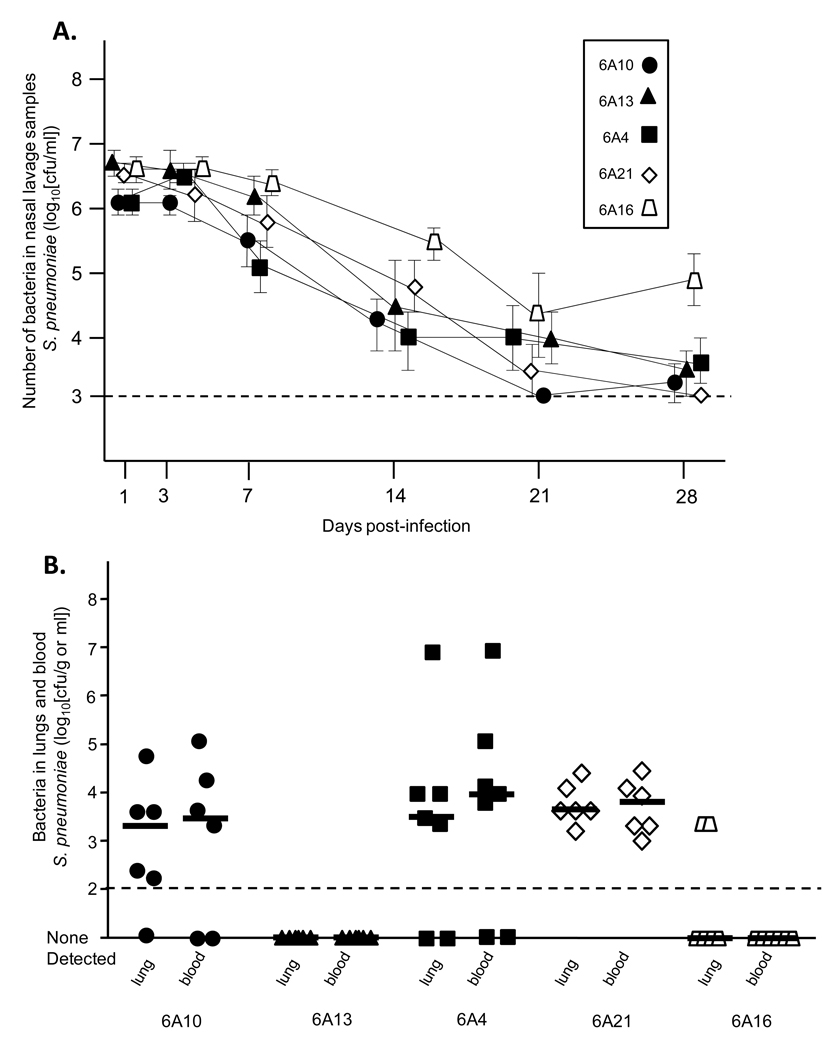
To more robustly confirm the observed disconnect between biofilm formation and colonization and invasion, we tested the ability of the 3 highest (6A4, 6A10, 6A13) and 2 lowest (6A16, 6A21) biofilm producers of serotype 6A to colonize the nasopharynx in a long-term model (Figure 3A) and following intratracheal challenge cause pneumonia (Figure 3B). Following intranasal challenge of mice, we observed no significant differences between levels of the 5 isolates on days 1, 3, 7, 14, 21 and 28. Thus the highest biofilm producers colonized the nasopharynx equally to those that formed little to no biofilms. Similarly, no differences were observed in the lungs of mice infected intratracheally with 6A4, 6A10, and 6A21. Of note, mice infected with 6A13 had no bacteria detectable in the lungs, as did the majority of mice infected with the low biofilm producing strain 6A16. Thus, in separate experiments, we confirmed that early in vitro biofilm forming capability was not correlated with the ability to colonize or cause pneumococcal disease. Finally, we also tested the reverse; whether early biofilm formation was affected by the deletion of established virulence determinants known to be required in the nasopharynx and lungs [19]. Isogenic mutants deficient in the toxin pneumolysin, the adhesin CbpA, and hydrogen peroxide synthesis, all formed biofilms comparable to the parent strain TIGR4 (T4 WT CV540 = 0.849 ± 0.121; T4 Δpln CV540 = 0.964 ± 0.124; T4 ΔcbpA CV540 = 0.737 ± 0.072; T4 ΔspxB CV540 = 1.093 ± 0.132). Thus early biofilm formation was determined not to be associated with these virulence determinants.
Figure 3. Ability of high and low serotype 6A biofilm producers to colonize the nasopharynx long-term and cause IPD.
A) Average bacterial titers in nasal lavage samples collected from mice colonized with the high biofilm producing strains (black shapes) 6A10, 6A13 and 6A21 as well as the low biofilm producing strains (white shapes) 6A21 and 6A16. Mice were challenged with 105 CFU in 10 µl PBS and bacterial titers in the nasopharynx determined on days 1, 3, 7, 14, 21 and 28. No statistical differences were observed between the groups using One-Way ANOVA. Error bars indicate ± SEM. B) Bacterial titers in the lungs and blood 48 hours after intratracheal challenge of mice with the same 6A isolates. Mice were infected with 105 CFU in 100 µl PBS. Individual shapes represent distinct mice. Horizontal bars designate the median bacterial value. Statistical analyses between the groups were performed using One-Way ANOVA.
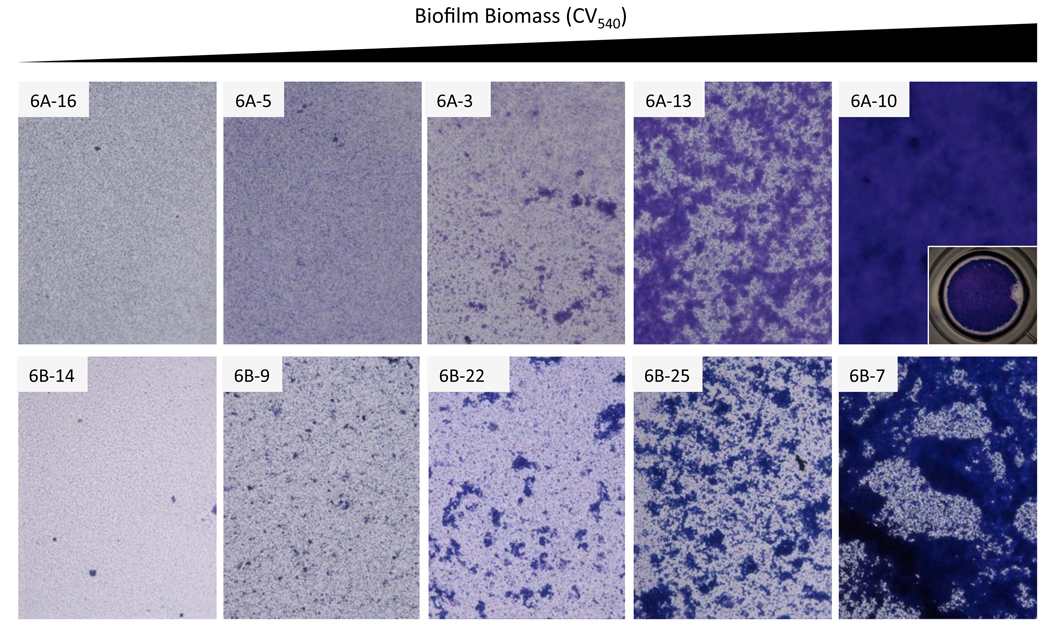
Bacteria aggregation contributes to early biofilm biomass
Incidentally, microscopic inspection of the surface attached biofilm biomass determined that the ability to form robust surface attached communities was associated with the formation of large aggregates on the polystyrene surface (Figure 4). Low early biofilm producers (e.g. 6A16, 6B14) formed only a thin layer on the microtiter well bottom composed primarily of individual diplococci, while mid level biofilm producers formed a similar bacterial “carpet” accentuated by the presence of small bacterial aggregates (e.g. 6A3, 6B22). As the respective CV540 values increased, the size of the bacterial aggregates also increased, ultimately coalescing into microcolonies ranging in size between 100 and 500 µm (e.g. 6A13, 6B25). Finally in the highest early biofilm producers, the microcolonies converged forming a thick confluent layer of bacteria along the bottom of the well (e.g. 6A10, 6B7). Thus, increased CV540 values were associated with an incremental ability to form bacterial aggregates following initial attachment to the plastic surface.
Figure 4. Increased biofilm biomass corresponds to the formation of increasingly larger bacterial aggregates.
Representative micrographs of pneumococcal biofilms forming on the bottom of the polystyrene microtiter plate wells. Images were taken with at 200X with an inverted microscope. For 6A10 we include an insert of a well taken at 15X magnification. This is to demonstrate that the 6A10 biofilm has completely covered the well bottom. Strains were selected based on their increasing CV540 value.
DISCUSSION
Because biofilm formation is a mechanism for the pneumococcus to evade the host-defense and resist antimicrobials, it is reasonable to hypothesize that strains better able to form biofilms are more virulent. To test this hypothesis, we examined the ability of 32 clinical isolates from individuals with IPD and 20 isolates from healthy asymptomatic carriers to form biofilms in vitro as well as cause IPD in mice. If our hypothesis were true, we would have predicted that the clinical isolates that caused invasive disease in humans, as well as in mice, were those that best formed biofilms in vitro. In fact, this was not observed and instead we determined that the ability of serotype 6A and 6B isolates to form biofilms in vitro was not correlated with the disease condition of the host (i.e. invasive or non-invasive), the ability to colonize the nasopharynx, or the ability to cause invasive disease in mice. Thus our experimental results suggested that the ability to form early biofilms was not important for IPD.
This study has important limitations. Foremost, we examined the ability of S. pneumoniae to form biofilms in vitro and thus our findings regarding virulence were correlative. Biofilm formation in vivo occurs under selective pressure and environmental signals that are distinct from in vitro. Environmental signals such as limited nutrients, the presence of antimicrobial host factors, and bacteria density may serve as signals for the expression of biofilm related genes. Previously we have shown that gene expression during bacterial growth in media is distinct from that which occurs in vivo, and from bacteria attached to epithelial cells in vitro [21]. Thus it is possible that biofilm factors are differentially expressed in vitro and that biofilm formation occurs differently in vivo. Furthermore, studies have shown that differences in the richness of growth media, bacteria seeding, and strain capsulation, affect biofilm formation on microtiter plates [10, 12–14, 16]. Had we used different media or altered our growth conditions we may have observed distinct results. Finally, we only tested serotype 6 clinical isolates. Capsule types 6A and 6B are near identical but the linkage between a component rhamnose and ribitol is 1→3 for serotype 6A and 1→4 for serotype 6B [22]. Perhaps for other serotypes, biofilm production under these conditions would have been positively correlated with invasive disease potential.
Currently two model systems are used to grow biofilms in vitro, a static microtiter plate system and a continuous flow system. Static systems such as the microtiter plate system permit growth of the bacteria in a vessel without replacement of media. Advantages of this model system are that it is easily amendable to high through-put screens, particularly when using a 96-well polystyrene plates. However, this model is short-term as nutrients are depleted and metabolic wastes accumulate; typically growth of the biofilm stops between 8 to 16 hours followed by loss of adhered bacteria presumably due to autolytic properties [13]. Recent findings also suggest the accumulation of a biofilm dispersion signal, cis-2-decenoic acid [23]; thus, without frequent exchanges of media, the static system does not allow for the formation of mature biofilms. Importantly, differences between the microtiter plate system and continuous flow reactor in pneumococcal biofilm formation have been reported. Studies by Allegruci et al., have shown that the inability to form early biofilms in a microtiter plate is not correlated with the ability to form biofilms in a flow-through reactor over an extended period [24]. Similar observations have been made for various other biofilm forming bacteria including Gram-negative pili mutants. While pili-deficient mutants have been shown to be defective in initial attachment to non-coated abiotic surfaces, most retain the capability to form biofilms (although with altered biofilm architecture compared to wild type) following several days of growth under flowing conditions [27–31]. Thus the continuous flow through model emphasizes distinct physiological properties and may have distinct in vivo correlates. It is therefore possible that had we used the continuous flow through reactor we would have observed distinct results. This possibility highlights the arbitrary nature of the current in vitro biofilm models.
Despite these considerations, our finding that the capability to initiate surface attached growth on microtiter plates was not correlated with disease suggests that for at least serotype 6A and 6B the ability to form early biofilms does not model pneumococcal events necessary for the development of invasive disease in humans or mice. Importantly, biofilm formation on polystyrene [or: in microtiter plates] with media would be distinct from events in vivo due to the fact that the bacteria would not be interacting with host components such as mucin, fibronectin, or laminin which may act as bridging molecules between individual bacteria and eukaryotic cells. Of note, finding that TIGR4 mutants deficient in pneumolysin, CbpA, and hydrogen peroxide synthesis were able to form biofilms was consistent with previous findings by Munoz-Elias et al. which showed that only deletion of lytC, a gene encoding a murein hydrolase, and cps4E, a capsule synthesis enzyme, affected early biofilm production following a screen of 6,500 TIGR4 mutants [10].
Interestingly, Munoz-Elias found 49 genes including cbpA that contributed to biofilm production in an acapsular strain. One explanation provided for the discrepancy between capsulated and unencapsulated mutants in biofilm production was increased bacterial interactions in the absence of capsule. Of note, 23 of the 49 genes identified by Munoz-Elias were subsequently shown to be important for nasopharyngeal colonization in TIGR4 [10]. Thus, when using unencapsulated pneumococci and a mutant with its isogenic parent, the early biofilm model provided important information regarding bacteria to bacteria interactions that were pertinent to the disease process. One important caveat is that clinical isolates of S. pneumoniae are almost always encapsulated, the exception being those collected from individuals with conjunctivitis [34]. Also that we observed contradictory results, strains unable to form dense biofilms colonized normally and in some instances were able to cause disease in mice. Thus these strains lacked biofilm determinants that were not required in vivo; alternatively, the presence of capsule alters the role of surface expressed proteins.
Despite the absence of an invasive disease correlation, our studies were the first to show that strains within a single serotype have a diverse ability to form early biofilms. While numerous studies have shown that the ability to form biofilms is nutrient dependent and enhanced by the absence of capsule [10, 13, 15, 35], our studies are the first to rigorously show that bacterial components other than capsule play important roles. The finding that phylogenetically similar strains (i.e. clonal derivatives), as determined by genetic content using microarrays, have diverse early biofilm forming abilities suggests that the ability to form biofilms may be regulated at the transcriptional level and not due to the presence or absence of genes [17]. Importantly, this study does not attempt to identify which genes are responsible for altered biofilm formation or correlate the serotype 6 clinical isolates with virulence gene expression. Our studies also corroborate earlier work by Hall-Stoodly et al. showing that the formation of microcolonies occurs and that they are important for biofilm biomass accumulation [14]. Examination of biofilm images revealed that early pneumococcal biofilm formation as determined by CV540 stating intensity coincided with the formation of bacterial aggregates superimposed on a bacterial “lawn”; suggesting that the formation of pneumococcal aggregates on top of a bacterial layer is a critical event during early biofilm formation.
In summary, it is clear that the ability to form early biofilms in vitro under the growth conditions tested were not positively correlated with the ability of clinical isolates to cause invasive disease. This suggests that the ability to form biofilms is not important for invasive disease. However, the discussed limitations in the model system and the fact that these studies are correlative, still leaves open the possibility that biofilm formation in vivo is important for invasive disease. These findings emphasize a need for future studies examining biofilm formation in animals during invasive disease.
ACKNOWLDEGEMENTS
This work was supported by National Institute of Health Grant AI078972.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infection. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 3.Hoa M, Tomovic S, Nistico L, Hall-Stoodley L, Stoodley P, Sachdeva L, et al. Identification of adenoid biofilms with middle ear pathogens in otitis-prone children utilizing SEM and FISH. Int J Pediatr Otorhinolaryngol. 2009 doi: 10.1016/j.ijporl.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Coates H, Thornton R, Langlands J, Filion P, Keil AD, Vijayasekaran S, et al. The role of chronic infection in children with otitis media with effusion: evidence for intracellular persistence of bacteria. Otolaryngol Head Neck Surg. 2008;138:778–781. doi: 10.1016/j.otohns.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta AJ, Lee JC, Stevens GR, Antonelli PJ. Opening plugged tympanostomy tubes: effect of biofilm formation. Otolaryngol Head Neck Surg. 2006;134:121–125. doi: 10.1016/j.otohns.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Reid SD, Hong W, Dew KE, Winn DR, Pang B, Watt J, et al. Streptococcus pneumoniae forms surface-attached communities in the middle ear of experimentally infected chinchillas. J Infect Dis. 2009;199:786–794. doi: 10.1086/597042. [DOI] [PubMed] [Google Scholar]
- 8.Hoa M, Syamal M, Sachdeva L, Berk R, Coticchia J. Demonstration of nasopharyngeal and middle ear mucosal biofilms in an animal model of acute otitis media. Ann Otol Rhinol Laryngol. 2009;118:292–298. doi: 10.1177/000348940911800410. [DOI] [PubMed] [Google Scholar]
- 9.Sanderson AR, Leid JG, Hunsaker D. Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis. Laryngoscope. 2006;116:1121–1126. doi: 10.1097/01.mlg.0000221954.05467.54. [DOI] [PubMed] [Google Scholar]
- 10.Munoz-Elias EJ, Marcano J, Camilli A. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect Immun. 2008;76:5049–5061. doi: 10.1128/IAI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trappetti C, Kadioglu A, Carter M, Hayre J, Iannelli F, Pozzi G, et al. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J Infect Dis. 2009;199:1497–1505. doi: 10.1086/598483. [DOI] [PubMed] [Google Scholar]
- 12.Oggioni MR, Trappetti C, Kadioglu A, Cassone M, Iannelli F, Ricci S, et al. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol Microbiol. 2006;61:1196–1210. doi: 10.1111/j.1365-2958.2006.05310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moscoso M, Garcia E, Lopez R. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol. 2006;188:7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon WJ, et al. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 2008;8:173. doi: 10.1186/1471-2180-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allegrucci M, Sauer K. Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J Bacteriol. 2008;190:6330–6339. doi: 10.1128/JB.00707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allegrucci M, Sauer K. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J Bacteriol. 2007;189:2030–2038. doi: 10.1128/JB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obert C, Sublett J, Kaushal D, Hinojosa E, Barton T, Tuomanen EI, et al. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun. 2006;74:4766–4777. doi: 10.1128/IAI.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 19.Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis. 2004;190:1661–1669. doi: 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- 20.Embry A, Hinojosa E, Orihuela CJ. Regions of Diversity 8, 9 and 13 contribute to Streptococcus pneumoniae virulence. BMC Microbiol. 2007;7:80. doi: 10.1186/1471-2180-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, Tuomanen EI. Microarray analysis of pneumococcal gene expression during invasive disease. Infect Immun. 2004;72:5582–5596. doi: 10.1128/IAI.72.10.5582-5596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mavroidi A, Godoy D, Aanensen DM, Robinson DA, Hollingshead SK, Spratt BG. Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J Bacteriol. 2004;186:8181–8192. doi: 10.1128/JB.186.24.8181-8192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies DG, Marques CNH. A Fatty Acid Messenger Is Responsible for Inducing Dispersion in Microbial Biofilms. J. Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allegrucci M, Hu FZ, Shen K, Hayes J, Ehrlich GD, Post JC, et al. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J Bacteriol. 2006;188:2325–2335. doi: 10.1128/JB.188.7.2325-2335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donlan RM, Piede JA, Heyes CD, Sanii L, Murga R, Edmonds P, et al. Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl Environ Microbiol. 2004;70:4980–4988. doi: 10.1128/AEM.70.8.4980-4988.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Budhani RK, Struthers JK. Interaction of Streptococcus pneumoniae and Moraxella catarrhalis: investigation of the indirect pathogenic role of beta-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob Agents Chemother. 1998;42:2521–2526. doi: 10.1128/aac.42.10.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shime-Hattori A, Iida T, Arita M, Park K-S, Kodama T, Honda T. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiology Letters. 2006;264:89–97. doi: 10.1111/j.1574-6968.2006.00438.x. [DOI] [PubMed] [Google Scholar]
- 28.Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. BdlA, a Chemotaxis Regulator Essential for Biofilm Dispersion in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Molecular Microbiology. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 30.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Molecular Microbiology. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 31.Gohl O, Friedrich A, Hoppert M, Averhoff B. The Thin Pili of Acinetobacter sp. Strain BD413 Mediate Adhesion to Biotic and Abiotic Surfaces. Appl. Environ. Microbiol. 2006;72:1394–1401. doi: 10.1128/AEM.72.2.1394-1401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orihuela CJ, Mahdavi J, Thornton J, Mann B, Wooldridge KG, Abouseada N, et al. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J Clin Invest. 2009;119:1638–1646. doi: 10.1172/JCI36759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes AR, McNab R, Millsap KW, Rohde M, Hammerschmidt S, Mawdsley JL, et al. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol Microbiol. 2001;41:1395–1408. doi: 10.1046/j.1365-2958.2001.02610.x. [DOI] [PubMed] [Google Scholar]
- 34.Martin M, Turco JH, Zegans ME, Facklam RR, Sodha S, Elliott JA, et al. An outbreak of conjunctivitis due to atypical Streptococcus pneumoniae. N Engl J Med. 2003;348:1112–1121. doi: 10.1056/NEJMoa022521. [DOI] [PubMed] [Google Scholar]
- 35.McEllistrem MC, Ransford JV, Khan SA. Characterization of in vitro biofilm-associated pneumococcal phase variants of a clinically relevant serotype 3 clone. J Clin Microbiol. 2007;45:97–101. doi: 10.1128/JCM.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]