Abstract
The transforming growth factor beta (TGF-β) has been studied with regard to the regulation of cell behavior for over three decades. A large body of research has been devoted to the regulation of epithelial cell and derivative carcinoma cell populations in vitro and in vivo. TGF-β has been shown to inhibit epithelial cell cycle progression and promote apoptosis that together significantly contribute to the tumor suppressive role for TGF-β during carcinoma initiation and progression. However, TGF-β is also able to promote an epithelial to mesenchymal transition that has been associated with increased tumor cell motility, invasion and metastasis. However, it has now been shown that loss of carcinoma cell responsiveness to TGF-β stimulation can also promote metastasis. Interestingly, the enhanced metastasis in the absence of a carcinoma cell response to TGF-β stimulation has been shown to involve increased chemokine production resulting in recruitment of pro-metastatic myeloid derived suppressor cell (MDSC) populations to the tumor microenvironment at the leading invasive edge. When present, MDSCs enhance angiogenesis, promote immune tolerance and provide matrix degrading enzymes that promote tumor progression and metastasis. Further, the recruitment of MDSC populations in this context likely enhances the classic role for TGF-β in immune suppression since the MDSCs are an abundant source of TGF-β production. Importantly, it is now clear that carcinoma-immune cell cross-talk initiated by TGF-β signaling within the carcinoma cell is a significant determinant worth consideration when designing therapeutic strategies to manage tumor progression and metastasis.
1. TGF-β signaling and cancer
1.1 Introduction
Transforming growth factor beta (TGF-β) is a secreted ligand that has been intimately linked to the regulation of tumor initiation, progression and metastasis. The regulation of tumorigenesis by TGF-β signaling is dependent upon the ability to regulate the behavior of tumor and host cell populations. TGF-β was initially identified as a functional mediator of epithelial and fibroblast cell proliferation and transformation over two decades ago. Shortly thereafter, it was also shown that TGF-β could significantly regulate immune cell populations [1–5]. In the time since its initial characterization, TGF-β has been linked to the control of tumor progression through modification of carcinoma cell behavior and importantly the interaction between carcinoma cells and adjacent cell populations within the tumor microenvironment. As a result, it is now generally accepted that TGF-β has a profound effect on nearly every cell type that has been shown to influence carcinoma initiation, progression and metastasis.
In recent years, carcinoma stromal cell populations including fibroblasts, endothelial cells and a diverse repertoire of immune cells have been the focus of a renewed effort to explore heterotypic interactions that regulate tumor initiation, progression and metastasis. One of the predominant stromal-epithelial axes associated with the regulation of cancer progression involves carcinoma-immune cell interactions within the tumor microenvironment [6, 7]. Specifically, TGF-β has been shown to suppress the anti-tumor activity of T-cells, NK cells, neutrophils, monocytes and macrophages that are known to have a significant role in the regulation of tumor progression [8–10]. Together, these cell populations have the ability to promote or suppress tumor progression depending on the context of each interaction. Importantly, the balance between suppression and promotion of tumor progression by the immune system is regulated by TGF-β in vivo.
1.2 Simplified overview of TGF-β signaling
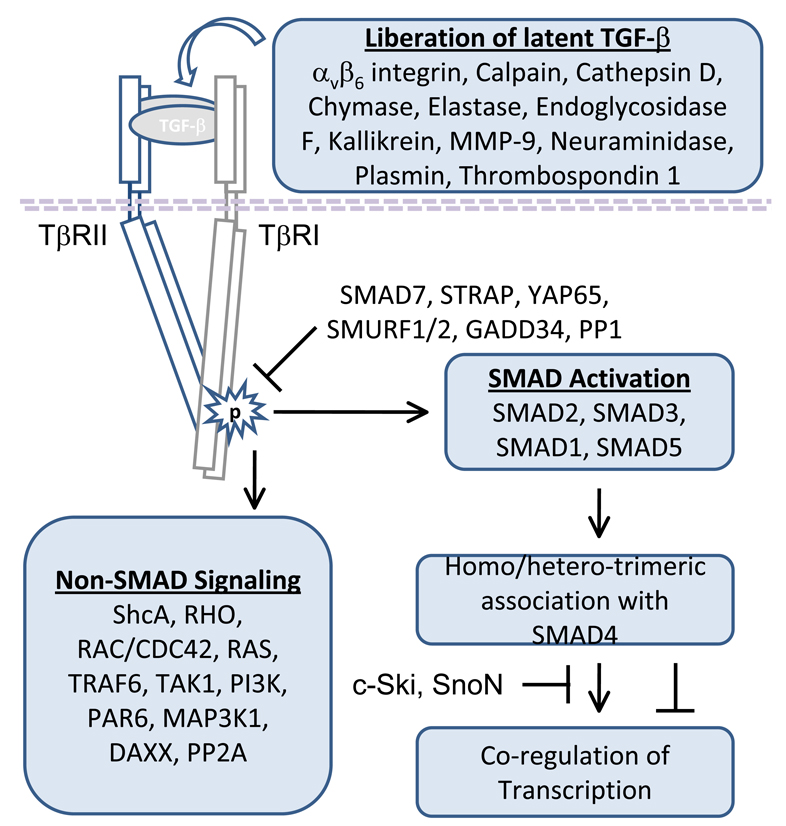
The transforming growth factor beta ligands (TGF-β1, TGF-β2 and TGF-β3) have been widely studied in the immune system and in association with the regulation of tumor progression [8, 9, 11, 12]. In cancer, TGF-β1 was shown to be more abundant that TGF-β2 or TGF-β3 and has therefore been the focal point for many of the mechanistic studies conducted in vitro and in vivo [13, 14]. Importantly, TGF-β1 expression is often upregulated in human cancer [15, 16]. TGF-β when expressed and secreted in vivo, is sequestered in the extra-cellular matrix (ECM) as part of an inactive complex until subsequent activation [17]. TGF-β activation can be induced by a number of mechanisms including the expression of αvβ6 integrin, calpain, cathepsin D, chymase, elastase, endoglycosidase F, kallikrein, MMP-9, neuraminidase, plasmin and thrombospondin-1 (TSP-1) [18–26]. In addition, reactive oxygen free radicals have been shown to activate latent TGF-β [27]. Once activated, TGF-β binds the type II TGF-β receptor (TβRII; TGFBR2) and the ligand bound TβRII is then able to efficiently trans-activate the type I TGF-β receptor (TβRI; TGFBR1) [28, 29]. The activation of signaling derived from TGF-β stimulation occurs primarily through serine threonine kinase activity associated with TβRI via a glycine and serine rich region termed the GS domain [29]. In addition to the serine-threonine kinase activity, it has now been shown that TGF-β signaling can directly promote tyrosine phosphorylation [30]. Downstream signaling includes canonical SMAD dependent and SMAD independent pathways [12, 29]. The SMAD proteins are transcription factors, and TGF-β signaling has been shown to robustly activate SMAD2 and SMAD3 (receptor associated SMAD family members; R-Smads) through phosphorylation of C-terminal regulatory residues [12, 29, 31]. Once phosphorylated, SMAD2 and SMAD3 are able to change confirmation thereby allowing the assembly of hetero- and homo-oligomeric complexes with SMAD4 (common mediator Smad; co-Smad) to initiate downstream signaling [12, 29, 31]. Interestingly, in addition to SMAD2 and SMAD3, it has now been shown that TGF-β can activate SMAD1 and SMAD5 (R-Smads) in epithelial cells, epithelial cell derived tumor cells and fibroblasts [32]. Although a majority of the Smad dependent processes associated with TGF-β signaling can be directly linked to SMAD2/SMAD3/SMAD4 complexes it is now clear that some processes, exemplified by TGF-β induced anchorage independent growth, may require activated SMAD1 or SMAD5 in association with SMAD2 or SMAD3 in a tertiary signaling complex with SMAD4 [32]. Importantly, R-SMAD activity that is known to be an important part of TGF-β signaling, can be modified by a number of mechanisms [12, 29, 31]. In addition to SMAD dependent signaling, SMAD independent pathways are also important for a response to stimulation by the TGF-β ligand. At present, the SMAD independent pathways are known to include ShcA, RHO, RAC/CDC42, RAS, TRAF6, TAK1, PI3K, PAR6, MAP3K1, DAXX and PP2A [12, 29–31, 33, 34]. Together, the net activation of SMAD dependent and independent pathways will determine the response to ligand stimulation in vitro or in vivo (Figure 1).
Figure 1. General overview of TGF-β signaling.
TGF-β can be liberated from latent complexes by αvβ6 integrin, calpain, cathepsin D, chymase, elastase, endoglycosidase F, kallikrein, matrix metalloproteinase 9 (MMP-9), neuraminidase, plasmin or thrombospondin 1. Once activated TGF-β is able to bind type II TGF-β receptor homo-dimers (TβRII) thereby permitting the efficient transactivation of type I TGF-β receptor homo-dimers (TβRI). As a result of receptor activation by TGF-β, downstream SMAD dependent and SMAD independent signaling is initiated. However, the level of downstream pathway activation is dependent the abundance of receptor associated TGF-β signaling repressors such as SMAD7, STRAP, YAP65, SMURF1, SMURF2, GADD34 and PP1. SMAD dependent signaling has been primarily associated with activation of SMAD2 or SMAD3, however it has now been shown that SMAD1 and SMAD5 can be activated by TGF-β. Once SMAD1, SMAD2, SMAD3 or SMAD5 are activated they are able to associate with SMAD4 to co-activate or co-repress transcription. Importantly, SMAD dependent signaling can also be repressed by complex association with other transcription factors, co-activators or co-repressors such the c-Ski or SnoN proto-oncogenes. The SMAD independent pathways are known to include ShcA, RHO, RAC/CDC42, RAS, TRAF6, TAK1, PI3K, PAR6, MAP3K1, DAXX and PP2A. Together, the balance of SMAD dependent signaling, SMAD independent signaling and the presence or absence of signaling repressors ultimately determines the response to TGF-β in vitro or in vivo.
1.3 Altered TGF-β signaling in human cancer
At present, substantial correlative data has been produced to demonstrate that that TGF-β signaling components including TGFBR1, TGFBR2, SMAD2 and SMAD4 are often lost in human cancer [16, 35]. The TGF-β1 ligand has been shown to be upregulated in breast, colon, esophageal, gastric, liver, lung pancreatic and prostate cancer [16, 35]. However, mutations and loss of type I and Type II TGF-β receptor expression have been detected in most types of common cancer including those that occur in the biliary tract, bladder, breast, colon, esophagus, stomach, brain, liver, lung, ovary, pancreas and prostate [16, 35]. In the case of TGFBR1, recent evidence suggest that alteration in a single allele is can result in an increased risk of cancer [36, 37]. In contrast with TGFBRI, TGFBR2 mutation was shown to be particularly frequent in micro-satellite instable (MSI+) tumors of the biliary, colon, gastric, brain and lung tissues [16, 35, 38]. The increased rate of mutation in the MSI+ tumor tissue was often associated with errors in a long adenine (10bp; Poly(A)10) repeat region within the TGFBR2 gene [38]. SMAD signaling was also shown to be frequently lost in human cancer. SMAD4 mutation, deletion and loss of expression has been reported in biliary, bladder, breast, cervical, colon, liver, intestine, esophagus, lung, ovary and pancreatic cancers [16, 35, 38]. In addition, loss of SMAD4 has been putatively linked to a causal role in juvenile polyposis-associated carcinoma initiation [39]. Further, mutation or loss of SMAD2 has been detected in cervical, colon, lung and liver cancer [16, 35, 38]. However, SMAD3 was maintained in most common cancers [16].
Interestingly, epigenetic alterations have also impact on the TGF-β pathway in human cancer. Recently, it has been shown that silencing of the TGFBR2 gene could occur through DNA methylation in human breast carcinoma cells [40]. Importantly, transcriptional repression and DNA methylation of TGFBR1 and TGFBR2 have been shown to occur in human cancer [41, 42]. TGFBR2 repression is particularly important, since it has been suggested that this mode of regulation may be responsible for most of the tumor associated TGF-β resistance observed in vivo [42]. In addition, it has been shown recently that TSP-1 suppression in colon cancer can be attributed to DNA methylation [43]. Notably, the reduced TSP-1 expression has been correlated with a decrease in latent TGF-β activation that could be reversed upon TSP-1 DNA de-methylation [43]. It has also been shown that SMAD4 suppression in advanced prostate cancer could be associated with promoter DNA methylation [44]. Notably, in human mammary epithelial cells and human mammary carcinoma cell lines it has been shown that TGF-β2, TGFBR1, TGFBR2 and TSP-1 expression was concurrently suppressed by DNA methylation and these genes could be coordinately re-induced upon de-methylation [45].
Mutation, loss of heterozygosity and epigenetic alterations are not the only factors that have been shown to regulate the TGF-β pathway in cancer. Functional suppressors of TGF-β signaling have also been demonstrated as important modifiers of TGF-β responsiveness in vitro and in vivo (Figure 1). SMAD activation downstream of TGF-β is highly regulated and can be attenuated through a number of distinct mechanisms. The inhibitory SMAD7 (I-SMAD) protein can associate with TβRI and effectively attenuate SMAD2/3 signaling [46, 47]. SMAD7 has been shown to promote recruitment of E3 ubiquitin ligases including SMAD ubiquitin regulatory factor 1 (SMURF1), and SMURF2 to the receptor complex [48, 49]. This E3 ubiquitin ligase recruitment results in ubiquitylation of the TGF-β receptors followed by proteasome mediated degradation. Binding of SMAD7 and SMURF proteins to the receptor complex also results in competitive inhibition of Smad2/3 binding to TβRI [50]. Further, it has been shown that SMAD7 can associate with GADD34 (growth arrest and DNA damage protein 34) to target protein phosphatase 1 (PP1) to TβRI resulting in dephosphorylation of the TβRI receptor [51]. Importantly, expression of other proteins, such as STRAP or YAP65 can interact with SMAD7 to facilitate receptor binding and thereby attenuate TGF-β signaling [52, 53]. The c-Ski and SnoN proto-oncoproteins are also known to repress SMAD activity and function as negative regulators of TGF-β signaling [54, 55]. In addition, increased expression of c-myc, cyclin D1 or Ras can attenuate the cytostatic ability of TGF-β signaling [12]. Other alterations that are common in cancer such as loss or mutation of p107, p15INK4b or p16INK4a can further contribute to the attenuation of TGF-β dependent growth inhibition [12]. Due to the frequency of mutation, loss and downregulation in human cancer the TGF-β pathway is a major tumor suppressor pathway and has been heavily studied to determine the role for this signaling network during tumor initiation, progression and metastasis.
2. Carcinoma cell specific response to TGF-β signaling and the link to inflammation
2.1 TGF-β dependent regulation of epithelial cell populations
TGF-β was initially described as a ligand that had the ability to promote or suppress cellular proliferation depending on the cell type and context of stimulation. In non-transformed or carcinoma-associated epithelial cells, TGF-β is well known for its ability to inhibit cell proliferation and promote an epithelial to mesenchymal transition (EMT) that has been associated with increased motility and invasiveness [56, 57]. In the years since discovery of TGF-β, an enormous volume of data has been produced in association with TGF-β dependent regulation of tumor progression [11, 12].
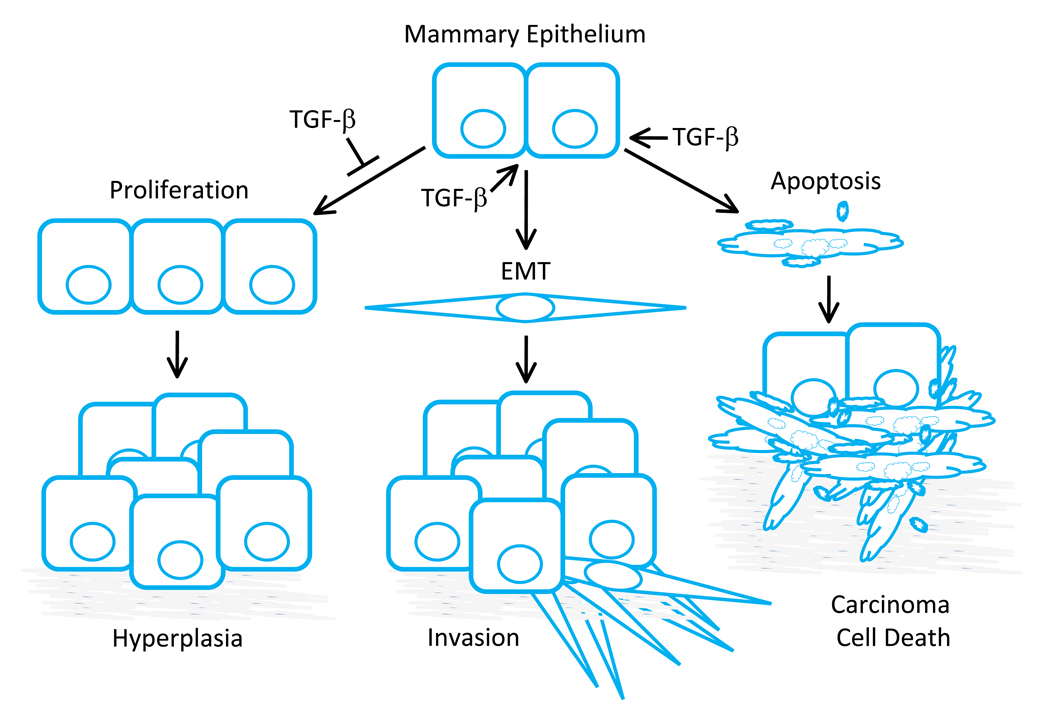
To determine the functional role for carcinoma cell specific responses to TGF-β signaling mouse models have been particularly useful. Although significant results demonstrating an essential role for carcinoma cell response to TGF-β signaling have been produced in several model systems, many of the general features associated with the known roles for TGF-β in this context have been illustrated using mammary epithelium and derivative mammary carcinomas. One of the predominant roles for TGF-β signaling is the regulation of epithelial cell growth (Figure 2). In early developmental studies it was noted that increased TGF-β expression could lead to inhibition of mammary epithelial outgrowth in vivo [58–65]. Conversely, expression of a dominant negative TβRII in the epithelium during mammary development resulted in mammary hyperplasia and precocious differentiation [66, 67]. In addition to suppressing cell cycle progression, TGF-β was also shown to promote apoptosis and epithelial to mesenchymal transition (EMT) in epithelial cells depending upon the cell type and context of stimulation [12, 56, 57]. Together, the early studies suggested that stimulation of epithelium by TGF-β would result in tumor suppression through enhanced growth arrest, apoptosis while stimulating carcinoma cell motility as a result of EMT.
Figure 2. Classic roles for TGF-β in the regulation of mammary epithelial cell growth and tumorigenesis.
TGF-β signaling has been shown to regulate many epithelial cell types, however many of the common features of signaling through this pathway have been demonstrated using mammary epithelium. TGF-β has been shown to inhibit epithelial cell proliferation predominantly through Smad dependent regulation of p107 (Rb) activity via regulation of p15INK4B, p16INK4A, p21Cip1 and p27Kip1 expression. In the presence of TGF-β, these cyclin dependent kinase inhibitors are upregulated and Rb remains hypo-phosphorylated thereby blocking cell cycle progression. TGF-β is also known to suppress c-myc expression that would otherwise provide an additional proliferative signal for the epithelium. However, one or more of these mediators are often mis-regulated during tumor progression and this can prevent the cytostatic regulation associated with TGF-β stimulation. In addition to cell cycle regulation, TGF-β is able to promote an epithelial to mesenchymal transition (EMT) that has been associated with increased motility and enhanced carcinoma cell invasion. Further, epithelial and carcinoma cell survival has also been associated with TGF-β signaling. Apoptosis induced by TGF-β can provide an additional level of regulation in favor of tumor suppression. The precise role for TGF-β in each epithelial cell population depends on the specific cell type and context of stimulation. Although non-transformed cells from a given organ under clearly defined conditions will often respond to TGF-β in a similar manner, due to the complex nature of parallel intrinsic and extrinsic signaling unique to the cancer evolution in each patient, it is often difficult to predict the proliferative, EMT, or apoptotic impact of TGF-β signaling in a carcinoma cell population a priori.
2.2 TGF-β regulates carcinoma initiation, progression and metastasis
TGF-β has a profound effect on mammary carcinoma initiation, progression and metastasis. The transgenic expression of TGF-β by mammary epithelium led to a significant increase in the latency associated with mammary tumor growth in TGF-α transgenic mice or those treated with the chemical carcinogen DMBA [68]. TGF-β expression by mammary epithelium also resulted in a decreased incidence of tumorigenesis induced by infection with the mouse mammary tumor virus [69]. Conversely, in the presence of TGF-α expression from mammary epithelium it was shown that attenuation of the carcinoma cell response to TGF-β by a dominant-negative type II receptor transgene (dnTβRII) significantly reduced tumor latency [66]. In addition to the difference in tumor latency, a decrease in carcinoma cell invasion was also observed in this model. Interestingly, the invasive lesions that did occur were shown to be negative for transgenic dnTβRII expression [66]. This suggested that in vivo, carcinoma cell specific TGF-β signaling was able to promote tumor cell invasion. These results correlated well with other studies in which expression of dnTβRII was shown to decrease extravascular pulmonary metastases in the MMTV-c-Neu mouse model [70]. Further, it was shown in several mouse models that mammary epithelial cell specific expression of an activated TGF-β ligand or expression of a constitutively active TβRI could enhance breast cancer associated lung metastases in vivo [70–72]. In support of a pro-metastatic role for TGF-β, systemic attenuation of TGF-β signaling through expression of a Fc-TβRII was also shown to prevent metastasis in the MMTV-c-neu mouse model or in mice that had been injected with malignant melanoma cells through the tail vein [73]. Despite variability in observed growth responses associated with TGF-β signaling during tumorigenesis in vivo, the enhanced invasion and metastasis in response to TGF-β stimulation was consistent [66, 70–73]. Together, the results suggested that TGF-β could suppress primary tumor growth while promoting metastasis through EMT of the responding carcinoma cells.
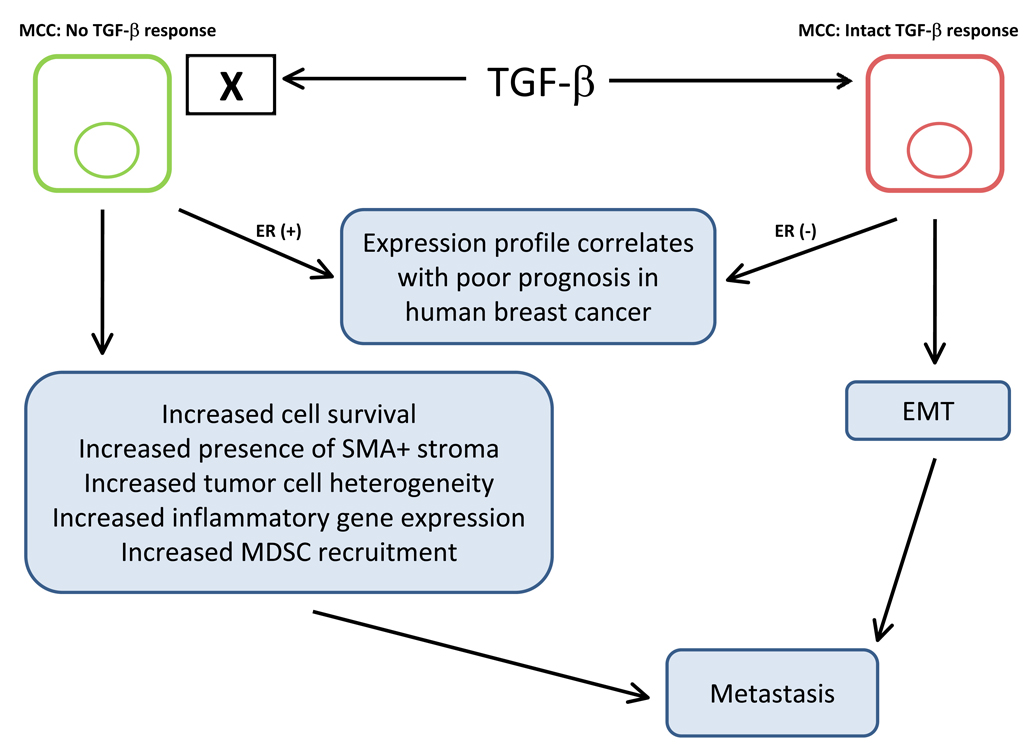
Importantly, upon complete ablation of TGF-β signaling from mammary epithelium another significant interaction was uncovered. In a mouse model of mammary carcinoma, with complete ablation of TGF-β response in mammary epithelium, a decrease in tumor latency was observed as predicted [74]. Notably, a striking increase in pulmonary metastasis was also clearly demonstrated [74–76]. This latter result was not expected due to the consistency of previous work demonstrating that TGF-β enhanced metastasis. In this model it was also observed that loss of TGF-β signaling in the mammary carcinoma cells increased the abundance of smooth muscle actin positive stroma, tumor cell heterogeneity and tumor cell survival. Further, it was shown that TGF-β regulated chemokine expression and resulting carcinoma-immune cell interactions could in part explain the seemingly divergent observations related to mammary carcinoma cell metastasis [75–77]. Specifically, the TGF-β response by carcinoma cells could suppress chemokine expression that regulates recruitment of pro-metastatic GR1+ CD11b+ and F4/80+ bone marrow derived cells to the tumor microenvironment [75–77]. In addition, a gene signature associated with the loss of carcinoma cell specific response to TGF-β was correlated with a poor patient prognosis in human breast cancer. Using a panel of 1,319 human breast cancer gene expression signatures it was shown that the loss of TGF-β signaling in human ER+ breast cancer correlated with an increased probability of relapse [77]. Conversely, a gene signature derived from breast cancer cells that had been stimulated with TGF-β was correlated with a poor prognosis in human ER− breast cancer [77, 78]. Importantly, in the TGF-β signaling deficient gene expression signature several chemokines were consistently upregulated whereas the same chemokines were consistently downregulated in the carcinoma cells that had been stimulated with TGF-β. The correlation with human breast cancer prognosis, regulation of chemokine expression and recruitment of tumor promoting myeloid cell populations suggested that the cross-talk between adjacent cell populations mediated by TGF-β is a significant factor worth consideration when designing strategies to manage human breast cancer progression (Figure 3).
Figure 3. Gain or loss of TGF-β signaling in mammary carcinoma cells (MCC) can promote metastasis.
Gene expression profiles representing gain or loss of TGF-β signaling in mammary carcinoma cells have been shown to correlate with poor prognosis in human breast cancer. Importantly, the results suggest that the difference may be correlated with the status of estrogen receptor (ER) expression. In the presence of ER, a gene signature representing complete abrogation of TGF-β signaling in mammary carcinoma cells correlates with poor clinical prognosis whereas a signature associated with intact TGF-β signaling in ER negative breast cancer correlates with poor clinical prognosis. In the case of intact TGF-β signaling, many studies have suggested that TGF-β dependent EMT promotes carcinoma invasion that initiates and promotes the seeding of distant metastases. In the absence of carcinoma cell specific TGF-β responsiveness, a number of interactions including increased carcinoma cell survival, the increased abundance of adjacent smooth muscle actin positive (SMA+) fibrovascular stroma, increased tumor cell heterogeneity, increased inflammatory gene expression and increased MDSC recruitment have been suggested to promote metastatic progression. Importantly, in a mixed population of carcinoma cells that differ in the ability to respond to TGF-β, it is possible that both types of pro-metastatic progression can co-exist in a single tumor microenvironment.
3. The TGF-β response in carcinoma cells can suppress recruitment of myeloid cells that promote invasion and metastasis
3.1 Immature bone marrow derived cells promotes invasion and metastasis
In many of the early studies, it was suggested that the loss of TGF-β signaling in carcinoma cells could lead to enhanced growth of primary tumors and metastases. However, it had also been suggested that TGF-β signaling was necessary for promoting an epithelial to mesenchymal transition that was thought to be necessary for metastatic dissemination. This latter point was difficult to reconcile with data demonstrating that increased TGF-β signaling or complete abrogation of TGF-β signaling in mammary carcinoma cells could both lead to a significantly increased rate of metastasis. However, the recently identified carcinoma-immune cell interactions mediated by TGF-β may help to explain these results. It is well known that recruitment of tumor associated macrophages, monocytes and neutrophils can promote tumor progression [6, 7], however a number of recent reports now support a significant role for immature myeloid cells as promoters of tumor progression and metastasis [79]. This tumor promoting immature bone marrow derived cell population within the tumor microenvironment has been referred to as myeloid derived suppressor cells (MDSCs), myeloid immune suppressor cells (MDSCs) and immature myeloid cells (iMCs). These names all refer to a family of bone marrow derived cells with characteristics similar to monocyte, macrophage, granulocyte and dendritic cell precursors at different stages of differentiation [79]. In mouse models, these cell populations are often identified by their cell surface expression of GR-1 and CD11b proteins. Within the GR-1+ CD11b+ cell population it has been shown that the GR-1high populations enriches for the polymorphonuclear cells whereas the GR1int/low population enriches for the mononuclear cells. Importantly, it has been suggested that the mononuclear fraction is better able to suppress CD8+ T-cell mediated immunity than the polymorphonuclear fraction [80]. Further, it has been suggested that a subset of GR1+ CD11b+ cells that also express F4/80 and IL4-Ra can differentiate into tumor associated macrophages or contribute to M2 type activation of tumor-associated macrophages [79, 81, 82].
3.2 Loss of TGF-β signaling in carcinoma cells can enhance recruitment of MDSCs
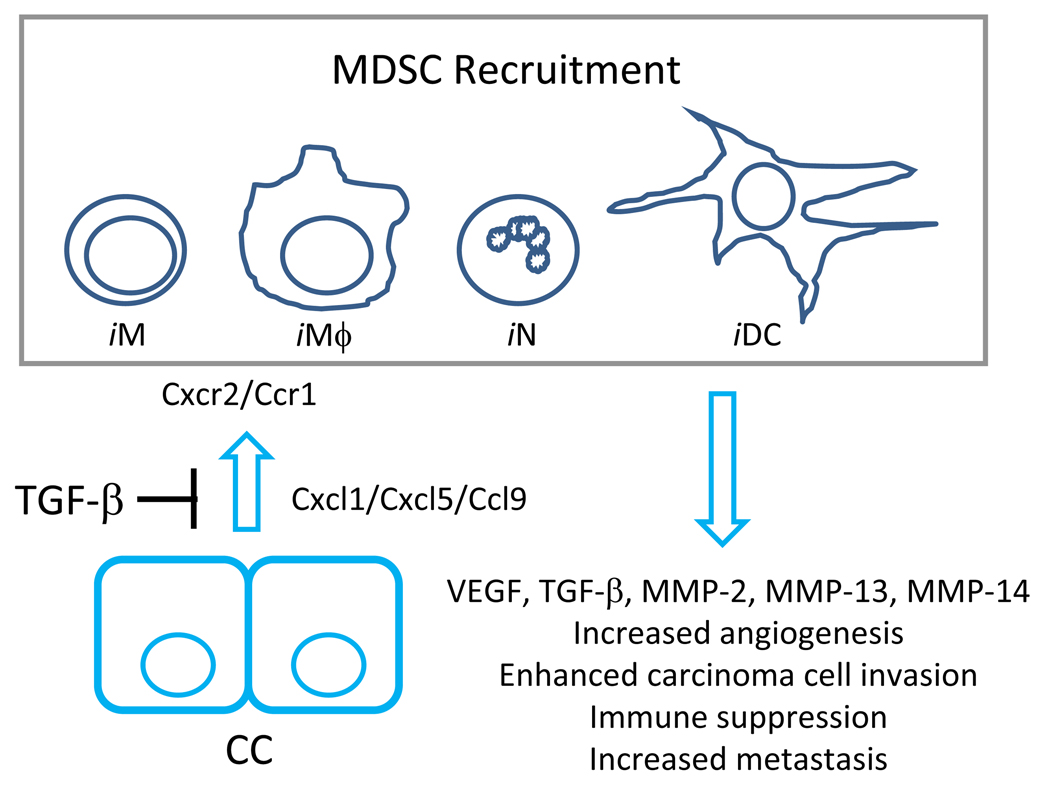
Mammary carcinoma cell specific ablation of TGF-β signaling led to enhanced metastasis and was associated with an increased myeloid cell infiltrate in mice [74–76]. The inflammation involved GR1+ CD11b+ and F4/80+ myeloid cells that were preferentially recruited to the leading edge of tumors exhibiting a carcinoma cell specific ablation of TGF-β responsiveness [75, 76]. The recruitment was correlated with increased expression of Cxcl1 and Cxcl5 in TGF-β signaling deficient tissues in vitro and in vivo [75–77] (Figure 4). Cxcl1 and Cxcl5 signal through the Cxcr2 receptor and it has been shown that the increased signaling through Cxcr2 signaling in MDSC populations could provide a motogenic signal for MDSC cells in vitro [76]. In vivo, it was shown that Cxcr2 signaling significantly contributed to enhanced metastasis observed from the TGF-β signaling deficient mammary carcinoma cell population when compared with the control mammary carcinoma cells [76]. Importantly, MDSC conditioned medium was shown to promote enhanced invasion of mammary carcinoma cells through an artificial basement membrane in vitro [76]. The enhanced invasion was MMP dependent, however it was not clear if MMPs necessary for the enhanced invasion were produced by MDSCs or by carcinoma cells in response to stimulation with MDSC conditioned medium [76]. However, MDSCs derived from tumor bearing hosts were shown to produce a relatively high level of VEGF, MMP-2, MMP-13 and MMP-14 when compared with those isolated from non-tumor bearing hosts [76], suggesting that at least some of the observed invasive ability associated with the MDSC conditioned medium may have come from MDSC derived MMP secretion. In addition to the enhanced MMP associated invasion, it was observed that MDSCs derived from tumor bearing hosts were a significant source of TGF-β production at the leading invasive tumor edge [76]. Importantly, recent results produced using a mouse model of colon cancer suggest the regulation of chemokine dependent MDSC recruitment by TGF-β may be operative in other types of cancer [83] (Figure 4). In this study it was shown that loss Smad4 on a single allele in the presence of an APC mutation resulted in colon carcinoma with enhanced Ccl9 dependent recruitment of Ccr1+ immature myeloid cells that significantly enhanced tumor progression [83].
Figure 4. TGF-β signaling significantly regulates recruitment of tumor promoting myeloid derived suppressor cell (MDSC) populations.
MDSC recruitment significantly enhances tumor progression and metastasis. This cell population is a heterogenous combination of myeloid derived cell populations that have characteristics associated with immature monocytes (iM), macrophages (iMϕ), neutrophils (iN) and dendritic cells (iDC). TGF-β signaling in non-transformed and carcinoma-associated epithelial cells (CC) has been shown to suppress the production of chemokines including Cxcl1, Cxcl5 and Ccl9 that when present have the ability to recruit MDSC populations. Cxcl1 and Cxcl5 signal through the Cxcr2 receptor and Ccl9 is known to signal through Ccr1. Abrogation of TGF-β signaling in carcinoma cells relieve the repression of these chemokines and thereby promotes Cxcr2 and Ccr1 dependent recruitment of MDSCs. The recruited MDSC populations are known to be an abundant source of VEGF, TGF-β, MMP-2, MMP-13 and MMP-14 that likely contribute to the MDSC-associated increase in angiogenesis, carcinoma cell invasion, immune suppression and metastasis.
3.3 MDSCs are present at the invasive front of human carcinomas
In human cancer, it has been shown that analogous MDSC populations were abundant and selectively localized within the leading invasive edge of breast ductal adenocarcinomas [76]. In addition to breast cancer, the increased production of antigen specific T-cell suppressing MDSC cells (predominantly CD34+ CD33+ CD15−) has been associated with head and neck and non-small cell lung cancer [84]. CD14+ ARG+ cells that also have the ability to impair T-cell function have been described in head and neck patients as well as those with multiple myeloma [85]. In metastatic melanoma patients, a population of circulating CD11b+ CD14+ myeloid cells was shown to be present that had the ability to suppress T-cell proliferation and effector functions [86]. CD11b+ CD15+ CD14− cells have also been detected in peripheral blood from metastatic renal cell carcinoma patients and these cells were also shown to suppress T-cell activity [87]. Importantly, it was recently shown that infiltrating CD34+ CCR1+ leukocytes are present at the invasive front of human colorectal cancers that had a mutation in the Poly(A)10 region of TGFBR2 [83].
4. TGF-β inhibits T-cell mediated rejection of carcinoma cells
4.1 T-cells are significantly regulated by TGF-β
The impact of TGF-β signaling in the immune system is well documented. Importantly, TGF-β is present in the carcinoma extra-cellular matrix regardless of the carcinoma cell specific ability to bind the activated ligand. Further, in tumors where the carcinoma cells were unable to respond to TGF-β, recent results suggest that stromal TGF-β was more abundant [76]. Thus, in carcinomas where the carcinoma cell specific response to TGF-β is attenuated or ablated there remains an abundant supply of TGF-β available for stimulation of adjacent cell populations including immune cell infiltrates. As a result, the presence of bioactive TGF-β in the tumor microenvironment can have a significant impact upon anti-tumor activity of T-cells. In early studies, it was shown through the production of TGF-β1 null mice, that TGF-β had a central role in autoimmune regulation [88, 89]. In the absence of global TGF-β1, multifocal autoimmune disease and a high rate of mortality were observed [88, 89]. Later it was shown that attenuation of TGF-β signaling specifically in T-cells, through expression of a dominant negative TβRII transgene in vivo, resulted in autoimmune disease and enhanced spontaneous effector T-cell differentiation [90]. TGF-β has also been shown to suppress T-cell proliferation through both IL-2 dependent and independent mechanisms [1]. The regulation of IL-2 dependent T-cell signaling by TGF-β has been reported to involve suppression of IL-2 production by Smad3, however the response to IL-2 induced proliferation (unlike TCR induced proliferation) was not directly dependent upon Smad3 activation [91]. TGF-β was also shown to regulate T-cell growth arrest in the presence of exogenous IL-2 and IL-4 that would normally promote proliferation [92]. In support of a direct effect of TGF-β stimulation on T-cell growth inhibition, p21Cip1 and p27Kip1 are known TGF-β targets that have been shown to significantly suppress the IL-2 dependent proliferative T-cell response [93]. Interestingly, it has been shown that in naïve CD4+ and CD8+ cell populations (CD44low) TGF-β stimulation significantly induced arrest of cell cycle progression in G1, whereas in the memory effector T-cell pool (CD44high), the impact of TGF-β signaling was less pronounced in the presence or absence of p21Cip1 and p27Kip1 [93]. These results were similar to those in other cell types wherein the context of TGF-β signaling is an important determinant for the outcome associated TGF-β stimulation. Importantly, TGF-β has been shown to functionally regulate the differentiation of helper and effector T-cell sub-populations in vitro and in vivo [8, 9, 94]. Specifically, TGF-β has been shown to inhibit Th1 and Th2 T-cell differentiation while promoting regulatory T-cell production and suppression of cytotoxic T-cell activity [8] (Figure 5A).
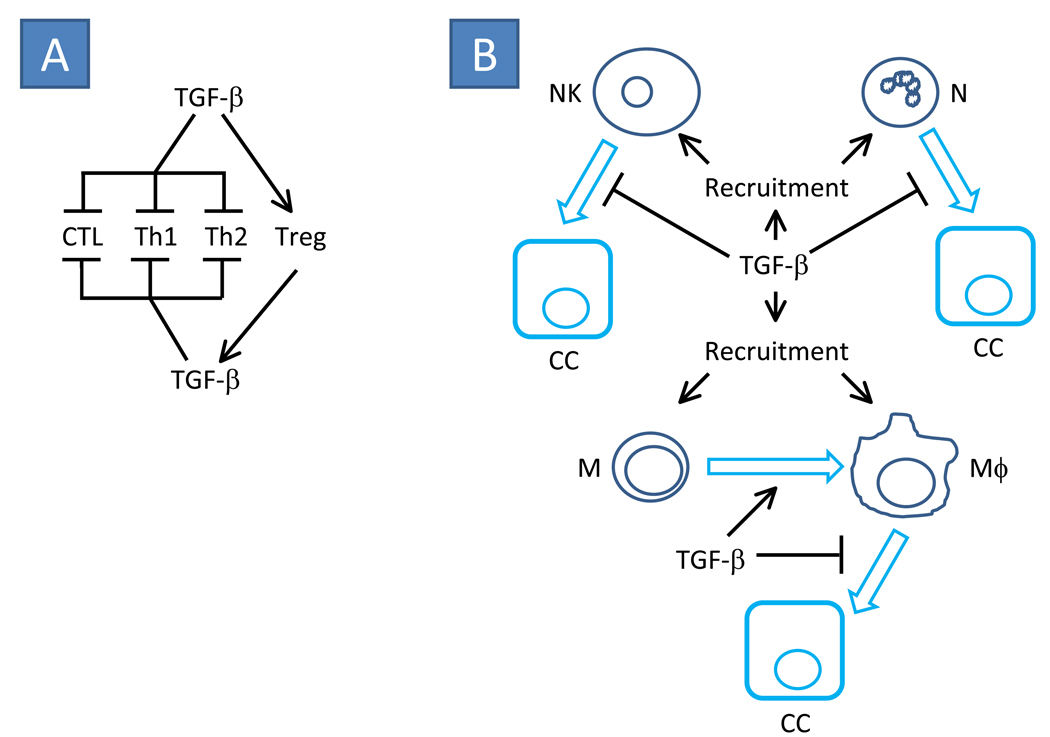
Figure 5. Classic roles for TGF-β signaling in immune suppression often associated with carcinoma-associated tumor progression.
(A) TGF-β is known to suppress the activity of cytotoxic T-lymphocytes (CTL) in addition to differentiation of helper Th1 and Th2 T-cell populations. However, TGF-β promotes the differentiation of regulatory T-cells (Treg) that are known to be an abundant source of TGF-β production that may further support suppression of CTL, Th1 and Th2 cells while promoting Treg expansion. (B) TGF-β is a potent chemoattractant for natural killer (NK), neutrophil (N), monocyte (M) and macrophage (Mϕ) cells. TGF-β has been shown to enhance M to Mϕ differentiation. However when present, TGF-β has been shown to prevent NK-, N- and Mϕ-associated carcinoma cell (CC) death. Together, the recruitment of immune cell populations and suppression of immune effector function can result in a tumor microenvironment that is rich in immune cell derived growth factors, morphogens, mitogens, and additional chemokines that further promote tumor progression and metastasis.
4.2 TGF-β regulates T-cell mediated tumor rejection
TGF-β is known to suppress T-cell mediated tumor rejection. When TGF-β signaling was attenuated in CD4+ and CD8+ T-cells, it was shown that the predominant tumor suppression mediated by TGF-β was associated with the CD8+ cell population [95]. In this study it was shown that the CD4+ cells were necessary for immune suppression, however it was TGF-β signaling in the CD8+ cell population that suppressed tumor rejection [95]. Although the CD4+ cells do not directly mediate tumor rejection they are important for evasion of immune surveillance and TGF-β can cause host macrophages to become suppressors of CD4+ T-cell proliferation [96]. Although there are many sources of TGF-β in the tumor microenvironment, it has recently been shown that the CD4+CD25+ regulatory T-cell population can provide a significant source of TGF-β that is responsible for attenuation of tumor antigen expanded CD8+ CTLs [97]. Importantly, in this study it was shown that TGF-β did not inhibit the expansion of the CD8+ CTL population; rather it had an impact on the effector function of the CTLs [97]. Further, the regulation by TGF-β was shown to be selective for ligand primed T-cell populations. The specificity of inhibition was demonstrated in experiments that involved the expression of a dominant negative TβRII (dnTβRII) in T-cells isolated from mice after priming with TRAMP-C2 prostate carcinoma cells [98]. In this model it was shown that adoptive transfer of dnTβRII expressing CD8+ T-cells significantly reduced TRAMP-C2 tumor growth and metastasis. This effect was shown to be selective for the tumor cell line used for priming since the TRAMP-C2 primed CD8+ T-cells were relatively ineffective when the transfers were performed in the presence of B16-F10 melanoma cells [98].
At present a number of studies have clearly demonstrated that TGF-β can suppress cytotoxic T-cell differentiation and cytotoxic T-cell mediated lysis of carcinoma cells [98–102]. In addition to the effect on differentiation, some of the molecular mechanisms have been described that help to explain the functional observations. It has been shown that TGF-β was able to suppress the pore forming protein (PFP) expression by CD8+ cytotoxic T-cells and prevent their cytolytic activity in vitro [103]. The regulation of PFP expression and cytotoxic potential was dose dependent and shown to be independent of the proliferative responses to TGF-β within the T-cells [103]. In addition, it has now been shown that TGF-β also prevents the expression of granzyme A, granzyme B, perforin, FasL and interferon-gamma that together promote CTL cytotoxicity [102, 104, 105]. The expression of granzyme B and interferon-gamma was directly linked to SMAD and ATF1 transcription factors providing a direct link to TGF-β mediated signal transduction within the CTLs [104]. Importantly, systemic attenuation of TGF-β signaling has been shown to result in enhanced clearance of carcinoma cells in vivo and it has been suggested that this outcome is mediated in part by the CTL cell response to TGF-β stimulation [104, 106].
5. TGF-β inhibits natural killer (NK) cell and neutrophil associated tumor rejection
5.1 TGF-β attenuates NK-cell associated tumor rejection
TGF-β inhibits NK cell and neutrophil effector functions and thereby contributes to a permissive microenvironment for tumor progression (Figure 5B). In early studies, it was shown that TGF-β stimulation resulted in suppression of natural killer (NK) cell activity. In vivo, it was shown that growth of MDA-MB-231 mammary carcinomas and derivative metastases could be significantly reduced when systemic TGF-β inhibition was performed [107]. This effect was not observed in beige nude mice that were NK cell deficient [107]. Interestingly, it has been shown that TGF-β could regulate the effect of tamoxifen treatment in vivo through an interaction involving NK cell populations [108]. In this context, it was shown the human breast cancer LCC2 cell line was resistant to tamoxifen in the absence of systemic TGF-β inhibition, however significant tamoxifen sensitivity was observed upon systemic administration of TGF-β neutralizing antibodies in mice. It was shown that the effect of systemic inhibition was not associated with carcinoma cell proliferation, but tumor cell production of TGF-β2 resulted in attenuation of NK cell mediated cytotoxicity. Further, the altered sensitivity to tamoxifen was not observed in NK deficient nude beige mice demonstrating a mechanistic link between the response to hormone therapy and NK cell activity in vivo [108].
Mechanistically the TGF-β associated NK-cell link may be explained in part by early observations demonstrating that TGF-β suppresses natural killer cell cytolytic activity and interferon responsiveness [3]. In addition, TGF-β has been shown to suppress MHC I and MHC II expression in a number of cell populations [94, 109–112]. Importantly, the TGF-β dependent decrease of MHC I expression in tumor cells has been shown to result in reduced tumor cell lysis by natural killer cells [109]. Recently, it has been shown that CD4+CD25+ regulatory T-cell production of TGF-β can inhibit NKG2D-mediated NK cell cytotoxicity thereby enhancing tumor growth and metastasis [113, 114]. TGF-β also attenuates the expression of triggering receptors including NKp30 and NKG2D that are important for NK cell mediated killing of dendritic cells and susceptible populations of tumor cells [115]. Interestingly, an inverse correlation has been identified between TGF-β1 secretion and NKG2D expression in cancer patients [116]. The reduced NKG2D receptor expression may contribute to the reduced NK-cell associated cytotoxicity that has been observed in association with disease progression [116]. At present the current literature suggests that increased TGF-β abundance in the tumor microenvironment can lead to reduced NK-cell cytotoxic activity thereby contributing to enhanced tumor progression and metastasis.
5.2 TGF-β can suppress neutrophil associated tumor rejection
TGF-β is a chemoattractant for neutrophils that also inhibits their ability to suppress tumorigenesis [10, 117] (Figure 5B). In early studies, it was determined that TGF-β is one of the most potent known chemoattractants for human peripheral blood neutrophils [117]. In fact, TGF-β was shown to promote directed migration of neutrophils at levels in the femtomolar range which is quite low compared with other conventional mitogens that require concentrations in the nanomolar range. It was also demonstrated that the migration invoked by TGF-β stimulation was dependent upon new protein and mRNA synthesis. Further, it was shown that TGF-β stimulation was associated with the polymerization of actin rather than alteration of GTPase or Ca2+ signaling that is common for other neutrophil mitogens [117]. In the context of cancer, TGF-β is an important factor that potently regulates the interaction between neutrophils and other cell populations within the tumor microenvironment. One of the predominant tumor suppressing functions for neutrophils in the tumor microenvironment involves the recognition and destruction of Fas ligand (FasL) expressing carcinoma cells. FasL expression is a significant component of tumor suppression that acts through initiating death signals in Fas receptor (CD95) expressing cells [118]. With regard to the mechanism, it has been shown that within a neutrophil, TGF-β inhibits the response to FasL via p38 MAPK [10]. Importantly, FasL expression promotes cell death in tumor infiltrating T-lymphocytes that could otherwise target and eliminate carcinoma cells [118]. However, in the presence of TGF-β neutrophils exhibit a decreased ability to eliminate FasL expressing cells and thereby foster a permissive microenvironment for tumor growth [10].
6. TGF-β regulates monocyte and macrophage activity
Monocytes and macrophages have been studied in great detail with regard to their contribution to tumor progression. In general, the recruitment of monocytes and macrophages to the tumor microenvironment has been associated with enhanced tumor progression. TGF-β promotes recruitment of monocytes and it has been suggested that TGF-β can promote monocyte to macrophage differentiation [119, 120] (Figure 5B). Early work suggested that in the case of macrophages that have the potential to be anti-tumorigenic, TGF-β inhibits the acquisition of effector functions [121]. In vitro, it was shown that TGF-β blocked both the priming by interferon-γ and triggering by lipopolysaccharide (LPS) of macrophages that were necessary for the ability of macrophages to efficiently kill tumor cells [121]. However, it was shown that the tumor necrosis factor associated cytotoxic ability of the macrophages did not depend upon macrophage response to TGF-β. Further, TGF-β did not have an impact on the cytostatic ability of activated macrophages upon the carcinoma cells. Interestingly, TGF-β pre-treatment of carcinoma cells attenuated both the cytotoxicity and cytostatic ability of macrophages in vitro [121]. In addition to direct effects upon the cytotoxic and cytostatic responses, TGF-β stimulation of macrophages has been shown to attenuate macrophage-associated suppression of CD4+ T-cell proliferation [96].
Importantly, many of the observed functional responses may be attributed to the regulation of gene expression in monocytes and macrophages that respond to TGF-β stimulation. In monocytes, TGF-β has been shown to promote expression of pro-inflammatory mediators including interleukin-1 (IL-1) and interleukin-6 (IL-6) while suppressing oxygen free radical production [120]. In macrophages, TGF-β has been shown to suppress MIP-1α, MIP-2, CXCL1, IL-1β, IL-8, GM-CSF and IL-10 expression [122, 123]. However production of other secreted factors, exemplified by tumor necrosis factor alpha (TNF-α) and MCP-1, may be alternatively regulated by TGF-β depending on the context of stimulation [120–125]. In addition, TGF-β is able to enhance the response to parallel chemotactic signals that are known to be abundant within the tumor microenvironment as illustrated in the case of SDF-1/CXCL12 ligand and CXCR4 receptor signaling [126]. It was shown that in monocytes and macrophages, stimulation with TGF-β could increase the expression of CXCR4 [126]. The enhanced receptor expression is likely significant since SDF-1, which is abundantly expressed by carcinoma associated fibroblasts, significantly contributes to tumor progression [127]. Together the results suggest that TGF-β response within monocytes and macrophages regulates their recruitment, differentiation, activation, gene expression profile and response to external stimulus that can significantly impact adjacent tumor progression.
7. Summary
The loss of TGF-β signaling in many common human carcinomas has been well documented [16]. Abundant literature dedicated to the impact of TGF-β signaling in non-transformed and carcinoma associated epithelium has focused primarily on the regulation of carcinoma cell behavior. Clearly the regulation of carcinoma cell survival, proliferation and epithelial to mesenchymal transition are significant factors that contribute to TGF-β mediated regulation of tumor progression. The current literature suggests that either gain or loss of TGF-β signaling in carcinoma epithelium can promote metastasis (Figure 3). During advanced stages of tumor progression many carcinoma cell populations find ways to circumvent the cytostatic and apoptotic responses attributed to TGF-β stimulation. This likely results in clonal expansion of carcinoma cells that are able to subvert these responses. In addition, TGF-β associated carcinoma cell EMT can be induced to promote migration and invasion while adjacent immune effectors are prevented from destroying the invasive carcinoma cells in response to TGF-β stimulation. However, it has now been shown that the loss of TGF-β signaling in carcinoma cells can have a profound impact on cross-talk with the adjacent tumor microenvironment including immune cell populations. When carcinoma cells lose the ability to respond to TGF-β they can upregulate pro-inflammatory cytokines including CXCL1 and CXCL5 that promote recruitment of tumor promoting MDSCs (Figure 4). MDSCs are known to be a significant source of TGF-β and the increased MDSC recruitment has been shown to correlate with increased focal TGF-β production in vivo. Importantly, the immature myeloid cells when present significantly contribute to tumor progression.
Despite the documented loss of TGF-β response by carcinoma cells, TGF-β is abundant in the tumor microenvironment. In early studies designed to investigate the role for TGF-β during tumor initiation, progression and metastasis two basic model systems have been widely utilized in vivo: 1) overexpression of an activated TGF-β1 ligand or 2) expression of a dominant negative receptor in tumor-associated epithelium. Importantly, the first approach (overexpression of a secreted bioactive TGF-β ligand) resulted in stimulation of all cell populations within the tumor microenvironment including carcinoma cells, adjacent fibroblasts, myofibroblasts, endothelial cells and infiltrating or resident immune cell populations. Importantly, TGF-β in the tumor microenvironment directly promotes immune suppression (Figure 5). TGF-β enhances motility and stimulates the recruitment of monocytes, macrophages, NK cells, dendritic cells and T-cells while directly inhibiting their anti-tumor effector functions [8, 9, 128, 129]. As a result, TGF-β associated inflammation can promote tumorigenesis due to secretion of growth-factors, cytokines, chemokines, proteases and extra-cellular matrix modifying enzymes from the recruited cell populations that stimulate carcinoma cell growth, motility and invasion. Together the current experimental evidence suggests, in addition to the direct impact of TGF-β upon the carcinoma cells, carcinoma-immune cell interactions regulated by TGF-β must be considered when designing relevant therapeutic approaches to manage human cancer progression and metastasis.
In recent years, it has become increasingly clear that cross-talk between cell populations within the tumor microenvironment can significantly regulate disease progression. TGF-β is able to significantly regulate the cross-talk between adjacent cell populations in the tumor microenvironment. However, many of the observations demonstrating regulation of paracrine signaling within each unique tumor-associated cell population were described in unique sub-populations and a different context of stimulation. In particular, the immature myeloid cells or mature derivatives such as neutrophils and macrophages have not been systematically stratified with response to their relative regulation by TGF-β. At present, it is not known how the regulation by TGF-β differs when comparing alternate immature myeloid subsets such as CD34+ CD33+ CD15−, CD14+ ARG+, CD11b+ CD14+, CD11b+ CD15+ CD14− or CD34+ CCR1+ cells that have been suggested as significant regulators of cancer progression. At present, it is not known how the responses in these immature cell populations compare with the mature neutrophil, monocyte or macrophage responses. Further, it is not known how primary normal or tumor-associated myeloid cells respond when compared to established immortal cell populations that were used for many of the current mechanistic studies. In addition, it is not clear if the location of the primary tumor has an impact on the TGF-β dependent regulation of myeloid cell recruitment or function responses in vivo. Clearly, significant progress has been made on all fronts of TGF-β regulation of tumor progression, however many essential questions remain. It is likely, that these interactions will be further explored and the carcinoma-immune cell interactions mediated by TGF-β will be delineated with increased fidelity. Clinically, it will be important to understand the details associated with the local and global carcinoma-immune cell interactions in order to design effective strategies for the management human cancer progression.
Biographies
Harold L. Moses, M.D. is the Hortense B. Ingram Professor of Molecular Oncology and Professor of Cancer Biology, Medicine and Pathology at Vanderbilt University School of Medicine. Dr. Moses was the founding director of the Vanderbilt-Ingram Cancer Center, which he led for twelve years; he is now director emeritus. Trained as a pathologist, Dr. Moses has devoted much of his career to basic research on growth factors and tumor suppressor genes, particularly in the TGF-β pathway, and has received many awards for his research. He has served as president of the American Association for Cancer Research, president of the Association of American Cancer Institutes, chair of the NIH Chemical Pathology Study Section, chair of the Molecular Oncogenesis Study Section, and chair of the National Cancer Institute Cancer Centers review panel. He is a member of the Institute of Medicine of the National Academies and currently chairs the National Cancer Policy Forum of the Institute of Medicine. He also currently co-chairs the Program Steering Committee for the NCI’s Tumor Microenvironment Network.

Brian Bierie, Ph.D. performed his undergraduate work at Colorado State University in his home town of Fort Collins, Colorado. After finishing his undergraduate work Brian spent time working in the area of analytical chemistry for a large pesticide production plant. During this time Brian realized that some of the mutagens were so abundant in the soil and waste water produced by the production facilities that he began to think about dedicating his time to studies that could help people rather than contributing to the production of industrial mutagenic compounds. As a result, Brian switched to the study mammary gland biology and neoplasia mentored by Dr. Lothar Hennighausen, Ph.D. at the NIH in Bethesda, Maryland. After a three year tour at NIH Brian initiated his Ph.D. studies mentored by Dr. Harold L. Moses, M.D. at Vanderbilt University in Nashville, Tennessee. Brian obtained his PhD in Cancer Biology and is now working as a postdoc mentored by Dr. Robert Weinberg, Ph.D. at the Whitehead Institute for Biomedical Research in Boston, Massachusetts.

Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kehrl JH, et al. Production of transforming growth factor beta by human T lymphocytes and its potential role in the regulation of T cell growth. J Exp Med. 1986;163(5):1037–1050. doi: 10.1084/jem.163.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kehrl JH, et al. Transforming growth factor beta is an important immunomodulatory protein for human B lymphocytes. J Immunol. 1986;137(12):3855–3860. [PubMed] [Google Scholar]
- 3.Rook AH, et al. Effects of transforming growth factor beta on the functions of natural killer cells: depressed cytolytic activity and blunting of interferon responsiveness. J Immunol. 1986;136(10):3916–3920. [PubMed] [Google Scholar]
- 4.Fontana A, et al. Expression of TGF-beta 2 in human glioblastoma: a role in resistance to immune rejection? Ciba Found Symp. 1991;157:232–238. doi: 10.1002/9780470514061.ch15. discussion 238-41. [DOI] [PubMed] [Google Scholar]
- 5.Mule JJ, et al. Transforming growth factor-beta inhibits the in vitro generation of lymphokine-activated killer cells and cytotoxic T cells. Cancer Immunol Immunother. 1988;26(2):95–100. doi: 10.1007/BF00205600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6(1):24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 8.Li MO, et al. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 9.Wrzesinski SH, Wan YY, Flavell RA. Transforming growth factor-beta and the immune response: implications for anticancer therapy. Clin Cancer Res. 2007;13(18 Pt 1):5262–5270. doi: 10.1158/1078-0432.CCR-07-1157. [DOI] [PubMed] [Google Scholar]
- 10.Chen JJ, Sun Y, Nabel GJ. Regulation of the proinflammatory effects of Fas ligand (CD95L) Science. 1998;282(5394):1714–1717. doi: 10.1126/science.282.5394.1714. [DOI] [PubMed] [Google Scholar]
- 11.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6(7):506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 12.Massague J. TGFbeta in Cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derynck R, et al. Synthesis of messenger RNAs for transforming growth factors alpha and beta and the epidermal growth factor receptor by human tumors. Cancer Res. 1987;47(3):707–712. [PubMed] [Google Scholar]
- 14.Dickson RB, et al. Activation of growth factor secretion in tumorigenic states of breast cancer induced by 17 beta-estradiol or v-Ha-ras oncogene. Proc Natl Acad Sci U S A. 1987;84(3):837–841. doi: 10.1073/pnas.84.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wojtowicz-Praga S. Reversal of tumor-induced immunosuppression by TGF-beta inhibitors. Invest New Drugs. 2003;21(1):21–32. doi: 10.1023/a:1022951824806. [DOI] [PubMed] [Google Scholar]
- 16.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17(1–2):41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Stover DG, Bierie B, Moses HL. A delicate balance: TGF-beta and the tumor microenvironment. J Cell Biochem. 2007 doi: 10.1002/jcb.21149. [DOI] [PubMed] [Google Scholar]
- 18.Abe M, Oda N, Sato Y. Cell-associated activation of latent transforming growth factor-beta by calpain. J Cell Physiol. 1998;174(2):186–193. doi: 10.1002/(SICI)1097-4652(199802)174:2<186::AID-JCP6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 19.Munger JS, et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96(3):319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 20.Lyons RM, Keski-Oja J, Moses HL. Proteolytic activation of latent transforming growth factor-beta from fibroblast-conditioned medium. J Cell Biol. 1988;106(5):1659–1665. doi: 10.1083/jcb.106.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akita K, et al. Impaired liver regeneration in mice by lipopolysaccharide via TNF-alpha/kallikrein-mediated activation of latent TGF-beta. Gastroenterology. 2002;123(1):352–364. doi: 10.1053/gast.2002.34234. [DOI] [PubMed] [Google Scholar]
- 22.Taipale J, et al. Human mast cell chymase and leukocyte elastase release latent transforming growth factor-beta 1 from the extracellular matrix of cultured human epithelial and endothelial cells. J Biol Chem. 1995;270(9):4689–4696. doi: 10.1074/jbc.270.9.4689. [DOI] [PubMed] [Google Scholar]
- 23.Miyazono K, Heldin CH. Role for carbohydrate structures in TGF-beta 1 latency. Nature. 1989;338(6211):158–160. doi: 10.1038/338158a0. [DOI] [PubMed] [Google Scholar]
- 24.Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14(2):163–176. [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz-Cherry S, Hinshaw VS. Influenza virus neuraminidase activates latent transforming growth factor beta. J Virol. 1996;70(12):8624–8629. doi: 10.1128/jvi.70.12.8624-8629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz-Cherry S, Murphy-Ullrich JE. Thrombospondin causes activation of latent transforming growth factor-beta secreted by endothelial cells by a novel mechanism. J Cell Biol. 1993;122(4):923–932. doi: 10.1083/jcb.122.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barcellos-Hoff MH, Dix TA. Redox-mediated activation of latent transforming growth factor-beta 1. Mol Endocrinol. 1996;10(9):1077–1083. doi: 10.1210/mend.10.9.8885242. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 29.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 30.Lee MK, et al. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. Embo J. 2007;26(17):3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng XH, Derynck R. Specificity and Versatility in TGF- Signaling Through Smads. Annu Rev Cell Dev Biol. 2005 doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 32.Daly AC, Randall RA, Hill CS. Transforming growth factor beta-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Mol Cell Biol. 2008;28(22):6889–6902. doi: 10.1128/MCB.01192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98(10):1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita M, et al. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell. 2008;31(6):918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17(1–2):29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Zeng Q, et al. Tgfbr1 haploinsufficiency is a potent modifier of colorectal cancer development. Cancer Res. 2009;69(2):678–686. doi: 10.1158/0008-5472.CAN-08-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valle L, et al. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science. 2008;321(5894):1361–1365. doi: 10.1126/science.1159397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101–128. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 39.Kim BG, et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006;441(7096):1015–1019. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- 40.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 41.Kang SH, et al. Transcriptional repression of the transforming growth factor-beta type I receptor gene by DNA methylation results in the development of TGF-beta resistance in human gastric cancer. Oncogene. 1999;18(51):7280–7286. doi: 10.1038/sj.onc.1203146. [DOI] [PubMed] [Google Scholar]
- 42.Kim SJ, et al. Molecular mechanisms of inactivation of TGF-beta receptors during carcinogenesis. Cytokine Growth Factor Rev. 2000;11(1–2):159–168. doi: 10.1016/s1359-6101(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 43.Rojas A, et al. The aberrant methylation of TSP1 suppresses TGF-beta1 activation in colorectal cancer. Int J Cancer. 2008;123(1):14–21. doi: 10.1002/ijc.23608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aitchison AA, et al. Promoter methylation correlates with reduced Smad4 expression in advanced prostate cancer. Prostate. (6) 2008;68:661–674. doi: 10.1002/pros.20730. [DOI] [PubMed] [Google Scholar]
- 45.Hinshelwood RA, et al. Concordant epigenetic silencing of transforming growth factor-beta signaling pathway genes occurs early in breast carcinogenesis. Cancer Res. 2007;67(24):11517–11527. doi: 10.1158/0008-5472.CAN-07-1284. [DOI] [PubMed] [Google Scholar]
- 46.Nakao A, et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389(6651):631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi H, et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89(7):1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 48.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6(6):1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 49.Ebisawa T, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276(16):12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 50.Inoue Y, Imamura T. Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci. 2008;99(11):2107–2112. doi: 10.1111/j.1349-7006.2008.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi W, et al. GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta type I receptor. J Cell Biol. 2004;164(2):291–300. doi: 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Datta PK, Moses HL. STRAP and Smad7 synergize in the inhibition of transforming growth factor beta signaling. Mol Cell Biol. 2000;20(9):3157–3167. doi: 10.1128/mcb.20.9.3157-3167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferrigno O, et al. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene. 2002;21(32):4879–4884. doi: 10.1038/sj.onc.1205623. [DOI] [PubMed] [Google Scholar]
- 54.Stroschein SL, et al. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science. 1999;286(5440):771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 55.Luo K, et al. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 1999;13(17):2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3(11):807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 57.Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21(2):166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 58.Daniel CW, Robinson SD. Regulation of mammary growth and function by TGF-beta. Mol Reprod Dev. 1992;32(2):145–151. doi: 10.1002/mrd.1080320210. [DOI] [PubMed] [Google Scholar]
- 59.Daniel CW, et al. TGF-beta 1-induced inhibition of mouse mammary ductal growth: developmental specificity and characterization. Dev Biol. 1989;135(1):20–30. doi: 10.1016/0012-1606(89)90154-1. [DOI] [PubMed] [Google Scholar]
- 60.Silberstein GB, Daniel CW. Reversible inhibition of mammary gland growth by transforming growth factor-beta. Science. 1987;237(4812):291–293. doi: 10.1126/science.3474783. [DOI] [PubMed] [Google Scholar]
- 61.Pierce DF, Jr, et al. Inhibition of mammary duct development but not alveolar outgrowth during pregnancy in transgenic mice expressing active TGF-beta 1. Genes Dev. 1993;7(12A):2308–2317. doi: 10.1101/gad.7.12a.2308. [DOI] [PubMed] [Google Scholar]
- 62.Boulanger CA, Wagner KU, Smith GH. Parity-induced mouse mammary epithelial cells are pluripotent, self-renewing and sensitive to TGF-beta1 expression. Oncogene. 2005;24(4):552–560. doi: 10.1038/sj.onc.1208185. [DOI] [PubMed] [Google Scholar]
- 63.Jhappan C, et al. Targeting expression of a transforming growth factor beta 1 transgene to the pregnant mammary gland inhibits alveolar development and lactation. Embo J. 1993;12(5):1835–1845. doi: 10.1002/j.1460-2075.1993.tb05832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kordon EC, et al. Ectopic TGF beta 1 expression in the secretory mammary epithelium induces early senescence of the epithelial stem cell population. Dev Biol. 1995;168(1):47–61. doi: 10.1006/dbio.1995.1060. [DOI] [PubMed] [Google Scholar]
- 65.Ewan KB, et al. Latent transforming growth factor-beta activation in mammary gland: regulation by ovarian hormones affects ductal and alveolar proliferation. Am J Pathol. 2002;160(6):2081–2093. doi: 10.1016/s0002-9440(10)61158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gorska AE, et al. Transgenic mice expressing a dominant-negative mutant type II transforming growth factor-beta receptor exhibit impaired mammary development and enhanced mammary tumor formation. Am J Pathol. 2003;163(4):1539–1549. doi: 10.1016/s0002-9440(10)63510-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lenferink AE, et al. Expression of TGF-beta type II receptor antisense RNA impairs TGF-beta signaling in vitro and promotes mammary gland differentiation in vivo. Int J Cancer. 2003;107(6):919–928. doi: 10.1002/ijc.11494. [DOI] [PubMed] [Google Scholar]
- 68.Pierce DF, Jr, et al. Mammary tumor suppression by transforming growth factor beta 1 transgene expression. Proc Natl Acad Sci U S A. 1995;92(10):4254–4258. doi: 10.1073/pnas.92.10.4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boulanger CA, Smith GH. Reducing mammary cancer risk through premature stem cell senescence. Oncogene. 2001;20(18):2264–2272. doi: 10.1038/sj.onc.1204312. [DOI] [PubMed] [Google Scholar]
- 70.Siegel PM, et al. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc Natl Acad Sci U S A. 2003;100(14):8430–8435. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muraoka-Cook RS, et al. Conditional overexpression of active transforming growth factor beta1 in vivo accelerates metastases of transgenic mammary tumors. Cancer Res. 2004;64(24):9002–9011. doi: 10.1158/0008-5472.CAN-04-2111. [DOI] [PubMed] [Google Scholar]
- 72.Muraoka-Cook RS, et al. Activated type I TGFbeta receptor kinase enhances the survival of mammary epithelial cells and accelerates tumor progression. Oncogene. 2006;25(24):3408–3423. doi: 10.1038/sj.onc.1208964. [DOI] [PubMed] [Google Scholar]
- 73.Yang YA, et al. Lifetime exposure to a soluble TGF-beta antagonist protects mice against metastasis without adverse side effects. J Clin Invest. 2002;109(12):1607–1615. doi: 10.1172/JCI15333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Forrester E, et al. Effect of conditional knockout of the type II TGF-beta receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer Res. 2005;65(6):2296–2302. doi: 10.1158/0008-5472.CAN-04-3272. [DOI] [PubMed] [Google Scholar]
- 75.Bierie B, et al. Transforming growth factor-beta regulates mammary carcinoma cell survival and interaction with the adjacent microenvironment. Cancer Res. 2008;68(6):1809–1819. doi: 10.1158/0008-5472.CAN-07-5597. [DOI] [PubMed] [Google Scholar]
- 76.Yang L, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13(1):23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bierie B, et al. Abrogation of TGF-beta signaling enhances chemokine production and correlates with prognosis in human breast cancer. J Clin Invest. 2009;119(6):1571–1582. doi: 10.1172/JCI37480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Padua D, et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133(1):66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marigo I, et al. Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol Rev. 2008;222:162–179. doi: 10.1111/j.1600-065X.2008.00602.x. [DOI] [PubMed] [Google Scholar]
- 80.Gallina G, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J Clin Invest. 2006;116(10):2777–2790. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sinha P, et al. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J Immunol. 2007;179(2):977–983. doi: 10.4049/jimmunol.179.2.977. [DOI] [PubMed] [Google Scholar]
- 82.Sica A, Bronte V. Altered macrophage differentiation and immune dysfunction in tumor development. J Clin Invest. 2007;117(5):1155–1166. doi: 10.1172/JCI31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kitamura T, et al. SMAD4-deficient intestinal tumors recruit CCR1+ myeloid cells that promote invasion. Nat Genet. 2007;39(4):467–475. doi: 10.1038/ng1997. [DOI] [PubMed] [Google Scholar]
- 84.Almand B, et al. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J Immunol. 2001;166(1):678–689. doi: 10.4049/jimmunol.166.1.678. [DOI] [PubMed] [Google Scholar]
- 85.Serafini P, et al. Phosphodiesterase-5 inhibition augments endogenous antitumor immunity by reducing myeloid-derived suppressor cell function. J Exp Med. 2006;203(12):2691–2702. doi: 10.1084/jem.20061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Filipazzi P, et al. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol. 2007;25(18):2546–2553. doi: 10.1200/JCO.2006.08.5829. [DOI] [PubMed] [Google Scholar]
- 87.Zea AH, et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 2005;65(8):3044–3048. doi: 10.1158/0008-5472.CAN-04-4505. [DOI] [PubMed] [Google Scholar]
- 88.Kulkarni AB, et al. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci U S A. 1993;90(2):770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shull MM, et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359(6397):693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gorelik L, Flavell RA. Abrogation of TGFbeta signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 2000;12(2):171–181. doi: 10.1016/s1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- 91.McKarns SC, Schwartz RH, Kaminski NE. Smad3 is essential for TGF-beta 1 to suppress IL-2 production and TCR-induced proliferation, but not IL-2-induced proliferation. J Immunol. 2004;172(7):4275–4284. doi: 10.4049/jimmunol.172.7.4275. [DOI] [PubMed] [Google Scholar]
- 92.Ruegemer JJ, et al. Regulatory effects of transforming growth factor-beta on IL-2- and IL-4-dependent T cell-cycle progression. J Immunol. 1990;144(5):1767–1776. [PubMed] [Google Scholar]
- 93.Wolfraim LA, et al. p21Cip1 and p27Kip1 act in synergy to alter the sensitivity of naive T cells to TGF-beta-mediated G1 arrest through modulation of IL-2 responsiveness. J Immunol. 2004;173(5):3093–3102. doi: 10.4049/jimmunol.173.5.3093. [DOI] [PubMed] [Google Scholar]
- 94.Gorelik L, Flavell RA. Transforming growth factor-beta in T-cell biology. Nat Rev Immunol. 2002;2(1):46–53. doi: 10.1038/nri704. [DOI] [PubMed] [Google Scholar]
- 95.Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-beta signaling in T cells. Nat Med. 2001;7(10):1118–1122. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- 96.Alleva DG, Walker TM, Elgert KD. Induction of macrophage suppressor activity by fibrosarcoma-derived transforming growth factor-beta 1: contrasting effects on resting and activated macrophages. J Leukoc Biol. 1995;57(6):919–928. doi: 10.1002/jlb.57.6.919. [DOI] [PubMed] [Google Scholar]
- 97.Chen ML, et al. Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-beta signals in vivo. Proc Natl Acad Sci U S A. 2005;102(2):419–424. doi: 10.1073/pnas.0408197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Q, et al. Adoptive transfer of tumor-reactive transforming growth factor-betainsensitive CD8+ T cells: eradication of autologous mouse prostate cancer. Cancer Res. 2005;65(5):1761–1769. doi: 10.1158/0008-5472.CAN-04-3169. [DOI] [PubMed] [Google Scholar]
- 99.Park JA, et al. Expression of an antisense transforming growth factor-beta1 transgene reduces tumorigenicity of EMT6 mammary tumor cells. Cancer Gene Ther. 1997;4(1):42–50. [PubMed] [Google Scholar]
- 100.Mukherjee P, et al. MUC1-specific CTLs are non-functional within a pancreatic tumor microenvironment. Glycoconj J. 2001;18(11–12):931–942. doi: 10.1023/a:1022260711583. [DOI] [PubMed] [Google Scholar]
- 101.Ranges GE, et al. Inhibition of cytotoxic T cell development by transforming growth factor beta and reversal by recombinant tumor necrosis factor alpha. J Exp Med. 1987;166(4):991–998. doi: 10.1084/jem.166.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ahmadzadeh M, Rosenberg SA. TGF-beta 1 attenuates the acquisition and expression of effector function by tumor antigen-specific human memory CD8 T cells. J Immunol. 2005;174(9):5215–5223. doi: 10.4049/jimmunol.174.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smyth MJ, et al. Regulation of lymphokine-activated killer activity and pore-forming protein gene expression in human peripheral blood CD8+ T lymphocytes. Inhibition by transforming growth factor-beta. J Immunol. 1991;146(10):3289–3697. [PubMed] [Google Scholar]
- 104.Thomas DA, Massague J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8(5):369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 105.Bonig H, et al. Transforming growth factor-beta1 suppresses interleukin-15-mediated interferon-gamma production in human T lymphocytes. Scand J Immunol. 1999;50(6):612–618. doi: 10.1046/j.1365-3083.1999.00635.x. [DOI] [PubMed] [Google Scholar]
- 106.Kontani K, et al. Spontaneous elicitation of potent antitumor immunity and eradication of established tumors by administration of DNA encoding soluble transforming growth factor-beta II receptor without active antigen-sensitization. Cancer Immunol Immunother. 2005:1–9. doi: 10.1007/s00262-005-0044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Arteaga CL, et al. Anti-transforming growth factor (TGF)-beta antibodies inhibit breast cancer cell tumorigenicity and increase mouse spleen natural killer cell activity. Implications for a possible role of tumor cell/host TGF-beta interactions in human breast cancer progression. J Clin Invest. 1993;92(6):2569–2576. doi: 10.1172/JCI116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arteaga CL, et al. Reversal of tamoxifen resistance of human breast carcinomas in vivo by neutralizing antibodies to transforming growth factor-beta. J Natl Cancer Inst. 1999;91(1):46–53. doi: 10.1093/jnci/91.1.46. [DOI] [PubMed] [Google Scholar]
- 109.Ma D, Niederkorn JY. Transforming growth factor-beta down-regulates major histocompatibility complex class I antigen expression and increases the susceptibility of uveal melanoma cells to natural killer cell-mediated cytolysis. Immunology. 1995;86(2):263–269. [PMC free article] [PubMed] [Google Scholar]
- 110.Lee YJ, et al. TGF-beta suppresses IFN-gamma induction of class II MHC gene expression by inhibiting class II transactivator messenger RNA expression. J Immunol. 1997;158(5):2065–2075. [PubMed] [Google Scholar]
- 111.Geiser AG, et al. Transforming growth factor beta 1 (TGF-beta 1) controls expression of major histocompatibility genes in the postnatal mouse: aberrant histocompatibility antigen expression in the pathogenesis of the TGF-beta 1 null mouse phenotype. Proc Natl Acad Sci U S A. 1993;90(21):9944–9948. doi: 10.1073/pnas.90.21.9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Johns LD, et al. Transforming growth factor-beta 1 differentially regulates proliferation and MHC class-II antigen expression in forebrain and brainstem astrocyte primary cultures. Brain Res. 1992;585(1–2):229–236. doi: 10.1016/0006-8993(92)91211-v. [DOI] [PubMed] [Google Scholar]
- 113.Ghiringhelli F, et al. CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-beta-dependent manner. J Exp Med. 2005;202(8):1075–1085. doi: 10.1084/jem.20051511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smyth MJ, et al. CD4+CD25+ T Regulatory Cells Suppress NK Cell-Mediated Immunotherapy of Cancer. J Immunol. 2006;176(3):1582–1587. doi: 10.4049/jimmunol.176.3.1582. [DOI] [PubMed] [Google Scholar]
- 115.Castriconi R, et al. Transforming growth factor beta 1 inhibits expression of NKp30 and NKG2D receptors: consequences for the NK-mediated killing of dendritic cells. Proc Natl Acad Sci U S A. 2003;100(7):4120–4125. doi: 10.1073/pnas.0730640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee JC, et al. Elevated TGF-beta1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J Immunol. 2004;172(12):7335–7340. doi: 10.4049/jimmunol.172.12.7335. [DOI] [PubMed] [Google Scholar]
- 117.Reibman J, et al. Transforming growth factor beta 1, a potent chemoattractant for human neutrophils, bypasses classic signal-transduction pathways. Proc Natl Acad Sci U S A. 1991;88(15):6805–6809. doi: 10.1073/pnas.88.15.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hahne M, et al. Melanoma cell expression of Fas(Apo-1/CD95) ligand: implications for tumor immune escape. Science. 1996;274(5291):1363–1366. doi: 10.1126/science.274.5291.1363. [DOI] [PubMed] [Google Scholar]
- 119.Bombara C, Ignotz RA. TGF-beta inhibits proliferation of and promotes differentiation of human promonocytic leukemia cells. J Cell Physiol. 1992;153(1):30–37. doi: 10.1002/jcp.1041530106. [DOI] [PubMed] [Google Scholar]
- 120.Fontana A, et al. Modulation of the immune response by transforming growth factor beta. Int Arch Allergy Immunol. 1992;99(1):1–7. doi: 10.1159/000236328. [DOI] [PubMed] [Google Scholar]
- 121.Haak-Frendscho M, et al. Transforming growth factor-beta 1 inhibits activation of macrophage cell line RAW 264.7 for cell killing. Clin Exp Immunol. 1990;82(2):404–410. doi: 10.1111/j.1365-2249.1990.tb05461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McDonald PP, et al. Transcriptional and translational regulation of inflammatory mediator production by endogenous TGF-beta in macrophages that have ingested apoptotic cells. J Immunol. 1999;163(11):6164–6172. [PubMed] [Google Scholar]
- 123.Fadok VA, et al. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest. 1998;101(4):890–898. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bogdan C, Nathan C. Modulation of macrophage function by transforming growth factor beta, interleukin-4, and interleukin-10. Ann N Y Acad Sci. 1993;685:713–739. doi: 10.1111/j.1749-6632.1993.tb35934.x. [DOI] [PubMed] [Google Scholar]
- 125.Feinberg MW, et al. Essential role for Smad3 in regulating MCP-1 expression and vascular inflammation. Circ Res. 2004;94(5):601–608. doi: 10.1161/01.RES.0000119170.70818.4F. [DOI] [PubMed] [Google Scholar]
- 126.Wang J, et al. Role of tyrosine phosphorylation in ligand-independent sequestration of CXCR4 in human primary monocytes-macrophages. J Biol Chem. 2001;276(52):49236–49243. doi: 10.1074/jbc.M108523200. [DOI] [PubMed] [Google Scholar]
- 127.Orimo A, et al. Stromal Fibroblasts Present in Invasive Human Breast Carcinomas Promote Tumor Growth and Angiogenesis through Elevated SDF-1/CXCL12 Secretion. Cell. 2005;121(3):335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 128.Gruber BL, Marchese MJ, Kew RR. Transforming growth factor-beta 1 mediates mast cell chemotaxis. J Immunol. 1994;152(12):5860–5867. [PubMed] [Google Scholar]
- 129.Maghazachi AA, al-Aoukaty A. Transforming growth factor-beta 1 is chemotactic for interleukin-2-activated natural killer cells. Nat Immun. 1993;12(2):57–65. [PubMed] [Google Scholar]