Abstract
Defects in apoptosis are not only a hallmark of cancer initiation and progression but can also underlie the development of chemoresistance. How the tightly regulated cascade of protein-protein interactions between members of three competing protein families regulating the apoptotic cascade is subverted in tumor cells is incompletely understood. Here, we show that KLF6-SV1, whose overexpression is associated with poor survival in several different cancers and is an alternatively spliced isoform of the Krüppel-like tumor suppressor KLF6, is a critical prosurvival/antiapoptotic protein. KLF6-SV1 binds the proapoptotic BH3-only protein NOXA, which results in their mutual HDM2-dependent degradation. In turn, this increases the intracellular concentration of the prosurvival binding partner of NOXA, Mcl-1, and effectively blocks apoptosis. In an ovarian cancer model, systemically delivered small interfering RNA against KLF6-SV1 induces spontaneous apoptosis of tumor cells, decreases tumor burden, and restores cisplatin sensitivity in vivo. Moreover, i.p. delivery of siKLF6-SV1 RNA halts ovarian tumor progression and improves median and overall survival (progression-free for >15 months; P < 0.0002) in mice in a dose-dependent manner. Thus, KLF6-SV1 represents a novel regulator of protein interactions in the apoptotic cascade and a therapeutically targetable control point.
Introduction
Apoptosis is an evolutionary conserved program in diverse biological systems (1) and an important mediator of the cytotoxic action of chemotherapeutic agents (2). The developmental and physiologic cues that trigger programmed cell death are controlled by specific competing protein-protein interactions between members of three protein families, two acting to promote cell death and the third to block this effect (3). So critical is this control that defects in apoptosis result in several pathologic disorders and are considered a hallmark of cancer initiation, progression, and metastasis (4). Overcoming these defects and exploiting selective interactions within the apoptotic pathway therefore represent an appealing therapeutic opportunity (5). For example, mimetics targeting the proapoptotic, BH3-only protein family, which directly respond to cytotoxic stresses, would represent a desirable strategy owing to the ability of the family members to act independently of the p53 status of a tumor cell (6, 7), specificity of Bcl-2 family member interaction (8), restricted activity in a particular tumor type (9), and potential as an adjuvant treatment in conventional chemotherapy (10).
Ovarian cancer is the fifth most common form of cancer in women in the United States, and with an estimated 15,520 deaths from among 21,650 newly diagnosed cases, it is the most lethal of all gynecologic cancers (11). Most patients present with advanced-stage disease, and although initially responsive to platinum-based chemotherapy, the majority will succumb to recurrence and chemoresistance (12). Recently, decreased levels of the tumor suppressor KLF6 and increased levels of its alternatively spliced isoform KLF6-SV1 have been linked to ovarian cancer progression and chemoresistance (13, 14). Intriguingly, although its function is unknown, KLF6-SV1 was originally identified and its overexpression is linked to a single nucleotide polymorphism associated with an increased lifetime risk of prostate cancer (15–18). Although present in both normal and cancerous cells, expression of this cytoplasmic isoform is significantly up-regulated in multiple cancers (13, 15, 17, 19) and its overexpression is associated with decreased survival in prostate and lung cancers (20, 21).
Given the known overexpression of KLF6-SV1 in ovarian tumors and the critical limitations associated with ovarian cancer treatment and recurrence, we directly investigated its function and potential therapeutic value. Here, we show that systemic administration of chemically modified KLF6-SV1 small interfering RNA (siRNA) molecules results in long-term silencing in tumor cells, restores cisplatin sensitivity to increase apoptosis, and in a dose-dependent manner provides long-term survival in mice harboring disseminated i.p. ovarian cancer. In accord with previous hypotheses suggesting the potential therapeutic opportunity in targeting BH3-only family members, we show that KLF6-SV1 is a prosurvival/antiapoptotic molecule that directly interacts with and regulates NOXA, targeting them both for HDM2-mediated degradation.
Materials and Methods
Animal models
For the i.p. model of ovarian cancer dissemination, 6- to 8-wk-old female BALB/c nu/nu mice were injected with 1 × 107 SKOV3-Luc cells (a kind gift from Achim Aigner, Philipps-University School of Medicine, Marburg, Germany) and whole-body bioluminescence was measured biweekly until mice were euthanized at day 50 (Fig. 1) or until the mice became moribund and displayed features of distress (Fig. 2). On sacrificing the mice, tumors as well as any ascitic fluid were harvested. All animal work and protocols were approved by the Mount Sinai School of Medicine Institutional Animal Care and Use Committee.
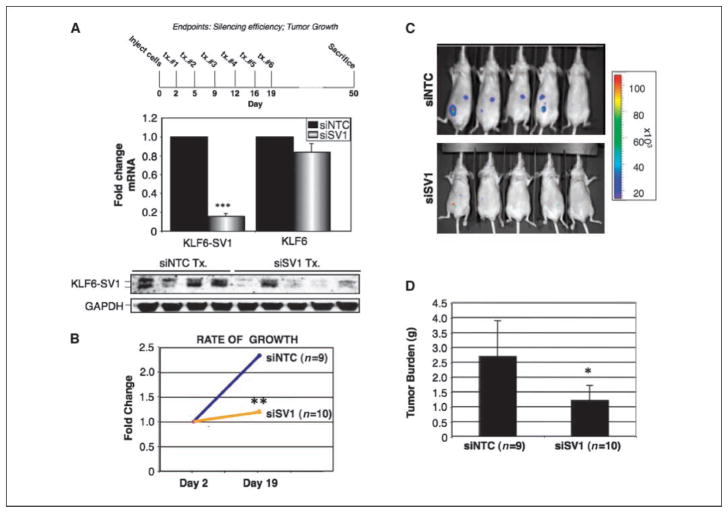
Figure 1.
In vivo antitumor effects of KLF6-SV1 inhibition. A, top, treatment regimen; middle, quantitative real-time PCR for KLF6-SV1 and KLF6 expression levels in i.p. tumors; bottom, Western blot analysis of KLF6-SV1 protein levels in i.p. tumors after treatment with siNTC or siSV1. B, rate of growth of tumors treated with siNTC or siSV1, measured by in vivo molecular imaging. C, total bioluminescent signal from the abdominal region of treated mice at day 19 after the final dose of siSV1 or siNTC. D, total tumor mass. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
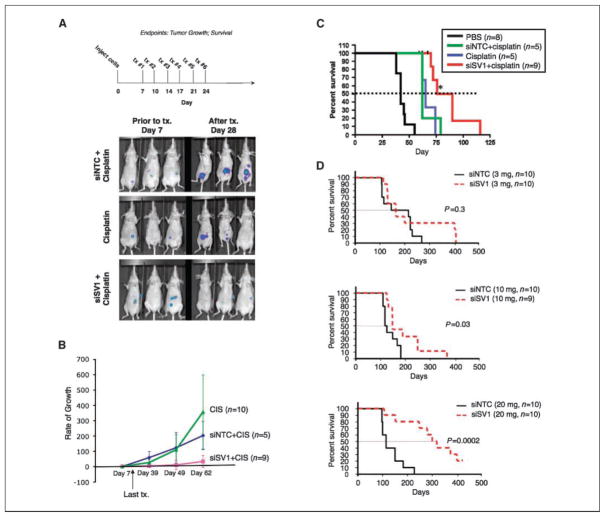
Figure 2.
KLF6-SV1 inhibition increases survival in mice bearing i.p. tumors. A, top, treatment regimen; bottom, in vivo whole-body bioluminescence imaging of a subset of mice before (day 7) and after treatment (day 28) with either siNTC (3 mg/kg) plus cisplatin (5 mg/kg), siSV1 (3 mg/kg) plus cisplatin (5 mg/kg), or cisplatin (5 mg/kg) alone. B, rate of tumor growth of all three groups (cisplatin, n = 10; siNTC + cisplatin, n = 5; siSV1 + cisplatin, n = 9) using whole-body bioluminescence imaging values compared with the initial bioluminescent signal before treatment (day 7). C, Kaplan-Meier survival curves comparing PBS-treated (n = 8), siNTC plus cisplatin–treated (n = 5), siSV1 plus cisplatin–treated (n = 9), or cisplatin alone–treated (n = 5) mice. D, Kaplan-Meier survival curves comparing siNTC and siSV1 at escalating doses. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
In vivo bioluminescent imaging
Bioluminescent imaging was performed with an IVIS-200 Imaging System (Xenogen). Ten to 15 min before in vivo imaging, animals received the substrate D-luciferin (Biosynth) at 150 mg/kg in PBS by i.p. injection and were anesthetized using 1% to 3% isoflurane (Abbott Laboratories). Imaging times ranged from 30 s to 20 min depending on the intensity of tumor nodules. The light emitted from the abdominal area of each mouse was quantified as photon/second using the Living Image software (Xenogen).
Flow cytometry
Following treatment with siNTC, siSV1, or cisplatin, cells were collected by trypsinization (Life Technologies) and centrifuged at 1,000 rpm for 5 min. The cell pellet was resuspended in 200 μL PBS, fixed in 70% ethanol, and then stored at 4°C overnight before analysis. Fixed cells were centrifuged at 1,000 rpm for 5 min and resuspended in 1 mL PBS containing 5 μg/mL propidium iodide (Boehringer Mannheim) and 50 μL of 10 mg/mL RNase (Roche). Cells were analyzed on FACSort flow cytometer (BD Biosciences) at 488-nm absorbance using CellQuest software. Propidium iodide staining was used for measuring the distribution of cells within the cell cycle. Sub-G1 peaks on DNA histograms from hypodiploid DNA represent dead cells, which may indicate apoptosis but may also contain some necrotic cells.
Western blot analysis
Cell extracts for Western blotting were harvested in radioimmunoprecipitation assay buffer (Santa Cruz Biotechnology standard protocol). Tumor tissue extracts were harvested and prepared in the T-PER reagent (Pierce). Equal amounts of protein (50 μg) as determined by the Bio-Rad detergent-compatible protein quantification assay were loaded and separated by PAGE and transferred to nitrocellulose membranes. Western blotting was performed using antibodies for actin and KLF6 (Santa Cruz Biotechnology), caspase-3 and PUMA (Cell Signaling), caspase-8 and NOXA (Calbiochem), Flag M2 (Sigma), V5 (Invitrogen), and BAX (Upstate).
siRNA-mediated inhibition
For the in vitro studies, all cell lines were seeded in 12-well culture plates at 60% to 70% confluence. The next day, Lipofectamine 2000 (Invitrogen) was used to transfect cells with siSTABLE-A4 siSV1 (5′-CAGGGAAGGAGAAAAGCCUUU) or siNTC (inverted β-galactosidase sequence; 5′-UAGCGACUAAACACAUCAAUU) at a final siRNA concentration of 100 nmol/L. For the in vivo study, Accell (Dharmacon) chemically synthesized siRNAs to either siSV1 (5′-CAGGGAAGGAGAAAAGCCUUU) or siNTC (inverted β-galactosidase sequence; 5′-UAGCGACUAAACACAUCAAUU) were injected i.p. at prespecified time points and concentrations as outlined in Figs. 1 and 2.
Statistical analyses
We conducted statistical analysis by ANOVA with the Student’s t test. A P value of ≤0.05 was considered significant. Kaplan-Meier survival curves and log-rank test were performed using GraphPad Prism. To confirm that all our treatment studies obtained sufficient power, we performed post hoc power analysis using G*Power software.
Results
In vivo inhibition of KLF6-SV1 prolongs survival
To investigate the potential in vivo antitumor effects of systemically delivered KLF6-SV1 siRNA, we used a well-characterized model of disseminated ovarian cancer (22) coupled with a novel chemically modified siRNA (Accell). SKOV3 cells stably expressing luciferase (SKOV3-Luc) were implanted into the peritoneal cavity of nude mice and imaged after 48 hours to confirm establishment of disseminated tumors. Three distinct sets of experiments were defined. As a test of the efficacy and stability of the siRNA, we first administered by peritoneal injection a relatively low dose of siRNA (3 mg/kg) targeted against either a control (siNTC) or KLF6-SV1 (siSV1) every third day for a total of six doses (Fig. 1A). All mice were sacrificed 1 month following the last dose (day 50) and i.p. tumor nodules were removed for analysis. siSV1 inhibition remained highly effective (~80% knockdown) and specific for KLF6-SV1 even 30 days following the last treatment both on the RNA and protein level (Fig. 1A). Even at this low dose (3 mg/kg) of siSV1, tumor growth was decreased ~2.5-fold in siSV1 mice (Fig. 1B–D).
Given that a recent study has reported that nonspecific stimulation of the TLR3 pathway may be an unintended consequence of siRNA dosing, we explored whether the effect of siSV1 on tumor burden was not a result of a similar cytokine induction (23). No significant differences in serum levels of IFNγ, tumor necrosis factor (TNF) α, or interleukin (IL)-1b, IL-2, IL-4, IL-5, IL-6, IL-10, and IL-12 were noted between control and treated mice (Supplementary Fig. S1). In addition, and to confirm uptake of siRNA into tumors, we also administered two doses of Cy3-labeled siSV1 molecules (10 mg/kg) via the i.p. route in parallel experiments. All disseminated tumors revealed strong uptake (Supplementary Fig. S2).
Ovarian cancer is clinically managed by surgical debulking in combination with platinum-based chemotherapy (24). Therefore, we next investigated if siSV1 combined with cisplatin would more effectively inhibit tumor growth and increase survival when compared against cisplatin alone (Fig. 2A). Mice were treated with either cisplatin (5 mg/kg) or cisplatin (5 mg/kg) combined with siNTC (3 mg/kg) or siSV1 (3 mg/kg) and then monitored for disease progression (Fig. 2A). Mice receiving siSV1 plus cisplatin had the greatest antitumor benefits. Mice receiving cisplatin alone or siNTC plus cisplatin had tumor growth rates that were at least five times greater (Fig. 2B). Moreover, the median survival of siSV1 plus cisplatin–treated mice (86 days) was increased >35% compared with mice treated with either siNTC plus cisplatin (63 days) or cisplatin alone (62 days; P < 0.018) and more than doubled when compared with PBS controls (42 days; P < 0.0001; Fig. 2C). Beyond this, the addition of siSV1 to cisplatin significantly increased overall survival by ~50% when compared with cisplatin alone (116 versus 79 days) or cisplatin with siNTC (74 days) and was twice that of PBS-treated mice (55 days; P < 0.02; Fig. 2C).
Finally, we tested the effectiveness of siSV1 as a single agent with survival as the final outcome. Median and overall survival were markedly enhanced and directly dose dependent (Fig. 2D). Mice were treated 1 week after tumor cell implantation with escalating i.p. doses (3, 10, or 20 mg/kg) of either siSV1 or siNTC every 3 days for a total of six treatments. The group of mice treated with 20 mg/kg siSV1 showed the most dramatic effects in both median and overall survival: median survival was tripled (114 versus 311 days) and overall survival was doubled [225 versus 456 days; P = 0.0002; 95% confidence interval (95% CI), 3.2–39.4]. Two mice achieved complete remission (Fig. 2D). Mice treated with 10 mg/kg siSV1 also showed significant median (110 versus 136 days) and overall survival differences (P = 0.03; 95% CI, 1.1–10.2; Fig. 2D). Although there were significant survival differences in the groups treated with siSV1, there were some mice that overcame siSV1 treatment and we are examining modes of acquired resistance in this subset.
KLF6-SV1 inhibition induces apoptosis and restores chemo-sensitivity
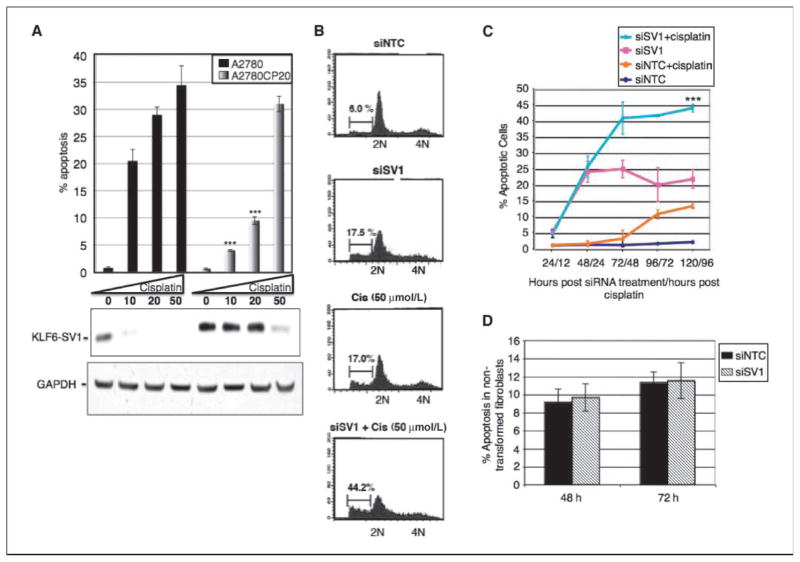
Given that the antitumor effect of siSV1 was amplified in the presence of cisplatin (Fig. 2B and C), we examined the effect of cisplatin on endogenous KLF6-SV1 expression levels in cisplatin-sensitive (A2780) and cisplatin-resistant (A2780CP) isogenic tumor cell lines. Over the range of cisplatin tested (0–50 μmol/L), A2780 cells were susceptible to cisplatin-induced apoptosis and there was a concomitant decrease in KLF6-SV1 protein levels (Fig. 3A). In contrast, the A2780CP-resistant cell line did not undergo apoptosis at either the 10 or 20 μmol/L dosing of cisplatin and KLF6-SV1 protein levels, which were much greater than in the A2780 line, were unaffected. Only at 50 μmol/L cisplatin did the A2780CP cells undergo apoptosis. At this dosage level, KLF6-SV1 protein was degraded (Fig. 3A).
Figure 3.
KLF6-SV1 inhibition induces apoptosis and synergizes with cisplatin in cancer cells. A, top, percentage of apoptosis in A2780 or A2780CP20 cells following treatment with increasing doses of cisplatin (1–50 μmol/L); bottom, Western blot analysis of A2780 or A2780CP20 cells after treatment with cisplatin (1–50 μmol/L). B, fluorescence-activated cell sorting of cells after treatment with siNTC, siSV1, cisplatin (50 μmol/L), or siSV1 plus cisplatin (50 μmol/L). C, SKOV3 cells were treated with the indicated agents and the percentage of apoptosis was determined. D, KLF6-SV1 inhibition does not induce apoptosis in noncancerous 293T fibroblasts.
Based on these findings that KLF6-SV1 overexpression may confer resistance to cisplatin and the fact that KLF6-SV1 levels are increased in several late-stage cancers (13, 15, 19), we explored the possibility that the link of KLF6-SV1 to tumorigenesis, progression, and chemoresistance may be through apoptosis. Therefore, we examined the effect of KLF6-SV1 inhibition on tumor cell death both in the presence and absence of cisplatin. KLF6-SV1 inhibition alone induced spontaneous apoptosis to the same level as cisplatin alone (~17.0%). When inhibited in combination with cisplatin treatment, the degree of apoptosis more than doubled (~44.2%; Fig. 3B). Isobologram analysis revealed that the effects of the two drugs on cell death were additive, not synergistic, in this model (Supplementary Fig. S3). Intriguingly, this effect increased over time. At 120 hours, nearly three times as many cells (45%) treated with siSV1 plus cisplatin had apoptosed compared with controls (16%; Fig. 3C). Unlike cisplatin, however, siSV1 activity seemed cancer cell specific. Cell death was not induced in the noncancerous fibroblast cell line 293T (Fig. 3D) but present in all cancer cell lines tested, including ovarian [SKOV3 (Fig. 3), A2780 (Supplementary Fig. S5), and 53S (Supplementary Fig. S4A)], lung (A549; ref. 21), prostate (PC3, PC3-M, LNCaP, and DU145; ref. 20), colon (HCT116; Supplementary Fig. S4A), and breast (MDA-435; Supplementary Fig. S4A).
KLF6-SV1 inhibition activates both extrinsic and intrinsic apoptotic cascades
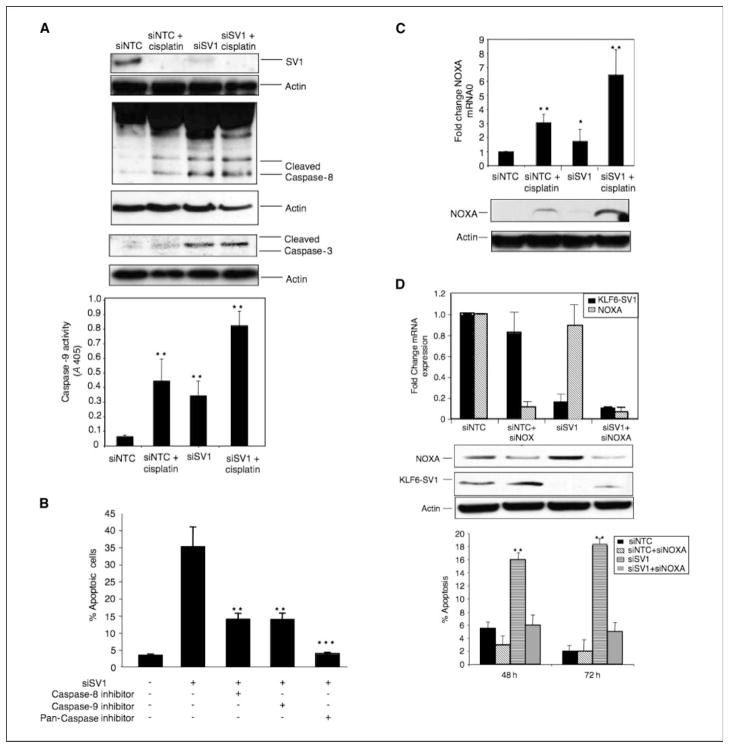
To confirm the cell cycle analysis and determine whether the intrinsic or extrinsic pathway of the apoptotic cascade was activated, we analyzed the activity of caspase-3, caspase-8, and caspase-9. KLF6-SV1 inhibition resulted in a significant increase in the amount of all active caspases (Fig. 4A). In addition, the combination of siSV1 and cisplatin resulted in the greatest degree of caspase activation (Fig. 4A). In all cases, both caspase-8 (extrinsic marker) and caspase-9 (intrinsic marker) were activated by siSV1, implying that both pathways may be activated (Fig. 4A). SKOV3 cells were then cotreated with siSV1 and either a broad-spectrum caspase inhibitor (Z-VAD-FMK), which blocks caspase-1, caspase-3, caspase-7, caspase-8, and caspase-9; a caspase-8–specific inhibitor (Z-IETD-FMK); or a caspase-9–specific inhibitor (Z-LEHD-FMK). All inhibitors repressed apoptosis induced by siSV1 (Fig. 4B). Blocking with the pan-caspase inhibitor abolished siSV1-dependent apoptosis (P = 0.01). Interestingly, specifically inhibiting caspase-8 or caspase-9 only blunted cell death by ~50% (P = 0.03; Fig. 4B). Collectively, these results suggest that multiple caspases are involved in siSV1-triggered apoptosis and that both the intrinsic and extrinsic pathways may be contributing to its function.
Figure 4.
KLF6-SV1 inhibition induces apoptosis through a NOXA-dependent mechanism. A, top, Western blot analysis of SKOV3 cell lysates following treatment with siNTC, siNTC plus cisplatin, siSV1, or siSV1 plus cisplatin; bottom, caspase-9 activity assay. B, suppression of siSV1-dependent apoptosis by caspase inhibitors. Following treatment of siSV1 with either a pan-caspase inhibitor, a caspase-8–specific (extrinsic marker) inhibitor, or caspase-9–specific (intrinsic marker) inhibitor, the percentage of apoptotic cells was measured. C, NOXA RNA and protein levels in SKOV3 cells following siNTC, siNTC plus cisplatin, siSV1, or siSV1 plus cisplatin treatment. D, quantitative reverse transcription-PCR (qRT-PCR) and Western blot analysis of NOXA and KLF6-SV1 following RNA interference (RNAi) transfection. Bottom, percentage of apoptosis followed by inhibition of NOXA, KLF6-SV1, or KLF6-SV1 in combination with NOXA. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
KLF6-SV1–mediated effects on apoptosis are NOXA dependent but p53 independent
A hallmark of both tumorigenesis and chemoresistance is the selection and outgrowth of cells resistant to apoptosis (4). We therefore compared expression levels of several apoptotic-associated genes following KLF6-SV1 inhibition, including members of the prosurvival Bcl-2 family (Bcl-2 and Mcl-1), BH3-only proteins (PUMA, Bad, NOXA), and the proapoptotic protein BAX. NOXA, which is induced by p53-dependent or p53-independent apoptotic stimuli (6, 25, 26), was the most up-regulated: ~4-fold following siSV1 treatment and ~2-fold with cisplatin treatment. When siSV1 and cisplatin were combined, NOXA mRNA was induced ~7-fold (Fig. 4C). These effects on NOXA in cultured cells were also present in tumors harvested from siRNA-treated mice (Supplementary Fig. S4B). NOXA levels were increased on average >5-fold in all tumors analyzed (Supplementary Fig. S4B).
The up-regulation of NOXA in the SKOV3 cell line, reported to be p53 null, suggested that siSV1 can induce NOXA by a p53-independent mechanism. To conclusively show this p53 independence, we repeated these experiments using the isogenic ovarian cancer cells A2780 (p53 wild-type) and A2780CP20 (p53 mutant). Again, and consistent with p53 independence, NOXA was up-regulated in both and the same degree of apoptosis was observed in both cell lines (Supplementary Fig. S5).
Finally, we examined the effect of combined NOXA and KLF6-SV1 inhibition on apoptosis. KLF6-SV1–mediated apoptosis was completely abrogated following NOXA silencing (Fig. 4D). This loss was evident across all tumor cell lines tested, including ovarian (SKOV3; Fig. 4D) and prostate (PC3, LNCaP, and DU145; data not shown) cancers. This response was NOXA specific because, and as an adjunct to siRNA-mediated inhibition, we also tested the apoptotic response of BAX wild-type and null isogenic derivatives of HCT116 cells. The presence or absence of BAX had no effect on siSV1-induced apoptosis (data not shown).
KLF6-SV1 binds and targets NOXA for degradation with resulting Mcl-1 accumulation
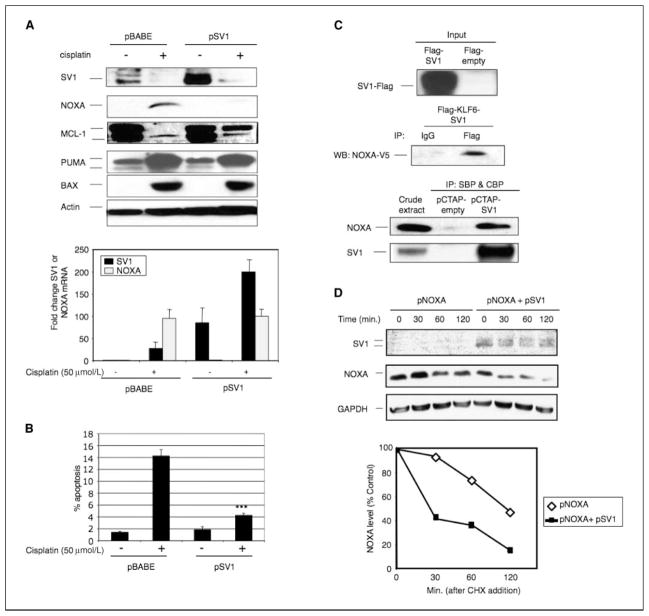
Because NOXA plays a role in DNA damage–induced apoptosis (25, 26), we next examined NOXA induction and stability in response to DNA damage in cells engineered to stably overexpress KLF6-SV1 (pSV1). NOXA RNA and protein levels were induced ~100-fold in control cells following cisplatin treatment (Fig. 5A). In marked contrast, NOXA protein was undetectable in cells overexpressing KLF6-SV1 despite NOXA RNA levels being increased ~100-fold. In addition, both endogenous and overexpressed (lane 4) KLF6-SV1 protein was also degraded following cisplatin treatment (Fig. 5A). In contrast, KLF6-SV1 overexpression had no effect on two other proapoptotic proteins, PUMA and BAX. Finally, and in further support of the antiapoptotic function of KLF6-SV1 and its ability to confer resistance to cisplatin-induced cell death, cells overexpressing KLF6-SV1 were decidedly less susceptible to cisplatin-induced apoptosis (Fig. 5B).
Figure 5.
KLF6-SV1 binds and regulates NOXA. A, top, Western blot analysis of KLF6, KLF6-SV1, NOXA, Mcl-1, PUMA, BAX, and actin in cell lysates from cells stably expressing pBABE (empty vector control) or pSV1 (KLF6-SV1) and in the presence or absence of cisplatin (50 μmol/L); bottom, qRT-PCR analysis of KLF6-SV1 and NOXA in pBABE or pSV1 cells treated with cisplatin (50 μmol/L). B, percentage of apoptosis in cells stably expressing pBABE or pSV1 in the presence or absence of cisplatin (50 μmol/L). C, top, coimmunoprecipitation assay of NH2-terminal flag-tagged KLF6-SV1 (SV1-flag) and COOH-terminal V5-tagged NOXA (NOXA-V5); bottom, TAP assay of COOH-terminal streptavidin-tagged (SBP) and calmodulin-tagged (CBP) KLF6-SV1 (pCTAP-SV1) or pCTAP-empty. D, half-life studies examining the effect of KLF6-SV1 overexpression on NOXA stability. Cells overexpressing NOXA alone or in combination with KLF6-SV1 were treated with cycloheximide (CHX) and harvested at various time points (0, 30, 60, and 120 min). ***, P < 0.0005.
Owing to its alternative splicing, KLF6-SV1 lacks the KLF6 zinc finger DNA binding domain while retaining the majority of the KLF6 NH2-terminal activation domain (17). The KLF activation domain, in general, mediates a broad range of protein-protein interactions and accounts for the diverse biological activities of the KLF family (27). Therefore, we hypothesized that KLF6-SV1 complexes with NOXA in part through this domain and this physical interaction results in their mutual and rapid degradation. This KLF6-SV1-NOXA neutralization model would in essence mimic the highly specific, physical interactions between BH3 ligands and Bcl-2 homologues required for initiation of apoptosis (8).
Two independent techniques confirmed our hypothesis. First, co-expressed tagged forms of NOXA and KLF6-SV1 were shown to physically interact by coimmunoprecipation in SKOV3 cells (Fig. 5C). Second, and in a second cell line, endogenous NOXA in PC3 cell lysates was shown to bind COOH-terminal, dual-tagged (streptavidin binding peptide/calmodulin binding peptide) KLF6-SV1 through use of tandem affinity purification (TAP; Fig. 5C, bottom).
Given these results, we first tested whether KLF6-SV1 binding had an effect on NOXA stability. The half-life of transfected NOXA alone was ~120 minutes, whereas in the presence of KLF6-SV1 half-life was decreased to ~25 minutes (Fig. 5D). Thus, KLF6-SV1 overexpression increases NOXA degradation.
NOXA binds Mcl-1, an antiapoptotic Bcl-2 family member (8, 28–31), and antagonizes its prosurvival function (8, 32). Therefore, we examined the indirect effects that KLF6-SV1 might have on Mcl-1 protein levels. In general, following cisplatin-induced DNA damage, Mcl-1 protein is rapidly degraded by the proteasome, and in turn, NOXA is released and induces apoptosis (32). In our studies, and in contrast to control cells, Mcl-1 evaded complete degradation in cells overexpressing KLF6-SV1 despite cisplatin treatment (Fig. 5A). Conversely, targeted inhibition of KLF6-SV1 resulted in decreased endogenous Mcl-1 levels, further supporting a regulatory effect of KLF6-SV1 on Mcl-1 levels (Supplementary Fig. S6).
KLF6-SV1 is ubiquitinated and its degradation is HDM2 mediated
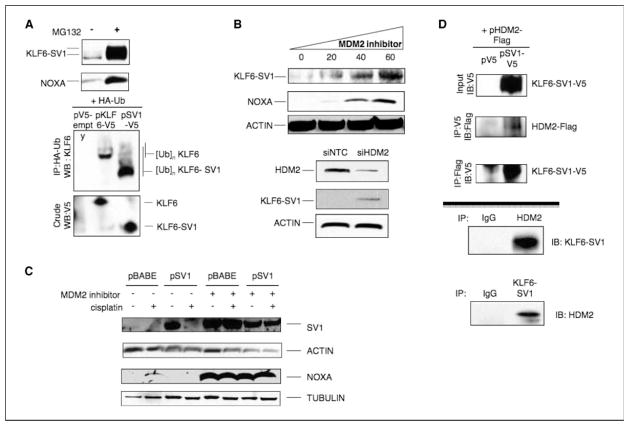
How does KLF6-SV1 binding to NOXA triggers their mutual degradation? We first ascertained whether the degradation was proteasome mediated because KLF6 has been previously shown to be ubiquitinated (33, 34). Treatment of SKOV3 cells with the potent and selective proteasome inhibitor MG132 resulted in dramatic increases in KLF6-SV1 and NOXA (Fig. 6A). We therefore hypothesized that KLF6-SV1 may serve as a substrate for ubiquitin modification. To test this, we coexpressed V5-tagged KLF6-SV1 and HA-tagged ubiquitin in 293T cells and coimmunoprecipitated the cell lysates. Immunoblotting of these lysates revealed that KLF6-SV1 is ubquitinated (Fig. 6A, bottom).
Figure 6.
HDM2 mediates the degradation of KLF6-SV1 and NOXA following cisplatin. A, top, KLF6-SV1 and NOXA accumulation following MG132 treatment (proteasome inhibitor). KLF6-SV1 is ubiquitinated. 293T cells were transiently transfected with pV5-empty, pSV1-V5, and HA-tagged ubiquitin (HA-Ub) expression vectors and then treated with MG132 for 6 h. Bottom, equal protein amounts were immunoprecipitated with anti-HA antibody and Western blotted with anti-KLF6 antibody. B, MDM2 inhibition leads to KLF6-SV1 and NOXA accumulation. Cells were treated with either increasing amounts of a synthetic inhibitor of MDM2 (EMD) for 6 h or RNAi targeting nontargeting control or HDM2 and cell lysates were immunoblotted with KLF6-SV1, NOXA, and actin. C, suppression of MDM2 abolished cisplatin-induced degradation of KLF6-SV1 and NOXA. Cells were simultaneously treated with cisplatin (50 μmol/L) and the MDM2 inhibitor (40 μmol/L) for 12 h and for protein analysis. D, HDM2 coimmunoprecipitates with KLF6-SV1. Reciprocal coimmunoprecipitations were performed using 293T cell extracts overexpressing pSV1-V5 and pMDM2-flag or pSV1-V5 alone. Top, soluble protein extracts were immunoprecipitated (IP) with anti-V5 or anti-flag antibodies and analyzed by immunoblotting. Endogenous immunoprecipitation using SKOV3 cell extracts. Bottom, soluble protein extracts were immunoprecipitated using anti-MDM2 antibody and immunoblotted with an anti-KLF6 antibody or an anti-KLF6 antibody and immunoblotted with anti-MDM2 antibody. IgG immunoprecipitation was used as control.
To identify the regulator of their proteasome-mediated degradation, we used a candidate gene approach. We first tested whether HDM2, a well-characterized E3 ubiquitin ligase that interacts with several key cancer genes, including p53 (35, 36), and targets them for degradation, may be involved. HDM2 overexpression markedly reduced KLF6-SV1 protein levels, but not RNA levels (data not shown), and this effect was abolished with the addition of MG132 (Supplementary Fig. S7A). The effect of suppressing HDM2 activity, using a synthetic compound that binds strongly to HDM2 and irreversibly blocks the formation of the HDM2/p53 protein complex (37), on KLF6-SV1 and NOXA protein levels was then tested. Escalating doses of the inhibitor resulted in an accumulation of both KLF6-SV1 and NOXA in both SKOV3 cells (Fig. 6B) and HeLa cells (Supplementary Fig. S7B). HDM2 inhibition also blocked cisplatin-induced degradation of KLF6-SV1 and NOXA (Fig. 6C).
Having identified this HDM2 requirement for cisplatin-induced degradation, we further characterized this finding by testing whether HDM2 and KLF6-SV1 can interact in a complex. The interaction between these two proteins was shown first by reciprocal coimmunoprecipitation assays in SKOV3 cells using HDM2-flag–tagged and KLF6-SV1-V5–tagged constructs (Fig. 6D, top) and then by reciprocal coimmunoprecipitation assays of endogenous proteins using 293HEK cellular extracts (Fig. 6D, bottom).
Discussion
In this study, we initially sought to explore the potential therapeutic efficacy of siRNA-mediated inhibition of KLF6-SV1 in ovarian cancer. KLF6-SV1 inhibition not only induced spontaneous apoptosis of tumor cells in culture and in vivo but also restored their chemosensitivity to cisplatin. Currently, ovarian cancer treatment comprises a dual approach based on tumor debulking through surgery combined with adjuvant chemotherapy using a combination of platinum- and taxane-based agents. Nonetheless, treatment failure occurs in the majority of patients because of late presentation and the development of chemoresistance (12). In an effort to overcome therapeutic failure, efforts have been made to optimize available cytotoxic agents by changing the route of administration or combining agents (38). I.p. delivery of chemotherapeutic agents would theoretically target the principal site of ovarian disease while minimizing some of the systemic toxicities associated with treatment. Our results suggest that siRNA-mediated treatment against KLF6-SV1 offers a novel, mechanism-based approach, which induces spontaneous apoptosis in tumor cells and restores cisplatin sensitivity. A particularly relevant aspect of our findings, given that most late-stage ovarian tumors are p53 deficient (39), is that this effect is p53 independent.
An intriguing similarity offered by the genetic relationship between KLF6 and KLF6-SV1 and other apoptosis family members is the fact that several proteins involved in the apoptotic pathway are themselves regulated by alternative splicing (40). This results in proteins generated from the same gene but with antagonistic functions or tissue altered expression patterns. With this in mind, it is noteworthy that our results identified a physical interaction between KLF6-SV1, which retains the wild-type KLF6 NH2-terminal activation domain, and NOXA, which results in their mutual and rapid HDM2-mediated degradation. Interestingly, KLF6 also binds NOXA and HDM2 but this pairing does not result in NOXA degradation (results not shown), further highlighting the specificity of the KLF6-SV1/NOXA interaction. The proposed KLF6-SV1/NOXA neutralization model, albeit providing a new layer of complexity to the apoptotic cascade, would parallel the highly specific, physical interactions between BH3 ligands and Bcl-2 homologues already known to be required for initiation of apoptosis (8).
The downstream effect of this interaction between KLF6-SV1 and NOXA is the stabilization of Mcl-1. Without this interaction, Mcl-1 is normally rapidly degraded in response to cell death signals due to its displacement from the BAK/Mcl-1 complex triggered by NOXA induction (8, 33, 41). Interestingly, elevated Mcl-1 expression has been previously correlated with resistance to chemotherapeutic agents (42), and its transcriptional repression by c-myc or by a multikinase inhibitor sensitizes cells to TNF-related apoptosis-inducing ligand–induced apoptosis (43).
Thus, KLF6-SV1 is a prosurvival/antiapoptotic protein that functions through the degradation of a proapoptotic/BH3-only family member. This suggests a novel “fourth” level of regulation in the apoptotic cascade. Interestingly, the existence of such a tier has already been defined. Chlamydiae, obligate intracellular bacteria, are known to escape destruction by averting apoptosis in infected human cells (44). They do so by broadly degrading BH3-only proteins, including NOXA. Most recently, a chlamydia-secreted protease factor was shown to be responsible for the degrading activity (45). Identification of KLF6-SV1 as a protein that can target a proapoptotic BH3-only molecule for destruction establishes a paradigm for the existence of other host-derived factors and intrinsic regulatory pathways important in cancer. Based on our findings, these host-encoded factors, including KLF6-SV1, may represent future therapeutic targets for cancers, which by definition have subverted apoptotic control mechanisms.
Supplementary Material
Acknowledgments
Grant support: Funded in part by NIH grant RO1 CA122332 (J.A. Martignetti) and a Howard Hughes Physician-Scientist Early Career Award (G. Narla).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–84. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- 2.Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108:153–64. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 3.Youle RJ. Cell biology. Cellular demolition and the rules of engagement. Science. 2007;315:776–7. doi: 10.1126/science.1138870. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson DW. From bench to clinic with apoptosis-based therapeutic agents. Nature. 2000;407:810–6. doi: 10.1038/35037747. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez Y, Verhaegen M, Miller TP, et al. Differential regulation of NOXA in normal melanocytes and melanoma cells by proteasome inhibition: therapeutic implications. Cancer Res. 2005;65:6294–304. doi: 10.1158/0008-5472.CAN-05-0686. [DOI] [PubMed] [Google Scholar]
- 7.Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 8.Willis SN, Fletcher JI, Kaufmann T, et al. Apoptosis initiated when BH3 ligands engage multiple BCL-2 homologs, not Bax or Bak. Science. 2007;315:856–9. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 9.Konopleva M, Contractor R, Tsao T, et al. Mechanism of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–88. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Adams JM, Cory S. The BCL-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–7. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 12.Ozols RF, Bookman MA, Connolly DC, et al. Focus on epithelial ovarian cancer. Cancer Cell. 2004;5:19–24. doi: 10.1016/s1535-6108(04)00002-9. [DOI] [PubMed] [Google Scholar]
- 13.DiFeo A, Narla G, Hirshfeld J, et al. Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res. 2006;12:3730–9. doi: 10.1158/1078-0432.CCR-06-0054. [DOI] [PubMed] [Google Scholar]
- 14.Macleod K, Mullen P, Sewell J, et al. Altered ErbB receptor signaling and gene expression in cisplatin-resistant ovarian cancer. Cancer Res. 2005;65:6789–800. doi: 10.1158/0008-5472.CAN-04-2684. [DOI] [PubMed] [Google Scholar]
- 15.Narla G, DiFeo A, Yao S, et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005;65:5761–8. doi: 10.1158/0008-5472.CAN-05-0217. [DOI] [PubMed] [Google Scholar]
- 16.Narla G, Heath KE, Reeves HL, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–6. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 17.Narla G, DiFeo A, Reeves HL, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1207–12. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser J. American Society of Human Genetics meeting. New prostate cancer genetic link. Science. 2004;306:1285. doi: 10.1126/science.306.5700.1285. [DOI] [PubMed] [Google Scholar]
- 19.Camacho-Vanegas O, Narla G, Teixeira MS, et al. Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int J Cancer. 2007;121:1390–5. doi: 10.1002/ijc.22809. [DOI] [PubMed] [Google Scholar]
- 20.Narla G, DiFeo A, Fernandez Y, et al. KLF6-SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. J Clin Invest. 2008;118:2711–21. doi: 10.1172/JCI34780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiFeo A, Feld L, Rodriguez E, et al. A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res. 2008;68:965–70. doi: 10.1158/0008-5472.CAN-07-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mesiano S, Ferrara N, Jaffe RB. Role of vascular endothelial growth factor in ovarian cancer: inhibition of ascites formation by immunoneutralization. Am J Pathol. 1998;153:1249–56. doi: 10.1016/S0002-9440(10)65669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinman ME, Yamada K, Takeda A, et al. Sequence-and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–7. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2007;18:ii12. doi: 10.1093/annonc/mdm017. [DOI] [PubMed] [Google Scholar]
- 25.Oda E, Ohki R, Murasawa H, et al. NOXA, a BH3-only member of the BCL-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–8. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 26.Villunger A, Michalak EM, Coultas, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and NOXA. Science. 2003;302:1036–8. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 27.Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–8. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 28.Letai A, Bassik MC, Walensky LD, et al. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–92. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 29.Kuwana T, Bouchier-Hayes L, Chipuk JE, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–35. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Willis SN, Wei A, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 31.Mei Y, Xie C, Xie W, et al. Noxa/Mcl-1 balance regulates susceptibility of cells to camptothecin-induced apoptosis. Neoplasia. 2007;9:871–81. doi: 10.1593/neo.07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H, Rafiuddin-Shah M, Tu HC, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 2006;8:1348–58. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 33.Banck MS, Beaven SW, Narla G, et al. KLF6 degradation after apoptotic DNA damage. FEBS Lett. 2006;580:6981–6. doi: 10.1016/j.febslet.2006.10.077. [DOI] [PubMed] [Google Scholar]
- 34.Slavin DA, Koritschoner NP, Prieto CC, et al. A new role for the Kruppel-like transcription factor KLF6 as an inhibitor of c-Jun proto-oncoprotein function. Oncogene. 2004;23:8196–205. doi: 10.1038/sj.onc.1208020. [DOI] [PubMed] [Google Scholar]
- 35.Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 36.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 37.Kumar SK, Hager E, Pettit C, et al. Design, synthesis and evaluation of novel boronic-chalcone derivatives as antitumor agents. J Med Chem. 2003;46:2813–3. doi: 10.1021/jm030213+. [DOI] [PubMed] [Google Scholar]
- 38.Agarwal A, Linch M, Kaye SB. Novel therapeutic agents in ovarian cancer. Eur J Surg Oncol. 2006;32:875–86. doi: 10.1016/j.ejso.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 39.Aunoble B, Sanches R, Didier E, Bignon YJ. Major oncogenes and tumor suppressor genes involved in epithelial ovarian cancer (review) Int J Oncol. 2000;16:567–76. doi: 10.3892/ijo.16.3.567. [DOI] [PubMed] [Google Scholar]
- 40.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 41.Lindsten T, Ross AJ, King A, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6:1389–99. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaufmann SH, Karp JE, Svingen PA, et al. Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood. 1998;91:991–1000. [PubMed] [Google Scholar]
- 43.Ricci MS, Kim SH, Ogi K, et al. Reduction of TRAIL-induced Mcl-1 and cIAP2 by c-Myc or sorafenib sensitizes resistant human cancer cells to TRAIL-induced death. Cancer Cell. 2007;12:66–80. doi: 10.1016/j.ccr.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 44.Fan T, Lu H, Hu H, et al. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J Exp Med. 1998;187:487–96. doi: 10.1084/jem.187.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pirbhai M, Dong F, Zhong Y, Pan KZ, Zhong G. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J Biol Chem. 2006;281:31495–5. doi: 10.1074/jbc.M602796200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.