Abstract
TRAF6 mediates Lys63 (K63)-linked polyubiquitination for NF-κB activation via its N-terminal RING and zinc finger domains. Here we report the crystal structures of TRAF6 and its complex with the ubiquitin conjugating enzyme (E2) Ubc13. The RING and zinc fingers of TRAF6 assume a rigid, strikingly elongated structure. Interaction of TRAF6 with Ubc13 involves direct contacts of the RING and the preceding residues while the first zinc finger plays a structural role. Surprisingly, this region of TRAF6 is dimeric both in the crystal and in solution, different from the trimeric C-terminal TRAF domain. Structure-based mutagenesis reveals that TRAF6 dimerization is critical for polyubiquitin synthesis and auto-ubiquitination. Fluorescence energy transfer analysis shows that TRAF6 dimerization induces higher order oligomerization of full-length TRAF6. The mismatch of dimeric and trimeric symmetry may provide a mode of infinite oligomerization that facilitates ligand-dependent signal transduction of many immune receptors.
Tumor necrosis factor (TNF) receptor associated factors (TRAFs) play important roles in intracellular signal transduction of many receptor families such as the TNF receptor superfamily, the IL-1 receptors (IL-1R), the Toll-like receptors (TLR), T-cell receptors (TCR) and B-cell receptors (BCR) 1,2. Upon receptor activation, TRAFs are directly or indirectly recruited to the intracellular domains of these receptors. They subsequently engage other signaling proteins to activate the inhibitor of κB (IκB) kinase (IKK) and MAP kinases, leading ultimately to activation of transcription factors such as NF-κB and AP-1 to induce immune and inflammatory responses and confer protection from apoptosis.
Most TRAFs contain an N-terminal domain with RING (really interesting gene) and a variable number of zinc fingers and a C-terminal TRAF domain that comprises a coiled coil domain and a conserved TRAF-C domain (Fig. 1a). Previous biochemical and structural studies have revealed that the TRAF domain forms a mushroom-shaped trimeric structure with the TRAF-C domain as the head for interaction with receptors and adaptor proteins and the coiled coil domain as the stalk for trimerization 3–5. Remarkably, TRAF6 is uniquely pleiotropic in participating in the signal transduction of many receptor systems while TRAF2, TRAF3 and TRAF5 appear to signal only within the TNF receptor superfamily 5.
Figure 1.
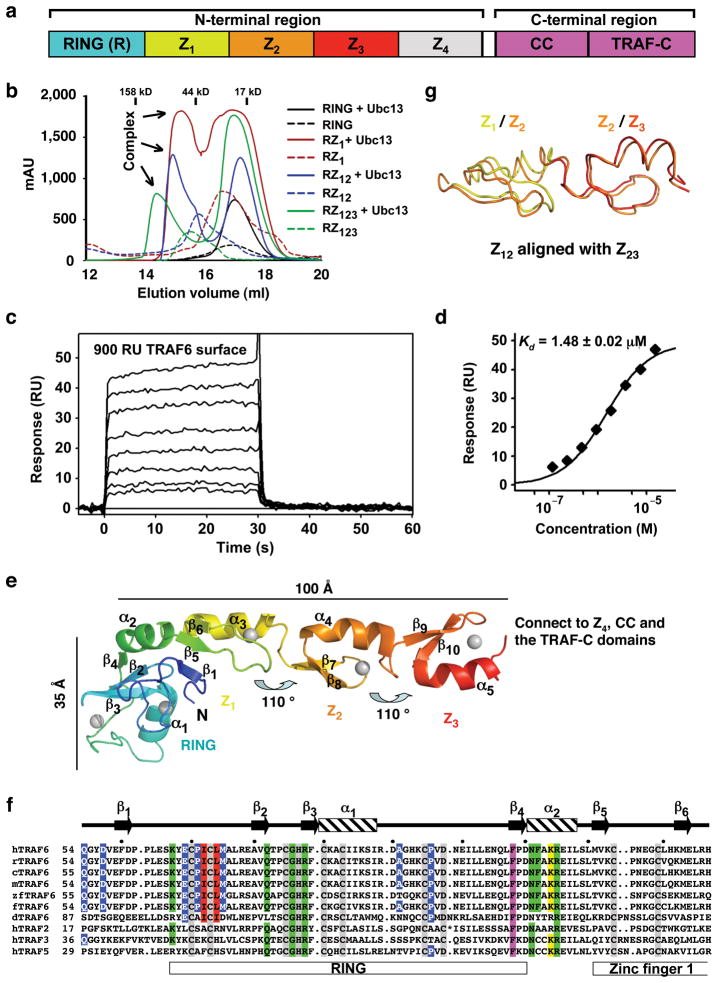
Biochemical characterization of the TRAF6/Ubc13 interaction and structure of the N-terminal region of TRAF6. (a) Domain organization of TRAF6. Z1-Z4: zinc fingers 1–4. CC: coiled coil. (b) Superimposed gel filtration profiles of TRAF6/Ubc13 mixtures to show that TRAF6 RZ1 is both necessary and sufficient for Ubc13 interaction. Approximate elution positions of molecular weight standards are shown. (c) Surface plasmon resonance measurement for the binding of Ubc13 to surface coupled TRAF6 RZ123 at different Ubc13 concentrations. (d) Plot of the binding response as a function of Ubc13 concentration to derive the dissociation constant (Kd). ± refers to the standard error of the mean of the fitting. (e) Ribbon diagram of the monomer structure of TRAF6 RZ123. The different domains are labeled and the rotational relationships between the zinc fingers are shown. (f) Sequence alignment of TRAF6 from different species and with TRAF2, TRAF3, and TRAF5. Domain boundaries and secondary structures are labeled. Zinc-coordinating residues are shaded in gray. Residues at the Ubc13 interface are highlighted in red for those that bury more than 60 Å2 surface areas and in blue for those with surface area burials of 20–60 Å2. Residues at the dimerization interface are highlighted in magenta for those that bury more than 100 Å2 surface areas and in green for those with surface area burials of 40–100 Å2. The major TRAF6 auto-ubiquitination site at K124 is highlighted in yellow. “*“ in the TRAF2 sequence represents an insertion of “ VHEGIYEEG”. h: human; r: rat; c: cow; m: mouse; zf: zebra fish; f: fowl; d: drosophila. (g) Superposition of the zinc finger 1 and 2 structure to the zinc finger 2 and 3 structure.
The downstream signaling mechanism of TRAFs was first revealed from biochemical and cellular studies of TRAF6 to show the involvement of K63-linked polyubiquitination 6–8. Ubiquitination is one of the most prevalent post-translational modifications 9. It is accomplished in three steps, 1) ATP-dependent attachment of ubiquitin (Ub) via a thioester bond to a Ub activating enzyme (E1), 2) transfer of Ub from E1 to the active site Cys of a Ub conjugating enzyme (E2), and 3) transfer of Ub from the E2 active site to Lys residues of substrates (including other molecules of Ub) with the aid of a Ub ligase (E3) 10–12. There are two types of well characterized E3s. The HECT domain-containing E3s harbor an essential catalytic Cys residue and promote substrate polyubiquitination via an E3 intermediate with a thioester linked Ub. The RING domain-containing E3s do not appear to exhibit catalytic activity but provide a bridge between E2s and substrates. TRAF6 is a RING-type E3 that facilitates K63-linked polyubiquitination. Unlike K48-linked polyubiquitin (poly-Ub) chains that are hallmarks for proteasomal degradation, the K63 linkage is non-degradative and has been discovered to function as a signaling moiety in DNA damage repair processes and innate immunity pathways 10,13.
Upon activation by the relevant signaling pathways after ligand stimulation, TRAF6 promotes K63-linked polyubiquitination of itself and downstream signaling proteins, a process that requires the heterodimeric E2 of Ubc13 and the ubiquitin E2 variant (Uev) known as Uev1A 10. Crystal structure of the complex between Ubc13 covalently bound to donor Ub and a Uev known as Mms2 has elegantly revealed that Uev possesses an acceptor Ub binding site and orientates the acceptor Ub for K63-linkage with the donor Ub 14. The K63-linked poly-Ub chains function as anchors to recruit the TAK1 kinase complex and IKK to activate both the MAP kinase pathway and the NF-κB pathway 15. TAK1 directly phosphorylates MAP kinases while IKK-mediated phosphorylation of IκB leads to its degradation to free NF-κB for transcription.
Despite extensive studies, how TRAF6 as well as other E3s promote polyubiquitination is poorly understood. Here we report biochemical, structural and cell biological studies on the N-terminal region of human TRAF6, which reveal both specificity and mechanism of TRAF6-mediated polyubiquitination. We show that the RING domain of TRAF6 does not function alone; instead, residues preceding the RING directly interact with Ubc13 and the first zinc finger plays a structural role. Surprisingly, we show that the N-terminal region of TRAF6 is dimeric, in contrast to the trimeric symmetry of its C-terminal region. Investigation on the functional consequence of this specific TRAF6 dimerization reveals unforeseen aspects of its E3 activity and mode of signal transduction.
RESULTS
Interaction with Ubc13 requires RING and zinc finger 1 (RZ1)
TRAF6-mediated K63-linked polyubiquitination requires the heterodimeric E2 complex of Ubc13 and Uev1A 10. While Ubc13 mediates direct interaction with an E3, Uev1A provides the linkage specificity 14,16–18. To understand how TRAF6 interacts with Ubc13, we constructed a number of deletion constructs of human TRAF6 containing the RING alone (residues 50-120), RZ1 (residues 50-159), RZ12 (residues 50-187) and RZ123 (residues 50-211). Surprisingly, gel filtration chromatography analysis of complex formation showed that the RING domain alone was not sufficient for Ubc13 interaction (Fig. 1b). This is in contrast to many other RING domains which are necessary and sufficient for E2 interaction 19. Instead, TRAF6 RZ1 (residues 50-159) is the shortest construct that is necessary and sufficient for formation of a complex with Ubc13 (Fig. 1b). The first 49 residues of TRAF6 do not possess any recognizable domains and are dispensable for poly-Ub synthesis (see below). They caused severe aggregation when included in any TRAF6 constructs.
We used surface plasmon resonance (SPR) to quantitatively measure the interaction between TRAF6 and Ubc13. An average dissociation constant of approximately 1.6 μM was obtained for the binding of Ubc13 to two coupling densities of TRAF6 RZ123 (Fig. 1c, 1d, Supplementary Fig. 1). This modest affinity is compatible with the necessity of an E2 to shuttle between its E1 and E3 20,21. As shown from the TRAF6/Ubc13 complex structure (below), substantial contributions to the TRAF6/Ubc13 interaction are afforded by additional interactions from residues preceding the RING and by a structural role of the first zinc finger domain. The RING domain per se of TRAF6 exhibits weak affinity to Ubc13 as shown from NMR studies of the TRAF6 RING domain comprising residues 67-124 (KD ≈ 2 mM) 22.
Elongated structure of RING and zinc fingers 1–3 (RZ123) of TRAF6
The structure of human TRAF6 RZ123 was determined at 2.6 Å resolution by single wavelength anomalous diffraction and refined at 2.2 Å resolution (Table 1, Fig. 1e, Supplementary Fig. 2). The monomer structure is elongated and resembles the shape of a golf club with the RING domain as the head of approximately 35 Å in length and the three zinc fingers as the shaft of approximately 100 Å in length (Fig. 1e). Surprisingly, instead of beads on a string, the structure is rigid as exemplified in the small RMSD of 0.5 Å between the two independent, dimerically related TRAF6 molecules in the crystallographic asymmetric unit. The zinc fingers align in a linear fashion along the long axis of the molecule with rotations of approximately 110° between the successive fingers (Fig. 1e). The fixed relationship between the successive fingers is at least partly due to the fixed sequence spacing between them. There are always three residues between the last Cys residue of the previous finger to the first β-strand of the next finger (Fig. 1f, Supplementary Fig. 3). Together with the last Cys in the previous finger, these three residues form a classical type I β-turn, which interacts with both zinc fingers to join them together. The first and second zinc finger region (Z12) of TRAF6 is superimposable to the second and third zinc finger region (Z23) with an RMSD of 0.9 Å (Fig. 1g). The fourth zinc finger (Z4) may be modeled based on its relationship with the previous zinc finger. This sequence spacing is conserved in different species of TRAF6 and in TRAF2, TRAF3 and TRAF5 (Supplementary Fig. 3), suggesting that it represents a conserved feature of the zinc finger arrangement in TRAFs.
Table 1.
Data collection, phasing and refinement statistics
| SeMet-TRAF6 | TRAF6 | TRAF6/Ubc13 | TRAF6/Ubc13 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | C2 | C2 | P1 | C2 |
| Cell dimensions | ||||
| a, b, c (Å) | 124.1, 81.1, 50.5 | 123.7, 80.8, 50.7 | 39.0, 42.8, 49.1 | 130.5, 41.4, 123.7 |
| α, β, γ(°) | 90.0, 91.5, 90.0 | 90.0, 91.4, 90.0 | 104.6, 99.6,108.6 | 90.0, 116.2, 90.0 |
| Resolution (Å) | 50-2.6 (2.69-2.6)* | 30-2.2 (2.28-2.2)* | 50-2.1 (2.18-2.1)* | 50-2.6 (2.69-2.6)* |
| Rsym | 0.074 (0.36) | 0.074 (0.48) | 0.050 (0.14) | 0.046 (0.39) |
| I/σI | 45.2 (6.9) | 28.7 (2.9) | 45.5 (9.3) | 55.3 (5.1) |
| Completeness | 0.97 (0.88) | 0.98 (0.82) | 0.96 (0.82) | 0.99 (0.94) |
| Redundancy | 5.7 (4.6) | 6.4 (4.1) | 3.9 (3.5) | 6.8 (5.9) |
| Refinement | ||||
| Resolution (Å) | 30-2.2 | 50-2.1 | 50-2.6 | |
| No. reflections | 23,334 | 15,563 | 17,678 | |
| Rwork/Rfree | 0.228/0.269 | 0.210/0.251 | 0.258/0.333 | |
| No. atoms | ||||
| Protein | 2,520 | 2,018 | 4,062 | |
| Water | 224 | 198 | 39 | |
| Average B-factors | ||||
| Protein (Å2) | 51.2 | 47.1 | 80.6 | |
| Water (Å2) | 68.7 | 57.3 | 63.4 | |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.006 | 0.006 | 0.008 | |
| Bond angles (°) | 1.39 | 1.20 | 1.47 | |
Values in parenthesis are for highest resolution shell. One crystal was used for each data set.
Structure of the TRAF6 RZ1/Ubc13 complex
The structures of the human TRAF6 RZ1/Ubc13 complex were solved independently in 2 crystal forms at 2.6 Å and 2.1 Å resolutions, respectively, by molecular replacement using the RZ1 model from the TRAF6 RZ123 structure and the previously determined Ubc13 structure 16 (Table 1, Fig. 2a). Since the structures are similar with pair wise RMSDs of 0.8 Å, the description below is based on the higher resolution structure. The TRAF6/Ubc13 complex buries approximately 1,000 Å2 surface areas, most of which are hydrophobic (Fig. 1f, 2a, 2b). The architecture of the interaction is similar to the RING/E2 interaction in the c-Cbl/UbcH7 complex 23 and to the U-box/E2 interaction in the CHIP/Ubc13 complex 24,25. RING and U-box domains share similar folds but the latter do not coordinate any metal ions 26.
Figure 2.
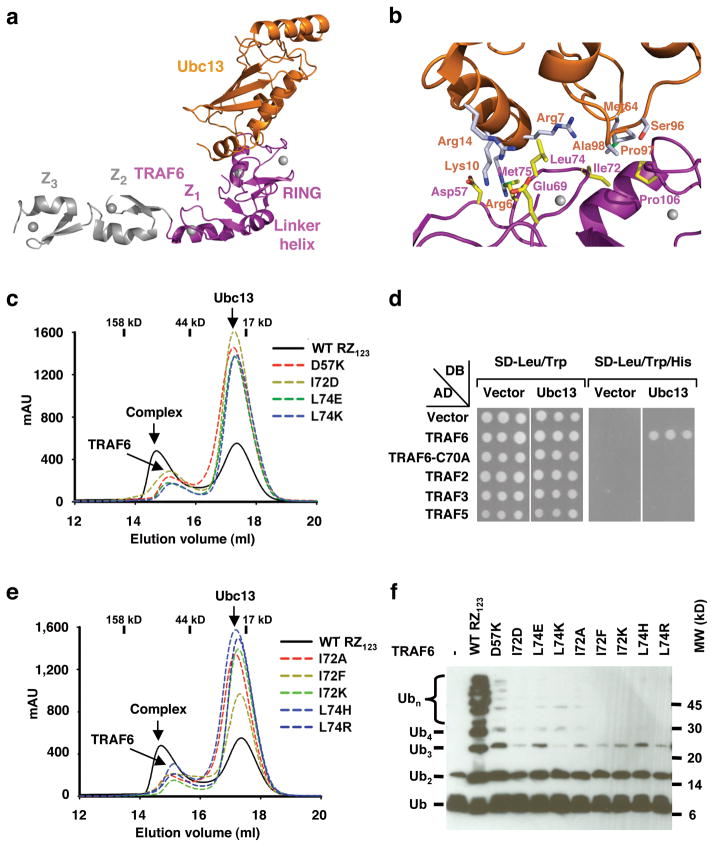
Structural analysis of the TRAF6/Ubc13 interaction. (a) Ribbon diagram of the TRAF6 RZ1/Ubc13 complex. Z2 and Z3 domains are modeled based on superposition of the TRAF6 RZ1/Ubc13 complex with the TRAF6 RZ123 structure and are shown in gray. (b) Detailed interaction between TRAF6 and Ubc13. TRAF6 is shown in magenta with the carbon atoms of its side chains in yellow. Ubc13 is shown in orange with the carbon atoms of its side chains in gray. (c) Superimposed gel filtration profiles of Ubc13 mixed with wild type or mutant TRAF6 RZ123 designed to disrupt the interaction. Approximate elution positions of molecular weight standards are shown. (d) Yeast two hybrid experiments on the interaction between Ubc13 and full-length TRAF2, TRAF3, TRAF5, TRAF6 (positive control) and its RING mutant C70A (negative control). (e) Superimposed gel filtration profiles of Ubc13 mixed with wild type or mutant TRAF6 RZ123 with interface residues switched to the corresponding sequences in other TRAFs: I72A (mutation to the corresponding TRAF2 sequence), I72K (TRAF3), I72F (TRAF5), L74H (TRAF3 and 5), and L74R (TRAF2). Approximate elution positions of molecular weight standards are shown. (f) Promotion of poly-Ub chain synthesis by wild type and mutant TRAF6 RZ123 in the presence of the E2 complex Ubc13/Uev1A and E1.
Despite the general resemblance, a structural comparison showed that there is marked structural plasticity at these E3/E2 interfaces. This is true even when comparing the TRAF6/Ubc13 interaction with the CHIP/Ubc13 interaction in which Ubc13 is the E2 in both complexes. When TRAF6 is aligned with CHIP, Ubc13 molecules in the two complexes exhibit rotational differences of approximately 10°. Some residues such as R6 of Ubc13 at the interface with TRAF6 show Cα position differences of up to 2 Å with the Ubc13 in complex with CHIP. Larger differences are also seen elsewhere in the Ubc13 structure. In addition, the interfacial residues exhibit considerable side chain conformational variability (Supplementary Fig. 4, Supplementary Fig. 5).
In the TRAF6/Ubc13 complex, seven residues within the RING domain of TRAF6, Glu69, Pro71, Ile72, Leu74, Met75, Ala101 and Pro106, form the major contact site with Ubc13 (Fig. 1f, 2b). Among these interactions, Ile72 and Leu74 are completely buried at the interface and contribute the most surface areas. Residues Ile72 and Leu74 of TRAF6 correspond to Ile236 and Phe238 of CHIP, respectively. Glu69 of TRAF6, which is Cys233 in CHIP, forms salt bridge with Arg14 of Ubc13. In the CHIP/Ubc13 complex, the same Arg14 residue is hydrogen bonded with Asp230 of CHIP (Supplementary Fig. 5).
Surprisingly, in addition to the RING of TRAF6, residues preceding it (residues 54-66) contribute further to the Ubc13 interaction, which may be the reason for an enhanced affinity in comparison with the RING domain per se. At least two residues in this region, Gln54 and Asp57, form direct interactions with Ubc13, with Asp57 in salt bridges with Arg6 and Lys10 of Ubc13 (Fig. 2b). These interactions are completely absent in the CHIP/Ubc13 complex and Arg6 of Ubc13 points to opposite directions in the two complexes. Involvement of Gln54 and Asp57 preceding the RING domain in Ubc13 interaction revealed an indirect structural role of the first zinc finger in Ubc13 interaction. Residues 59-61 in the sequence preceding the RING domain form a β-strand (β1) that interacts with the β-hairpin (β5 and β6) in the first zinc finger (Fig. 1e, 1f). This interaction is important for maintaining the proper conformation of the region for residues such as Gln54 and Asp57 to interact with Ubc13. The TRAF6 RING domain protein containing the preceding sequence but without the first zinc finger (RING, residues 50-120) did not form a complex with Ubc13 on gel filtration chromatography (Fig. 1b). Participation of residues preceding the RING in interaction with other E2s has also been observed in the c-Cbl complex with UbcH7 23, in which the linker helix of c-Cbl contacts the similar N-terminal region of the E2.
Structure based mutations confirmed the TRAF6/Ubc13 interaction
We designed structure-based mutations on TRAF6 residues Ile72 and Leu74, which bury the most surface areas, and on Asp57, a residue preceding the RING and contributing polar contacts with Ubc13. Mutations D57K, I72D, L74E, and L74K were generated with the goal of disrupting the TRAF6/Ubc13 interaction. Gel filtration analysis using the TRAF6 RZ123 protein showed that all mutants no longer interacted with Ubc13 (Fig. 2c). These results were further confirmed by yeast two hybrid assays using full length TRAF6 (Supplementary Methods and Supplementary Fig. 6), demonstrating the importance of these residues in Ubc13 interaction.
Other TRAFs exhibit undetectable interactions with Ubc13
While all interfacial residues with Ubc13 are conserved among TRAF6 sequences from different species, most are not conserved in TRAF2, TRAF3 and TRAF5 (Fig. 1f). This is surprising because at least TRAF2 and TRAF5 are known to mediate NF-κB activation through K63-linked polyubiquitination2,27–29. Indeed, using yeast two hybrid assays, we showed that full length TRAF2, TRAF3 and TRAF5 did not interact with Ubc13 while TRAF6 interacted well with Ubc13 (Fig. 2d).
To understand the molecular basis for the lack of interactions, we generated mutations on TRAF6 that switch to the corresponding sequences in these TRAFs, I72A (TRAF2), I72K (TRAF3), I72F (TRAF5), L74H (TRAF3 and 5) and L74R (TRAF2) (Fig. 1f). Using gel filtration and yeast two hybrid analyses, all these mutants were shown to be defective in their interactions with Ubc13 in comparison with the wild type (Fig. 2e, Supplementary Fig. 6), confirming that these substitutions could underlie the failure of these TRAFs to interact with Ubc13. Therefore, despite being in the same family of signaling proteins, TRAF2, TRAF3 and TRAF5 do not use the dimeric E2 Ubc13/Uev1A. Evolutionarily, TRAF6 is the oldest TRAF family member. It is likely that an initial ability to interact with Ubc13 becomes lost. One way that this loss is compensated may be through association with a Ubc13-interacting E3. In this regard, it is known that TRAF2 is constitutively associated with RING-containing proteins cIAP1 and cIAP2 30. Sequence analysis predicts that cIAPs can interact with Ubc13 (Supplementary Fig. 3) and in vitro they promote polyubiquitination in the presence of Ubc13 (data not shown). These observations are consistent with recent reports showing the role of cIAPs in formation of the TNF receptor signaling complex 31 and the critical roles of cIAPs in TNF-mediated NF-κB activation32.
Poly-Ub synthesis, auto-ubiquitination and NF-κB activation
Because many E3s promote poly-Ub synthesis 11, we tested the ability of TRAF6 to stimulate poly-Ub chain assembly by the E2 complex Ubc13/Uev1A in vitro. TRAF6 RZ123 (residues 50-211), as well as RZ1 (residues 50-159) and RZ1234 (residues 50-279), strongly promoted poly-Ub chain synthesis in the presence of E1, E2, Ub and ATP (Fig. 2f and data not shown). The interaction between TRAF6 and Ubc13 is required as TRAF6 mutants generated to disrupt the interaction or to mimic other TRAFs were all defective in promoting poly-Ub chain synthesis (Fig. 2f).
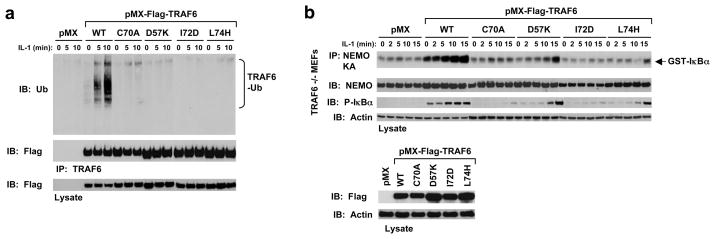
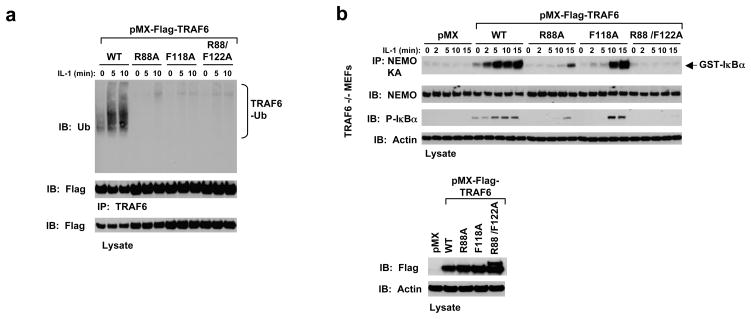
TRAF6 undergoes auto-ubiquitination upon receptor stimulation by ligands such as IL-1 and this auto-ubiquitination is required for NF-κB activation 8. To determine whether the observed TRAF6/Ubc13 interaction is also required for TRAF6 auto-ubiquitination and NF-κB activation upon stimulation, we used wild type and mutant TRAF6 to rescue TRAF6-deficient mouse embryonic fibroblasts (MEFs). Retroviral infection of wild type TRAF6 rescued TRAF6 auto-ubiquitination upon IL-1 treatment (Fig. 3a). In contrast to the wild type, TRAF6 with disruptive mutations for Ubc13 interaction failed to rescue TRAF6 auto-ubiquitination in response to IL-1 (Fig. 3a). Furthermore, we determined whether IKK was activated in these cells by pulling down IKK using antibody against the IKK regulatory subunit NEMO. In vitro kinase assay in phosphorylating purified GST-IκBα showed that IKK was active only in MEFs infected with wild type TRAF6, and not Ubc13-binding defective TRAF6 mutants (Fig. 3b). These data support a critical role of the observed Ubc13 interaction in TRAF6 function in cells.
Figure 3.
Cellular effects of TRAF6 mutants that fail to interact with Ubc13. (a) TRAF6 mutants defective in Ubc13 interaction failed to rescue IL-1-induced TRAF6 auto-ubiquitination in TRAF6-deficient MEFs. The indicated stable cells lines were treated with IL-1 (1 ng/ml) for the indicated times and the clarified lysates were immunoblotted with the indicated antibodies. (b) TRAF6 mutants defective in Ubc13 interaction failed to rescue IL-1-induced IKK activation and IκB phosphorylation.
TRAF6 dimerization in the crystal and in solution
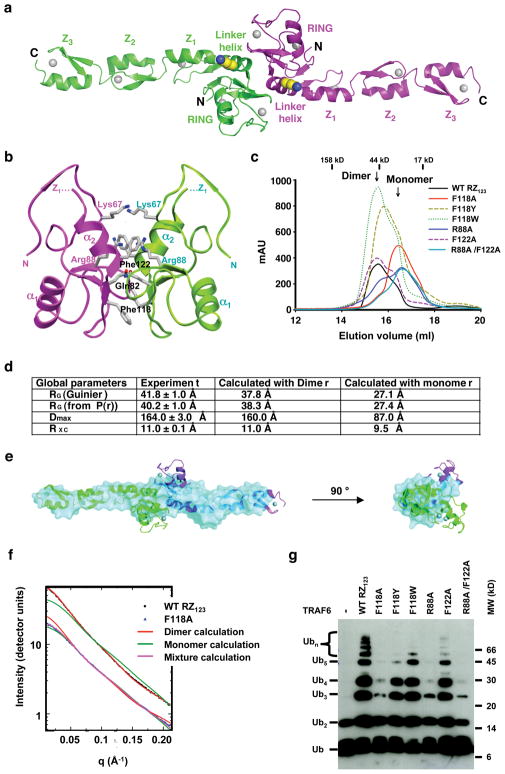
The N-terminal region of TRAF6 forms a strikingly elongated non-crystallographic dimer in the crystal (Fig. 4a). We did not anticipate this because the C-terminal coiled coil and TRAF-C domains form trimeric structures 3,4,6,7. The dimeric N-terminal domain of TRAF6 causes a symmetry mismatch in the full-length TRAF6 structure. The dimerization interface is formed via the RING and part of the linker helix α2, and buries a total of 1,270 Å2 surface areas, most of which are hydrophobic (Fig. 1f, 4a, 4b). Along the 2-fold axis, the solvent exposed hydrophobic side chains of Phe118 of both monomers stack against each other and likely form the core of the interface. At the adjacent region, Lys67, Gln82, Arg88 and Phe122 form another patch of the interface.
Figure 4.
TRAF6 dimerization is crucial for its ability to promote poly-Ub chain synthesis. (a) Dimeric structure of TRAF6 RZ123, shown with the two fold axis perpendicular to the page. The major auto-ubiquitination residue K124 is shown in ball-and-stick model. (b) Detailed interactions at the TRAF6 dimerization interface. (c) Superimposed gel filtration profiles of wild type and dimerization mutants of TRAF6 RZ123. (d) Global shape parameters derived from SAXS of wild type TRAF6 RZ123 and in comparison with those calculated from monomeric and dimeric TRAF6 crystal structure. ± refers to the standard error of the mean of the fitting. (e) Solution structure from SAXS data on TRAF6 Z123. The averaged SAXS envelope (cyan) generated from 10 independent runs of DAMMIN 44 is overlaid on the crystallographic dimer shown in a ribbon representation. DAMMIN fits the scattering data directly and does not require the number of amino acids as an input. In addition no assumptions on symmetry were applied. (f) Experimental and calculated scattering profiles from the wild type and the F118A mutant of TRAF6 RZ123. The experimental scattering profile from the wild type TRAF6 (black circles) is well fit by the scattering profile calculated from the dimer found in the crystal structure (red) and poorly fit by the monomer (green). The experimental scattering profile from the F118A mutant (blue triangles, and artificially scaled down for clarity) is best fit by a mixture of monomers and dimers (magenta). (g) Promotion of polyubiquitin chain synthesis by wild type and dimerization mutants of TRAF6 RZ123.
We generated potentially disruptive TRAF6 mutations F118A, F122A, R88A and the double mutation R88A/F122A, and the more conservative substitutions F118Y and F118W on TRAF6 RZ123. In comparison with wild type RZ123, the gel filtration elution positions of F118A, R88A and the R88A/F122A double mutants of TRAF6 RZ123 shifted towards lower molecular weight (Fig. 4c). This suggests that the F118A, R88A and the R88A/F122A double mutations disrupted TRAF6 dimerization. The elution position of F118Y shifted slightly towards lower molecular weight, suggesting that it is partially defective in dimerization.
To determine if the TRAF6 dimer in solution is the elongated dimer we observed in the crystal (Fig. 4a), we performed small angle X-ray scattering (SAXS) on the wild type RZ123 protein of TRAF6 and its F118A mutant. The scattering profile from wild type TRAF6 was used to derive its global shape parameters such as the overall radius of gyration, maximum dimension, and the radius of gyration of the cross section. These model-independent, experimentally derived parameters showed excellent agreement with those calculated from the dimeric, but not the monomeric, TRAF6 crystal structure (Fig. 4d). Low resolution shape reconstruction from the SAXS data of the wild type TRAF6 showed an elongated molecular envelope that superimposes well with the dimeric TRAF6 structure (Fig. 4e). The only region of TRAF6 that protrudes outside the molecular envelope corresponds to the α1-β4 loop, a region with high B factors in the crystal.
We analyzed the scattering profiles measured from wild type and the F118A mutant TRAF6 by comparing them against those calculated from the crystal structures (Fig. 4f). The χ2 value, as defined in the program CRYSOL 33, was used as a metric of agreement. For wild type TRAF6, the scattering profile matched that calculated from the dimeric crystal structure with a good χ2 agreement of 1.41 while that from a monomer gave a poor χ2 agreement of 11.2. The SAXS curve of the F118A mutant fit to a mixture of 44% monomer and 56% dimer with a good χ2 agreement of 1.30. As compared with that from the gel filtration profile, this apparent higher dimeric proportion may be due to the much higher protein concentration used in the SAXS measurement. These data demonstrated that TRAF6 exists as the elongated dimer in solution.
TRAF6 dimerization is critical for its biological functions
Given the dimeric structure of TRAF6, we were wondering if this dimerization is important for TRAF6 function. Interestingly, residues at the dimerization interface are much more conserved than residues for Ubc13 interaction (Fig. 1f). They are conserved not only among the different species of TRAF6 but also among TRAF2, TRAF3 and TRAF5. Remarkably, TRAF6 RZ123 mutants with defective dimerization were impaired in their ability to promote poly-Ub chain synthesis in vitro (Fig. 4g). The three TRAF6 mutants that were most defective in dimerization, F118A, R88A and R88A/F122A, were severely impaired in poly-Ub synthesis with R88A/F122A being the most impaired. F118Y was partially impaired in its ability to promote poly-Ub formation, consistent with its partial defectiveness in dimerization. Therefore, a high correlation was present in the dimerization tendencies of wild type and mutant TRAF6 and their E3 activities. The TRAF6 dimerization interface is away from the Ubc13 interaction interface and we confirmed the ability of the dimerization mutants in interacting with Ubc13 (Supplementary Fig. 7).
TRAF6 is both an E3 and a substrate that undergoes functionally important auto-ubiquitination 8. Full-length TRAF6 defective in dimerization also failed to undergo auto-ubiquitination (Fig. 5a). The major auto-ubiquitination site in TRAF6 has been mapped to Lys124 8, which resides on the linker helix between the RING domain and the first zinc finger domain (Fig. 1f). Although Lys124 resides on the same linker helix involved in dimerization, its side chain protrudes away from the interface (Fig. 4a). In fact, the K124R mutant with defective TRAF6 auto-ubiquitination 8 is still dimeric and interacts with Ubc13 (data not shown). Consistent with the defective auto-ubiquitination, dimerization mutants of TRAF6 also exhibited impaired abilities to restore IKK activation in TRAF6−/− MEFs (Fig. 5b). The R88A/F122A mutant was the most defective, with R88A and F118A showing residual and delayed IKK activation, consistent with the severity of impairment in poly-Ub synthesis in vitro (Fig. 4g).
Figure 5.
Cellular effects of TRAF6 mutants that fail to dimerize. (a) TRAF6 dimerization mutants failed to rescue IL-1-induced TRAF6 auto-ubiquitination in TRAF6-deficient MEFs. The indicated stable cells lines were treated with IL-1 (1 ng/ml) for the indicated times and the clarified lysates were immunoblotted with the indicated antibodies. (b) TRAF6 dimerization mutants failed to rescue IL-1-induced IKK activation and IκB phosphorylation.
TRAF6 dimerization induces higher order oligomerization
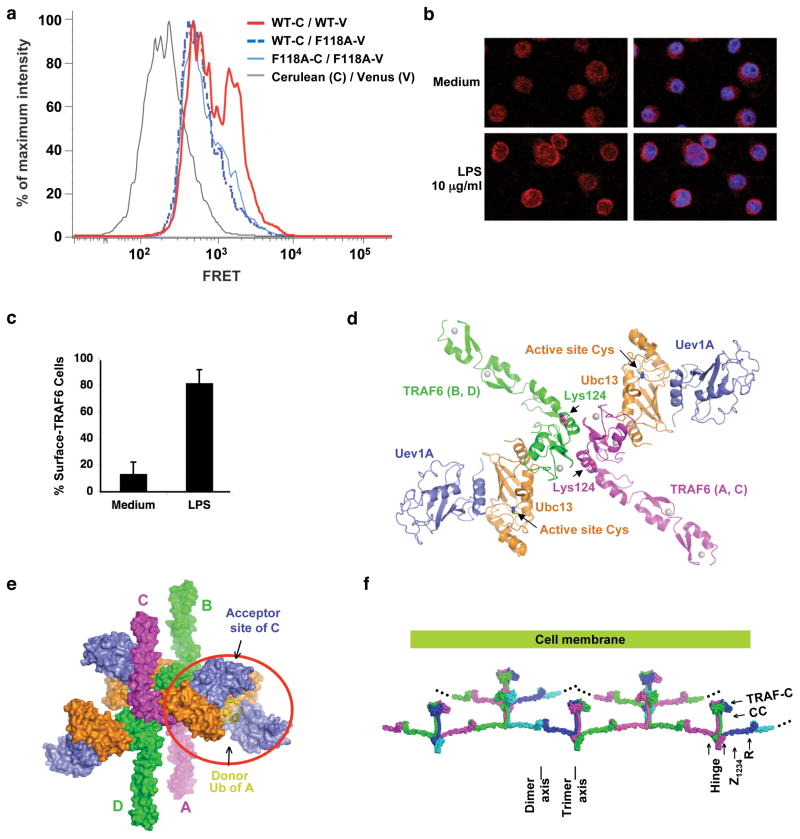
Because the C-terminal region of TRAF6 is trimeric, we wondered what kind of oligomer is full-length TRAF6 and what consequence disrupting dimerization would lead to. To determine this, we fused TRAF6 to the N-terminus of Cerulean or Venus and performed fluorescence energy transfer (FRET) experiments. When BJAB lymphoma cells were co-transfected with wild type TRAF6-Cerulean and TRAF6-Venus fusion constructs, intense FRET signals were observed (Fig. 6a). In contrast, when the F118A-Venus fusion construct was co-transfected with either wild type TRAF6-Cerulean or F118A-Cerulean, much less FRET signals were observed (Fig. 6a). The larger FRET signals correlated with a spontaneous aggregation seen under confocal microscopy of wild type TRAF6 in BJAB cells upon expression of the fusion constructs (data not shown). In contrast, the F118A mutant fusion constructs only gave a smeared expression pattern, suggesting that TRAF6 dimerization is also critical for higher order oligomerization of TRAF6.
Figure 6.
TRAF6 oligomerization. (a) Measurement of TRAF6 self-association by FRET. BJAB lymphoma cells were co-transfected with Cerulean- (C) and Venus- (V) fusion constructs for expressing wild type or F118A mutant TRAF6 fusion proteins as indicated. The intensity of FRET signals were determined by flow cytometry, gating on live Cerulean positive cells that had been excited by a 407nm laser and measuring emission post a 550/50nm band-pass filter. (b) Endogenous TRAF6 aggregation at the cell surface during signaling. Distribution of intracellular TRAF6 (red) in BJAB cells changes from diffused toward membranous condensed patterns upon LPS stimulation for 2 hours. Hoechst 33342 (blue) specifies nuclei. Error bars refer to the standard deviations of the measurements. (c) The percentages of cells having surface TRAF6 in unstimulated and LPS stimulated samples are shown. Data represents more than 10 images from 3 independent experiments. (d) A dimeric TRAF6/Ubc13/Uev1A complex built based on the dimeric TRAF6 structure, the dimeric TRAF6/Ubc13 structure and the Ubc13/Mms2 structure 14. TRAF6 auto-ubiquitination sites at K124 and the active site locations of Ubc13 are shown. (e) A potential tetrameric assembly of the TRAF6/Ubc13/Uev1A complex mediated by the interaction between a loaded Ub on Ubc13 (yellow, stick model) of one dimeric complex and an acceptor site on Uev1A of another dimeric complex. (f) A model of oligomerized full-length TRAF6 in the activated state, showing the potential for infinite aggregation. TRAF6 is shown in 4 alternative colors (magenta, green, cyan and blue) and is modeled by piecing together the N-terminal region structure and the C-terminal region structure. Zinc finger 4 is modeled based on its predicted relationship with zinc finger 3. The linker between zinc finger 4 and the coiled coil may act as a flexible hinge.
To determine whether spontaneous aggregation is also observed upon receptor stimulation under endogenous conditions, we used lipopolysaccharides (LPS) to stimulate Toll-like receptors on BJAB cells. Endogenous TRAF6 was detected using anti-TRAF6 antibody followed by visualization with Alexa Fluor 568-coupled secondary antibody. Comparison of TRAF6 distribution before and after LPS treatment showed a clear coalescence of TRAF6 in discernable clusters to the cell surface upon activation (Fig. 6b, 6c). The fact that the coalescence is visible microscopically suggests that a large number of individual TRAF6 molecules participate in this signaling induced aggregation, similar to the spontaneous aggregation observed under TRAF6 over-expression.
DISCUSSION
RING domains and their variants may comprise the largest family of ubiquitin ligases 19. The established concept is that the RING interacts with E2 to bring the substrate to the proximity of E2 for ubiquitination. Our studies show that the RING of TRAF6 does not function alone, but function together with the neighboring sequences in TRAF6 including residues preceding the RING and a structural role of the first zinc finger. Despite the availability of several E2/E3 complex structures 26,27, these details in the interaction of TRAF6 with Ubc13 could not have been predicted. Surprisingly, this interaction is specific for TRAF6, but not other members of the TRAF family, perhaps highlighting the unique critical biological roles of TRAF6 in multiple receptor signaling pathways.
Despite the trimeric symmetry of the C-terminal domain of TRAF6, the N-terminal domain of TRAF6 is dimerized via its RING domain and the linker helix to enable the formation of a dimeric TRAF6/Ubc13/Uev1A complex (Fig. 6d). While RING dimerization has been observed before 34,35, its functional consequence has not been fully investigated. In the case of TRAF6, dimerization is important for its ability to promote poly-Ub synthesis, auto-ubiquitination and NF-κB activation and correlates with its higher order oligomerization. However, how TRAF6 dimerization facilitates these activities remains to be elucidated. For synthesizing long poly-Ub chains, it is crucial to preferentially re-use poly-Ub intermediates in the following cycle of ubiquitination. Therefore, a simplest explanation might be that TRAF6 dimerization results in increased local concentration of the Ubc13/Uev1A complex to trap locally generated poly-Ub intermediates. Heterologous dimerization has been shown to activate the E2 cdc34 E2 directly in the absence of E3 36. In addition, when Ubc13 is loaded with donor Ub, the E2/E3 complex dimers may further tetramerize through the low affinity interaction between Uev1A and the Ub 14 (Fig. 6e). The tendency to form oligomers for Ub-loaded E2 UbcH5 has been shown and is required for processive BRCA1-directed ubiquitination 37.
For TRAF6 auto-ubiquitination, it is possible that dimerization of the TRAF6 N-terminal region is required because it promotes higher order aggregation. We have shown that the N-terminal region per se is not sufficient for TRAF6 auto-ubiquitination in cells; instead, the trimeric coiled coil region is also needed (data not shown). On the molecular level, the observed higher order aggregation of TRAF6 is consistent with an infinite expandable model of full-length TRAF6 in its activated state as a consequence of the symmetry mismatch between its N- and C-terminal regions. In this model, the dimeric and trimeric symmetry axes are roughly parallel to each other and both perpendicular to the membrane surface to enable multiple engagements of receptor trimers (Fig. 6f). The size of the aggregate may depend on TRAF6 concentration, receptor engagement and interaction with other signaling proteins in the cell. Both dimerization and trimerization would cooperate in this model to form the lattice of interactions and the expanded lattice would in turn strengthen dimerization and trimerization. It is likely that the lattice of interactions, as well as the inherent flexibility at the N- and C-terminal region junctions, provides the necessary geometry required for transfer of poly-Ub chains, in trans, from one E2 to the TRAF6 auto-ubiquitination site at K124 of another TRAF6/E2 complex.
In the context of this model, the difference in FRET signals between wild type TRAF6 and the F118A mutant may have been resulted from stabilization of TRAF6 trimerization in the expanded lattice of interactions. The distance between the C-terminus of TRAF6 in the protomers of the trimer is approximately 26 Å in the crystal structure of the C-terminal region of TRAF6 3,5, while the estimated distance between dimerically related C-termini is more than 200 Å. Alternatively, because the linker region between the end of zinc finger 4 and the beginning of the coiled coil is predicted to lack secondary structures and therefore may be flexible, it is also possible that different C-terminal trimers may be brought into close proximity as well in this higher order aggregation, leading to enhanced FRET signals.
Therefore, our studies have not only unveiled interesting aspects of TRAF6 E2 interaction and dimerization, but also revealed an unexpected platform of oligomerization in mediating TRAF6 function. The massive increase in local concentrations of the associated signaling proteins in this aggregation platform likely acts as a factory to promote poly-Ub synthesis, auto-ubiquitination and recruitment of downstream proteins such as the TAK1 complex and the IKK complex. In keeping with this concept, recent studies suggest that the critical role of oligomerization in ubiquitination in general may be more prevalent than previously anticipated 38–40. Interestingly, the proposed model of TRAF6 aggregation bears unexpected similarity to assembly of the death receptor signaling complex upon activation of Fas, a death receptor in the TNF receptor superfamily 41–43. The aggregations in both systems could provide an elegant scaffold to facilitate proximity-induced caspase activation, ubiquitination or kinase activation. On a conceptual level, these observations signify a convergence of the signaling mechanisms of caspase mediated death pathways and the TRAF mediated survival pathways in immune receptor signal transduction.
METHODS
Protein expression, purification and mutagenesis
We constructed human TRAF6 RING (residues 50-120), RZ1 (residues 50-159), RZ12 (residues 50-187), RZ123 (residues 50-211) and RZ1234 (residues 50-279) with C-terminal polyhistidine tags and human Ubc13 and Uev1A with N-terminal polyhistidine tags. We expressed these proteins in BL21-CodonPlus®(DE3) cells and purified them by Ni-affinity chromatography (Qiagen) and gel filtration chromatography (Superdex 200, GE Healthcare). We removed the N-terminal polyhistidine tags by thrombin (GE Healthcare) cleavage. To obtain the Ubc13/Uev1A complex, we mixed equimolar purified tagless Ubc13 and Uev1A and passed the mixture through the gel filtration column. For complex formation with Ubc13, we incubated a given TRAF6 construct (concentrations from 0.16 to 0.6 mM) with excess Ubc13 (molar ratios from 1:1.2 to 1:2) for gel filtration chromatography. Injection volumes ranged from 450μl to 735 μl. We performed mutagenesis using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene).
Crystallization and structure determination
We crystallized TRAF6 RZ123 and the RZ1/Ubc13 complex using hanging drop vapor diffusion at 20 °C. The crystallization condition for RZ123 is 1.19–1.33 M (NH4)2SO4 and 0.1 M CHES (pH 9.4–9.5). Crystals formed in space group C2 with two molecules per crystallographic asymmetric unit. The crystallization condition for the RZ1/Ubc13 complex in both the P1 and the C2 space groups is 8 % (w/v) PEG 4000. The P1 and C2 crystals contain one and two complexes per crystallographic asymmetric unit, respectively (Table 1). We collected all diffraction data at beam line X4A of Brookhaven National Lab and processed them using HKL2000 45.
We determined the RZ123 structure by single wavelength anomalous diffraction (SAD) 46 from the intrinsic zinc atoms using the program SOLVE and RESOLVE 47. We performed iterative model building in WinCoot 48 and refinement in CNS 1.2 49. We used molecular replacement to solve the RZ1/Ubc13 structure using the CCP4 suite 50. We generated all structural presentations using Pymol (DeLano Scientific) and Setor 51. The final atomic models of RZ123 in the C2 space group and RZ1/Ubc13 in the P1 and C2 space groups contain 86.8%, 83.5% and 79.2% residues, respectively, in the most favored regions and 100%, 100% and 99.5% residues, respectively, in the allowed regions of the Ramachandran Plots.
Ubiquitination assays
We incubated 100 nM mouse E1, 200 nM E2, 2μM wild type or mutant RZ123, RZ1 or RZ1234, and 20 μM Ub at 37 °C for 45 min in reaction buffer containing 25 mM Tris-HCl at pH 7.5, 2.5 mM MgCl2, 0.1 mM DTT, 2 mM ATP, 5 mM creatine phosphate, 0.6 units ml−1 creatine kinase and 0.6 units ml−1 inorganic pyrophosphatase. We quenched the reactions using SDS loading buffer and resolved them on 15 % (v/v) SDS-PAGE. We transferred the protein bands to PVDF membranes using a Tran-Blot® SD Semi-Dry Transfer Cell (BioRad). We probed the membranes with anti-Ub primary antibody (Santa Cruz Biotechnology) in 1:1000 dilution, washed them and then incubated them with HRP-linked anti-mouse secondary IgG (Cell Signaling) in 1:4000 dilution. We used the SuperSignal West Pico Trial Kit (Pierce) to visualize the reactive bands as instructed by the manufacturer.
Surface plasmon resonance (SPR)
We assayed the interaction between TRAF6 RZ123 and Ubc13 at 25 °C using a Biacore 2000 optical biosensor equipped with a CM4 sensor chip and equilibrated with 20 mM Tris-HCl at pH 7.5, 100 mM NaCl, 5 mM DTT, 0.005 % (v/v) Tween-20, and 0.1 mg ml−1 BSA. We immobilized TRAF6 RZ123 using amine coupling chemistry to sensor chips at 580 and 900 RU respectively in running buffer containing 20 mM Tris-HCl at pH 7.5, 100 mM NaCl, and 5 mM DTT. We test binding of Ubc13 to the surface-tethered TRAF6 in two-fold dilution series of 0.12, 0.23, 0.47, 0.94, 1.88, 3.75, 7.5, and 15.0 μM. We double referenced the binding responses 52 and fit them to a simple binding isotherm to determine the affinities using the program Scrubber 2 (BioLogic Software Ltd., Campbell, Australia).
Small angle X-ray scattering (SAXS)
We collected SAXS data at the SIBYLS beam line (12.3.1) at the Advanced Light Source in Lawrence Berkeley National Laboratory. The incident wavelength used in the experiment was 1.54 Å with a q range of 0.011 – 0.21 Å−1 (q = 4π sin (θ/2)/λ where θ is the scattering angle and λ is the wavelength). The detector was a MAR 165 CCD area detector. We used a 1 mm thick cuvette with sample volumes of 15 μL. We collected data sets at concentrations of approximately 10 mg ml−1 for both the wild type and the F118A mutant and processed them similarly as described previously 53. Briefly, we used a Guinier plot from scattering curves extrapolated to zero concentration to determine the radius of gyration RG (Guinier) using the freeware program PRIMUS 54. We subjected the scattering curves to an indirect Fourier transform using the GNOM program 55 to yield the pair distribution function P(r), from which we derived the maximum dimension and the radius of gyration RG (P(r)) 56. We also generated the low resolution ab initio model from the values of GNOM output using the dummy atom approach as implemented in GASBOR 57. Ten runs of GASBOR with and without 2 fold symmetry yielded similar results. Results from each independent run agreed well with one another as judged by the program DAMAVER 58. We calculated the scattering profiles from PDB files using the program CRYSOL 33 with an option which best fits the data by adjusting properties of the hydration shell. We used the algorithm for elongated proteins to extract the radius of gyration of cross-section (Rxc) 59.
Supplementary Material
Acknowledgments
We thank Drs. Tongpil Min and Jee Y. Chung for their earlier work on the project, Dr. Xuejun Jiang and Dr. Xinjiang Wang of the Sloan-Kettering Institute for purified E1, Dr. Zhijian (James) Chen of University of Texas Southwestern Medical School for the expression constructs of Ubc13 and Uev1A, Randy Abramowitz and John Schwanof of X4A of NSLS for data collection, and Jin Wu for maintaining our X-ray and computer equipment. This work was supported by National Institute of Health (RO1 AI045937 to HW and RO1 AR053540 to BGD), Department of Defense (DOE Contract DE-AC02-05CH11231 for GH), the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (to LZ and MJL) and institutional start-up funds to BGD. SCL and YCL were postdoctoral fellows of the Cancer Research Institute and ML was a postdoctoral fellow of the American Heart Association.
Footnotes
Accession codes. Protein Data Bank: Coordinates for TRAF6 and the TRAF6/Ubc13 complex in two crystal forms have been deposited with accession code XXXX, XXXX and XXXX, respectively.
References
- 1.Wu H. Assembly of post-receptor signaling complexes for the tumor necrosis factor receptor superfamily. Adv Protein Chem. 2004;68:225–79. doi: 10.1016/S0065-3233(04)68007-7. [DOI] [PubMed] [Google Scholar]
- 2.Pineda G, Ea CK, Chen ZJ. Ubiquitination and TRAF signaling. Adv Exp Med Biol. 2007;597:80–92. doi: 10.1007/978-0-387-70630-6_7. [DOI] [PubMed] [Google Scholar]
- 3.Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398:533–8. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- 4.McWhirter SM, et al. Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc Natl Acad Sci U S A. 1999;96:8408–13. doi: 10.1073/pnas.96.15.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye H, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–7. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 6.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 8.Lamothe B, et al. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem. 2007;282:4102–12. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 10.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 12.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–50. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 13.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–20. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 15.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–65. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105:711–20. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 17.Moraes TF, et al. Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13. Nat Struct Biol. 2001;8:669–73. doi: 10.1038/90373. [DOI] [PubMed] [Google Scholar]
- 18.McKenna S, et al. Noncovalent interaction between ubiquitin and the human DNA repair protein Mms2 is required for Ubc13-mediated polyubiquitination. J Biol Chem. 2001;276:40120–6. doi: 10.1074/jbc.M102858200. [DOI] [PubMed] [Google Scholar]
- 19.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 20.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–4. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 21.Huang DT, et al. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol Cell. 2005;17:341–50. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 22.Mercier P, et al. Structure, interactions, and dynamics of the RING domain from human TRAF6. Protein Sci. 2007;16:602–14. doi: 10.1110/ps.062358007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–9. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M, et al. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–38. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Xu Z, et al. Structure and interactions of the helical and U-box domains of CHIP, the C terminus of HSP70 interacting protein. Biochemistry. 2006;45:4749–59. doi: 10.1021/bi0601508. [DOI] [PubMed] [Google Scholar]
- 26.Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–5. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothe M, Wong SC, Henzel WJ, Goeddel DV. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–92. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 28.Nakano H, et al. TRAF5, an activator of NF-kappaB and putative signal transducer for the lymphotoxin-beta receptor. J Biol Chem. 1996;271:14661–4. doi: 10.1074/jbc.271.25.14661. [DOI] [PubMed] [Google Scholar]
- 29.Ishida TK, et al. TRAF5, a novel tumor necrosis factor receptor-associated factor family protein, mediates CD40 signaling. Proc Natl Acad Sci U S A. 1996;93:9437–9442. doi: 10.1073/pnas.93.18.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–52. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 31.Santoro MM, Samuel T, Mitchell T, Reed JC, Stainier DY. Birc2 (cIap1) regulates endothelial cell integrity and blood vessel homeostasis. Nat Genet. 2007;39:1397–402. doi: 10.1038/ng.2007.8. [DOI] [PubMed] [Google Scholar]
- 32.Mahoney DJ, et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105:11778–83. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svergun D, Baraberato C, Koch MH. CRYSOL - a Program to evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 34.Kostic M, Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J Mol Biol. 2006;363:433–50. doi: 10.1016/j.jmb.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 35.Knipscheer P, Sixma TK. Protein-protein interactions regulate Ubl conjugation. Curr Opin Struct Biol. 2007;17:665–73. doi: 10.1016/j.sbi.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Gazdoiu S, et al. Proximity-induced activation of human Cdc34 through heterologous dimerization. Proc Natl Acad Sci U S A. 2005;102:15053–8. doi: 10.1073/pnas.0507646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–80. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–43. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 39.Tang X, et al. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–76. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 40.Peschard P, et al. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol Cell. 2007;27:474–85. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Yang JK, et al. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol Cell. 2005;20:939–49. doi: 10.1016/j.molcel.2005.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrington PE, et al. The structure of FADD and its mode of interaction with procaspase-8. Mol Cell. 2006;22:599–610. doi: 10.1016/j.molcel.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 43.Siegel RM, et al. SPOTS: signaling protein oligomeric transduction structures are early mediators of death receptor-induced apoptosis at the plasma membrane. J Cell Biol. 2004;167:735–44. doi: 10.1083/jcb.200406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–86. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Hendrickson WA. Analysis of protein structures from diffraction measurements at multiple wavelengths. Trans Am Crystallogr Assoc. 1985;21:11. [Google Scholar]
- 47.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 50.Collaborative Computational Project, N. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 51.Evans SV. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J Mol Graph. 1993;11:134–8. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 52.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–84. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 53.Iyer RR, et al. The MutSalpha -PCNA interaction in human DNA mismatch repair. J Biol Chem. 2008 doi: 10.1074/jbc.M800606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konarev PV, Volkov VV, Sokolova AV, MHJK, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
- 55.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
- 56.Nagar B, et al. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol Cell. 2006;21:787–98. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 57.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–53. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Cryst. 2001;34:33–41. [Google Scholar]
- 59.Glater O, Kratky O. Small Angle X-ray Scattering. Academic Press; London: 1982. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.