Abstract
Recent advances in mouse genomics have revealed considerable variation in the form of single-nucleotide polymorphisms (SNPs) among common inbred strains. This has made it possible to characterize closely related strains and to identify genes that differ; such genes may be causal for quantitative phenotypes. The mouse strains DBA/1J and DBA/2J differ by just 5.6% at the SNP level. These strains exhibit differences in a number of metabolic and lipid phenotypes, such as plasma levels of triglycerides (TGs) and HDL. A cross between these strains revealed multiple quantitative trait loci (QTLs) in 294 progeny. We identified significant TG QTLs on chromosomes (Chrs) 1, 2, 3, 4, 8, 9, 10, 11, 12, 13, 14, 16, and 19, and significant HDL QTLs on Chrs 3, 9, and 16. Some QTLs mapped to chromosomes with limited variability between the two strains, thus facilitating the identification of candidate genes. We suggest that Tshr is the QTL gene for Chr 12 TG and HDL levels and that Ihh may account for the TG QTL on Chr 1. This cross highlights the advantage of crossing closely related strains for subsequent identification of QTL genes.
Keywords: quantitative trait loci, triglycerides, body weight, Tshr, Villin, Cyp27a1, Ihh
The leading cause of mortality in developed nations is ischemic cardiovascular disease, and the pathological basis is atherosclerosis. Major risk factors for atherosclerosis are high plasma levels of triglycerides (TGs) (1) and LDL, as well as low levels of HDL cholesterol (2). High levels of plasma HDL provide protection against heart disease, as shown in both human (3–5) and animal studies (6–11). Many genes and pathways controlling LDL levels are known; however, those that control TG and HDL levels are less well characterized. A successful route toward identifying genes that affect quantitative phenotypes is through the use of inbred mouse strains and quantitative trait locus (QTL) analysis. A combination of genetic tools and databases, along with improved mapping techniques (12), are helping to identify these QTL genes (13–15).
When mapping QTLs in mouse models, it is important to use strains that differ in the phenotype of interest and to capture a large proportion of the genetic variation present in the inbred strains (16). However, in this study we used two mouse strains that differ in phenotype but are genetically closely related, demonstrating that QTL genes can be identified by using models of limited genetic variability. The strains used are DBA/1J (D1) and DBA/2J (D2).
The DBA strain was developed by Clarence Cook Little in 1909 and is the oldest of all inbred strains of mice. In 1929–30, crosses were made between substrains, and several new substrains were established, including D1 and D2. Differences between the substrains are too large to be accounted for by mutation and probably result from residual heterozygosity following the crosses between substrains (www.informatics.jax.org/external/festing/search_form.cgi). D1 and D2 have been assayed for ∼140,000 single-nucleotide polymorphisms (SNPs) (17). Less than 6% of these differed between D1 and D2, and regions that are not identical by descent (IBD) contained just 1,697 known or predicted genes. Genes that are in polymorphic locations between D1 and D2 and underlie a QTL would be regarded as candidate genes.
The D1 and D2 strains exhibit significant differences in circulating plasma levels of TG and HDL, as well as other metabolic factors such as insulin, leptin, body weight, and percent body weight as fat (18). We report the QTL found in a D1 × D2 cross for TG, HDL, and body weight and propose some candidate genes.
MATERIALS AND METHODS
Mice
DBA/1J (D1) and DBA/2J (D2) mice, and the strain BALB/cBy.RF-Tshrhyt/+/J (henceforth referred to Tshrhyt/+), carrying a mutation in the thyroid-stimulating hormone receptor were obtained from The Jackson Laboratory (Bar Harbor, ME) and maintained on a 14 h light/10 h dark cycle. Mice were housed in individually pressurized cages (Thoren Caging Systems) containing pine shaving bedding, topped with a polyester filter, and allowed ad libitum access to acidified water and a chow diet containing 6% fat by weight (LabDiet 5K52; LabDiet, Scott Distributing, Hudson, NH). Animal protocols were reviewed and approved by the Animal Care and Use Committee at The Jackson Laboratory. Tshrhyt was originally identified as a spontaneous hypothyroid mutation in an RF/J mouse and was subsequently backcrossed into a BALB/cByJ background (N10) and maintained through heterozygous × wild-type sib mating, because homozygotes are infertile (19, 20). A strain deficient in villin1 (Vil1), a protein expressed in the brush border of the intestine, was generated at the Institut Curie (21) by villin1 gene targeting in 129S2/SvPas ES cells followed by injection into C57BL6 blastocysts; chimeric mice were subsequently intercrossed with DBA/2 mice to generate homozygous Vil1−/− mice on a mixed 129S2, B6, and D2 background. This strain is named STOCK Vil1tm1Syr.
D1 and D2 mice were reciprocally mated to generate 40 mice, 20 (D1×D2) F1 and 20 (D2×D1) F1. The F1 males were mated in trios with one D1 and one D2 female; females were separated before giving birth. F1 females were paired and mated to D1 or D2 males. A total of 294 backcross mice were generated with N = 200 originating from D1×D2 grandparents and N = 94 from D2×D1 grandparents.
Phenotyping
Blood samples from mice fasted for 4 h in the morning were collected in tubes containing EDTA, centrifuged at 9,000 rpm for 5 min, and plasma was frozen at −20°C until analyzed. Plasma samples were thawed, vortexed, and analyzed within a week of being collected. Concentrations of TG, HDL, and total cholesterol were measured directly using enzymatic Reagent Kits (#650207, #467825, and #445850; Beckman Coulter, Inc.) used according to the manufacturer's recommendations on the Synchron CX Delta System (Beckman Coulter, Inc.). In addition, thyroxine (T4) was measured in male backcross plasma samples that had been frozen at −80°C (Beckman assay #445995).
D1 by D2 mice
Mice were weighed at weaning (21 days) and at each blood sampling time point: 8, 11, and 16 weeks of age. After the 8 week sample, mice were fed a high-fat (HF) diet for the remainder of the experiment. This diet contained 15% (by weight) dairy fat, 1% cholesterol, and 0.5% cholic acid and has been previously described (22).
Tshrhyt colony
Plasma from chow-fed mice was taken at 8 weeks of age and T4 as well as lipids were measured. Controls were littermate Tshr+/+ and Tshrhyt/+ mice from heterozygous matings.
Vil1-deficient colony
Plasma from chow-fed mice was taken at 12 weeks of age; controls were B6×D2 F1 mice.
Genotyping
SNP genotyping was performed on genomic DNA isolated from tails. A total of 40 markers (see supplementary Table I) were chosen from a panel designed to facilitate genotyping in inbred mouse strains (23) and was performed by the Allele Typing Service at The Jackson Laboratory in conjunction with KBiosciences (Herts, UK). Markers were chosen at evenly spaced intervals where possible. However, on many chromosomes, D1 and D2 differ at only small regions and are hence only represented by markers where polymorphisms existed. For this reason, the 95% confidence intervals of the QTLs are not meaningful and are not provided. More recently, the Broad Institute released additional SNPs that indicated polymorphic regions between D1 and D2 (www.broad.mit.edu/personal/claire/MouseHapMap) that were not represented by the Jax SNP panel. At these locations, assays were developed based on the sequencing data available from the Broad Institute.
QTL and statistical analyses
QTL analysis was performed using Pseudomarker 2.03 (http://www.jax.org/staff/churchill/labsite/software/pseudomarker) as described previously (24). Approximate cM coordinates for the SNP markers for QTL analysis were obtained by dividing Millions of base pair (Mbp) positions (mouse genome build 36) by a factor of 2 except for chromosome (Chr) 19, where we used the Mb/1.04. The validity of this approximation was confirmed by comparison to estimated map positions in Pseudomarker and also from previous cM to Mb comparisons in the mouse (25). A QTL was named if it was significant or if it was suggestive but confirmed a previously reported QTL.
Phenotypes were normally distributed and thus not transformed. Interval mapping was performed with sex as an additive covariate and also sex as an interactive covariate, which provides a test for a sex-by-QTL interaction. Genome-wide significance was determined using 1,000 permutations with stratification by sex (26). A pairscan analysis was performed to detect gene interactions; however, no significant interactions were identified. Other statistical analyses were performed using JMP version 6.0 (SAS Institute); data are presented as means ± SEM unless otherwise noted. Paired Student's t-tests were used for comparisons.
SNP and haplotype analysis
A total of 140,000 SNPs were downloaded from the Mouse Phenome Database (www.jax.org/phenome) for the strains DBA/1J and DBA/2J for direct SNP and haplotype comparisons. In addition, SNPs from C57BL/6J, 129S/SvImJ, and other strains were also used for combining haplotypes with concordant QTLs found in previously published crosses (27). Gene lists for each chromosome were obtained from build 36 of the Ensembl database (http://mouse.ensembl.org). We have implemented a function in the R computing environment (http://www.r-project.org/) to identify lists of genes in regions differing at the haplotype level between D1 and D2. Those gene lists were further reduced according to differing haplotypes of strain-pairs that gave rise to concordant QTLs.
RNA isolation, cDNA synthesis, and real-time quantitative PCR
Samples were collected from 4 hr morning-fasted male mice under conditions suitable for RNA extraction and stored in RNAlater (Ambion) overnight before being frozen at −80°C. RNA was extracted from frozen samples using TRIzol® following the manufacturer's recommendations (Invitrogen). Quality of the RNA was assessed using a 2100 Bioanalyzer instrument and RNA 6000 Nano LabChip assay (Agilent Technologies). The First-Strand cDNA synthesis kit (GE Healthcare/Amersham Biosciences) was used for first-strand cDNA synthesis.
Power SYBR® Green PCR Master Mix (Applied Biosystems) was used with a 7300 Real-time PCR system (Applied Biosystems) with accompanied software (Sequence Detector v2.0). Reactions were carried out on 96-well MicroAmp optical plates with optical adhesive tape (Applied Biosystems). For each reaction, three identical (systematic) replicates were carried out on four biological replicates. For each gene examined, identical reactions were processed on the same 96-well plate with primers for mouse Actb to normalize for variation in loading between samples (ΔCT).
Real-time quantitative PCR (RT-PCR) was performed on mouse Tshr, Vil1, Ihh, and Cyp27a1. Tshr was examined in adipose and liver mRNA, although we could not detect Tshr in the liver. Cyp27a1 was examined in liver mRNA, Vil1 was examined in intestines, and Ihh was examined in liver and intestines. Log dilution comparisons were performed between the genes under investigation and the control (Actb) to establish the working concentration at which comparable amplification is linear. Multiple primers were tested for each gene, and primer pairs with linear ΔCT amplification over a 64-fold window were selected for RT-PCR.
Design of primers was carried out using Primer3, available online at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi, using sequences from the Mouse Genome Sequencing Consortium Assembly build 37. The rodent mispriming library was chosen as a constraint for improved primer design. Tshr: F-TGCTCAAGTTTCTTGGCATTT, R-GTTTTCAGGGACCGAAGTCA; Vil1: F-TGGGACCAGGTCTTCTTCTG, R-CTCAAGGTCTCGGTTTCCAG; Ihh: F-ACCGTGACCGAAATAAGTATGG, R-GCCGAATGCTCAGACTTGAC; Cyp27a1: F-TCTGGCTACCTGCACTTCCT, R-GTGTGTTGGATGTCGTGTCC; and Actb: F-TTGGGTATGGAATCCTGTGG, R-CTTCTGCATCCTGTCAGCAA.
Sequencing
Genes were sequenced using primers designed from genomic C57BL/6J sequence to sequence the exons of each gene with 50 nucleotides of adjacent introns. Purified PCR products were sequenced by the Jackson Laboratory DNA Sequencing Service using capillary-based sequencing machines.
RESULTS
Differences between D1 and D2
Phenotypic
D1 and D2 differ in their plasma levels of TG for females fed a chow diet and for males fed either chow or a HF diet for 8 weeks (Table 1). These strains also differed in HDL for males fed the HF diet. In each case, D2 has the higher phenotypic value. Interestingly, F1 mice fed a chow diet have significantly higher levels of circulating lipids than either parent strain (Table 1). The strains also differ in insulin (D1 = 3.2 ± 0.5, D2 = 4.6 ± 0.6 ng/ml, P < 0.05), leptin (D1 = 10.7 ± 1.2, D2 = 22.1 ± 3.6 ng/ml, P < 0.005), and percent body fat (D1 = 22.5 ± 1.7, D2 = 29.1 ± 1.4 ng/ml, P < 0.005) (18).
TABLE 1.
Plasma lipid differences between DBA/1J and DBA/2J
| Female |
Male |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chow |
8 Week HF |
Chow |
8 Week HF |
|||||||
| Strain | Trait | Mean | P | Mean | P | Mean | P | Mean | P | |
| D1 | TG | 80.4 | 126.6 | 103.6 | 130.3 | |||||
| D2 | 172.1 | <0.0001a | 128.0 | 0.93 | 218.6 | <0.0001 | 190.8 | <0.0001a | ||
| F1 | D1×D2 | 257.0 | <0.0001 | 134.5 | 0.71 | 220.2 | <0.0001 | 162.7 | 0.07 | |
| F1 | D2×D1 | 198.9 | <0.0001 | 130.3 | 0.84 | 247.1 | <0.0001 | 194.4 | <0.0001a | |
| D1 | HDL | 39.3 | 74.9 | 73.7 | 118.6 | |||||
| D2 | 43.5 | 0.015 | 78.6 | 0.48 | 72.7 | 0.73 | 147.2 | <0.0001a | ||
| F1 | D1×D2 | 64.8 | <0.0001a | 86.6 | 0.07 | 92.8 | <0.0001 | 133.2 | 0.03 | |
| F1 | D2×D1 | 58.2 | <0.0001a | 84.7 | 0.11 | 90.2 | <0.0001 | 147.9 | <0.0001a | |
| D1 | TChol | 59.3 | 160.5 | 96.5 | 232.0 | |||||
| D2 | 65.4 | 0.030 | 173.8 | 0.26 | 98.7 | 0.53 | 234.8 | 0.77 | ||
| F1 | D1×D2 | 85.0 | <0.0001a | 155.0 | 0.70 | 105.4 | 0.02 | 184.6 | <0.0001a | |
| F1 | D2×D1 | 76.9 | <0.0001a | 140.0 | 0.14 | 106.2 | 0.01 | 200.6 | 0.005 | |
HF, high-fat diet. TChol, total cholesterol; TG, triglyceride. P-value is the t-test comparison within sex-diet to D1.
A Bonferroni correction for multiple testing requires a P < 0.001 for significance.
Genotypic
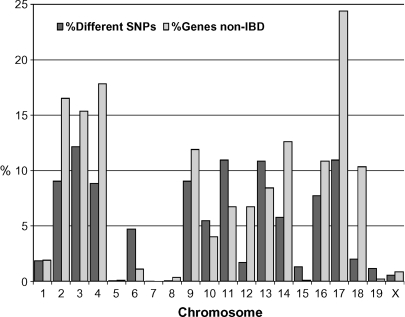
Overall, D1 and D2 differ by 5.6% when examining approximately 140,000 SNPs generated by the Broad Institute as well as several other sources (www.jax.org/phenome). This translates to 6,949 observable polymorphic SNPs representing 1,697 known or predicted genes in regions that are not IBD (6.6%). Some chromosomes, such as Chrs 5, 7, 8, 15, 19, and X are almost identical between the two strains, having between 0 and 9 genes in non-IBD regions. Chrs 2, 3, 4, and 17 have the greatest number of genes in differing haplotypes, with 15–25% of all genes located in polymorphic regions (Fig. 1).
Fig. 1.
Genotypic differences between DBA/1J and DBA/2J by chromosome. The 140,000 single-nucleotide polymorphisms (SNPs) used in this analysis were generated from multiple sources and are available through www.jax.org/phenome. IBD, identical by descent.
QTL analysis
QTL analysis was performed for each lipid trait at 8 weeks of age on a chow diet, at 11 weeks of age, after the mice had been on a HF diet for 3 weeks, and at 16 weeks of age, after the mice had been on the high fat diet for 8 weeks. If QTLs mapped within 20 cM of each other for all three diet conditions, we assigned one QTL name to represent all QTLs; however, if the distance was greater than 20 cM or there were differences in sex effects, we assigned a separate QTL name (Table 2).
TABLE 2.
TG and HDL QTL between DBA/1J and DBA/2J
| 8 Week (Chow Diet) |
11 Week (+3 Week HF) |
16 Week (+8 Week HF) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Chr | cM | LOD | Chr | cM | LOD | Chr | cM | LOD | |||
| TG | 1 | 34 | 2.6 | 1 | 34 | 2.6 | 1 | 34 | 4.1 | Tgq9 | ||
| 2 | 3 | 4.5 | Tgq10 | 2 | 27 | 5.1 | Tgq11 | |||||
| 2 | 75 | 2.9 | Tgq12a | |||||||||
| 3 | 18 | 4.7 | Tgq13 | 3 | 41 | 4.8 | Tgq14 | |||||
| 3 | 71 | 5.2 | Tgq15 | 3 | 73 | 3.9 | ||||||
| 4 | 45 | 3.5 | Tgq16 | 4 | 24 | 2.9 | ||||||
| 4 | 68 | 4.4 | Tgq17 | |||||||||
| 5 | 26 | 3.1 | 5 | 26 | 2.6 | |||||||
| 7 | 55 | 2.7 | 7 | 53 | 3.5 | |||||||
| 8 | 40 | 4.5 | Tgq18 | 8 | 41 | 2.7 | 8 | 40 | 2.9 | |||
| 9 | 11 | 3.7 | 9 | 18 | 4.1 | Tgq19 | 9 | 6 | 4.7 | Tgq19 | ||
| 10 | 52 | 4.7 | Tgq20 | 10 | 45 | 2.8 | 10 | 60 | 5.5 | Tgq21 | ||
| 11 | 25 | 4.3 | Tgq22 | 11 | 19 | 5.4 | Tgq22 | |||||
| 12 | 45 | 5.6 | Tgq23 | 12 | 45 | 3.6 | 12 | 45 | 3.7 | |||
| 13 | 60 | 3.7 | 13 | 58 | 4.3 | Tgq24 | ||||||
| 14 | 48 | 4.4 | Tgq25 | 14 | 22 | 3.5 | 14 | 25 | 4.6 | Tgq26 | ||
| 15 | 31 | 3.8 | Tgq27a | |||||||||
| 16 | 40 | 3.5 | 16 | 40 | 5.5 | Tgq28 | 16 | 40 | 3.9 | |||
| 17 | 14 | 3.6 | 17 | 16 | 3.2 | |||||||
| 18 | 7 | 3.3 | 18 | 17 | 2.5 | |||||||
| 19 | 51 | 4.8 | Tgq29 | 19 | 51 | 3.3 | 19 | 51 | 2.7 | |||
| HDL | 2 | 21 | 3.6 | 2 | 8 | 3.3 | ||||||
| 3 | 2 | 4.0 | Hdlq61 | |||||||||
| 4 | 10 | 2.5 | ||||||||||
| 4 | 60 | 2.5 | 4 | 64 | 2.9 | Hdlq64 | ||||||
| 9 | 1 | 4.1 | Hdlq62 | |||||||||
| 10 | 42 | 2.5 | 10 | 37 | 3.3 | |||||||
| 12 | 45 | 2.5 | Hdlq63 | 12 | 45 | 2.5 | Hdlq63 | |||||
| 14 | 26 | 2.6 | ||||||||||
| 16 | 39 | 3.9 | Hdlq65 | 16 | 30 | 2.5 | Hdlq65 | |||||
Chr, chromosome; LOD, logarithm of the odds ratio; TG, triglyceride. The 95% confidence limits for quantitative trait loci (QTLs) are not given, because they are not meaningful in a cross for which the parents are identical for most of the genome. All QTLs are suggestive at >0.37 or significant at >0.95 (given in boldface). Suggestive/significance thresholds (determined after 1,000 permutations) for chow, HF 3 weeks, and HF 8 weeks are 2.2/3.8, 2.3/3.8, and 2.4/4.1 for TG and 2.3/3.9, 2.4/4.0, and 2.6/4.1 for HDL.
Suggestive QTLs on Chr 2@75 and Chr 15@31 cM for TG, and Chr 12@41 and Chr 14@26 cM for HDL were named because they had been previously found in other crosses (see Table 3). QTLs for total cholesterol, body weight, and thyroxine (T4) can be found in supplementary Table II.
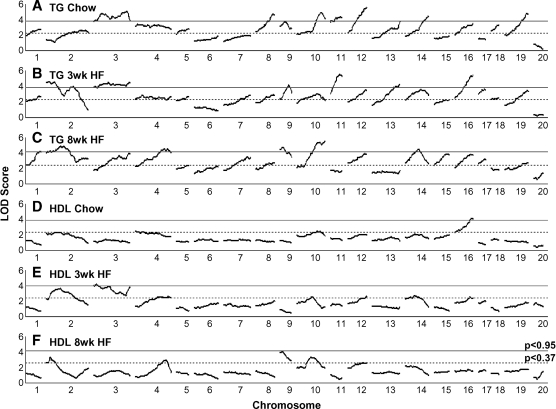
Triglycerides
Significant or suggestive QTLs for TG were found on nearly all chromosomes except Chrs 6 and X (Fig. 2A–C, Table 2). Eight QTLs are significant on the chow diet, 6 on the 3 week HF and 6 on the 8 week HF diet (Table 2). All significant QTLs, except for Tgq17 (Ch4 @68cM), are significant or suggestive in at least a second diet/age condition, but none are significant for all three conditions. Seven loci are at least suggestive across all three diet conditions for TG: Chr 1@34 cM; Chr 8@40 cM; Chr 9@6 to18 cM; Chr 10@45–60 cM; Chr 12@45 cM; Chr 16 @40 cM, and Chr 19@51 cM. Chr 3 is complex, with at least 2 QTLs present for the chow diet and a third one on the 3 week HF diet; these are further complicated by sex interactions. Many of the QTLs observed for TG were either sex specific or were strongly affected by sex, as shown by a change in logarithm of the odds ratio (LOD) score >2 when comparing the genome scans using sex as additive or interactive covariate; in all such cases, males had the higher TG. Only 2 of the QTLs in mice fed a chow diet were influenced by sex; however, 13 of the 18 QTLs for the 3 week HF diet and 7 of 14 QTLs for the 8 week HF diet had significant sex interactions. Many QTLs reported were significant in both sexes; however, for 2 TG QTLs, the effect was greater in males (M+,F), and an additional 6 QTLs were specific to males (M/) (Table 3).
Fig. 2.
Genome scan for triglyceride (TG) (A–C) and HDL (D–F). Chow diet at 8 weeks of age (A, D), at 11 weeks of age after being on a high-fat (HF) diet for 3 weeks (B, E), and at 16 weeks of age after being on a HF diet for 8 weeks (C, F). Sex is fitted as an additive and interactive covariate (Y = QTL + Sex + Sex*QTL). QTL, quantitative trait locus.
TABLE 3.
Summary of genome-wide significant TG QTL (updated from (27) with new QTL indicated in bold)
| Chr | Cross | Peak Genetic Marker | cM | CI | LODe | Sexd | High Allele | QTL | Ref |
|---|---|---|---|---|---|---|---|---|---|
| 1 | D1×D2 BC | rs3683684 | 34 | b | (4.1) | F, M | DBA/2 | Tgq9 | This report |
| NZB×SM | D1Nds2 | 62 | (3.5) | F, M | (49) | ||||
| MRL×SJL | D1Mir43 | 76 | 55–78 | 3.8 | F | MRL | Tglq1 | (50) | |
| B×Aa | rs3724524 | 79 | 77–90 | 5.4 | M/ | A/J | (24) | ||
| (NZO×NON)NONa | D1Mit37 | 89 | 73–92 | 2.9 | M | NON | (51) | ||
| B×C3H | D1Mit270 | 92 | 78–97 | 3.2 | F | C3H | (52) | ||
| B6×RR | D1Mit206 | 96 | 85–112 | 4.4 | F | RR | Trglyd | (53) | |
| 2 | D1×D2 BC | rs13476339 | 3 | 0–30 | (4.5) | F, M | DBA/2 | Tgq10 | This report |
| D1×D2 BC | rs13476339 | 27 | 0–30 | (4.5) | F, M | DBA/2 | Tgq11 | This report | |
| (NZO×NON)NONa | D2Mit109 | 73 | 66–80 | 3.7 | M | NON | (51) | ||
| B6×DBA | D2Mit495 | 79 | 3.0 | F | DBA/2 | (54) | |||
| B×C3H | D2MIT285 | 80 | 64–87 | (4.7) | F | C3H | Stylianou et al., in prep | ||
| 3 | D1×D2 BC | rs3700620c | 18 | 10–74 | 4.7 | F, M | DBA/2 | Tgq13 | This report |
| D1×D2 BC | rs3668158c | 41 | 14–68 | (4.8) | M/ | DBA/2 | Tgq14 | This report | |
| B×C3H | D3MIT57 | 58 | 37–65 | (2.5) | F | C3H | Stylianou et al., in prep | ||
| D1×D2 BC | rs3668158 | 70 | 18–71 | 5.2 | F, M | DBA/2 | Tgq15 | This report | |
| 4 | PERA×B6-Ldlr | D4Mit143 | 43 | (3.6) | M/ | PERA | (55) | ||
| D1×D2 BC | rs3708285 | 68 | 40–75 | (4.4) | M/ | DBA/2 | Tgq17 | This report | |
| B6×129 | D4Mit308 | 58 | 30–84f | F | 129 | Tgq3 | (56) | ||
| KK×BALB/c | D4Mit336 | 59 | 50–75 | 3.2 | M | BALB | Tgls1 | (57) | |
| MRL×BALB/c | D4Mit54 | 66 | 2.9 | (58) | |||||
| 5 | B6×DBA | D5Mit1 | 13 | 4.3 | F | DBA/2 | (54) | ||
| MRL×SJL | nr | 55 | F | (50) | |||||
| NZB×SM | D5Mit65 | 68 | (3.2) | F, M | (49) | ||||
| 7 | NZB×SM | D7Mit55 | 15 | (5.1) | F, M | (49) | |||
| NZB×NZW | rs3670069 | 50 | 29–58 | 3.1 | F+M | NZW | (59) | ||
| 8 | NZB×NZW | rs3691954 | 10 | 0–25 | 3.2 | F+M | NZW | (59) | |
| KK×RR | D8Mit205 | 30 | 4.7 | F | KK | Trigq2 | (60) | ||
| D1×D2 BC | rs13479852b | 40 | b | 4.5 | F, M | DBA/2 | Tgq18 | This report | |
| B×C3H | D8Mit41 | 41 | 33–51 | (3.8) | F | C3H | (52) | ||
| KK×BALB/c | D8Mit166 | 56 | 44–71 | 4.8 | M | KK | Tgl1 | (57) | |
| 9 | B6×KK | D9Mit163 | 33 | 20–57 | 4.2 | F, M | Trigq1 | (61) | |
| D1×D2 BC | rs3023961 | 6 | 0–20 | 4.0 (4.7) | F, M | DBA/2 | Tgq19 | This report | |
| B6×129 | D9Mit281 | 66 | 44–68 | (2.2)f | F | B6 | Tgq2 | (56) | |
| 10 | D1×D2 BC | rs13480749 | 52 | 42–62 | 4.7 | F, M | DBA/2 | Tgq20 | This report |
| D1×D2 BC | rs3653850 | 60 | 37–62 | 5.5 | M/ | DBA/2 | Tgq21 | This report | |
| 11 | LG×SM | D11Mit71–173 | 10 | (nr) | F, M | SM | Tgyl1 | (62) | |
| D1×D2 BC | rs3023254 | 19 | 11–26 | 4.3(5.4) | M+,F | DBA/2 | Tgq22 | This report | |
| B6×DBA | D11Mit54 | 44 | 3.6 | F | DBA/2 | (54) | |||
| 12 | NZB×RF | D12Mit182 | 8 | 0–30 | 8.7 | F | NZB | (63) | |
| MRL×SJL | D12Mit201 | 26 | 0–31 | 4.1 | F | SJL | Tglq2 | (50) | |
| D1×D2 BC | rs3711162 | 45 | b | 5.6 | M+,F | DBA/2 | Tgq23 | This report | |
| 13 | D1×D2 BC | rs3710370 | 58 | b | 4.3 | M/ | DBA/2 | Tgq24 | This report |
| 14 | D1×D2 BC | rs3090594 | 25 | 7–36 | (4.6) | M/ | DBA/2 | Tgq25 | This report |
| D1×D2 BC | rs3686670 | 48 | 22–48 | 4.4 | F, M | DBA/2 | Tgq26 | This report | |
| 15 | NZB×RF | D15Mit13 | 2 | 0–25 | (3.5) | F | RF | (63) | |
| MRL×BALB/c | D15Mit37 | 32 | 2.1 (2.7) | (58) | |||||
| 16 | D1×D2 BC | rs4210264 | 40 | b | (5.5) | M/ | DBA/2 | Tgq28 | This report |
| 17 | MRL×SJL | nr | 30 | 2.6 | F | Tgq1 | (50) | ||
| 18 | NZB×NZW | Rs4231907 | 34 | 26–39 | 4.1 | F+M | NZB | (59) | |
| B×C3H | D18MIT9 | 39 | 19–42 | (3.2) | F | C3H | Stylianou et al., in prep | ||
| B6×129 | D18Mit50 | 42 | 37–44 | (3.2) | F | Het | (56) | ||
| 19 | MRL×BALB/c | D19Mit93 | 3 | (4.0) | (58) | ||||
| D1×D2 BC | rs30858740 | 51 | b | 4.8 | F, M | DBA/2 | Tgq29 | This report |
CI, confidence interval; nr, not reported. Only genome-wide-significant QTLs are given.
The cross is described, but the TG phenotype is unpublished.
Indicates QTL from this study represented by a single marker.
Broad peak spanning most of the polymorphic area.
Both sexes tested and are male (M/)-, or female (F/)-specific; M, F, affects both sexes, with a greater effect in males (M+,F) or females (M,F+).
HF diet indicated by parenthesis.
Interacting QTL, high allele homozygous 129 on Chr 4 interacting with homozygous B6 on Chr 9.
HDL
A total of 3 significant QTLs for HDL were identified, 1 on Chr 16@39 cM for the chow diet, 1 on Chr 3@2 cM for the 3 week HF diet, and 1 on Chr 9@1 cM for the 8 week HF diet. Additional suggestive QTLs were identified on Chrs 4 and 10 for chow diet, Chrs 2, 4, 12, 14, and 16 for 3 week HF diet, and Chrs 2, 4, 10, and 12 for the 8 week HF diet (Table 2, Fig. 2D–F). The significant QTL on Chr 3 at 6 cM identified on the 3 week HF diet appears to be transient, because the effect is not present for either the chow diet or the later 8 week HF diet time point. It is possible that the TG QTL on Chr 3 is also affecting HDL, especially on the chow diet, where HDL QTL maps to 18 cM. An additional significant QTL was found in the related trait for total cholesterol on Chr 16 (on the chow diet 85–95% of total cholesterol is HDL), and suggestive QTLs for total cholesterol were detected on Chrs 2, 3, 4, 8, 9, 10, 12, 13, and 14, which map to approximately the same locations as the HDL QTLs (see supplementary Table II and supplementary Fig. IA–C). Under all three age/diet conditions, D2 contributed the high allele for significant or suggestive QTLs.
Body weight and T4
Six significant and 6 suggestive QTLs were identified for body weight at 21 days, immediately prior to weaning (see supplementary Table II and supplementary Fig. ID–G). None of these QTLs were present at later time points. Overall, the effect was determined by unknown paternal effects. Progeny derived from F1 females (either D1×D2 or D2×D1) mated to D1 fathers (10.0 ± 0.2 g) were significantly larger than progeny from F1 females mated to D2 fathers (8.0 ± 0.2 g, P < 0.0001). Whether the grandsire was D1 or D2 did not have a significant effect on the grand-progeny (P = 0.77). T4 was measured in males on the 8 week HF diet (see supplementary Table II and supplementary Fig. IH). A significant QTL was identified on Chr 2@18 cM and a suggestive locus on Chr 14@28 cM.
Candidate gene identification
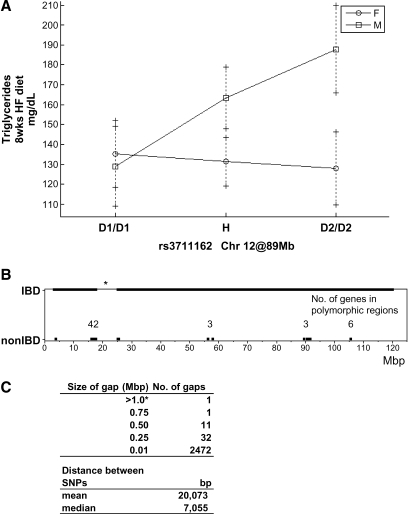
Chr 12@45 cM Tgq23/Hdlq63
There are 54 genes located in polymorphic regions between D1 and D2 on Chr 12: 42 are clustered between 17 and 26 Mb, 3 are located at ∼56 Mb, 3 between 89 and 91 Mb, and a further 6 at ∼105 Mb (Fig. 3B). SNP coverage to detect non-IBD regions between D1 and D2 is relatively good, with a median distance between SNPs of ∼7 kb, and only one large gap spanning 7.5 Mb with no SNPs (Fig. 3B, C).
Fig. 3.
Chromosome (Chr) 12 TG QTL. A: Effect plot for the SNP rs3711162 at 89 Mb. B: Diagram illustrating locations of IBD between D1 and D2. C: Table showing SNP coverage between D1 and D2. Note: there is one region on Chr 12 that is poorly covered with SNPs between D1 and D2. This region spans from 17 to 25 Mb with no SNP information.
The QTL on Chr 12, significant for TG and suggestive for HDL, has a peak marker located at 89 Mb. The QTL region close to the peak marker (89.9–91.8 Mb) contains three genes: neurexin 3 (Nrxn3), an uncharacterized Riken gene (4930534B04Rik), and thyroid-stimulating hormone receptor (Tshr). Nrxn3 is a well-characterized gene that is involved in synaptogenesis (28) and is not expressed in any tissues that would imply involvement in lipid metabolism (http://symatlas.gnf.org). The Riken clone, 4930534B04Rik, has no known function, although expression profiling suggests a possible function in the testis. The third gene, Tshr, is involved in controlling thyroid hormone levels, which have previously been associated with atherosclerosis, obesity, bone disease, and other metabolic diseases (29–31), making Tshr a probable candidate gene.
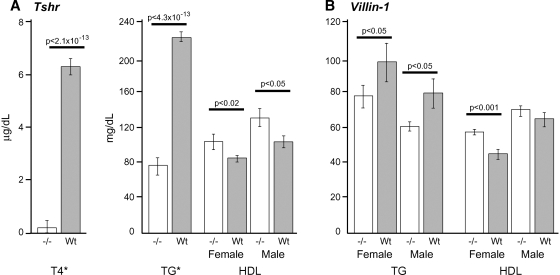
In humans, Tshr is primarily expressed in the thyroid but also to some extent in adipose tissue, a primary peripheral tissue for the storage of cholesterol (32). Similarly, in the mouse, Tshr is expressed at high levels in adipose tissue. Consequently, Tshr was selected as the most probable candidate gene for the HDL/TG QTL on Chr 12. A strain with a point mutation causing a natural knockout of Tshr, called hypothyroid (33), was obtained from The Jackson Laboratory. Tshr+/− heterozygotes were intercrossed, and littermates that included all three genotypes were compared with each other.
Originally, T4 was reported as undetectable in this strain (19), but with more-sensitive assays, we are able to detect the presence of T4 in Tshrhyt/hyt males at 0.40 ± 0.5 SE μg/dl, and a trace amount in females at 0.1 ± 0.7 SE μg/dl (detection threshold, 0.20 μg/dl). This low level could represent cross-reactivity of the antibody with T3 or it could be low levels of T4. At 8 weeks of age, TG is significantly lower in homozygous Tshrhyt/hyt (77 ± 6 mg/dL ±SD) versus heterozygous and wild-type mice (223 ± 10 mg/dL ±SD, P < 4.3 × 10−13), with no significant difference between males and females (Fig. 4A). However, HDL is significantly different between males and females (P < 0.001) and is thus analyzed separately. Nonetheless, both sexes of Tshrhyt/hyt mice show a significant increase in HDL, 20% in females (P < 0.02) and 30% in males (P < 0.05), mirroring the relative impact of the QTL effect observed between D1 and D2 for HDL and TG (Fig. 3A).
Fig. 4.
TG and HDL in Tshr- and Vil1-deficient mice. Means and SEM are given for each group maintained on a chow diet. A: Tshrhyt/hyt lipid testing. *There is no sex difference in T4 and TG between homozygous mutants versus wild-type (P = 0.4), thus data are presented sex-averaged. HDL is presented separately for each sex, because there was a significant difference between sexes (P < 0.01). B: Vil1−/− lipid testing. TG and HDL both had significant sex effects and thus are presented separately. TG (mg/dl), female Vil1−/− = 78 ± 7, Vil1+/+ = 97 ± 8 (P < 0.05); male Vil1−/− = 61 ± 9, Vil1+/+ = 80 ± 8 (P < 0.05); HDL (mg/dl), female Vil1−/− = 58 ± 2, Vil1+/+ = 45 ± 3 (P < 0.05); male, Vil1−/− = 70 ± 3, Vil1+/+ =65 ± 3 (P < 0.05). Male HDL values do not appear significant; however, in the context of a mixed model with sex as a fixed effect, TG and HDL are significantly different; P = 0.03, 0.006 for TG and HDL, respectively. N = 5 for each group.
Chr 1@34 cM Tgq9
The significant TG QTL on Chr 1 for an 8 week HF diet is suggestive for the other two diet conditions. The only identifiable region on Chr 1 that differs between D1 and D2 spans 67.9 to 74.9 Mb and contains 24 known or predicted genes (Table 4). Three genes are of particular interest because they are highly expressed in tissues related to lipid metabolism or have known functions related to lipid metabolism. These are cytochrome P450, family 27, subfamily a, polypeptide 1 (Cyp27a1); villin 1 (Vil1); and Indian hedgehog (Ihh).
TABLE 4.
Genes in non-IBD region between D1 and D2 on Chr 1
| Start of Gene (bp) | Gene Symbol | Known Function | Primary Tissue of Expressiona | Further Interest? |
|---|---|---|---|---|
| 67,973,387 | Erbb4 | Multiple | Uniform | |
| 69,760,212 | Spag16 | Male fertility | Testis | |
| 70,658,923 | A830006F12Rik | Unknown | Uniform | |
| 72,516,478 | Smarcal1 | Schimke immuno osseous dysplasia. | Various | |
| 72,576,187 | Ankar | Unknown | Uniform | |
| 72,644,879 | Rpl37a | Ribosomal/transcription | B-, T-cells | |
| 72,948,284 | Tnp1 | Male fertility | Testis | |
| 73,846,153 | Tns1 | Renal | Lung, kidney | |
| 74,058,748 | Rufy4 | Unknown | Uniform | |
| 74,087,201 | Il8rb | Immune response | Bone | |
| 74,342,615 | Vil1 | Microvilli morphogenesis | Intestine | Yes |
| 74,372,319 | Usp37 | Unknown | Unknown | |
| 74,438,458 | Rqcd1 | Cell differentiation | Embryonic | |
| 74,476,095 | Plcd4 | Gametogenesis | Uniform | |
| 74,499,649 | Zfp142 | Unknown | Pancreas | |
| 74,526,960 | Rnf25 | Unknown | Oocyte | |
| 74,534,662 | Stk36 | Unknown | Oocyte | |
| 74,594,961 | Ttll4 | Unknown | Testis | |
| 74,646,781 | Cyp27a1 | Degradation of cholesterol to bile acids | Liver | Yes |
| 74,814,716 | Fev | Unknown | Embryonic | |
| 74,823,151 | Cryba2 | Unknown | Retina | |
| 74,835,287 | B230363K08Rik | Unknown | Uniform | |
| 74,878,522 | Ihh | Development | Intestine | Yesb |
| 74,900,901 | Nhej1 | DNA repair | Uniform |
IBD, identical by descent.
Expression data are taken from C57BL6 mice from the SymAtlas database.
Ihh−/− is homozygous lethal.
Cyp27a1 is a likely candidate because it is almost exclusively expressed in the liver. The role of CYP27A1 is to convert cholesterol into bile acids for secretion, and TG and cholesterol levels are significantly increased in Cyp27a1−/− mice (34). Ihh was identified as a candidate because it is specifically expressed in the intestine, and inhibition of Ihh by antibodies significantly reduces the rate of TG absorption (35). Ihh−/− mice are homozygous lethal; heterozygotes were not available for testing. Vil1 also was identified as a candidate because it is specifically expressed in the intestines, which is an important route for lipid absorption and metabolism. We were able to obtain Vil1−/− mice (21) on a C57BL/6 (B6) and D2 mixed genetic background and compared these to B6D2 F1 mice for differences in TG and HDL (Fig. 4B). Lipid traits were significantly different between sexes (TG P = 0.05, HDL P < 0.001) and thus are presented separately. Triglyceride levels were significantly lower in both male and female Vil1−/− mice; HDL levels were lower only in female Vil1−/− mice (Fig. 4B).
Further testing of candidate genes
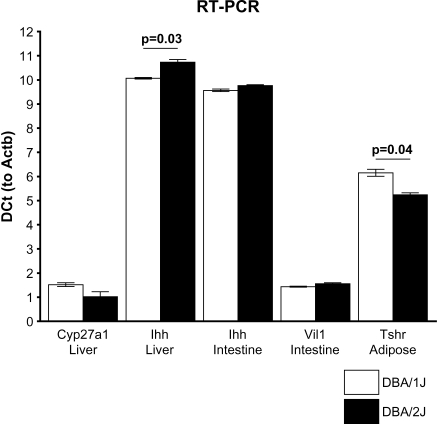
A QTL gene must have an expression difference between the parental strains or a coding region polymorphism that can change function. To further test the candidates Tshr, Vil1, Ihh, and Cyp27a1, we carried out RT-PCR and sequenced each gene. We found that Tshr is upregulated nearly 2-fold in DBA/2J adipose tissue relative to DBA/1J, requiring almost one less amplification cycle to reach the logarithmic threshold using Actb expression as control (Fig. 5). Tshr cDNA was not detectable in liver. Sequencing of the exons revealed no SNP differences between D1 and D2 indicating that promoter region differences may exist that affect transcript abundance leading to the QTL effect, rather than functional changes in the protein sequence.
Fig. 5.
RT-PCR comparisons of Tshr, Vil1, Ihh, and Cyp27a1. Comparisons were performed in relevant tissues in DBA/1J and DBA/2J male mice, N = 4 for each group with mean DCt and SEM controlled to Actb.
For the Chr 1 candidate genes, we found that Cyp27a1 showed a 40% upregulation in D2 liver compared with D1, but this failed to reach significance (P = 0.08), and Ihh showed a significant 60% increase in D1 liver compared with D2 (P = 0.03). Ihh and Vil1 were both examined in intestines because this is where these genes are principally expressed. However, neither Ihh nor Vil1 differed significantly in intestine expression between the two strains (Ihh, P = 0.12; Vil1, P = 0.32).
Sequencing of Ihh and Vil1 revealed that neither gene had coding region SNPs but that both have polymorphisms in the 3′ untranslated region (UTR); Vil1 has an adenosine trinucleotide insertion, whereas Ihh has four SNPs between D1 and D2 (Table 5). Sequencing of Cyp27a1 revealed no polymorphisms in the coding region.
TABLE 5.
Summary of polymorphisms identified between D1 and D2 for candidate genes Vil1, Ihh, and Cyp27a1 on Chr 1 and Tshr on Chr 12
| Polymorphisms |
||||||
|---|---|---|---|---|---|---|
| Chr | QTL | Gene | Bpa | D1 | D2 | Location |
| 1 | Tgq9 | Cyp27a1 | 0 | 0 | 0 | |
| Vil1 | 3,126 | ::: | AAA | 3′UTR | ||
| Ihh | 2,274 | C | T | 3′UTR | ||
| 2,307 | C | T | 3′UTR | |||
| 2,313 | C | G | 3′UTR | |||
| 2,425 | : | T | 3′UTR | |||
| 12 | Tgq23/Hdlq63 | Tshr | 0 | 0 | 0 | |
UTR, untranslated region. A colon indicates the absence of a base pair.
Bp position is relative to the start of the coding sequence.
Candidate gene identification for QTLs on other chromosomes
Of the significant TG QTLs identified, 11 are located in locations not previously reported as significant in mouse QTL studies. These are: Chr 1@34, 2@3, 2@27, 4@68, 9@6–18, 10@45–60, 12@40, 13@58, 14@25–48, 16@40, and 19@51 cM. The significant TG QTLs located on Chr 4@68, 8@40, and 11@19 cM, have been previously reported in other mouse QTL crosses (Table 3). Combining crosses or haplotyping did not help to narrow the QTLs significantly (data not shown). For HDL, the 3 significant QTLs identified in this study are located in regions not previously reported to be associated with HDL in mouse studies.
DISCUSSION
Increased levels of LDL and TG, as well as decreased levels of HDL, are predictors of atherosclerosis and cardiovascular disease. However, lipid levels are controlled by genes and pathways that largely remain unknown. Whereas mouse models have been extensively used to map QTLs for lipids, the focus has been to use strains that have extreme differences at the phenotype level and also at the genotype level (27, 36). Although this approach is useful in capturing maximal genetic variation between any two strains, the big differences at the genetic level between the initial cross strains mean that the confidence intervals that result from these QTL studies usually contain a prohibitive number of genes in terms of candidate analysis.
In this study, we limited the genetic variability by crossing the closely related strains DBA/1 and DBA/2. This allowed us to map multiple QTLs for lipids, and many of these QTLs mapped to regions containing a small number of genes. However, comparisons between closely related strains could be more susceptible to type 2 errors, given that polymorphisms could exist that are more difficult to detect than when comparing two more distantly related strains. Here we used over 140,000 SNPs with a median interval of ∼7 kb, which should limit such errors.
On the chow diet, QTL for TG affected males and females approximately equally; however, on a HF diet, many QTLs were male-specific. This is consistent with the observations of the parental phenotypes; D1 and D2 males differed significantly on both diets, but females differed only on a chow diet. Although D1 and D2 differed significantly in body weight at 8 weeks when fed chow and differed even more after consuming a HF diet for 8 more weeks, no significant obesity QTLs were observed beyond weaning age, indicating a possible lack of SNP coverage for a potential body weight locus.
Nonetheless, as demonstrated here, by using over 140,000 SNPs, we were able to capture sufficient variation to map numerous QTLs and identify candidate genes, especially for TG and HDL levels. By examining gene expression profiles from public databases, we were able to identify two novel candidates, Tshr and Ihh.
Chr12@45 cM (Tgq23) candidate gene
Tshr
Tshr was identified as one of only three genes in the Chr 12 QTL interval that could be responsible for the significant effect on TG and the suggestive effect on HDL. Testing a hypomorphic strain for Tshr revealed that this gene does indeed have a strong effect on both TG and HDL levels. The mutant Tshrhyt strain has a Pro556Leu mutation (19, 20) that makes it hypothyroid with decreased TG and increased HDL relative to controls. In our QTL, D2 alleles give rise to both increased TG and increased HDL, so the effect is not quite the same. However, the background of the Tshrhyt mutant (mixed BALB/cByJ and RF/J) is very different from the context of the D1 by D2 QTL analysis, and polymorphisms within each of these strains for the other thyroid hormone pathway genes may account for these differences. Sequencing revealed no coding region polymorphisms between D1 and D2 in Tshr, but mRNA was significantly upregulated in D2 relative to D1.
In humans, the thyroid hormone pathway has long been linked to dyslipidemia. The association of lipids to subclinical hypothyroidism is inconsistent with studies reporting that hypothyroid populations can have both decreased and increased LDL, HDL, and total cholesterol (37, 38). However, in general, people with overt or subclinical hypothyroidism have elevated LDL and total cholesterol, whereas HDL and TG remain unchanged (38–40). Treating hypothyroidism with T4 generally decreases total cholesterol and LDL, but does not change HDL and TG (41–44).
In conclusion, it appears that the thyroid hormone pathway is important for lipid metabolism in both mice and humans. However, treating hypothyroidism with T4, the end product of this pathway, is not effective for correcting TG and HDL levels. Rather, specific genes in this pathway could be targeted to modify lipids that would not directly affect T4 levels and would hence avoid adverse cardiac side effects. This has recently been demonstrated for an agonist of liver thyroid hormone receptor β, leading to lowered LDL but unchanged cardiac function (45). As shown here, Tshr could also be a valid target for influencing HDL and TG levels.
Chr 1@34 cM (Tgq9) candidate genes
Cyp27a1, Ihh and Vil1
This locus contains three genes of interest that were examined further. Cyp27a1 showed no significant expression difference and no coding region polymorphisms. It is true that mice lacking this gene have altered lipid metabolism, including altered TG levels (34), but we find no evidence that Cyp27a1 can account for the TG QTL in this cross. Vil1 is almost exclusively expressed in the mouse intestine, and its expression is so specific that the Vil1 promoter is frequently used in expression systems to upregulate target genes in the intestines. VILLIN1 is produced primarily in epithelial cells that develop the brush border responsible for nutrient uptake, and it has been characterized as a calcium-dependent actin binding protein (46). We identified a trinucleotide insertion in the 3′UTR; however Vil1 did not have any coding region differences between D1 and D2, nor were there any expression differences. We did show that TG was altered in a Vil1 knockout, but these altered TG levels may result from the true QTL gene in the mixed genetic background of the knockout, which included not only B6 and D2 (the predominant backgrounds of the Vil1 knockout strain) but also 129S2 (the genetic background of the original ES cell line). Ihh did show significantly different expression in the livers of D1 and D2 mice and had polymorphisms in the 3′ UTR, which might account for the expression difference. This leaves Ihh as the most likely candidate gene. Further studies potentially utilizing knockdown technologies performed in the DBA strains could provide more definitive proof that Ihh can affect lipid metabolism.
In summary, a total of 21 different QTLs were identified for TG, 12 of which were significant in at least one of the three diet conditions, and 6 of which were significant for two diet conditions. We conclude that diet strongly affects TG levels, and also that TG is a highly complex trait, even in the presence of a model with relatively limited genetic variability. Larger genetically heterogeneous mouse models are very powerful in identifying QTL genes (47–49); however, as demonstrated here, closely related mouse strains can also be used in a more trait-focused manner to identify complex trait genes.
Supplementary Material
Acknowledgments
The authors wish to thank Fred Rumill and Heather Sawick for technical assistance, and Edward Leiter and Luanne Peters for their helpful comments on the manuscript.
Published, JLR Papers in Press, May 23, 2008.
Footnotes
This work was supported by The American Heart Association, Grant 0525816T (I.M.S.), The National Institutes of Health, Grants HL-77796, HL-81162, and HL-74086 (B.P), and by CA34196, a Cancer Center Core grant to The Jackson Laboratory.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of a figure and two tables.
References
- 1.Assmann G., H. Schulte, and A. von Eckardstein. 1996. Hypertriglyceridemia and elevated lipoprotein(a) are risk factors for major coronary events in middle-aged men. Am. J. Cardiol. 77 1179–1184. [DOI] [PubMed] [Google Scholar]
- 2.Fruchart J. C., and P. Duriez. 2002. HDL and triglyceride as therapeutic targets. Curr. Opin. Lipidol. 13 605–616. [DOI] [PubMed] [Google Scholar]
- 3.Gordon D. J., J. L. Probstfield, R. J. Garrison, J. D. Neaton, W. P. Castelli, J. D. Knoke, D. R. Jacobs, Jr., S. Bangdiwala, and H. A. Tyroler. 1989. High-density lipoprotein cholesterol and cardiovascular disease. Four prospective American studies. Circulation. 79 8–15. [DOI] [PubMed] [Google Scholar]
- 4.Nissen S. E., T. Tsunoda, E. M. Tuzcu, P. Schoenhagen, C. J. Cooper, M. Yasin, G. M. Eaton, M. A. Lauer, W. S. Sheldon, C. L. Grines, et al. 2003. Effect of recombinant ApoA-I Milano on coronary atherosclerosis in patients with acute coronary syndromes: a randomized controlled trial. J. Am. Med. Assoc. 290 2292–2300. [DOI] [PubMed] [Google Scholar]
- 5.Wilson P. W., K. M. Anderson, T. Harris, W. B. Kannel, and W. P. Castelli. 1994. Determinants of change in total cholesterol and HDL-C with age: the Framingham Study. J. Gerontol. 49 M252–M257. [DOI] [PubMed] [Google Scholar]
- 6.Badimon J. J., L. Badimon, and V. Fuster. 1990. Regression of atherosclerotic lesions by high density lipoprotein plasma fraction in the cholesterol-fed rabbit. J. Clin. Invest. 85 1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okamoto H., F. Yonemori, K. Wakitani, T. Minowa, K. Maeda, and H. Shinkai. 2000. A cholesteryl ester transfer protein inhibitor attenuates atherosclerosis in rabbits. Nature. 406 203–207. [DOI] [PubMed] [Google Scholar]
- 8.Plump A. S., C. J. Scott, and J. L. Breslow. 1994. Human apolipoprotein A-I gene expression increases high density lipoprotein and suppresses atherosclerosis in the apolipoprotein E-deficient mouse. Proc. Natl. Acad. Sci. USA. 91 9607–9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rittershaus C. W., D. P. Miller, L. J. Thomas, M. D. Picard, C. M. Honan, C. D. Emmett, C. L. Pettey, H. Adari, R. A. Hammond, D. T. Beattie, et al. 2000. Vaccine-induced antibodies inhibit CETP activity in vivo and reduce aortic lesions in a rabbit model of atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 20 2106–2112. [DOI] [PubMed] [Google Scholar]
- 10.Rubin E. M., R. M. Krauss, E. A. Spangler, J. G. Verstuyft, and S. M. Clift. 1991. Inhibition of early atherogenesis in transgenic mice by human apolipoprotein AI. Nature. 353 265–267. [DOI] [PubMed] [Google Scholar]
- 11.Sugano M., N. Makino, S. Sawada, S. Otsuka, M. Watanabe, H. Okamoto, M. Kamada, and A. Mizushima. 1998. Effect of antisense oligonucleotides against cholesteryl ester transfer protein on the development of atherosclerosis in cholesterol-fed rabbits. J. Biol. Chem. 273 5033–5036. [DOI] [PubMed] [Google Scholar]
- 12.DiPetrillo K., X. Wang, I. M. Stylianou, and B. Paigen. 2005. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 21 683–692. [DOI] [PubMed] [Google Scholar]
- 13.Hillebrandt S., H. E. Wasmuth, R. Weiskirchen, C. Hellerbrand, H. Keppeler, A. Werth, R. Schirin-Sokhan, G. Wilkens, A. Geier, J. Lorenzen, et al. 2005. Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat. Genet. 37 835–843. [DOI] [PubMed] [Google Scholar]
- 14.Korstanje R., and B. Paigen. 2002. From QTL to gene: the harvest begins. Nat. Genet. 31 235–236. [DOI] [PubMed] [Google Scholar]
- 15.Wang X., M. Ria, P. M. Kelmenson, P. Eriksson, D. C. Higgins, A. Samnegard, C. Petros, J. Rollins, A. M. Bennet, B. Wiman, et al. 2005. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat. Genet. 37 365–372. [DOI] [PubMed] [Google Scholar]
- 16.Churchill G. A., D. C. Airey, H. Allayee, J. M. Angel, A. D. Attie, J. Beatty, W. D. Beavis, J. K. Belknap, B. Bennett, W. Berrettini, et al. 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36 1133–1137. [DOI] [PubMed] [Google Scholar]
- 17.Wade C. M., and M. J. Daly. 2005. Genetic variation in laboratory mice. Nat. Genet. 37 1175–1180. [DOI] [PubMed] [Google Scholar]
- 18.Svenson K. L., R. Von Smith, P. A. Magnani, H. R. Suetin, B. Paigen, J. K. Naggert, R. Li, G. A. Churchill, and L. L. Peters. 2007. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J. Appl. Physiol. 102 2369–2378. [DOI] [PubMed] [Google Scholar]
- 19.Beamer W. J., E. M. Eicher, L. J. Maltais, and J. L. Southard. 1981. Inherited primary hypothyroidism in mice. Science. 212 61–63. [DOI] [PubMed] [Google Scholar]
- 20.Gu W. X., G. G. Du, P. Kopp, A. Rentoumis, C. Albanese, L. D. Kohn, L. D. Madison, and J. L. Jameson. 1995. The thyrotropin (TSH) receptor transmembrane domain mutation (Pro556-Leu) in the hypothyroid hyt/hyt mouse results in plasma membrane targeting but defective TSH binding. Endocrinology. 136 3146–3153. [DOI] [PubMed] [Google Scholar]
- 21.Ferrary E., M. Cohen-Tannoudji, G. Pehau-Arnaudet, A. Lapillonne, R. Athman, T. Ruiz, L. Boulouha, F. El Marjou, A. Doye, J. J. Fontaine, et al. 1999. In vivo, villin is required for Ca(2+)-dependent F-actin disruption in intestinal brush borders. J. Cell Biol. 146 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishina P. M., J. Verstuyft, and B. Paigen. 1990. Synthetic low and high fat diets for the study of atherosclerosis in the mouse. J. Lipid Res. 31 859–869. [PubMed] [Google Scholar]
- 23.Petkov P. M., Y. Ding, M. A. Cassell, W. Zhang, G. Wagner, E. E. Sargent, S. Asquith, V. Crew, K. A. Johnson, P. Robinson, et al. 2004. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 14 1806–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stylianou I. M., S. W. Tsaih, K. DiPetrillo, N. Ishimori, R. Li, B. Paigen, and G. Churchill. 2006. Complex genetic architecture revealed by analysis of high-density lipoprotein cholesterol in chromosome substitution strains and F2 crosses. Genetics. 174 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen-Seaman M. I., T. S. Furey, B. A. Payseur, Y. Lu, K. M. Roskin, C. F. Chen, M. A. Thomas, D. Haussler, and H. J. Jacob. 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Churchill G. A., and R. W. Doerge. 1994. Empirical threshold values for quantitative trait mapping. Genetics. 138 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., and B. Paigen. 2005. Genome-wide search for new genes controlling plasma lipid concentrations in mice and humans. Curr. Opin. Lipidol. 16 127–137. [DOI] [PubMed] [Google Scholar]
- 28.Rowen L., J. Young, B. Birditt, A. Kaur, A. Madan, D. L. Philipps, S. Qin, P. Minx, R. K. Wilson, L. Hood, et al. 2002. Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics. 79 587–597. [DOI] [PubMed] [Google Scholar]
- 29.Cooper D. S. 2001. Clinical practice. Subclinical hypothyroidism. N. Engl. J. Med. 345 260–265. [DOI] [PubMed] [Google Scholar]
- 30.Knudsen N., P. Laurberg, L. B. Rasmussen, I. Bulow, H. Perrild, L. Ovesen, and T. Jorgensen. 2005. Small differences in thyroid function may be important for body mass index and the occurrence of obesity in the population. J. Clin. Endocrinol. Metab. 90 4019–4024. [DOI] [PubMed] [Google Scholar]
- 31.Toft A. D. 2001. Clinical practice. Subclinical hyperthyroidism. N. Engl. J. Med. 345 512–516. [DOI] [PubMed] [Google Scholar]
- 32.Bell A., A. Gagnon, L. Grunder, S. J. Parikh, T. J. Smith, and A. Sorisky. 2000. Functional TSH receptor in human abdominal preadipocytes and orbital fibroblasts. Am. J. Physiol. Cell Physiol. 279 C335–C340. [DOI] [PubMed] [Google Scholar]
- 33.Stein S. A., E. L. Oates, C. R. Hall, R. M. Grumbles, L. M. Fernandez, N. A. Taylor, D. Puett, and S. Jin. 1994. Identification of a point mutation in the thyrotropin receptor of the hyt/hyt hypothyroid mouse. Mol. Endocrinol. 8 129–138. [DOI] [PubMed] [Google Scholar]
- 34.Repa J. J., E. G. Lund, J. D. Horton, E. Leitersdorf, D. W. Russell, J. M. Dietschy, and S. D. Turley. 2000. Disruption of the sterol 27-hydroxylase gene in mice results in hepatomegaly and hypertriglyceridemia. Reversal by cholic acid feeding. J. Biol. Chem. 275 39685–39692. [DOI] [PubMed] [Google Scholar]
- 35.Buhman K. K., L. C. Wang, Y. Tang, E. A. Swietlicki, S. Kennedy, Y. Xie, Z. Y. Liu, L. C. Burkly, M. S. Levin, D. C. Rubin, et al. 2004. Inhibition of Hedgehog signaling protects adult mice from diet-induced weight gain. J. Nutr. 134 2979–2984. [DOI] [PubMed] [Google Scholar]
- 36.Wang X., and B. Paigen. 2005. Genetics of variation in HDL cholesterol in humans and mice. Circ. Res. 96 27–42. [DOI] [PubMed] [Google Scholar]
- 37.Boberg J., P. A. Dahlberg, B. Vessby, and H. Lithell. 1984. Serum lipoprotein and apolipoprotein concentrations in patients with hyperthyroidism and the effect of treatment with carbimazole. Acta Med. Scand. 215 453–459. [DOI] [PubMed] [Google Scholar]
- 38.Canaris G. J., N. R. Manowitz, G. Mayor, and E. C. Ridgway. 2000. The Colorado thyroid disease prevalence study. Arch. Intern. Med. 160 526–534. [DOI] [PubMed] [Google Scholar]
- 39.Kanaya A. M., F. Harris, S. Volpato, E. J. Perez-Stable, T. Harris, and D. C. Bauer. 2002. Association between thyroid dysfunction and total cholesterol level in an older biracial population: the health, aging and body composition study. Arch. Intern. Med. 162 773–779. [DOI] [PubMed] [Google Scholar]
- 40.Vierhapper H., A. Nardi, P. Grosser, W. Raber, and A. Gessl. 2000. Low-density lipoprotein cholesterol in subclinical hypothyroidism. Thyroid. 10 981–984. [DOI] [PubMed] [Google Scholar]
- 41.Danese M. D., P. W. Ladenson, C. L. Meinert, and N. R. Powe. 2000. Clinical review 115: effect of thyroxine therapy on serum lipoproteins in patients with mild thyroid failure: a quantitative review of the literature. J. Clin. Endocrinol. Metab. 85 2993–3001. [DOI] [PubMed] [Google Scholar]
- 42.Kong W. M., M. H. Sheikh, P. J. Lumb, R. P. Naoumova, D. B. Freedman, M. Crook, C. J. Dore, and N. Finer. 2002. A 6-month randomized trial of thyroxine treatment in women with mild subclinical hypothyroidism. Am. J. Med. 112 348–354. [DOI] [PubMed] [Google Scholar]
- 43.Meier C., J. J. Staub, C. B. Roth, M. Guglielmetti, M. Kunz, A. R. Miserez, J. Drewe, P. Huber, R. Herzog, and B. Muller. 2001. TSH-controlled L-thyroxine therapy reduces cholesterol levels and clinical symptoms in subclinical hypothyroidism: a double blind, placebo-controlled trial (Basel Thyroid Study). J. Clin. Endocrinol. Metab. 86 4860–4866. [DOI] [PubMed] [Google Scholar]
- 44.Tzotzas T., G. E. Krassas, T. Konstantinidis, and M. Bougoulia. 2000. Changes in lipoprotein(a) levels in overt and subclinical hypothyroidism before and during treatment. Thyroid. 10 803–808. [DOI] [PubMed] [Google Scholar]
- 45.Berkenstam A., J. Kristensen, K. Mellstrom, B. Carlsson, J. Malm, S. Rehnmark, N. Garg, C. M. Andersson, M. Rudling, F. Sjoberg, et al. 2008. The thyroid hormone mimetic compound KB2115 lowers plasma LDL cholesterol and stimulates bile acid synthesis without cardiac effects in humans. Proc. Natl. Acad. Sci. USA. 105 663–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friederich E., E. Pringault, M. Arpin, and D. Louvard. 1990. From the structure to the function of villin, an actin-binding protein of the brush border. Bioessays. 12 403–408. [DOI] [PubMed] [Google Scholar]
- 47.Valdar W., L. C. Solberg, D. Gauguier, S. Burnett, P. Klenerman, W. O. Cookson, M. S. Taylor, J. N. Rawlins, R. Mott, and J. Flint. 2006. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 38 879–887. [DOI] [PubMed] [Google Scholar]
- 48.Williams, R. W., K. W. Broman, J. M. Cheverud, G. A. Churchill, R. W. Hitzemann, K. W. Hunter, J. D. Mountz, P. Pomp, R. H. Reeves, L. C. Schalkwyk, et al. 2002. A collaborative cross for high-precision complex trait analysis. 1st Workshop Report of the Complex Trait Consortium, Memphis, TN.
- 49.Purcell-Huynh D. A., A. Weinreb, L. W. Castellani, M. Mehrabian, M. H. Doolittle, and A. J. Lusis. 1995. Genetic factors in lipoprotein metabolism. Analysis of a genetic cross between inbred mouse strains NZB/BINJ and SM/J using a complete linkage map approach. J. Clin. Invest. 96 1845–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srivastava A. K., S. Mohan, G. L. Masinde, H. Yu, and D. J. Baylink. 2006. Identification of quantitative trait loci that regulate obesity and serum lipid levels in MRL/MpJ x SJL/J inbred mice. J. Lipid Res. 47 123–133. [DOI] [PubMed] [Google Scholar]
- 51.Reifsnyder P. C., G. Churchill, and E. H. Leiter. 2000. Maternal environment and genotype interact to establish diabesity in mice. Genome Res. 10 1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su Z., Y. Li, J. C. James, M. McDuffie, A. H. Matsumoto, G. A. Helm, J. L. Weber, A. J. Lusis, and W. Shi. 2006. Quantitative trait locus analysis of atherosclerosis in an intercross between C57BL/6 and C3H mice carrying the mutant apolipoprotein E gene. Genetics. 172 1799–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suto J., Y. Takahashi, and K. Sekikawa. 2004. Quantitative trait locus analysis of plasma cholesterol and triglyceride levels in C57BL/6J x RR F2 mice. Biochem. Genet. 42 347–363. [DOI] [PubMed] [Google Scholar]
- 54.Colinayo V. V., J. H. Qiao, X. Wang, K. L. Krass, E. Schadt, A. J. Lusis, and T. A. Drake. 2003. Genetic loci for diet-induced atherosclerotic lesions and plasma lipids in mice. Mamm. Genome. 14 464–471. [DOI] [PubMed] [Google Scholar]
- 55.Seidelmann S. B., C. De Luca, R. L. Leibel, J. L. Breslow, A. R. Tall, and C. L. Welch. 2005. Quantitative trait locus mapping of genetic modifiers of metabolic syndrome and atherosclerosis in low-density lipoprotein receptor-deficient mice: identification of a locus for metabolic syndrome and increased atherosclerosis on chromosome 4. Arterioscler. Thromb. Vasc. Biol. 25 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishimori N., R. Li, P. M. Kelmenson, R. Korstanje, K. A. Walsh, G. A. Churchill, K. Forsman-Semb, and B. Paigen. 2004. Quantitative trait loci that determine plasma lipids and obesity in C57BL/6J and 129S1/SvImJ inbred mice. J. Lipid Res. 45 1624–1632. [DOI] [PubMed] [Google Scholar]
- 57.Shike T., S. Hirose, M. Kobayashi, K. Funabiki, T. Shirai, and Y. Tomino. 2001. Susceptibility and negative epistatic loci contributing to type 2 diabetes and related phenotypes in a KK/Ta mouse model. Diabetes. 50 1943–1948. [DOI] [PubMed] [Google Scholar]
- 58.Gu L., M. W. Johnson, and A. J. Lusis. 1999. Quantitative trait locus analysis of plasma lipoprotein levels in an autoimmune mouse model: interactions between lipoprotein metabolism, autoimmune disease, and atherogenesis. Arterioscler. Thromb. Vasc. Biol. 19 442–453. [DOI] [PubMed] [Google Scholar]
- 59.Su Z., S. W. Tsaih, J. Szatkiewicz, Y. Shen, and B. Paigen. 2008. Candidate genes for plasma triglyceride, free fatty acid, and glucose revealed from an intercross between inbred mouse strains NZB/B1NJ x NZW/LacJ. J. Lipid Res. 49 1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suto J., and K. Sekikawa. 2003. Quantitative trait locus analysis of plasma cholesterol and triglyceride levels in KK x RR F2 mice. Biochem. Genet. 41 325–341. [DOI] [PubMed] [Google Scholar]
- 61.Suto J., S. Matsuura, H. Yamanaka, and K. Sekikawa. 1999. Quantitative trait loci that regulate plasma lipid concentration in hereditary obese KK and KK-Ay mice. Biochim. Biophys. Acta. 1453 385–395. [DOI] [PubMed] [Google Scholar]
- 62.Cheverud J. M., T. H. Ehrich, T. Hrbek, J. P. Kenney, L. S. Pletscher, and C. F. Semenkovich. 2004. Quantitative trait loci for obesity- and diabetes-related traits and their dietary responses to high-fat feeding in LGXSM recombinant inbred mouse strains. Diabetes. 53 3328–3336. [DOI] [PubMed] [Google Scholar]
- 63.Wergedal J. E., C. L. Ackert-Bicknell, W. G. Beamer, S. Mohan, D. J. Baylink, and A. K. Srivastava. 2007. Mapping genetic loci that regulate lipid levels in a NZB/B1NJxRF/J intercross and a combined intercross involving NZB/B1NJ, RF/J, MRL/MpJ, and SJL/J mouse strains. J. Lipid Res. 48 1724–1734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.