Abstract
The steroid 20-hydroxyecdysone (20E) is the primary regulatory hormone that mediates developmental transitions in insects and other arthropods. 20E is produced from ecdysone (E) by the action of a P450 monooxygenase that hydroxylates E at carbon 20. The gene coding for this key enzyme of ecdysteroidogenesis has not been identified definitively in any insect. We show here that the Drosophila E-20-monooxygenase (E20MO) is the product of the shade (shd) locus (cytochrome p450, CYP314a1). When shd is transfected into Drosophila S2 cells, extensive conversion of E to 20E is observed, whereas in sorted homozygous shd embryos, no E20MO activity is apparent either in vivo or in vitro. Mutations in shd lead to severe disruptions in late embryonic morphogenesis and exhibit phenotypes identical to those seen in disembodied (dib) and shadow (sad) mutants, two other genes of the Halloween class that code for P450 enzymes that catalyze the final two steps in the synthesis of E from 2,22-dideoxyecdysone. Unlike dib and sad, shd is not expressed in the ring gland but is expressed in peripheral tissues such as the epidermis, midgut, Malpighian tubules, and fat body, i.e., tissues known to be major sites of E20MO activity in a variety of insects. However, the tissue in which shd is expressed does not appear to be important for developmental function because misexpression of shd in the embryonic mesoderm instead of the epidermis, the normal embryonic tissue in which shd is expressed, rescues embryonic lethality.
One principal reason for the success of insects is their rigid exoskeleton (cuticle), which in many cases protects them from desiccation and predators while also providing the substrate for the development of jointed legs and wings. Growth is ultimately restricted by the surface area of this cuticle, and the insect must synthesize a larger cuticle exterior to the old one. As this process occurs, the old cuticle is digested by specific enzymes, the products of which are recycled, and the insect finally sheds the remnants of the original cuticle (ecdysis). This molting process is therefore required for the use of this rigid exoskeleton and indirectly for the success of insects on this planet.
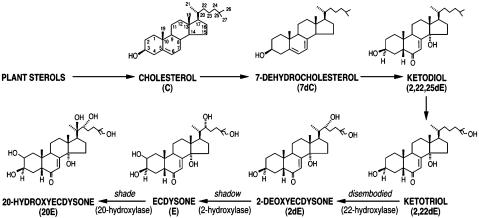
The trigger, or initiator of the molting process, is a polyhydroxylated steroid, 20-hydroxyecdysone (20E), the molting hormone of the vast majority of arthropods, which elicits not only molting but most of the morphogenetic processes that comprise insect growth and metamorphosis (1). That is, almost the entire insect is the target of 20E. 20E itself is the product of the hydroxylation of ecdysone (E) mediated by an E-20-monooxygenase (E20MO), i.e., ecdysone 20-hydroxylase, so E is a precursor of 20E, although it may have hormonal roles as well (2, 3). E is synthesized from cholesterol or plant sterols in the prothoracic gland cells of the ring gland (Fig. 1).
Fig. 1.
Scheme of 20E biosynthesis. The top portion represents a theoretical pathway in which plant sterols obtained in the diet are converted to the ketodiol (23). The bottom portion of the figure shows the final three steps in the Drosophila pathway that involve conversion of the ketotriol intermediate to 20-hydroxyecdysone by the activity of the P450 enzymes coded for by dib, sad, and shd (ref. 16 and this work).
Although it has been known for several decades that the E20MO is a P450 enzyme that is localized in the endoplasmic reticulum (4) and/or mitochondria (5, 6), depending on the insect, tissue, and developmental stage (7, 8), it has not been purified to homogeneity nor has the gene coding for this enzyme been cloned. Here, we report the cloning of shade (CYP314a1), a member of the Halloween gene family (9), and demonstrate that its gene product codes for the Drosophila E20MO.
Materials and Methods
Drosophila Strains. shd mutant strains were obtained from C. Nüsslein-Volhard (Max Planck Institute, Tubingen, Germany) and cultured on standard cornmeal/yeast extract/dextrose medium. The alleles were described by Jürgens et al. (10). twist-Gal4 line 2517 and armadillo-Gal4 line 1560 were obtained from the Bloomington Stock Center (http://flystocks.bio.indiana.edu).
Phenotypic Characterization of shd Embryos. Cuticle preparations, staining for spectrin (an actin-binding cytoskeletal component) and IMP-E1 (a 20E-inducible gene) expression in embryos have been described (9).
Gene Cloning and Identification of Mutations. The full-length cDNA sequence was amplified from a Drosophila embryonic cDNA library in pNB40 (11) by PCR using the MACH amplification protocol (12). Two pairs of primers were designed on the 5′ end (A + B) and 3′ end (C + D): ShdA 5′-AGGAGCGGCCGGAGGTAGATATC-3′; ShdB 5′-GAAGAACACGCTCCTTGAGGACTTC-3′; ShdC 5′-GCCTGGCTCGCAGTAGTTCG-3′; and ShdD 5′-GGGATCCGGATACACTGGTGG-3′. To identify the mutations in the shd mutants, genomic DNA from heterozygous animals was amplified by PCR using the primers 5′-ATAAGTGCCTCCAAAGCGGATC-3′ and 5′-AAACGCCTGAGGGTAGGCAC-3′ and sequenced by using the Thermosequenase cycle sequencing kit (United States Biochemical) according to the manufacturer's protocol. The mutant lesion was identified by the presence of two bases at a particular position that corresponds to either the mutant or balancer genomic sequences.
Tissue Expression of shd by RT-PCR. Total RNA was extracted from tissues of wandering third instar larvae (ring gland, brain, gut, fat body, salivary glands, and epidermis) and from adult males or females (head, gut, gonads, and carcass) by using the SV Total RNA Extraction kit (Promega). Single-stranded cDNAs for PCRs were synthesized from total RNAs (100 ng for all tissues) with M-MLV reverse transcriptase (Promega). Specific primers for shd (ShdUp1 5′-CGCTCTCCATCGGCACAAAT-3′ and ShdDo1 5′-AGCAGCACCACCTCCATTTC-3′) and rpL17A (L17Up1 5′-GTGATGAACTGTGCCGACAA-3′ and L17Do1 5′-CCTTCATTTCGCCCTTGTTG-3′) were used, giving 1.4-kb and 300-bp fragments. Thirty-five and 40 PCR cycles were carried out for rpL17A and shd amplification, respectively (94°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min 30 sec), followed by a 10-min extension period at 72°C.
shd in Situ Hybridization. The RNA hybridization and detection were done according to standard protocols (9). To obtain the shd RNA antisense probe, pNB40-shd was linearized with HindIII and transcribed with the T7 promoter. For the shd sense probe, pNB40-shd was linearized with NotI and transcribed with the SP6 promoter.
Construction of Upstream Activator Sequence (UAS) Transgenes and Rescue Crosses. The UAS-shd construct was generated by ligation of a BglII/NotI fragment, isolated from a pNB40 clone containing the full-length cDNA of shd, into a BglII/NotI site of the pUAST vector (13). Transformants were obtained by using standard protocols. Males carrying a tissue-specific Gal4 driver, either twist (14) or armadillo (15), and the mutant allele shdZ329/TM6B were crossed with virgin females carrying a UAS transgene and a different mutant allele, shdZ320/TM6B. Progeny were scored for the survival and fertility of shd-homozygous adults.
Endogenous Embryonic Ecdysteroid Titers and Composition. Dechorionated, wild-type embryos collected every 2 h or selected homozygous mutant shdZ320 embryos, mechanically sorted from heterozygous mutant shdZ320/TM3-armadillo-GFP embryos (both 10–14 h old) (16) by their lack of expression of GFP fluorescence (Copas Select, Harvard Bioscience, Holliston, MA), were homogenized and extracted exhaustively with methanol. Aliquots of the pooled extracts from replicate 20-mg samples (≈2,000 embryos) were subjected to RIA with the H22 antibody (16). Results are expressed in E equivalents. The replicate extracts were then pooled, evaporated to dryness, and subjected to reverse-phase (RP)-HPLC/TLC and differential RIA analysis with both the H22 and SHO3 antibodies to determine the composition of endogenous embryonic ecdysteroids (17). High specific activity 23,24-[3H]E (60 Ci/mmol; 1 Ci = 37 GBq) was purchased from NEN, E and 20E standards were purchased from Sigma-Aldrich, and 2-deoxyecdysone (2dE) was a gift from R. Lafont (Université Pierre et Marie Curie).
Transfection of S2 Cells. S2 cells were transfected with shd cDNA under the control of the actin 5C promoter or with a control construct constitutively expressing the GFP protein by using DDAB-mediated transfection (16, 18). shd cDNA was cloned as a HindIII/NotI fragment containing the full-length cDNA of shd into the StuI/NotI of the S2 expression vector pBRAcpA (19). Shd, Sad, and Dib proteins were epitope-tagged at the C terminus by introducing a NotI site after the last amino acid. A three-copy hemagglutinin (HA) epitope was dropped into the NotI site and the constructs were checked by sequencing.
In Vitro E20MO Activity. Conversion of [3H]E to [3H]20E was measured 3 days after the transfection of S2 cells with shd or GFP (control) expression constructs. The cells (8 × 106) were collected by centrifugation (1,200 × g for 5 min) and homogenized into medium (1 ml) containing [3H]E (1.0 μCi), nonradiolabeled E (1 μg, 2 μM), and NADPH (0.5 mM), and incubated at 25°C for 8 h. Alternatively, the cells were transferred to fresh medium (11 ml) containing [3H]E (1.0 μCi) and E (0–2 μM), but without NADPH, and incubated as before. Methanol extracts were analyzed by RP-HPLC, TLC, and electrospray ionization (ESI)-MS (16).
In a similar manner, to assay E20MO activity during Drosophila development, 20 mg of dechorionated wild-type embryos, 2,000 machine-sorted homozygous or heterozygous mutant shdZ320 embryos (10–14 h old), or whole bodies or selected tissues from yw, heterozygous, or homozygous (rescued) UAS-shd/twistGal4 wandering late third instar mutant larvae or 3-day-old adults were homogenized in Grace's medium (1 ml, pH 7.0) containing [3H]E (0.5 μCi) and NADPH (0.5 mM) and incubated at 25°C for 6 h. Samples were extracted repeatedly with methanol, the pooled solvents were evaporated, and the residues were subjected to RP-HPLC and TLC analysis along with added E and 20E standards (1 μg).
Subcellular Localization of Halloween Gene Products. S2 cells were cotransfected with HA-tagged versions of the protein of interest and an endoplasmic reticulum (mSpitz-GFP) marker (20). Three days after transfection the cells were plated on Con A-coated Lab-Tek II slides (21). After 2 h, the cells were washed, treated with 250 nM MitoTracker Red (Molecular Probes), fixed with 3.7% paraformaldehyde, and permeabilized with methanol. 12CA5 anti-HA (Roche) and goat anti-mouse Alexa 488 or 633 (Molecular Probes) were used to detect the HA-tagged proteins. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), mounted in PermaFluor (Thermo Shandon, Pittsburgh), and analyzed by confocal microscopy (Axioplan 2, Zeiss). Individual optical sections are shown.
Results
Confirmation of shade as CYP314a1. The Halloween genes dib and sad code for the ecdysteroid C22-monooxygenase (CYP302a1) and C2-monooxygenase (CYP315a1), respectively (Fig. 1 and ref. 16). Because both dib and sad proved to code for ecdysone biosynthetic pathway components, we analyzed other Halloween mutants characterized by similar abnormal cuticular patterning (9, 10). We determined that shade (shd), which mapped to 70D2-E8 (10), was in the vicinity of CYP314a, a P450 located at 70E4 (http://flybase.bio.indiana.edu; ref. 22). To determine whether shd corresponds to this P450, CYP314a genomic DNA was amplified by PCR from the appropriate heterozygous mutant stock and sequenced. We found that ShdZ320 has a stop codon at position 136 (a change in the first base from C to T), shdZ383 has a point mutation resulting in an amino acid change (glutamic acid to lysine) at position 225 (a change in the first base from G to A), and shdZ329 has a mutation in the acceptor site of intron 1 confirming that CYP314a is the product of the shd gene.
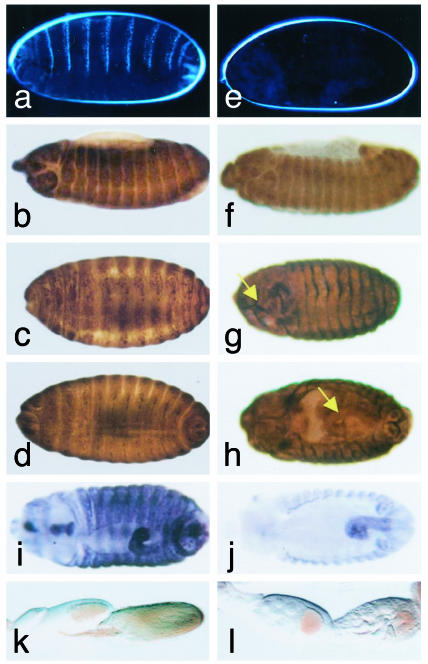
Phenotype of shd Mutants. Like other mutants of the Halloween class, shd mutants do not produce differentiated cuticle (Fig. 2 a vs. e). Spectrin staining (to highlight the general morphology of embryos) reveals that early embryonic development of shd mutants appears normal until approximately stage 14 (10.3–11.3 h) (Fig. 2 b vs. f). At stages 15–16 (11.3–16.0 h) (Fig. 2 c and d vs. g and h), abnormal morphogenetic movements become apparent that involve failure of head involution (indicated by an arrow in Fig. 2g), defects in dorsal closure, and aberrant gut looping (indicated by an arrow in Fig. 2h), a phenotype very similar to that of dib and sad mutants (9, 10). All shd alleles exhibit phenotypes equivalent to shdZ320, which we assume is a null allele because the stop codon eliminates the critical catalytic heme-binding domain. As with dib and sad mutants (9, 16), shd mutants lack epidermal expression of the ecdysone responsive gene IMP-E1 in stage 15 (11.3–13.0 h) embryos relative to wild type (Fig. 2 i and j), consistent with our hypothesis that this gene is also involved in ecdysteroid synthesis.
Fig. 2.
Phenotype of homozygous shd embryos and egg chambers. (a) Normal cuticle development in wild-type embryos. (b–d) Normal embryonic development of wild-type embryos. (e–h) Homozygous shd embryos. (e) Lack of cuticle differentiation in shd mutant embryos. (f) Normal embryonic development at stage 14. (g) Failure of head involution at embryonic stages 15–16 (see arrow). (h) Defect in dorsal closure and aberrant gut looping at embryonic stages 15–16 (see arrow). Embryos in b–d and f–h were stained with spectrin antibody. (i) Stage-15 wild-type embryo showing normal IMP-E1 expression. (j) Reduced IMP-E1 epidermal expression in stage 13 shd mutant embryos. The remaining gut staining is presumably under the influence of a nonecdysone response enhancer element (9, 16). (k) Wild-type egg chambers. (l) Egg chambers showing arrest and degeneration at stages 8–9 of oogenesis in rescued females of the genotype twist>gal4, shdz329/UAS-shd, shdz320.
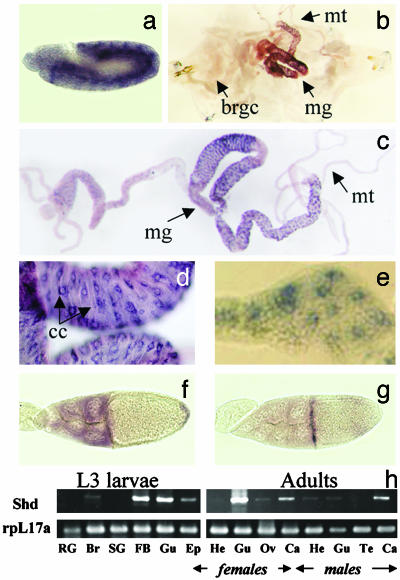
Expression of shd. To determine whether the expression pattern of shd is consistent with a role for shd in ecdysteroidogenesis, in situ hybridization of embryos, third instar larvae, and adult ovaries was performed. Unlike dib and sad, shd is not expressed early in embryogenesis (data not shown), but by the time of maximum germband extension (Fig. 3a, stage 10, 4:20–5:20 h), strong shd expression is observed in the epidermal cells. Expression of shd decreases significantly in older embryos but remains primarily epidermal (data not shown). In situ hybridization of third instar larvae reveals expression of the shd transcript in the midgut copper cells, Malpighian tubules, and fat body (Fig. 3 b–e). There is no shd expression in the brain-ring gland complex (Fig. 3b). Shd expression is also very low or absent in salivary glands and muscle tissue. In the adult ovaries, shd is expressed in both follicle and nurse cells (Fig. 3f), and prominent staining occurs in the centripetally migrating follicle cells (Fig. 3g). At present, we cannot exclude the possibility of a maternal contribution. Semiquantitative RT-PCR analysis of both larval and adult tissues is consistent with the in situ results (Fig. 3h).
Fig. 3.
In situ expression pattern of shd. (a) shd expression in the epidermis at the stage of germband extension (stage 10, 4:20–5:20 h). (b) shd expression in midgut (mg) and Malpighian tubules (mt) in third instar larval whole body but not in the brain-ring gland complex (brgc). (c) shd expression in third instar larval mg and mt. (d) Copper cells (cc) of the third instar larval midgut. (e) Third instar larval fat body. (f) Nurse cells of the adult ovary. (g) Centripetally migrating follicle cells. (h) RT-PCR study of shd tissue expression (Upper) and a control gene, rpL17A (Lower), in third instar larvae (Left) and adults (Right). RG, ring gland; Br, brain; SG, salivary glands; FB, fat body; Gu, gut; Ep, epidermis; He, head; Ov, ovary; Te, testis; Ca, carcass.
Subcellular Localization of Dib, Sad, and Shd Proteins. Numerous charged residues and other conserved motifs characteristic of mitochondrial P450 localization signals (23, 24) were found at the N terminus of Sad and Dib, suggesting that these gene products are likely mitochondrial (9, 16), in agreement with the previously reported subcellular localization of the 2- and 22-monooxygenase activities (25). To test this prediction experimentally, epitope-tagged versions of these proteins were cotransfected into S2 cells with a GFP-tagged marker for the endoplasmic reticulum, and mitochondria were labeled with MitoTracker Red. As shown in Fig. 4, both Sad (a–c) and Dib (d–f) colocalized with the mitochondrial marker, without a detectable signal in the endoplasmic reticulum (not shown), thus confirming their predicted localization. Analogous studies employing an epitope-tagged version of Shd revealed its colocalization with mitochondria as well (Fig. 4 g–i).
Fig. 4.
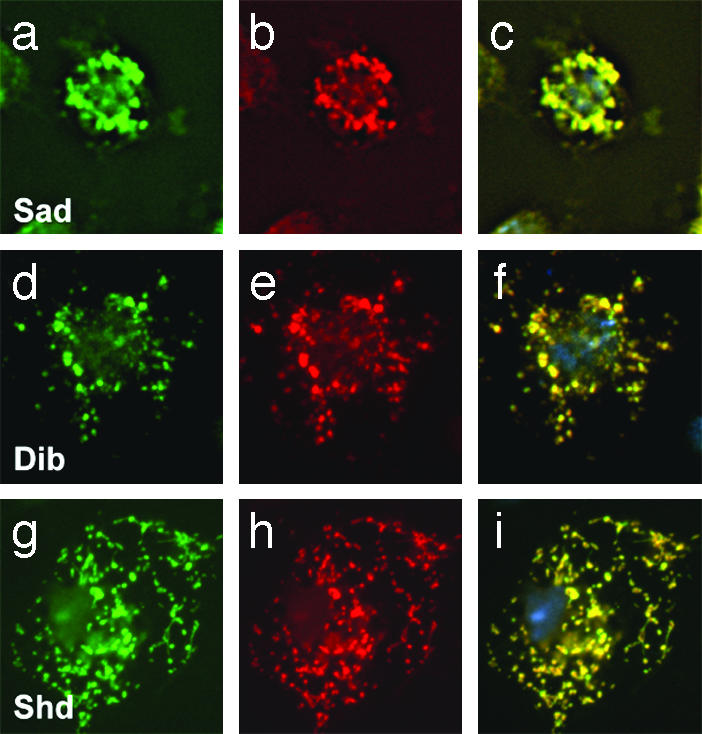
Immunolocalization of Halloween proteins. Confocal sections of S2 cells transfected with HA-tagged Sad (a–c), Dib (d–f), and Shd (g–i). HA immunoreactivity is shown in a, d, and g (green), MitoTracker Red signal is shown in b, e, and h (red), and the merge of HA and MitoTracker staining is shown in c, f, and i.
Transgenic Rescue of shade Mutants. Because shd shows strong expression in both the embryonic and larval midgut (Fig. 3 a–e), particularly in the copper cells, we expressed shd by using the Gal4-UAS expression system with twist or armadillo promoters. Twist is an embryonic mesodermal driver that is also expressed in the larval midgut in both copper cells and muscle nuclei, but not in fat body, Malpighian tubules, salivary glands, or epidermis (data not shown), and armadillo is a ubiquitous driver that is expressed throughout development (15). When shd was expressed by using either the twist or armadillo drivers, all shd mutant embryos developed into normal larvae and these larvae eclosed into adults [for twist>Gal4, shdz329/TM6B × UAS-shd, shdZ320, 248 total progeny were recovered, of which 88 were rescued animals (102% of expected); for arm>Gal4, shdz329/TM6B × UAS-shd, shdZ320, 145 total progeny were recovered, of which 38 were rescued animals (80% of expected)]. However, with the armadillo driver, the adults were fertile, whereas with the twist driver all of the females were sterile and ovaries stopped development at stages 8–9 (Fig. 2 k vs. l).
Embryonic Ecdysteroid Titers and Composition. Previously, very low ecdysteroid titers had been measured by RIA in sorted homozygous mutant dib and sad embryos relative to the titers in either sorted mutant heterozygous dib and sad or wild-type embryos of the same age (16). However, analysis of ecdysteroid titers in extracts from machine-sorted 10- to 14-h-old homozygous or heterozygous mutant shdZ320 embryos were identical to those of 10- to 14-h-old wild-type embryos, i.e., ≈50 pg/mg of wet weight. Nevertheless, subsequent chromatographic separation (RP-HPLC and TLC) and differential RIA analysis of the ecdysteroids in these sorted homozygous shdZ320 embryos revealed that E, together with its immediate precursor 2dE (26), were the major ecdysteroids present. No 20E was detected (Table 1). In contrast, in both sorted heterozygous shdZ320 and wild-type embryos of the same age, 20E and more polar products predominated and E and 2dE were low or absent (27). In fact, the ecdysteroid profile in these 10- to 14-h-old homozygous mutant shdZ320 embryos was very similar to that of early (4–6 h old) wild-type embryos (27), i.e., before the appearance at 6–8 h of either detectable endogenous 20E or significant E20MO activity (28) (see below).
Table 1. Endogenous embryonic ecdysteroid composition: Percent of total Ecdysteroids (average of two replicates).
| Wild type
|
Homozygous shdz320
|
Heterozygous shdz320
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age, h | VPP | 20E | E | 2dE | LPP | VPP | 20E | E | 2dE | LPP | VPP | 20E | E | 2dE | LPP |
| 0-2 | 0 | 0 | 3 | 97 | 0 | ||||||||||
| 2-4 | 0 | 0 | 20 | 80 | 0 | ||||||||||
| 4-6 | 0 | 0 | 50 | 50 | 0 | ||||||||||
| 6-8 | 5 | 10 | 72 | 10 | 3 | ||||||||||
| 10-12 | 85 | 5 | 0 | 0 | 10 | ||||||||||
| 12-14 | 87 | 3 | 0 | 0 | 10 | 4 | 0 | 45 | 50 | 1* | 85 | 8 | 0 | 0 | 7* |
Values were corrected for antibody cross-reactivity. In order of decreasing polarity: VPP, very polar products; 20E; E; 2dE; LPP, low-polarity products.
Ten to 14 h
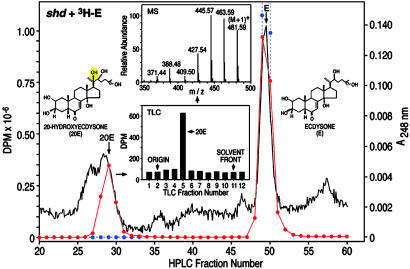
Characterization of E20MO Enzymatic Activity in S2 Cells Transfected with shd. Because the above RIA data suggested that shd encodes the E20MO, S2 cells transfected with shd or the GFP-control construct were homogenized into media containing both [3H]E and nonradiolabeled E for 8 h. The medium and cells were then extracted and analyzed by RP-HPLC/TLC/ESI-MS. The data revealed that these shd-transfected S2 cells hydroxylate E solely to 20E, whereas no such activity was observed in GFP-transfected cells. As shown in Fig. 5, 0.23 μg of 20E was recovered from 1 μg of E substrate, a yield of 23% at a substrate concentration of 2 μM. The characterization of this product as 20E was not only consistent with its UV spectrum (UVmax 248 nm; data not shown) but was also consistent with its specific UV absorption (E 12,400) and radiochemical analysis (Fig. 5). The identity of this product was confirmed by RP-HPLC (20% acetonitrile)/ESI-MS after additional normal-phase TLC purification (Fig. 5 Insets). Note the molecular ion (M + 1)+ at 481 and the sequential loss of four molecules of water at (M + 1)+-H2O (1–4), characteristic of 20E (29). Additional kinetic analysis with intact S2 cell cultures revealed a Km of 1.0 μM and a Vmax of 1 nmol/8 h for this enzyme (data not shown). We conclude from these data that shd codes for the E20MO.
Fig. 5.
RP-HPLC/TLC/ESI-MS analysis of ecdysteroids after shd- or GFP-transfected S2 cell homogenate incubation (8 h) with [3H]E containing E (1 μg) and NADPH (30–100% methanol gradient). Radioactivity was measured after incubations with shd-transfected (red circles) or GFP-transfected (blue circles) cell homogenates with substrate. UV absorption was measured at 248 nm (solid line). (Upper Inset) TLC (chloroform/ethanol) of RP-HPLC-purified 20-hydroxyecdysone (20E) product (1/1,000th of total sample). (Lower Inset) RP-HPLC/ESI-MS on a TSQ Quantum (Thermo Finnigan, San Jose, CA) of the TLC-purified 20E product.
Embryonic, Larval, and Adult in Vitro E20MO Enzymatic Activity. In a similar fashion, analysis of embryonic tissue homogenates (Table 2) revealed that both wild-type (10–12 and 12–14 h old) and heterozygous mutant shdZ320 embryos (10–14 h old) exhibited definitive conversion of the [3H]E substrate to [3H]20E (averages of 1.8% and 1.7%, respectively). In contrast, E20MO activity was undetectable in both 10- to 14-h-old homozygous shdZ320 embryos and in wild-type embryos before 6–8 h, i.e., the stage when significant E20MO activity (0.5%) is first observed (28) and shd expression first becomes apparent (Fig. 3)
Table 2. E20-MO activity in vitro: Percent conversion of [3H]E to [3H]20E.
| Embryo (average of two or more replicates)
|
|||
|---|---|---|---|
| Age | Wild type | Homozygous shdZ320 | Heterozygous shdZ320 |
| 0-2 | 0.0 | ||
| 2-4 | 0.0 | ||
| 4-6 | 0.0 | ||
| 6-8 | 0.5 | ||
| 10-12 | 1.5 | ||
| 12-14 | 2.0 | 0.0* | 1.7* |
Ten to 14 h.
Later, during the third instar (28), considerable E20MO activity was observed in wandering wild-type, yw, or heterozygous UAS-shd/twistGal4 larval homogenates (42.2%, 31.0%, and 19.2%, respectively), whereas very low activity (1.3%) was present in homozygous UAS-shd/twistGal4 (rescued) larvae (Table 3). Within individual larval tissues, the majority of activity was found in the fat body, i.e., wild-type, yw, and heterozygous UAS-shd/twistGal4 (26.7%, 12.8%, and 7.3%, respectively), whereas E20MO activity was undetectable in homozygous UAS-shd/twistGal4 (rescued) larval fat body. Significant E20MO activity was also observed in wild-type, yw, and heterozygous UAS-shd/twistGal4 Malpighian tubules and salivary glands, yet none was detected in these same tissues from homozygous UAS-shd/twistGal4 (rescued) larvae. In contrast, homogenates of the remaining carcass (mostly muscle and epidermis) and midgut tissues exhibited moderate E20MO activity in all types of larvae, whether wild-type or transgenic. However, no E20MO activity was found in larval brain ring-gland complexes.
Table 3. E20-MO activity in vitro: Percent conversion of [3H]E to [3H]20E.
| Wild type | yw | Homozygous UAS-shd/twistGal4 | Heterozygous UAS-shd/twistGal4 | |
|---|---|---|---|---|
| Larvae* | ||||
| Whole larvae | 42.2 | 31.0 | 1.3 | 19.2 |
| Fat body | 26.7 | 12.8 | 0.0 | 7.3 |
| Malphigian tubules | 5.6 | 4.6 | 0.0 | 3.6 |
| Salivary glands | 0.5 | 0.6 | 0.0 | 0.3 |
| Brain-ring gland complex | 0.0 | — | 0.0 | — |
| Midgut | 3.2 | 3.0 | 1.0 | 1.6 |
| Carcass | 1.2 | 3.1 | 0.5 | 2.3 |
| Adult* | ||||
| Whole fly | 1.8 | 16 | 0.09 | 1.1 |
| Ovaries | 1.7 | 2.1 | 0.0 | 1.0 |
| Carcass | 0.4 | 0.3 | 0.08 | 0.2 |
Fifteen larvae (wandering late third instar) or 15 adults (3-day-old females) or tissues from 15 larvae or adults.
E20MO activity could also be detected in whole fly homogenates of all 3-day-old females (Table 3), i.e., wild-type, yw, and heterozygous UAS-shd/twistGal4 (1.8%, 1.6%, and 1.1%, respectively), but it was much lower in homozygous UAS-shd/twistGal4 (rescued) adults (0.09%). Although the majority of this whole-body activity was found to be localized in the ovarian tissues of wild-type, yw, and heterozygous UAS-shd/twistGal4 (1.7%, 2.1%, and 1.0%, respectively) it was not present in the homozygous UAS-shd/twistGal4 (rescued) ovaries. Nevertheless, low but detectable E20MO activity was found in the carcass (comprising all of the remaining tissues) of all four fly lines.
Discussion
The identification of Dib as the ecdysteroid C22-monooxygenase and Sad as the ecdysteroid C2-monooxygenase demonstrated the advantage of using a combination of Drosophila molecular genetics and biochemistry to elucidate the details of the ecdysteroid biosynthetic pathway (16). Herein, this paradigm was used to identify the shd gene product (CYP314a1) as the ecdysone 20-monooxygenase responsible for mediating the conversion of E to the active molting hormone, 20E. The capacity of S2 cells transfected with shd to convert E to 20E, in addition to the absence of further metabolism of E to 20E in machine-sorted homozygous shd mutant embryos both in vitro (Table 2) and in vivo (Table 1), in contrast to wild-type or heterozygous shd mutant embryos, not only proves that Shd has E20MO activity but also that Shd is responsible for all of this activity during embryogenesis. Thus, unlike dib and sad homozygous mutants, which are “low E” embryos (9, 16), the homozygous shd embryos are “low 20E” mutants. This results in reduced IMP-E1 expression and embryonic developmental abnormalities similar to those observed in the “low E” Halloween mutants. The similarity in phenotype between these two types of mutants suggests that E, which is present in high levels in shd mutant embryos, has little functional activity at this stage.
The data in Fig. 3 and Tables 2 and 3 also indicate that Shd is responsible for all E20MO activity during larval and adult stages, because it is expressed in tissues (fat body, midgut, Malpighian tubules, epidermis, salivary glands, and ovaries) known to possess this enzyme activity in vitro during these stages, not only in Drosophila (30–34) but also in other Dipteran insects (35). In addition, shd is not expressed in tissues where such activity has been reported to be absent, such as in the ring glands, brain, ventral ganglion, and muscle of Diptera and other insects including Drosophila (5, 36–42). The complete absence of in vitro E20MO activity in the larval fat body, Malpighian tubules, salivary glands, and adult ovaries of rescued homozygous UAS-shd/twistGal4 flies relative to that observed in corresponding heterozygous UAS-shd/twistGal4, yw, or wild-type tissues is presented as proof that Shd is solely responsible for E20MO activity in these tissues.
In embryos, it seems probable that both E and 20E are highly mobile and that 20E is able to reach all target tissues irrespective of where it is made. This follows because expression of shd in embryos is normally confined to epidermal cells. Nevertheless, ectopic expression of shd in the embryonic mesoderm as well as in larval and (presumably) pupal gut and muscle by using the UAS-shd/twistGal4 system completely rescued embryonic, larval, and pupal development to the adult. That is, although the fat body is normally by far the most active tissue in the conversion of E to 20E during larval stages in wild-type, yw, and heterozygous UAS-shd/twistGal4 individuals (Tables 2 and 3), in the rescued homozygous UAS-shd/twistGal4 individuals, fat body activity is nonexistent. Instead, relatively low E20MO activity expressed under twist promoter control is only observed in the midgut and muscle tissues of the carcass. Yet, this activity is nevertheless sufficient for normal development to the adult. In addition, when shd expression is placed under the control of the ubiquitous armadillo promoter and so is expressed in all tissues at all times, the same result is observed. Thus, it follows that the appearance of active hormone (20E) is not the result of the precise developmental control of shd transcription but rather of the biosynthesis and release of E from the prothoracic gland cells of the ring gland. Circulating E is efficiently distributed to sites of Shd activity for conversion to 20E, which subsequently equilibrates throughout the embryo, larva, pupa, and developing adult.
However, in the homozygous UAS-shd/twistGal4 adult, no E20MO activity was detected in the ovaries (Tables 2 and 3) and severe degeneration of the egg chambers after stage 9 was observed (Fig. 2), even though some E20MO activity was found in the adult carcass. Apparently, in the adult, the ovary may be a closed system. That is, the ovary must not only synthesize ecdysone (43) but also must convert it to the active hormone 20E for normal ovarian development and fertility. Thus, armadillo, but not twist, is a suitable driver for necessary adult shd expression leading to fertility.
Another interesting aspect of the shd embryonic expression pattern is its delayed appearance relative to dib and sad, which are both first expressed at the blastoderm stage. The lag in shd expression likely accounts for the observations first made by Maróy et al. (27) and Mitchell and Smith (28), and confirmed here, that Drosophila embryos initially synthesize E from less-polar components (i.e., 2dE) during the first 6 h of development and only later (6–14 h) convert E to 20E and more-polar compounds. Indeed, it isonlyafter6hthatthe 20E-inducible cascade of early transcription factor genes E75A&B, DHR3, DHR4, DHR39, and βFTZ-F1 becomes apparent (April A. Sullivan and Carl S. Thummel, personal communication). Additional support comes from recent findings that E20MO activity, both in vivo and in vitro, increases dramatically after gastrulation in developing nondiapause eggs of the silk moth, Bombyx mori, relative to the very low activity measured in diapause-destined eggs, which cease further embryonic development at this time (44, 45).
Finally, in S2 cells, the Dib, Sad, and Shd enzymes are all localized to the mitochondria. For Dib and Sad, this is consistent with prior biochemical fractionation data showing that both the 22- and 2-monooxygenase activities are mitochondrial (29). Prior cell fractionation data for the E20MO is more ambiguous, with a microsomal and/or mitochondrial localization identified, depending on the insect, tissue, and developmental stage being examined (4, 5, 7, 46, 47). The N terminus of the Shd protein contains both hydrophobic signal-type sequences typical of microsomal P450s (48) as well as a charged segment containing sequences characteristic of typical mitochondrial enzymes (49). Thus, as a result of differential post translational modification, it is possible that Shd resides in either location depending on tissue type and developmental stage. However, the cofactors for supplying reducing equivalents are quite different for mitochondrial and microsomal P450 enzymes, as are the sequences that are thought to interact with these cofactors. Thus, it remains a possibility that E20MO activity has evolved independently in different insect species and so a microsomal location in other species might reflect a divergent P450 not related to Shd, as is suggested by recent data concerning the tentative identification of CYP6H1 as a E20MO in the Malpighian tubules of the locust (50, 51). The future unequivocal identification of ecdysone 20-monooxygenases from a variety of insects may reveal whether there was evolution of the enzyme itself or of subcellular localization signals for a common enzyme.
Acknowledgments
We thank Susan Whitfield for graphics, Beni Shilo for ER GFP marker, C. Nüsslein-Volhard for shd alleles, and Carl Thummel, Rene Feyereisen, and Stan Smith for constructive criticism of the manuscript. The University of North Carolina MS facility is supported in part by National Institutes of Health Grants P30 CA16086 and P30 ES10126. This work was supported by National Institutes of Health Training Grant HD33692 (to A.P.) and National Science Foundation Grant IBN0130825 (to L.I.G. and J.T.W.). M.B.O. is an Investigator of the Howard Hughes Medical Institute. C.D.-V. and J.-P.P. are supported by the Université Pierre et Marie Curie and the Ministère de la Recherche Scientifique.
Abbreviations: CYP, cytochrome P450; 2dE, 2-deoxyecdysone; E, ecdysone; E20MO, E-20-monooxygenase; ESI, electrospray ionization; 20E, 20-hydroxyecdysone; HA, hemagglutinin; RP, reverse-phase; UAS, upstream activator sequence.
Data deposition: The sequence of shade cDNA has been deposited in the GenBank database (accession no. AF484414).
References
- 1.Henrich, V. C., Rybczynski, R. & Gilbert, L. I. (1999) Vitam. Horm. 55, 73–125. [DOI] [PubMed] [Google Scholar]
- 2.Warren, J. T. & Gilbert, L. I. (1986) Insect Biochem. 16, 65–82. [Google Scholar]
- 3.Hiruma, K., Böcking, D., Lafont, R. & Riddiford, L. M. (1997) Gen. Comp. Endocrinol. 107, 84–97. [DOI] [PubMed] [Google Scholar]
- 4.Feyereisen, R. & Durst, F. (1978) Eur. J. Biochem. 88, 37–47. [DOI] [PubMed] [Google Scholar]
- 5.Bollenbacher, W. E., Smith, S., Wielgus, J. & Gilbert, L. I. (1977) Nature 268, 660–663. [Google Scholar]
- 6.Blais, C. & Lafont, R. (1986) Arch. Insect Biochem. Physiol. 3, 501–512. [DOI] [PubMed] [Google Scholar]
- 7.Smith, S., Bollenbacher, W., Cooper, D., Schleyer, H., Wielgus, J. & Gilbert, L. I. (1979) Mol. Cell. Endocrinol. 15, 111–133. [DOI] [PubMed] [Google Scholar]
- 8.Smith, S. L., Bollenbacher, W. E. & Gilbert, L. I. (1983) Mol. Cell. Endocrinol. 31, 227–251. [DOI] [PubMed] [Google Scholar]
- 9.Chávez, V. M., Marqués, G., Delbecque, J. P., Kobayashi, K., Hollingsworth, M., Burr, J., Natzle, J. E. & O'Connor, M. B. (2000) Development (Cambridge, U.K.) 127, 4115–4126. [DOI] [PubMed] [Google Scholar]
- 10.Jürgens, G., Wieschaus, E., Nüsslein-Volhard, C. & Kluding, H. (1984) Roux's Arch. Dev. Biol. 193, 283–295. [DOI] [PubMed] [Google Scholar]
- 11.Brown, N. & Kafatos, F. (1988) J. Mol. Biol. 203, 425–437. [DOI] [PubMed] [Google Scholar]
- 12.Haerry, T. E. & O'Connor, M. B. (2002) Gene 291, 85–93. [DOI] [PubMed] [Google Scholar]
- 13.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 14.Furlong, E. E. M., Anderson, E., Null, B., White, K. & Scott, M. (2001) Science 293, 1629–1633. [DOI] [PubMed] [Google Scholar]
- 15.Sanson, B., White, P. & Vincent, J. P. (1996) Nature 383, 627–630. [DOI] [PubMed] [Google Scholar]
- 16.Warren, J. T., Petryk, A., Marqués, G., Jarcho, M., Parvy, J.-P., Dauphin-Villemant, C., O'Connor, M. B. & Gilbert, L. I. (2002) Proc. Natl. Acad. Sci. USA 99, 11043–11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warren, J. T., Steiner, B., Dorn, A., Pak, M. & Gilbert, L. I. (1986) J. Liquid Chromatogr. 9, 1759–1782. [Google Scholar]
- 18.Han, K. (1996) Nucleic Acids Res. 24, 4362–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherbas, L., Moss, R. & Cherbas, P. (1994) Methods Cell Biol. 44, 161–179. [DOI] [PubMed] [Google Scholar]
- 20.Tsruya, R., Schlesinger, A., Reich, A., Gabay L., Sapir, A. & Shilo, B.-Z. (2002) Genes Dev. 16, 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers, S. L., Rogers G. C., Sharp, D. J. & Vale, R. D. (2002) J. Cell Biol. 158, 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams, M. D., Celniker, S., Holt, R., Evans, C., Gocayne, J., Amanatides, P., Scherer, S., Li, P., Hoskins, R., Galle, R., et al. (2000) Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 23.Feyereisen, R. (1999) Annu. Rev. Entomol. 44, 507–533. [DOI] [PubMed] [Google Scholar]
- 24.Tijet, N., Helvig, C. & Feyereisen, R. (2001) Gene 262, 189–198. [DOI] [PubMed] [Google Scholar]
- 25.Kappler, C., Kabbouh, M., Hetru, C., Durst, F. & Hoffmann, J. A. (1988) J. Steroid Biochem. 31, 891–898. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert, L. I., Rybczynski, R. & Warren, J. T. (2002) Annu. Rev. Entomol. 47, 883–916. [DOI] [PubMed] [Google Scholar]
- 27.Maróy, P., Kaufmann, G. & Dübendorfer, A. (1988) J. Insect Physiol. 34, 633–637. [Google Scholar]
- 28.Mitchell, M. J. & Smith, S. L. (1988) Gen. Comp. Endocrinol. 72, 467–470. [DOI] [PubMed] [Google Scholar]
- 29.Wainwright, G., Prescott, M., Lomas, L., Webster, S. & Rees, H. H. (1997) Arch. Insect. Biochem. Physiol. 35, 21–31. [Google Scholar]
- 30.Bownes, M., Dübendorfer, A. & Smith, T. (1984) J. Insect Physiol. 30, 823–830. [Google Scholar]
- 31.Dübendorfer, A. & Maróy, P. (1986) Insect Biochem. 16, 109–113. [Google Scholar]
- 32.Sommé-Martin, G., Colardeau, J. & Lafont, R. (1988) Insect Biochem. 18, 729–734. [DOI] [PubMed] [Google Scholar]
- 33.Sommé-Martin, G., Colardeau, J. & Lafont, R. (1988) Insect Biochem. 18, 735–742. [DOI] [PubMed] [Google Scholar]
- 34.Grau, V. & Lafont, R. (1994) J. Insect Physiol. 40, 87–96. [Google Scholar]
- 35.Smith, S. & Mitchell, M. (1986) Insect Biochem. 16, 49–55. [Google Scholar]
- 36.Bollenbacher, W. E., Goodman, W., Vedeckis, W. V. & Gilbert, L. I. (1976) Steroids 27, 309–324. [DOI] [PubMed] [Google Scholar]
- 37.Redfern, C. P. F. (1984) Proc. Natl. Acad. Sci. USA 81, 5643–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pak, M. & Gilbert, L. I. (1987) J. Liquid Chromatogr. 10, 2591–2611. [Google Scholar]
- 39.Henrich, V., Pak, M. & Gilbert, L. I. (1987) J. Comp. Physiol. 157, 543–549. [DOI] [PubMed] [Google Scholar]
- 40.Böcking, D., Dauphin-Villemant, C., Toullec, J. Y., Blais, C. & Lafont, R. (1994) C. R. Acad. Sci. Ser. III 317, 891–898. [Google Scholar]
- 41.Warren, J. T., Bachman, J., Dai, J. D. & Gilbert, L. I. (1996) Insect Biochem. Mol. Biol. 26, 931–943. [DOI] [PubMed] [Google Scholar]
- 42.Warren, J. T. & Gilbert, L. I. (1996) Insect Biochem. Mol. Biol. 26, 917–929. [DOI] [PubMed] [Google Scholar]
- 43.Kelly, T. J. (1994) in Perspectives in Comparative Endocrinology, eds. Davey, K. G., Peter, R. G. & Tobe, S. S. (Natl. Res. Council of Canada, Ottawa), pp. 282–290.
- 44.Horike, N. & Sonobe, H. (1999) Arch. Insect Biochem. Physiol. 41, 9–17. [DOI] [PubMed] [Google Scholar]
- 45.Sonobe, H., Tokushige, H., Makka, T., Tsutsumi, H., Hara, N. & Fujimoto, Y. (1999) Zool. Sci. 16, 935–943. [Google Scholar]
- 46.Mitchell, M. J. & Smith, S. L. (1986) Insect Biochem. 16, 525–537. [Google Scholar]
- 47.Smith, S. L. & Mitchell, M. J. (1986) Insect Biochem. 16, 49–55. [Google Scholar]
- 48.Van den Broek, P., Barroso, M. & Lechner, M. (1996) Experientia 52, 851–855. [DOI] [PubMed] [Google Scholar]
- 49.Omura, T. & Ito, A. (1991) Methods Enzymol. 206, 75–81. [DOI] [PubMed] [Google Scholar]
- 50.Winter, J., Bilbe, G., Richener, H., Sehringer, B. & Kayser, H. (1999) Biochem. Biophys. Res. Commun. 259, 305–310. [DOI] [PubMed] [Google Scholar]
- 51.Winter, J., Eckerskorn, C., Waditschatks, R. & Kayser, H. (2001) Biol. Chem. 382, 1541–1549. [DOI] [PubMed] [Google Scholar]