Abstract
Stable genetic modification of stem cells holds great promise for gene therapy and marking, but commonly used γ-retroviral vectors were found to influence growth/survival characteristics of hematopoietic stem cells (HSCs) by insertional mutagenesis. In this article, we show that promoter-deprived γ-retroviral self-inactivating (pd-SIN) vectors allow stable genetic marking of serially reconstituting murine HSC. In contrast to findings with γ-retroviral long terminal repeat (LTR) vectors, serial transplantation of pd-SIN-marked HSC in a sensitive mouse model was apparently not associated with induced clonal imbalance of gene-marked HSC. Furthermore, insertions of pd-SIN into protooncogenes, growth-promoting and signaling genes occurred significantly less frequent than in control experiments with LTR vectors. Also, transcriptional dysregulation of neighboring genes potentially caused by the pd-SIN insertion was rarely seen and comparatively weak. The integration pattern of promotor-deprived SIN vectors in reconstituting HSC seems to depend on the transcriptional activity of the respective gene loci reflecting the picture described for LTR vectors. In conclusion, our data strongly support the use of SIN vectors for gene-marking studies and suggest an increased therapeutic index for vectors lacking enhancers active in HSC.
Introduction
Because of their ability to stably integrate in their host cells' genomes, retroviral vectors have been successfully used for gene marking1,2,3 and gene therapy,4,5,6 primarily in the hematopoietic system. However, growing evidence from animal7,8 as well as clinical studies6,9,10 indicates a significant influence of γ-retroviral vector insertions in the vicinity of growth-regulatory genes (i.e., insertional mutagenesis) on the in vivo behavior of the affected clones potentially resulting in induced clonal dominance or leukemia development. The actual prevalence of those side effects may depend on additional factors, e.g., target cell population,11 transgene12 or even underlying disease.13,14 The latter has recently been demonstrated by several groups that performed large-scale analysis of insertion sites in several clinical gene therapy studies.6,15,16,17 Obviously, the potential impact of insertional mutagenesis has important implications for (i) the interpretation of marking experiments and (ii) the planning of clinical gene therapy studies which are based on this vector type.
Molecular analysis of retroviral vector–mediated malignant transformation has revealed activation of growth-promoting genes as the main mechanism of insertional mutagenesis.6,7,8,9,10,12,13,14,17,18 In fact, the strong enhancer and promotor sequences located in the retroviral long terminal repeats (LTRs) may cause transactivation of neighboring genes resulting in their aberrant expression. In accord, self-inactivating (SIN) vectors lacking the relevant sequences in the U3 region of their LTRs have been shown to be significantly less mutagenic both in vitro and in vivo.19,20 However, there is strong evidence that even γ-retroviral SIN vectors containing internal viral promotor sequences are eventually able to transform hematopoietic stem cells (HSCs) by insertional mutagenesis.19,20,21
We asked here whether gene marking of HSC would be feasible with a retroviral vector lacking any promotor/enhancer elements. We therefore constructed a promotor-deprived γ-retroviral SIN (pd-SIN) vector and used this for HSC marking in a serial bone marrow transplantation (BMT) assay, previously shown to represent a sensitive model for detecting insertional clonal imbalance and leukemogenesis.7,8,22
Results
Development and testing of pd-SIN
The promotor-deprived vector pd-SIN was generated by excising the internal SF-promotor/enhancer sequences from SINSFeGFP (Figure 1a), a SIN γ-retroviral vector described previously.20,23 To test whether pd-SIN is still able to express enhanced green fluorescent protein (eGFP), SC-1 cells were transduced in parallel with pd-SIN and SINSFeGFP at comparable efficiencies (59% for pd-SIN and 64% for SINSFeGFP). Surprisingly, in about one per three pd-SIN transduced cells, weak expression of eGFP was detectable by fluorescence-activated cell sorting (FACS) analysis, which was, however, reduced as compared to SINSFeGFP-transduced cells by a factor of >50, as determined by mean fluorescence intensities (data not shown).
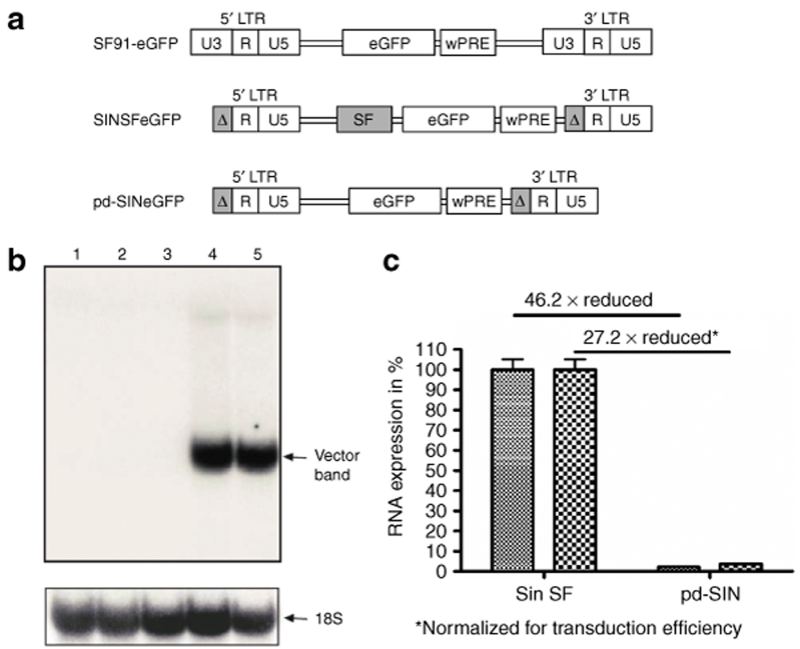
Figure 1.
Promoter-deprived γ-retroviral self-inactivating (pd-SIN) vector design and transcriptional activity. (a) Construction of the vector SF91-eGFP was described earlier.22 pd-SIN was derived from SINSFeGFP (refs. 19,23) by deleting the SF-promotor/enhancer sequences. It thus does not contain any promotor/enhancer element. (b) No enhanced green fluorescent protein (eGFP) transcription was detectable in murine SC-1 fibroblasts after transduction with pd-SIN (performed in duplicates) by northern blot analysis using an eGFP probe (lanes 2,3). In contrast, SC-1 cells transduced in parallel with SINSFeGFP (in duplicates) revealed strong eGFP transcription (arrow, lanes 4,5). Lane 1 shows a mock control. RNA load was controlled by hybridization with an 18S probe (lower blot, arrow). (c) Since pd-SIN-transduced cells were weakly eGFP-positive in fluorescence-activated cell sorting analysis, quantitative reverse transcription–PCR analyses were performed. As shown, normalized transcription levels were reduced by a factor of >25.
We next measured RNA levels for both pd-SIN and SINSFeGFP. In northern blot analysis no eGFP message was evident in pd-SIN as well as mock-transduced cells, whereas a strong signal was seen in SC-1 cell transduced with SINSFeGFP (Figure 1b). However, with a more sensitive quantitative reverse transcription–PCR technique,19 eGFP transcription was detectable in pd-SIN-transduced cells (Figure 1c). The low-level expression of eGFP observed in SC-1 cells, as well as in primary HSC probably indicates the presence of some cryptic promotor activity in the retroviral leader.
Serial transplantation of HSC marked with pd-SIN
Two independent mouse experiments (Figure 2) were subsequently performed, each with observation periods of ~1 year. In both experiments, the γ-retroviral pd-SIN vector was used for gene marking in the experimental group and a standard γ-retroviral vector (SF91-eGFP) bearing the strong SFFV U3 promotor/enhancer region in the control group.24 Earlier studies with the latter and similar LTR vectors have shown a significant mutagenic potential which manifested in induced clonal dominance or even malignant transformation.6,7,8,9,10,12,13,14,15,25
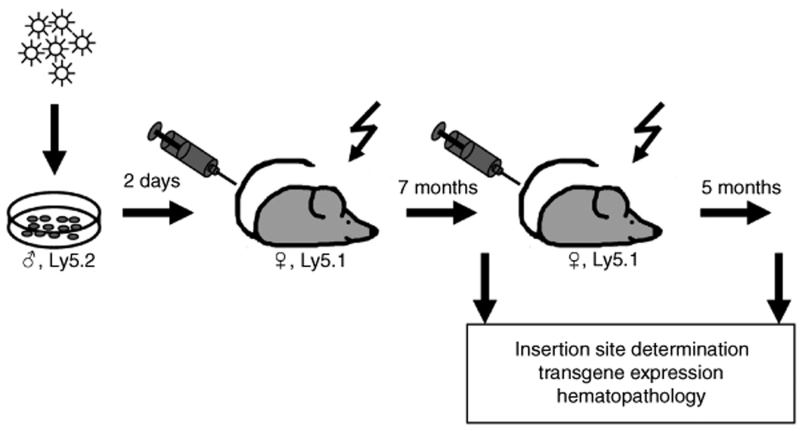
Figure 2.
Experimental setup. The design of the mouse study follows the well established, sex-mismatched diallelic (CD45.2/Ly5.2 into CD45.1/Ly5.1) serial bone marrow transplantation model.7,8,21 This model had enabled us to discover serious side effects of γ-retroviral gene transfer before their occurrence in clinical gene therapy studies, in particular the phenomenon of induced clonal dominance. Shortly, Lin– bone marrow cells from male donors were transduced with the promoter-deprived γ-retroviral self-inactivating vector and transplanted into female mice after TBI (10 Gy). Seven months later, hematopoietic organs of these mice were thoroughly investigated before secondary transplantation into irradiated mice took place. After a further observation period of 5 months, final analysis of hematopoietic organs, including molecular analysis of vector insertion sites, was performed.
Gene transfer efficiency into HSC for both pd-SIN and SF91-eGFP was in the range of 50% as estimated by flow cytometry and quantitative PCR (data not shown). Transduced HSC were serially transplanted into lethally irradiated mice,7,8,22 which were followed up for the indicated periods of time (Figure 2).
In the pd-SIN group, 14 animals from the first cohorts (Exp. I: 7; Exp. II: 7) and 13 from the second cohorts (Exp. I: 5; Exp. II: 8) survived and were available for detailed investigation. The corresponding numbers in the SF91-eGFP control group were 14 for the first cohorts (Exp. I: 7; Exp. II: 7) and 17 for the second cohorts (Exp. I: 8; Exp. II: 9).
FACS analysis22 showed hematopoietic donor chimerism in the peripheral blood (Materials and Methods) and normal cell counts for all blood lineages in all animals (n = 58); also, final pathological inspection of all 58 available animals did not reveal any abnormalities (data not shown).
Limited numbers of dominant clones in both the pd-SIN and the SF91-eGFP groups
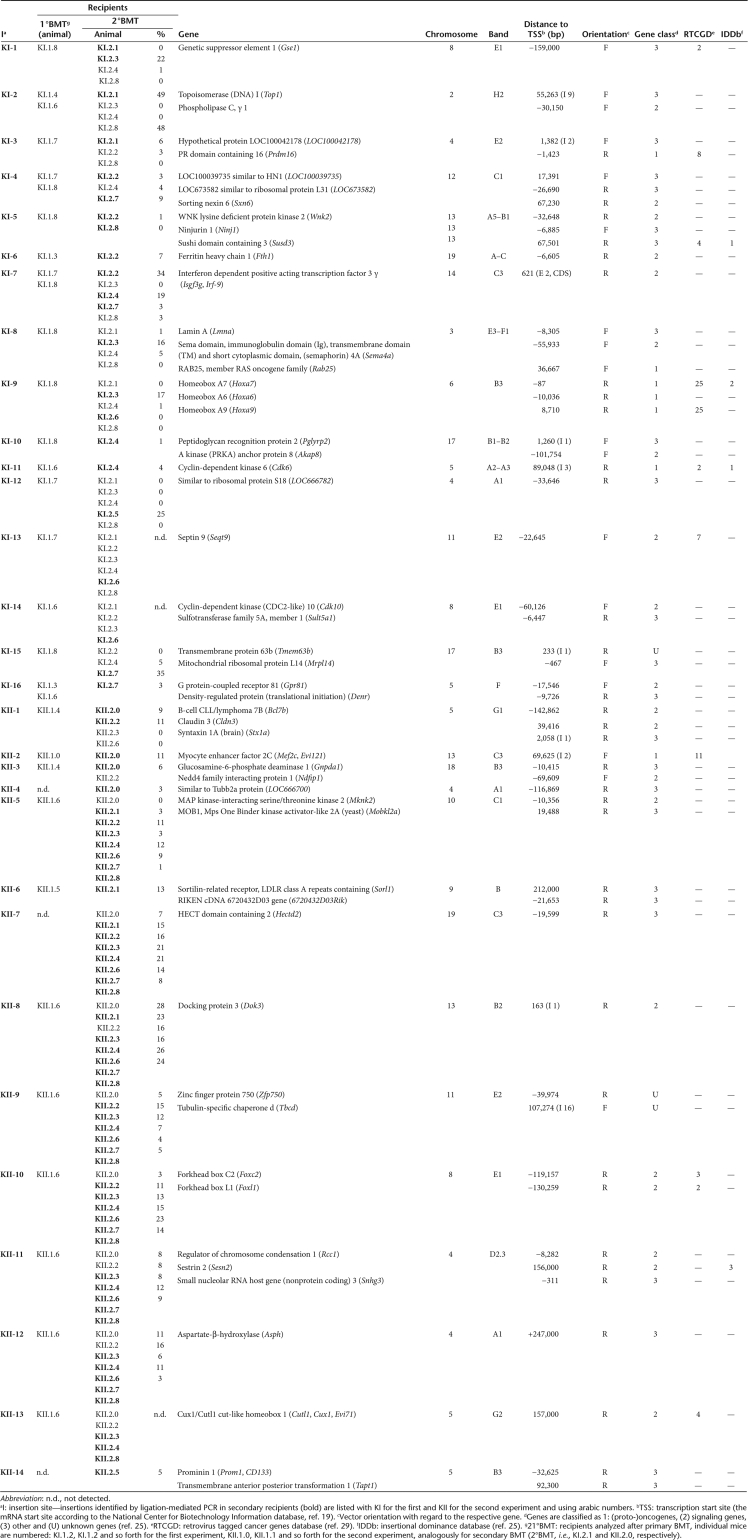
In order to preferentially identify insertion sites in clones dominating hematopoiesis we used a modified ligation-mediated PCR (LM-PCR) protocol.26 To concentrate on long-term HSC,27,28 we focused our analysis on clones dominating hematopoiesis after serial BMT (as illustrated for pd-SIN in Figure 3a).25,26 A retroviral vector insertion identified by LM-PCR was considered to represent a dominant clone only if the signal was retrieved in at least two independent LM-PCRs. Using this definition, we found a total of 25 “dominant” pd-SIN insertions in the 13 animals of the two second cohorts available for analysis (Exp. I: 12 insertions, 5 mice; Exp. II: 13 insertions, 8 mice; Table 1). For SF91-eGFP, 30 HSC clones were retrieved from 14 secondary transplanted animals (Table 2).
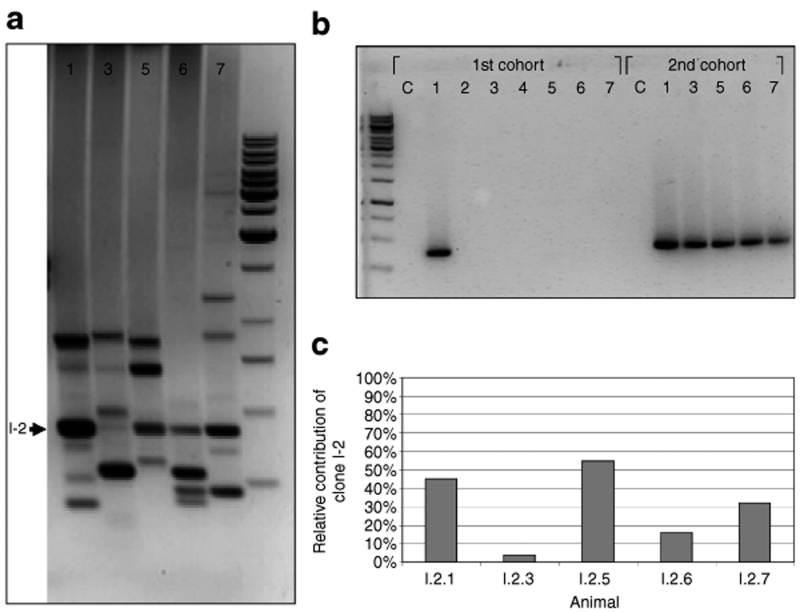
Figure 3.
Analysis of promoter-deprived γ-retroviral self-inactivating insertion sites by different PCR techniques. (a) A modified ligation-mediated PCR (LM-PCR) method26 was used to assess genomic locations of integrated vector copies in reconstituting hematopoietic cells (~12 months after initial transduction and 5 months after serial bone marrow transplantation). A representative agarose gel analysis showing the five secondary recipients from Exp. 1 is depicted. Twelve different insertions representing dominant clones could be identified. One insertion (I-2) was dominant in four out of five secondary animals (arrow). (b) Insertion site specific (is-PCR) revealed the distribution of hematopoietic clones bearing specific insertion in individual animals of the first and second cohorts of transplanted mice. As a representative example, is-PCR to detect the hematopoietic clone marked by insertion I-2 is shown. As illustrated, this clone took part in the reconstitution of hematopoiesis in all secondary animals. Insertion I-2 had occurred into the Efcab2 gene not known to be involved in hematopoietic stem cell (HSC) growth regulation. (c) Shot-gun cloning of LM-PCR products followed by sequencing of individual clones was performed to assess the relative contribution of specifically marked HSC clones to hematopoiesis in individual animals. Again, insertion I-2 has been selected as a representative example for this type of analysis. Quantitative data are in very good agreement with data from (a) LM-PCR and (b) is-PCR.
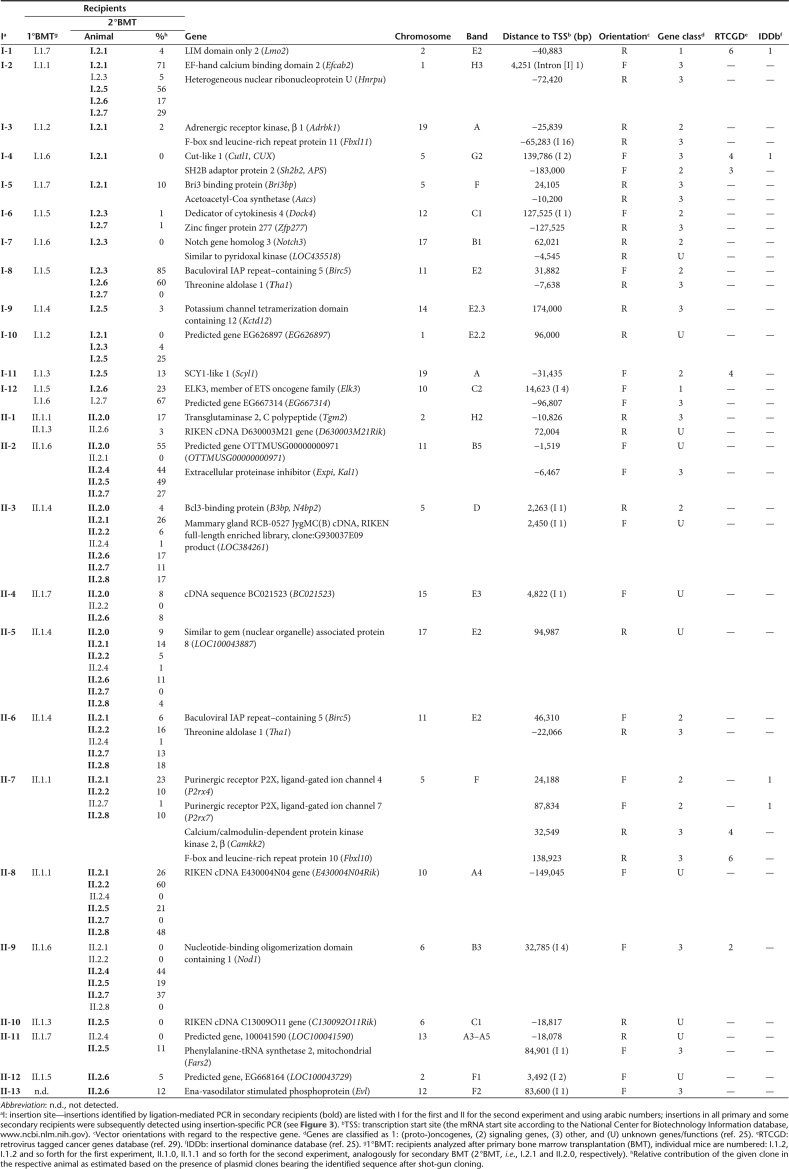
Table 1.
Insertion sites of pd-SIN in dominant clones
Table 2.
Insertion sites of SF91-eGFP in dominant clones
These limited numbers confirm the impact of serial transplantation on the establishment of clonal dominance,2,28 and may also reflect impaired survival of long-term repopulating HSC under our culture conditions. Additional selection steps seem to further reduce HSC clone number as noted in our previous study with mutagenic γ-retroviral marking LTR vectors, in which only 12 “dominant insertions” were retrieved from 16 secondary recipients.8
Using insertion-specific PCR (is-PCR) (Supplementary Materials and Methods) we were able to establish comprehensive figures of the distribution of HSC clones in primary and secondary recipients marked by pd-SIN (Figure 3b, Table 1) or SF91-eGFP (Table 2), respectively. For pd-SIN we found just two insertions (II.3 and II.5) that showed a fully identical distribution pattern by both LM-PCR and is-PCR analysis indicating that these two insertions had occurred within one and the same cell, thus representing only one dominant clone. For the control vector at least three clones apparently contained two insertion sites (KI-1 and KI-8, KII-5 and KII-7, KII-9 and KII-10) and one clone was marked by three insertions (KII-11, KII-12 and KII-13). It might be speculated that in HSC clones with multiple vector integrations only one insertion was involved in the establishment of induced clonal dominance (e.g., KII-10: Foxc2 or KII-11: Sesn2), whereas the other insertion(s) may just represent marking events or “bystander insertions.” This may lead to an overestimation of relative numbers of insertion sites in genes previously not associated with clonal dominance (gene class 3 in Figure 4).
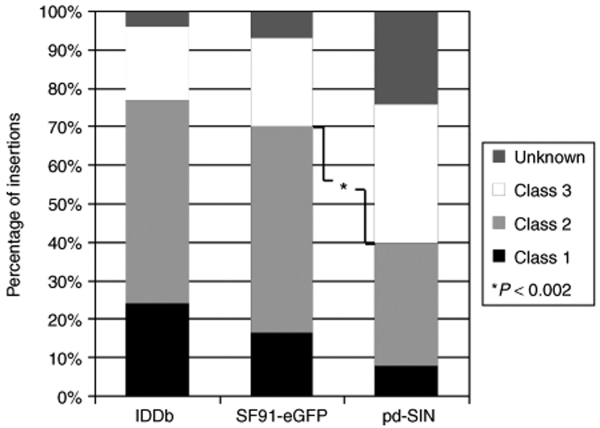
Figure 4.
Specific distribution of promoter-deprived γ-retroviral self-inactivating (pd-SIN) insertion sites in reconstituting hematopoietic stem cell. Insertion sites were categorized based on the genes present in a 150-kb window at both the 5′ and the 3′ sites of each individual insertion. Genes were classified into (putative) protooncogenes (class 1), signaling genes (class 2), other genes (class 3), and genes of unknown function as previously described.25 For control, respective data obtained in the two parallel control experiments with the γ-retroviral long terminal repeat (LTR) vector SF91-eGFP are shown. As evident, pd-SIN insertions show a strongly different insertion pattern as compared to γ-retroviral LTR vectors. Based on the frequency of hits into putative growth–promoting genes (classes 1 and 2), this difference is highly significant (P < 0.002; Fisher's exact test). In contrast, no significant differences were found between insertion patterns for SF91-eGFP (control group) and historical data from the IDDb.25
Notably, LM-PCR and is-PCR analysis, i.e., two independent techniques, led to fully identical results during analysis of insertion site patterns (Tables 1 and 2). Importantly, results for different insertions within one and the same clone were highly consistent. For instance, if a “dominant” clone contains two vector insertions, one would expect that both insertions will independently be detectable by our LM-PCR technique (focusing on “dominant insertions”). This was indeed the case for all respective clones and in all tested animals (compare, for instance, insertions II-3 and II-5, in animals II.2.0, II.2.1, II.2.2, II.2.6, II.2.7, II.2.8). Accordingly, if a clone contributed in a minor fashion to hematopoiesis, all insertions present in that clone were independently retrieved by the more sensitive is-PCR, but none of them by LM-PCR (compare the same insertions II-3 and II-5 in animals II.1.4 and II.2.4). Together these data confirm the validity of the used definition for clonal dominance.8,26
All other insertion sites revealed a distinct distribution pattern between primary and secondary animals which strongly suggests that they represented HSC clones marked by single integrations. Altogether, a total of 24 HSC clones (23 with one, and 1 with two insertions) marked by pd-SIN and 25 clones (21 with one, and 3 with two and 1 with three insertions) marked by SF91-eGFP were analyzed in this study.
Relative contribution of single clones to hematopoiesis
Next we aimed at determining the relative contribution of defined (dominant) clones to hematopoiesis in single animals. To do so, we established a high-throughput sequencing approach for insertion sites retrieved by LM-PCR based on shot-gun cloning of whole LM-PCR products. For each secondary recipient, 96 colonies were sequenced; thus 1,248 (13 × 96) additional sequences were obtained for the 13 secondary recipients in the two pd-SIN experiments. Nine hundred and ninety of these sequences represented pd-SIN insertion sites which were unambiguously locatable in the murine genome; the mean number of identifiable sequences was 76 ± 12 per mouse.
In Figure 3c, an example of such data is provided for one insertion (I-2) detected by LM-PCR in animals I.2.1; I.2.5, I.2.6, I.2.7 (compare Figure 3a) and additionally by is-PCR in animal I.2.3 (compare Figure 3b). Remarkably, the quantitative sequencing data are in excellent agreement with the LM-PCR data (Figure 3c). Indeed, only in those cases in which the given clone accounted for >15% of the total detectable transduced clones did it become a “dominant clone” in the LM-PCR analysis. In contrast, in animal I.2.3, where the same clone was only detectable by is-PCR, it constituted <5% of the transduced hematopoietic cells detectable with our assay (Figure 3a). Similar results were also obtained for the other insertions (Table 1), providing data with regard to the relative contribution of single clones to the hematopoiesis in a given animal.
An analogous analysis was performed for the SF91-eGFP control group. To avoid sequencing of internal vector control bands, those were excluded based on a newly developed PCR approach. A total of 1,632 sequences were investigated for 17 secondary BMT recipients, of which 1,229 represented SF91-eGFP insertions (72 ± 7 per mouse). The obtained clonal quantification data are presented in Table 2.
It is important to note, however, that the above quantification procedure has certain biases. Both the LM-PCR (influenced, for example, by the length of the amplified fragment) and the cloning (not all pieces of eukaryotic genomes are equally “clonable” in bacteria) steps are not reliably quantitative methods. This may particularly apply, if several clones are present in a given animal resulting in a competitive situation. Those restrictions may explain our observation that not all insertions that represented dominant clones in the gel analysis quantitatively contributed to shot-gun cloning (Table 1). This finding underlines the importance of directly cutting out and sequencing dominant bands from the agarose gel after LM-PCR in order to avoid loss of insertion sites during the cloning step.
Genomic locations of insertion sites
An important question was whether the genomic location pattern of pd-SIN insertion sites qualitatively differs from that found with LTR vectors in clones dominating hematopoiesis after serial BMT. To answer this question all identified insertions were blasted against the National Center for Biotechnology Information mouse build 37 genome database (www.ncbi.nlm.nih.gov); genes in a 150-kb window around the insertion were included into analysis (Tables 1 and 2).
Six of the 25 pd-SIN vector insertions found in reconstituting HSC were located in intergenic regions with the next (predicted) gene ~100 kb or further away. In the remaining 19 dominant clones, only two insertions were located close to or within established protooncogenes (Lmo2, Elk3), eight nearby signaling genes (including the putative growth–promoting gene Notch3 and the apoptosis inhibitor Birc5 which was found to be independently hit in both mouse experiments). Fifteen integrations had occurred in the proximity of “other,” unknown or predicted genes (Table 1; Figure 4). In line with that, only a minority of four clones (17%) was found to be dominant in more than half of the secondary recipients (Exp. I: I-2; Exp. II: II-3, II-5, II-8). Two of the latter (II-5, II-8) bore insertions which were located far away from the next gene (95 and 149 kb, respectively). The other two were located in the proximity of Efcab2 (5 kb downstream, 1st intron; I-2) and B3bp (2 kb downstream, 1st intron; II-3), two genes not known to be involved in growth regulation in HSC or other cell types. These data strongly argue against a role of insertional mutagenesis during establishment of clonal dominance of pd-SIN-marked clones.
In contrast, the distribution of insertion sites of the SF91-eGFP control vector well resembled earlier results from our and many other laboratories reported for this vector type25 (Figure 4). In fact, as in the IDDb (ref. 25), >70% of all insertions were found in signaling and/or protooncogenes. Five different protooncogenes (Table 2) were identified as insertion sites in dominant clones, four of them previously shown to be involved in hematopoietic malignancies. Most remarkably, one of them, Prdm16, is not only represented in the Retrovirus Tagged Cancer Gene Database (RTCGD),29 but its human counterpart has also been associated with induced clonal dominance in a clinical gene therapy study.6 In line, three other protooncogenes (HoxA7, Cdk6, Mef2c/Evi121) were identified before by the Copeland group as common retroviral insertion sites in retrovirus-induced malignancies.29 Moreover, HoxA7 and Cdk6 had already been linked to induced clonal dominance in the IDDb.25
Besides those protooncogenes, several other genes hit by SF91-eGFP were previously suggested to be involved in malignant transformation and/or induced clonal dominance (Table 2). These include the FoxL1 and FoxF1a genes (one genomic locus) listed in the RTCGD29 and IDDb,25 respectively. Another common retroviral insertion sites in both RTCGD and IDDb is Susd3, which is particularly interesting because its function is so far largely unknown. Two other genes hit by SF91-eGFP in dominant clones could be found in the RTCGD: Cux1/Evi71 (also an insertion site of pd-SIN, Table 1) and Sept9.
In summary, the overrepresentation of SF91-eGFP integrations in protooncogenes already shown to be able to induce benign and/or malignant clonal outgrowth after retroviral insertion–mediated upregulation (Prdm16, Hoxa7, Cdk6, Mef2c/Evi121) strongly argues for a potential role of insertional mutagenesis in the affected clones. The actual role of these, and also the other named candidate genes, in the establishment of induced clonal dominance is currently under investigation.
In conclusion, the insertion pattern observed for pd-SIN was strikingly different from that seen in our previous studies and with the control vector SF91-eGFP (Figure 4).8 In fact, based on Fisher's exact test, integrations of pd-SIN into oncogenes, growth-promoting and signaling genes (gene groups 1 and 2) occurred significantly (P < 0.002) less frequent than with the SF91-eGFP control vector.
No strong transcriptional dysregulation of genes at pd-SIN insertion sites
Although pd-SIN would not be expected to transactivate neighboring genes, its integration may still have resulted in transcriptional dysregulation which principally might have facilitated later dominance of an affected clone. The latter might particularly be expected with the insertions in the proximity of the protooncogenes (e.g., Lmo2, Elk3) or for those clones which in a dominant way contributed to hematopoiesis in more than half of the secondary recipients. As stated above, only for two (out of four) of those clones genes were detectable in the proximity of the vector integration (Efcab2, B3bp).
To address this question we measured transcription levels for 32 adjacent genes in splenocytes of affected animals using quantitative reverse transcription–PCR (Figure 5). Changes in transcription levels compared to control splenocytes for most genes analyzed (19/32, 59%) were below a factor of 2. For almost 90% (28/32), the changes were <3, a threshold often used for defining significant transcriptional upregulation in array analysis (Figure 5). In particular, transcriptional upregulation of Lmo2 was hardly detectable. In line, integration into the Lmo2 gene had occurred far away from the described negative regulatory element (data not shown).30
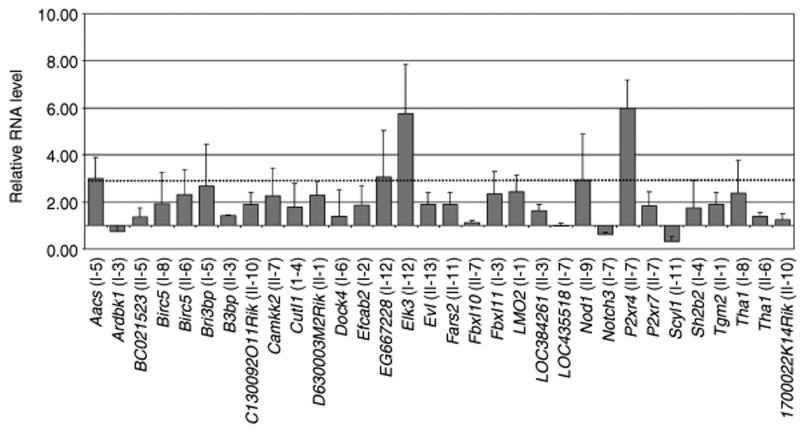
Figure 5.
Minor transcriptional dysregulation in gene loci targeted by promoter-deprived γ-retroviral self-inactivating (pd-SIN). Real-time reverse transcription–PCR using QuantiTect Primer Assays were used to assess transcription levels of genes adjacent to pd-SIN vector insertions. RNA levels as compared to healthy control mice splenocytes were established using the ΔΔCt method.8,49 Only comparatively slight changes in transcription levels were detected. In particular, genes adjacent to insertion sites found to mark clones dominant in more than half of the secondary recipients did not show relevant dysregulation of transcription (I-2: Efcab2; II-3: B3bp). Relative error bars indicate SDs from at least three independent assays.
Three of the genes with higher upregulation (Aacs: factor 3.0 ± 0.9; EG667228: 3.1 ± 2.0; P2xr4: 6.0 ± 1.2) have no described function in cell survival or hematopoiesis. Also, we found no large changes in the mean transcription of Efcab2 (1.9 ± 0.8) and B3bp (1.4 ± 0.1) in the secondary animals reconstituted by those two clones comprising a high reconstitution potential (see above) (Figure 5).
A comparatively strong transcriptional upregulation was found for Elk3, although the big SDs indicate large inter-animal variations in the transcription of this gene (Figure 5). Elk3 is a member of the Ets family of transcription factors. Because a potential role of Elk3, which represents an effector of mitogen-activated protein kinase pathways, in growth regulation of HSC could not be excluded, its upregulation and a potential contribution to clonal survival need further investigation. However, after serial BMT only one animal was reconstituted by a dominant Elk3 clone (Table 1) arguing against strong induced clonal dominance.
Integrations preferentially occur into active genomic loci
The above data indicate that pd-SIN marking of HSC is not associated with induced clonal dominance. Still, the distribution seems to be different from what would be expected in a pure chance setting. In fact, among the 25 vector insertions we found two insertions into protooncogenes (~8%) (Figure 4). On the other hand, such preference is well in line with the reported integration bias of γ-retroviral vectors in mixed cultures of HSC and progenitor cells.31
It has been suggested that this integration bias may be based on the expression levels of the targeted gene loci in HSC.31,32 To proof this possibility, we made use of an expression database for murine HSCs established by the Goodell lab.33 On the basis of that database, we were able to determine expression levels for 18 genes adjacent to the integration sites. Genes located 5′ from the insertion site were preferentially used since many insertions were found in introns thus obviously depending on the activity of the promoter located 5′. However, if there was no gene detectable in the 5′ proximity of an insertion, we used the next 3′ gene for analysis. In some cases, closely located genes were identified on both sites of the vector integration. It is remarkable that log2-transformed expression values for those genes were nearly identical, independent of the expression strength of the given locus (insertion I-11: Malat1 – 13.59 versus Ltbp3 – 12.82; II-2: Expi – 3.58 versus Ccl4 – 3.41). These data underline the validity of our approach.
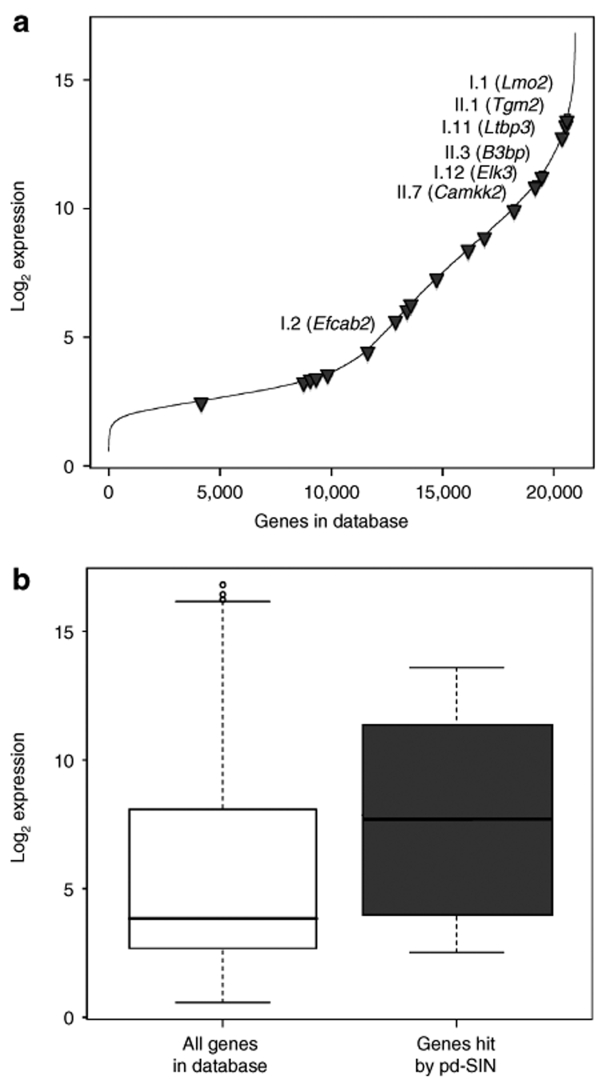
In order to assess transcriptional activity of the targeted gene loci in HSC we plotted the expression values of the hit genes against all genes in the Goodell database (Figure 6a). As evident, most targeted gene loci show intermediate-to-high-expression levels, whereas only one (II-13, Degs2) of the 18 analyzed genes (5.6%) is located in the lowest third. This is also reflected in the overall comparison depicted in Figure 6b clearly indicating comparatively higher expression in the targeted loci.
Figure 6.
Promoter-deprived γ-retroviral self-inactivating (pd-SIN) preferentially integrates into transcriptionally active regions. (a) Insertion sites of pd-SIN were correlated with transcription levels of the targeted gene loci. To do so we made use of an expression database for murine hematopoietic stem cells established by Goodell and colleagues.33 All genes contained in this database were plotted against their respective expression values. The shown line consists of >20,000 individual data points representing the highest expression values for each gene on the array. As shown, most genes hit by pd-SIN (gray triangles) were transcriptionally highly active in hematopoietic stem cell (HSC). (b) The relative transcriptional activity of genes targeted by pd-SIN was compared with that of all genes contained in the HSC expression database.29 Clearly, pd-SIN preferred actively transcribed gene loci for integration.
Remarkably, the two protooncogenes identified as insertion sites (Lmo2, Elk3) are amongst those gene loci showing the highest expression level. The same is true for B3bp, an insertion site found in one of the clones (II-3) dominant in more than half of the secondary HSCT recipients. Another dominantly repopulating clone (I-2) had an insertion in Efcab2, a gene which is characterized by intermediate expression in HSC.
Discussion
Different animal models and also several clinical trials have clearly shown the great potential of γ-retroviral vectors for gene marking and gene therapy.1,2,3,4,5,6,34,35,36,37 Gene marking has allowed investigation of the reconstitution dynamics of the hematopoietic system after HSCT,1,2 the impact of ex vivo culture and conditioning regimens on engraftment34,35 and the role of contaminating malignant cells for relapse development after autologous HSCT in clinical settings.3 At the same time, the successful treatment of inherited monogenic diseases of the hematopoietic system, such as severe combined immunodeficiencies (SCID-X1, ADA-SCID)4,5,36 or chronic granulomatous disease,6 using γ-retroviral vector–mediated gene correction has provided the long-awaited proof of principle for genetic therapy.
Unfortunately, further progress has been thwarted by the occurrence of severe side effects including induced dominance as well as malignant transformation of affected clones in both animal models7,8,37 and clinical studies.6,9,10 Thorough molecular analysis allowed attributing those side effects to insertional mutagenesis.6,7,8,9,10,12,13,14,37 Although insertional mutagenesis had been reported with replication-competent retroviruses decades ago,38 it was thought to be quite unlikely with nonreplicating vectors given the low probability of hitting a protooncogene in a susceptible (stem) cell. In accord, no evidence for insertional oncogenesis after retroviral vector–mediated gene transfer had been found in numerous animal studies.12,13,14,39
In most instances, insertional mutagenesis was shown to be conferred by transactivation of neighboring genes from the strong promotor/enhancer elements located in the U3 region of retroviral LTRs.6,7,8,9,10,11,12,13,14 However, even SIN vectors may still be mutagenic, given that they contain a sufficiently strong internal promotor. In fact, γ-retroviral SIN vectors containing internal retroviral promotors were still able to facilitate in vitro immortalization19 as well as leukemia development21 in suitable models, albeit at decreased incidences. In addition, lentiviral vectors equipped with the strong SFFV-LTR revealed genotoxicity in a tumor-prone mouse model.40 Moreover, even SIN lentivectors harboring physiological promotor/enhancer sequences derived from the β-globin locus control region were shown to cause insertional dysregulation of cellular genes in erythroid cells over long distances and at high frequencies.41
Given those results, we explored the usefulness of an almost totally “neutral” gene-marking vector lacking all promotor/enhancer elements. This pd-SIN vector would not be expected to directly transactivate neighboring genes. Using pd-SIN vectors to mark HSC in our sensitive serial BMT model,7,8 we were able to address a number of questions. First, we investigated whether pd-SIN vectors are still associated with induced clonal dominance. The significantly lower incidence (as compared to LTR vectors) of pd-SIN insertions into putative growth–promoting genes and the lack of evidence for very strong transcriptional upregulation of adjacent genes (in contrast to that observed with LTR vectors8) argue against this possibility. Indeed, most observed transcriptional changes were below factor 2. Moreover, given the high SDs, most differences seem to rather represent natural interindividual variations than induced upregulation or downregulation (compare SDs, Figure 5). However, the varying contribution of single clones to hematopoiesis of individual mice complicates reliable quantitative statements.
It should be taken into account that even a promotor-deprived vector is potentially genotoxic, because its integration may disrupt or truncate open-reading frames or gene-regulatory regions. Intriguingly, for the insertion into the Elk3 protooncogene we observed a sixfold upregulation (though with large inter-animal deviations). Although the affected clone reconstituted only one animal in a dominant fashion, we cannot fully rule out the involvement of insertional mutagenesis in its fate.
Still it is safe to conclude that the picture obtained with pd-SIN reflects mainly an unbiased insertion profile of γ-retroviral vectors in long-term reconstituting HSC. Indeed, our data relating the identified vector integration sites with the expression characteristics of the targeted loci (based on the expression profile of the best-characterized murine HSC population as established by the Goodell group33) suggest that integration site selection largely depended on the activity of a given gene locus. These data confirm the preference of γ-retroviral vectors for actively transcribed genomic regions42,43 and are well in line with earlier in vitro results for cultured progeny of human CD34+ cells.31 Other data44,45 suggest that the apparent preference of retroviral vectors for certain genomic loci (“hot spots”) may be caused by specific interactions of retroviral preintegration complexes with certain transcription factor binding sites. Interestingly, Recchia et al.45 have provided evidence that, in addition to the retroviral integrase, direct interactions of transcription factors with LTR enhancer sequences play a pivotal role in directing proviral integrations toward transcription factor binding site–rich genomic regions. To investigate this further, a large-scale insertion site database for the pd-SIN vector lacking any enhancers should be established (using novel sequencing techniques46) which will allow comparing its insertion site preferences with the data obtained for LTR vectors.15,16,17,31,43,44,45
The above findings lead to the assumption that the clonal dominance seen in our secondary transplanted animals represents a direct result of serial BMT and thus reflects intrinsic properties of serially transplanted HSC. This observation indicates that most conclusions of earlier marking studies with γ-retroviral LTR vectors1,2 still hold true. At the same time it underlines that the concept of using promotor-deprived SIN vectors for gene marking may overcome limitations of γ-retroviral LTR vectors, namely their strong mutagenicity leading to a rare, but unfortunately overriding phenotype of induced clonal dominance.6,8,25
In summary, to investigate gene marking with the virtually neutral pd-SIN, we have performed two independent long-term animal experiments with very similar results. Based thereon it seems safe to draw the following conclusions: (i) γ-retroviral, pd-SIN vectors ensure stable genetic marking of reconstituting HSC; (ii) the insertion pattern of pd-SIN vectors is significantly different from that of γ-retroviral LTR vectors, but shows a similar dependence on transcriptional activity of targeted loci. This might explain the slight preference for growth-regulatory genes;31 (iii) insertion of pd-SIN vectors has primarily a neutral affect on adjacent genes, i.e., major transcriptional dysregulation of adjacent genes seems to be a rare event; (iv) in accord, gene marking with γ-retroviral pd-SIN apparently usually does not result in induced clonal dominance.
Our observations may be useful for the design of future gene marking1,2 and competitive engraftment34,35 studies. Also, the use of vectors that do not influence clonal outgrowth will permit the analysis of the insertion bias of different vector types in various types of target cells. Finally, our results underline the great promise of using improved SIN vectors for gene therapy. Indeed, as seen here, the mutagenicity of SIN vectors containing tissue-specific promotors that are silent in stem cells significantly lower the likelihood of severe side effects of stem cell gene therapy.
Materials and Methods
Retroviral vector production. In the animal experiments we used the retroviral vectors pd-SIN (see below), and SF91-eGFP/wPRE (“SF91-eGFP”) containing the SFFV U3 promotor/enhancer region and a wPRE element.24 Cell-free supernatants containing ecotropic viral particles were generated by transient transfection of PhoenixGP cells (kindly provided by G Nolan) as described.47,48 Titers (as established on NIH3T3 cells) for pd-SIN were ~3 × 105 per ml (Exp. I) and 5 × 106 per ml (Exp. II). For SF91-eGFP titers were ~1 × 106 per ml in Exp. I and 4 × 106 per ml in Exp. II.
Serial BMT model. Two independent serial HSC transplantation experiments were set up according to our established C57Bl/6J mouse model (Figure 2).7,8,22 In brief, murine BM isolated from young adult male donors was lineage-depleted and taken in culture for ex vivo transduction.48 Lin− BM cells were transduced with vector-containing supernatant at a multiplicity of infection of three to obtain gene transfer of ~50–60%.48 Transduced cells (1 × 106 per recipient) were transplanted into female recipient mice that had received total body irradiation (9.5 Gy) (Exp. I: n = 8; Exp. II: n = 10). FACS analysis22 showed donor-derived hematopoiesis in the peripheral blood of all animals (pd-SIN – Exp. I: mean 87%, 78–93%; Exp. II: mean 31%, 9–69%; SF91-eGFP – Exp. I: mean 80%, 64–93%; Exp. II: mean 32%, 13–57%). In the eight transplanted groups (two experiments each including serial BMT, two vectors) a total of 13 mice were lost because of graft failure or unknown reasons. This is in line with our previous experience in the given model.
After 7 months, transplanted mice were killed. At autopsy gross evaluation of hematopoietic organs (spleen, thymus, lymph nodes) was performed before they were conserved. Cells from BM, PB, and thymus were submitted to detailed FACS analysis of different hematopoietic lineages. Splenocytes were used to isolate DNA and RNA for molecular analysis. Blood smears were investigated for presence of leukemic blasts.
Harvested BM was pooled and transplanted into a second cohort of female recipient mice (Exp. I: n = 8; Exp. II: n = 10) after total body irradiation (9.5 Gy) at 1 × 106 per mouse. Remaining BM cells, as well as cells isolated from other hematopoietic organs, were conserved for and/or directly applied to FACS and molecular analysis.
Mice from the second cohort were observed for at least 5 months before final analysis (as above) took place.
Molecular analysis of insertion sites in dominant clones. For PCR assays, DNA and RNA were isolated from hematopoietic organs as described.8 For northern blot analysis, RNA was isolated from SC-1 cultures by standard procedures.23 Northern blots were performed using a 32P-labeled eGFP probe.23
To identify insertion sites in dominant clones we made use of a modified LM-PCR protocol.26 Once “dominant insertion sites” were identified (see below), primers were designed for is-PCR (see Supplementary Materials and Methods and Supplementary Table S1).
Transcription levels of genes adjacent to pd-SIN vector insertions were comparatively analyzed for splenocytes from mice bearing those insertions (Table 1) versus control splenocytes (cells from nontransplanted mice and/or from secondary recipients that did not bear the clone of interest) using QuantiTect Primer Assays (Qiagen, Hilden, Germany). Differences were determined based on the ΔΔCt method.8,49
To quantitatively determine the contribution of dominant and nondominant hematopoietic clones, whole LM-PCR products were shot-gun cloned into pCRII-TOPO (Invitrogen, Karlsruhe, Germany) according to the manufacturer's protocol. For SF91-eGFP, bacterial colonies were first screened using a vector-specific PCR (primers in Supplementary Table S2) to exclude LM-PCR products representing the internal vector control from further analysis (there is no internal PCR signal with the LM-PCR primers used for pd-SIN).19 Ninety-six cloned insertion sites per individual animal were sequenced using the terminator dye approach (GATC, Heidelberg, Germany).
Expression levels of targeted gene loci in HSC. In order to assess expression levels of targeted gene loci in HSC we made use of a database established by Goodell and colleagues based on microarray technology.33 According to that work, murine HSC are defined as side population cells which are Sca-1+, c-Kit+, and Lin−.33 To evaluate the expression level of those loci where vector integrations had happened, we first identified the closest 5′ located gene for each single insertion site. If no 5′ gene was found to be present in the Goodell database, the closest 3′ gene was used instead. For all selected genes, log2-transformed expression values (representing the mean for two different HSC preparations) were retrieved from the database.33 If the database contained more than one entry, the highest value was included into further analyses. Those expression values were set into relation to the expression values of all other genes (again only the highest values were taken for each individual gene). Analysis and plotting were performed using R-2.7.0.50
Supplementary MaterialTable S1. Primer Sequences used for insertion-specific (is-)PCR.Table S2. Primer Sequences for vector-specific PCR.Materials and Methods.
Supplementary Material
Primer Sequences used for insertion-specific (is-)PCR.
Primer Sequences for vector-specific PCR.
Acknowledgments
We thank Gökhan Arman-Kalcek, and Melanie Engel for their expert technical assistance. We are grateful to Carol Stocking for critical reading of the manuscript. This work has been supported by the Deutsche Forschungsgemeinschaft DFG [FE568/9-1 (SPP1230) and BA1837/7-1 (excellence cluster REBIRTH)], the Roggenbuck-Stiftung and the Frankfurter Stiftung für Krebskranke Kinder. B.F.'s position is funded by the Deutsche Krebshilfe, A.S.'s by the Else-Kröner-Stiftung. This work is part of K.C.'s PhD thesis. The authors declare no competing financial interests.
REFERENCES
- Dick JE, Magli MC, Huszar D, Phillips RA., and , Bernstein A. Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell. 1985;42:71–79. doi: 10.1016/s0092-8674(85)80102-1. [DOI] [PubMed] [Google Scholar]
- Lemischka IR, Raulet DH., and , Mulligan RC. Developmental potential and dynamic behavior of hematopoietic stem cells. Cell. 1986;45:917–927. doi: 10.1016/0092-8674(86)90566-0. [DOI] [PubMed] [Google Scholar]
- Tey SK., and , Brenner MK. The continuing contribution of gene marking to cell and gene therapy. Mol Ther. 2007;15:666–676. doi: 10.1038/sj.mt.6300109. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Slavin S, Aker M, Ficara F, Deola S, Mortellaro A, et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Li Z, Düllmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Düllmann J, Kamino K, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newrzela S, Cornils K, Li Z, Baum C, Brugman MH, Hartmann M, et al. Resistance of mature T cells to oncogene transformation. Blood. 2008;112:2278–2286. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- Baum C, Düllmann J, Li Z, Fehse B, Meyer J, Williams DA, et al. Side effects of retroviral gene transfer into hematopoietic stem cells. Blood. 2003;101:2099–2114. doi: 10.1182/blood-2002-07-2314. [DOI] [PubMed] [Google Scholar]
- Baum C, Kustikova O, Modlich U, Li Z., and , Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum Gene Ther. 2006;17:253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- Baum C, von Kalle C, Staal FJ, Li Z, Fehse B, Schmidt M, et al. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol Ther. 2004;9:5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiuti A, Cassani B, Andolfi G, Mirolo M, Biasco L, Recchia A, et al. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J Clin Invest. 2007;117:2233–2240. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzwaelder K, Howe SJ, Schmidt M, Brugman MH, Deichmann A, Glimm H, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nienhuis AW, Dunbar CE., and , Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knöss S, Schambach A, et al. Cell culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2453. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- Modlich U, Schambach A, Brugman MH, Wicke DC, Knoess S, Li Z, et al. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia. 2008;8:1519–1528. doi: 10.1038/leu.2008.118. [DOI] [PubMed] [Google Scholar]
- Li Z, Fehse B, Schiedlmeier B, Düllmann J, Frank O, Zander AR, et al. Persisting multilineage transgene expression in the clonal progeny of a hematopoietic stem cell. Leukemia. 2002;16:1655–1663. doi: 10.1038/sj.leu.2402619. [DOI] [PubMed] [Google Scholar]
- Schambach A, Mueller D, Galla M, Verstegen MM, Wagemaker G, Loew R, et al. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 2006;13:1524–1533. doi: 10.1038/sj.gt.3302807. [DOI] [PubMed] [Google Scholar]
- Schambach A, Wodrich H, Hildinger M, Bohne J, Kräusslich HG., and , Baum C. Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol Ther. 2000;2:435–445. doi: 10.1006/mthe.2000.0191. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Geiger H, Li Z, Brugman MH, Chambers SM, Shaw CA, et al. Retroviral vector insertion sites associated with dominant hematopoietic clones mark “stemness” pathways. Blood. 2007;109:1897–1907. doi: 10.1182/blood-2006-08-044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustikova OS, Baum C., and , Fehse B. Retroviral integration site analysis in hematopoietic stem cells. Methods Mol Biol. 2008;430:255–267. doi: 10.1007/978-1-59745-182-6_18. [DOI] [PubMed] [Google Scholar]
- Purton LE., and , Scadden DT. Limiting factors in murine hematopoietic stem cell assays. Cell Stem Cell. 2007;1:263–270. doi: 10.1016/j.stem.2007.08.016. [DOI] [PubMed] [Google Scholar]
- McKenzie JL, Gan OI, Doedens M, Wang JC., and , Dick JE. Individual stem cells with highly variable proliferation and self-renewal properties comprise the human hematopoietic stem cell compartment. Nat Immunol. 2006;7:1225–1233. doi: 10.1038/ni1393. [DOI] [PubMed] [Google Scholar]
- Akagi K, Suzuki T, Stephens RM, Jenkins NA., and , Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Crable SC., and , Anderson KP. Negative regulatory elements are present in the human LMO2 oncogene and may contribute to its expression in leukemia. Leuk Res. 2005;29:89–97. doi: 10.1016/j.leukres.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Szilvassy S, Eaves C., and , Humphries R. Proliferation of totipotent hematopoietic stem cells in vitro with retention of long-term competitive in vivo reconstituting ability. Proc Natl Acad Sci USA. 1992;89:1968–1972. doi: 10.1073/pnas.89.5.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel WS, Yoder MC, Pech NK., and , Dinauer MC. Donor chimerism and stem cell function in a murine congenic transplantation model after low-dose radiation conditioning: effects of a retroviral-mediated gene transfer protocol and implications for gene therapy. Exp Hematol. 2002;30:1324–1332. doi: 10.1016/s0301-472x(02)00927-x. [DOI] [PubMed] [Google Scholar]
- Gaspar HB, Bjorkegren E, Parsley K, Gilmour KC, King D, Sinclair J, et al. Successful reconstitution of immunity in ADA-SCID by stem cell gene therapy following cessation of PEG-ADA and use of mild preconditioning. Mol Ther. 2006;14:505–513. doi: 10.1016/j.ymthe.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Putten H, Quint W, van Raaij J, Maandag ER, Verma IM., and , Berns A. M-MuLV-induced leukemogenesis: integration and structure of recombinant proviruses in tumors. Cell. 1981;24:729–739. doi: 10.1016/0092-8674(81)90099-4. [DOI] [PubMed] [Google Scholar]
- Kohn DB, Sadelain M, Dunbar C, Bodine D, Kiem HP, Candotti F, et al. American Society of Gene Therapy (ASGT) ad hoc subcommittee on retroviral-mediated gene transfer to hematopoietic stem cells. Mol Ther. 2003;8:180–187. doi: 10.1016/s1525-0016(03)00212-0. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. The genotoxic potential of integrative vectors is strongly modulated by vector design and the profile of integration site selection. Mol Ther. 2008;16 suppl. 1:S362. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove PW, Kepes S, Hanawa H, Obenauer JC, Pei D, Cheng C, et al. Globin lentiviral vector insertions can perturb the expression of endogenous genes in beta-thalassemic hematopoietic cells. Mol Ther. 2008;16:525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- Mooslehner K, Karls U., and , Harbers K. Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. J Virol. 1990;64:3056–3058. doi: 10.1128/jvi.64.6.3056-3058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and , Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J., and , Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia A, Cittaro D, Cattoglio C, Felice B, Miccio A, Ferrari G, et al. Integration of retroviral vectors into the human genome is biased by specific subsets of transcription factor binding sites. Mol Ther. 2008;16 suppl. 1:S360. [Google Scholar]
- Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer W, Westphal M, Ostertag W., and , von Laer D. Oncoretrovirus and lentivirus vectors pseudotyped with lymphocytic choriomeningitis virus glycoprotein: generation, concentration, and broad host range. J Virol. 2002;76:1488–1495. doi: 10.1128/JVI.76.3.1488-1495.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Schwieger M, Lange C, Kraunus J, Sun H, van den Akker E, et al. Predictable and efficient retroviral gene transfer into murine bone marrow repopulating cells using a defined vector dose. Exp Hematol. 2003;31:1206–1214. doi: 10.1016/j.exphem.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team R: a language and environment for statistical computing R Foundation for Statistical Computing 2008Vienna, Austria. ISBN 3-900051-07-0; , < http://www.R-project.org >. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer Sequences used for insertion-specific (is-)PCR.
Primer Sequences for vector-specific PCR.