Abstract
Precise manipulations of complex genomes by zinc-finger nucleases (ZFNs) depend on site-specific DNA cleavage, which requires two ZFN subunits to bind to two target half-sites separated by a spacer of 6 base pairs (bp). ZFN subunits consist of a specific DNA-binding domain and a nonspecific cleavage domain, connected by a short inter-domain linker. In this study, we conducted a systematic analysis of 11 candidate-based linkers using episomal and chromosomal targets in two human cell lines. We achieved gene targeting in up to 20% of transfected cells and identified linker variants that enforce DNA cleavage at narrowly defined spacer lengths and linkers that expand the repertoire of potential target sites. For instance, a nine amino acid (aa) linker induced efficient gene conversion at chromosomal sites with 7- or 16-bp spacers, whereas 4-aa linkers had activity optima at 5- and 6-bp spacers. Notably, single aa substitutions in the 4-aa linker affected the ZFN activity significantly, and both gene conversion and ZFN-associated toxicity depended on the linker/spacer combination and the cell type. In summary, both sequence and length of the inter-domain linker determine ZFN activity and target-site specificity, and are therefore important parameters to account for when designing ZFNs for genome editing.
Introduction
Methods of introducing precise and stable modifications in a mammalian genome hold great potential for the study of gene function and gene therapy. One particularly promising technology is based on the homologous recombination (HR) pathway and is known as gene targeting. Proof-of-concept studies with the rare cutting homing endonuclease I-SceI have shown that the low frequency of HR in mammalian cells (~1 HR event per 106 cells) can be overcome by introducing a site-specific DNA double-strand break (DSB) at the locus of interest.1,2 The DSB activates cellular DNA repair pathways and, as a consequence, stimulates HR between the target locus and an exogenous donor DNA 100- to 10,000-fold. To exploit such DSBs for therapeutic genome modifications, tailor-made nucleases such as zinc-finger nucleases (ZFNs)3 need to be generated. ZFNs have a simple and modular architecture. They are composed of an engineered Cys2-His2 zinc-finger domain and a nonspecific nuclease domain from the type IIs restriction enzyme FokI.4 A short inter-domain linker connects the two domains. Because the FokI cleavage domain is enzymatically active only as a dimer,5,6 the introduction of a DSB depends upon the dimerization of two ZFN subunits on the DNA target site.7,8 Accordingly, two ZFN subunits are designed to recognize the target sequence in a tail-to-tail conformation, with each monomer binding to “half-sites” separated by a “spacer” sequence (Figure 1a). On the basis that each zinc-finger domain recognizes ~3 bp of the DNA target sequence,9 a dimer of three-finger ZFNs is expected to bind to an 18-bp DNA target site, which statistically defines a unique sequence in the human genome.7
Figure 1.
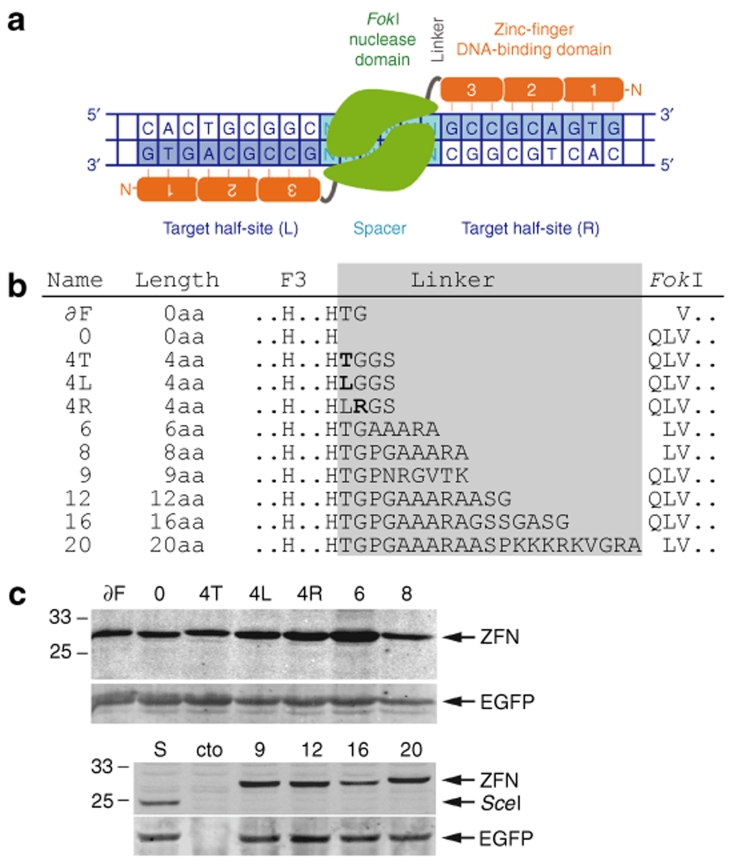
Zinc-finger nuclease (ZFN) linker variants. (a) Schematic of the EB2-N homodimer. All ZFN variants are based on EB2-N23 and contain an N-terminal HA epitope tag, three zinc-fingers (1, 2, 3), the inter-domain linker, and the FokI cleavage domain. The two target half-sites (L and R) are separated by a spacer sequence of defined length (see Supplementary Table S1). (b) Sequence of ZFN linker variants. The names, linker lengths, and the relevant amino acid sequences are indicated. The ZFN inter-domain linker length has been defined as the number of amino acids (aa) between the last conserved histidine in the third zinc-finger (F3) and the first residue of the FokI cleavage domain (QLV). Note that in some variants the first residues of the FokI domain were deleted and offset against the linker length. (c) Expression analysis of ZFN linker variants. HEK293T cells were transfected with an expression plasmid encoding a ZFN linker variant along with pEGFP as an internal transfection control. After 30 h, the cells were harvested and the lysates were probed simultaneously with antibodies against both the HA tag and EGFP. HA-tagged I-SceI and mock-transfected cells (cto) were included as controls. The positions of the various ZFNs, I-SceI, EGFP, and size markers are indicated.
Several articles have highlighted the tremendous potential of ZFNs for use in studying biological systems in mammalian cells10 or in whole organisms such as Drosophila,11,12 Caenorhabditis elegans,13 and plants.14,15 Moreover, gene conversion frequencies of up to 29% at the endogenous human IL2R-γ (interleukin-2 receptor γ-chain) locus in the absence of selection have demonstrated the usefulness of the ZFN technology in gene therapy.16,17 Despite these successes, ZFN-induced toxicity in human cells, which is likely caused by excessive ZFN cleavage at off-target sites, has remained an important issue.18,19,20,21,22,23 However, ZFN variants that combine high activity with significantly lower toxicity could be generated by improving the DNA-binding specificities of the zinc-finger domains23 and/or by regulating the DNA-cleavage activity of the nuclease domain through redesign of the FokI protein–protein interface.21,22
Evaluation of the modular ZFN architecture suggests a third parameter that is likely to affect target site selection and toxicity: the inter-domain linker (Figure 1a). ZFNs containing linkers of various lengths were initially tested in vitro and by microinjection of purified ZFNs into frog oocytes. These experiments showed that ZFNs with an 18-residue linker revealed cleavage activity at target half-sites separated by spacers in the range of 6–18 bp, with an optimum at 8 bp. ZFNs with shorter inter-domain linkers of 0 and 2 aa were more stable after microinjection and had an activity optimum at 6-bp spacers.24 Note that the 0, 2, and 18-aa linkers used in the prior study correspond to 4, 6, and 22 aa linkers as per the current definition (Figure 1b). On the basis of these results, ZFNs with short linkers of 4–7 aa have been used in the most recent studies and, depending on whether 3- or 4-finger ZFN subunits were used, the ZFNs were designed to bind to either 9-bp11,12,13,14,15,19,20,22,23,25,26 or 12-bp10,16,17,21,27 target half-sites separated by 5- or 6-bp spacer sequences.
The aa composition and the length of the inter-domain linker are probably major factors that determine target site selectivity and ZFN-associated toxicity. On the one hand, a promiscuous inter-domain linker will allow a ZFN dimer to bind to half-sites separated by spacers of different lengths. The consequence of such an ambiguity is that a pair of ZFNs will not only cut at sites with a spacer length of, say, 6 bp, but also at related sites with 5-bp and 7-bp spacers, thereby potentially increasing the number of off-target cleavage events. Therefore a linker that provides perfect specificity for a single spacer length should increase target site selectivity and, in turn, decrease ZFN off-target activity. On the other hand, the design of inter-domain linkers that will allow a ZFN dimer to bind to noncanonical target sites, i.e., target sites separated by spacer lengths other than 5 or 6 bp, would expand the repertoire of potential target sites. In the light of a recent publication indicating that currently used engineering methods for assembling zinc-finger arrays are likely to be successful in only a small fraction of all potentially targetable half-sites,26 this would seem highly desirable. In this study, we conducted a systematic analysis of 11 candidate-based linkers to identify both linker variants that enforce ZFN-mediated DNA cleavage at target sites with highly defined spacer lengths and linkers that will expand the repertoire of target sites beyond the canonical 5-/6-bp spacers.
Results
The ZFN inter-domain linker affects target site selection on episomal DNA
To identify ideal combinations of a specific ZFN inter-domain linker and the length of the spacer sequence in the target site, a repertoire of 11 ZFN linker variants was generated (Figure 1b, Supplementary Table S1). The linkers had a length ranging from 0 to 20 residues and were cloned into a previously described ZFN, known as EB2-N.23 As reported earlier, zinc-finger array EB2 encodes a highly specific DNA-binding domain28 which induced only moderate toxicity when overexpressed in the context of a ZFN.23 A quantitative expression analysis revealed that all ZFN variants were expressed to comparable steady-state levels, although some variants (4L, 4R, and 6) consistently revealed a slightly higher expression level, thereby suggesting higher stability of these proteins (Figure 1c).
A slightly modified version of a recently described episomal recombination assay19 was used for quantifying the activities of these 11 ZFN linker variants on 11 different target plasmids (Supplementary Table S2). The target plasmids harbored an entire lacZ gene which overlapped at its 3′-end with an out-of-frame EGFP gene (Figure 2a). The frameshift (fs) and the deletion of the initiator ATG ensured that no functional EGFP was expressed from the plasmid. The two target half-sites were placed as inverted repeats of the 9-bp binding sites into the 3′-end of the lacZ gene and separated by spacer lengths ranging from 4 to 18 bp. This setup allowed us to assess the activity of the ZFN linker variants as homodimers, thereby ensuring that the inter-domain linker was the only variable parameter affecting the ZFN's ability to stimulate gene targeting between the lacZ(fs)EGFP target plasmid and the donor DNA. The donor plasmid harbored a nontranscribed 5′-truncated lacZ-EGFP fusion gene and was designed to rescue EGFP expression by generating a lacZ-EGFP fusion gene through HR. Two days after co-transfection of HEK293T cells with the target plasmid, the donor DNA, and each of the various ZFN expression vectors, the number of EGFP-positive cells was determined by flow cytometry.
Figure 2.
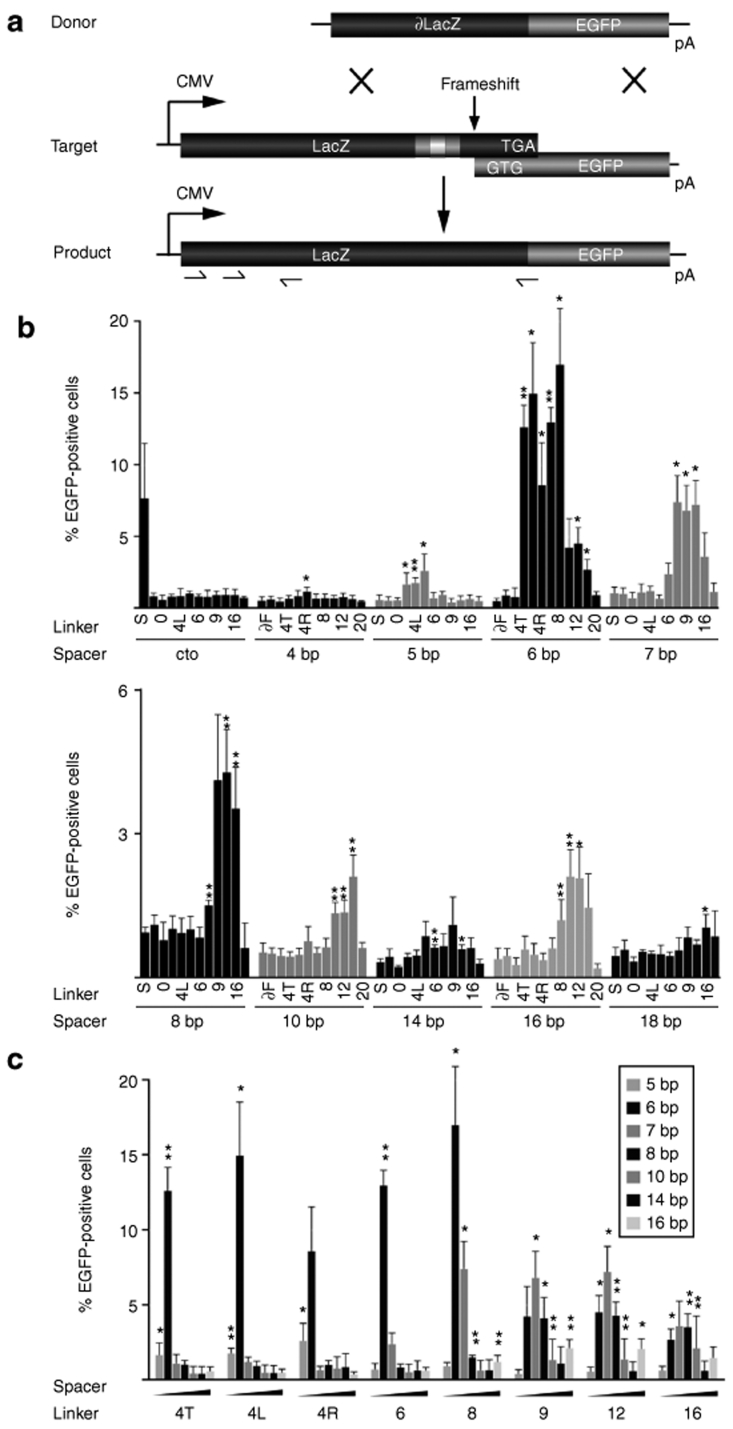
Activity profile of the zinc-finger nuclease (ZFN) linker variants on episomal targets. (a) Experimental setup. The donor and target plasmids are described in the text. The positions of the two binding half-sites for the homodimeric ZFN linker variants (gray boxes), the spacer sequence (white box), and the ‐1 frameshift are indicated in the target. TGA designates the stop codon of the lacZ gene while GTG represents the first triplet of the EGFP open reading frame. Arrows denote the positions of the primers used for genotyping. (b) Episomal recombination assay. HEK293T cells were transfected with plasmids encoding donor, target, and ZFN expression vectors. The columns designate the percentage of EGFP-positive cells 2 days after transfection, as determined by flow cytometry. The respective linker variants and spacer lengths are indicated. In cto, a target containing a recognition site for I-SceI but not for the ZFNs was used. For all other targets, I-SceI was included as a negative control to indicate the level of nonstimulated gene targeting. Note the change in scale between the first and second graph and that column labels alternate. (c) Activity profile of the ZFN linker variants. Data from b was replotted for all functional ZFN linker variants and itemized according to linker length to display target site selectivity. A statistically significant increase in gene targeting relative to nonstimulated HR is indicated by * (P < 0.05) or ** (P < 0.01).
For the control target plasmid (cto), containing a recognition site for I-SceI but not for the ZFNs, <1% of the transfected HEK293T cells turned green in the presence of the different ZFN linker variants, confirming that none of these ZFNs induced nonspecific gene targeting in our assay (Figure 2b). The 0.5–1% EGFP-positive cells we observed in all episomal HR assays represent the nonstimulated background recombination rate. Expression of I-SceI on the other hand, stimulated gene correction of the control target plasmid but not of any of the various target plasmids bearing a ZFN recognition site. In agreement with previous results,7,24 none of the ZFN linker variants we tested stimulated HR efficiently at a site bearing a 4-bp spacer (Figure 2b). On all other targets, at least one ZFN linker variant induced gene correction significantly (P < 0.05) above background. The peak activity of most linker variants was observed at a 6-bp spacer, with up to 17% of transfected cells expressing a functional lacZ-EGFP gene. While ZFNs with inter-domain linkers of 4 aa were also active on 5-bp spacers, ZFNs with linkers ranging in length from 8 to 16 aa supported ZFN-mediated activation of gene targeting on targets with spacer lengths of 7 to 16 bp. The gene targeting frequency steadily decreased with increasing spacer length, with the exception of a second weak optimum at 16 bp. Expression of linker variants ∂F, 0, and 20 did not stimulate HR at any spacer length, suggesting that very short and very long linkers did not allow the formation of active ZFN dimers on DNA target sites. However, linkers as long as 22 aa have been shown to work in previous studies7,24 and the lack of activity of our 20-aa ZFN linker variant might be attributable to the large number of positively charged residues in the linker sequence, which may interfere with DNA-binding.
Interestingly, although variants with 4- to 8-aa long linkers showed maximal activity at a 6-bp spacer, longer linkers of 9–16 residues revealed peak activity at a 7-bp spacer, with up to ~7% of cells expressing EGFP (Figure 2c). It also became evident that ZFNs with short linkers have a restricted activity profile, working either only at 5-/6-bp spacers for the 4-aa linkers or at 6-/7-bp spacers for 6- and 8-aa linkers, whereas longer 9–16-aa linkers were active on a wider array of spacer lengths ranging from 6- to 16-bp spacers.
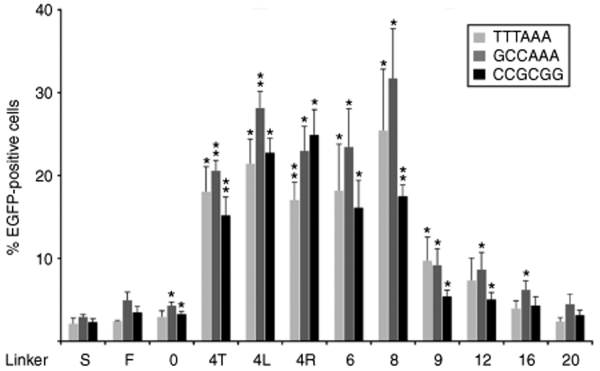
All the target plasmids assayed contained AT-rich spacers separating the two 5′-gccgcagtg half-sites. We next investigated whether the sequence composition of the spacer influences the activity of the ZFNs. To this end, novel target plasmids were generated, containing either a GC-rich or a balanced 6-bp spacer (Supplementary Table S2). A direct side-by-side comparison in the episomal recombination assay revealed that—with one exception—the spacer sequence was not important for ZFN activity (Figure 3). Interestingly, variant 4R revealed a significantly (P = 0.02) higher activity on the GC-rich spacer as compared to the activity on the AT-rich spacer. Together, these results suggest that the cleavage activity of ZFNs in human cells depends mainly on linker length, but that sequence composition of the ZFN inter-domain linker can have an effect on activity as well.
Figure 3.
Influence of spacer sequence on zinc-finger nuclease (ZFN) activity. HEK293T cells were transfected with plasmids encoding ZFN expression vectors, donor, and targets containing an AT-rich spacer, a GC-rich spacer, or a balanced 6-bp spacer. The bars designate the percentage of EGFP-positive cells 2 days after transfection as determined by flow cytometry. The respective linker variants and the sequence of the spacers are indicated. I-SceI was included as a negative control to indicate the level of nonstimulated gene targeting. A statistically significant increase in gene targeting relative to nonstimulated HR is indicated by * (P < 0.05) or ** (P < 0.01).
The ZFN inter-domain linker affects target site selection on chromosomal DNA
Previous studies have shown that the effects of the DNA target site spacer length and the ZFN inter-domain linker sequence on ZFN cleavage activities were more restrictive on episomal DNA in frog oocytes than in in vitro assays.7,24 However, to date, a systematic analysis on chromosomal targets has not been performed. To this end, HEK293 cells were transduced with lentiviral vectors to generate four polyclonal target cell lines containing integrated lacZ(fs)EGFP target loci with EB target half-sites separated by 5, 6, 7, and 16 bp. We chose to use polyclonal cell lines lest positional effects on the EGFP reporter affect the gene conversion frequency.
Flow cytometry was used to determine the extent of chromosomal gene targeting 5 days after co-transfection of the various ZFN expression vectors with an HR donor plasmid. In the control cell line 293LV31,29 which contains an I-SceI recognition site but no EB target sites, rescue of EGFP expression was observed in 9% of the cells transfected with an I-SceI expression plasmid, but was not observed in cells transfected with plasmids expressing any of the ZFN linker variants (Figure 4a). This finding confirmed that none of the ZFN linker variants had nonspecific activity on the target locus.
Figure 4.
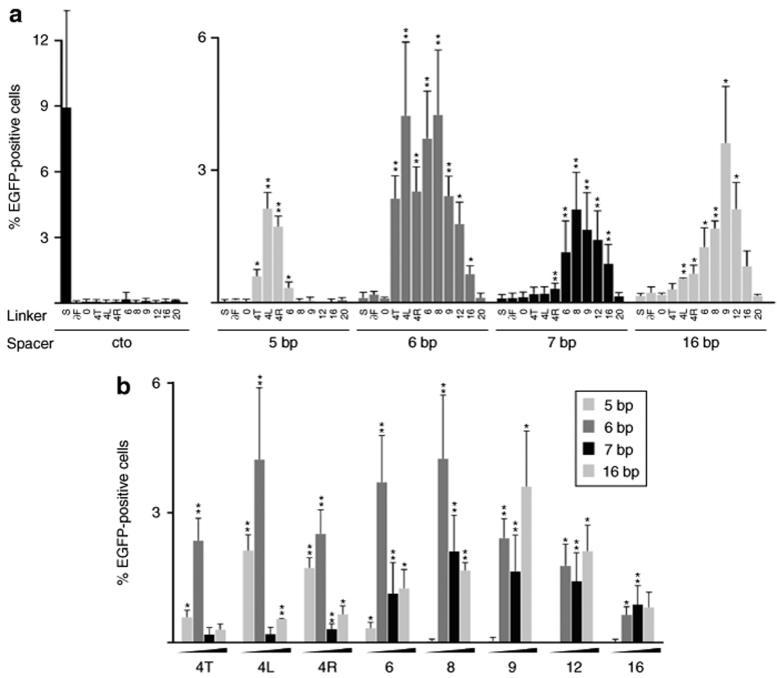
Activity profile of the zinc-finger nuclease (ZFN) linker variants on chromosomal targets. (a) Chromosomal recombination assay. HEK293-based cell lines that contain an integrated version of the target loci used in Figure 2 were transfected with the various ZFN expression vectors, a donor plasmid, and a DsRed-Express (REx) expression vector to normalize for transfection efficiency. The columns designate the percentage of EGFP-positive cells 5 days after transfection as determined by flow cytometry. The respective linker variants and spacer lengths are indicated. In cto, a cell line containing an integrated target locus with a recognition site for I-SceI but not for the ZFNs was used. For all other target cell lines, I-SceI was included as a negative control to indicate the level of nonstimulated gene targeting. (b) Activity profile of the ZFN linker variants. Data from a was replotted for all functional ZFN linker variants and itemized according to linker length to display target site selectivity of the linker variants. A statistically significant increase in gene targeting relative to nonstimulated HR is indicated by * (P < 0.05) or ** (P < 0.01).
In all four novel target cell lines, <0.2% of the transfected cells were scored as EGFP-positive upon expression of I-SceI, indicating the level of nonstimulated gene targeting (Figure 4a). Expression of linker variants ∂F, 0, and 20 did not stimulate gene correction significantly above background in any of these cell lines. In agreement with the episomal data, gene targeting was most efficient at a 6-bp spacer, reaching editing of the lacZ(6fs)EGFP locus in up to 4.3% of the transfected cells. Surprisingly, the gene targeting frequency at the 16-bp target locus was almost as high (3.6%), while rescue of EGFP expression was observed in ~2% of cells containing 5- or 7-bp spacers in the target site.
As seen on episomal targets, ZFNs with short linkers had a more restricted activity profile than those with longer linkers (Figure 4b). All 4-aa ZFN linker variants showed high activity only at 5- and 6-bp spacers, with variant 4L being the most active one. Variants 4T and 6 revealed the most restricted activity profile whereas the ZFN variant with the 9-aa linker turned out to be rather nonselective with regard to spacer length, stimulating gene targeting at 6-, 7-, and 16-bp spacers. Although the relative activities of the ZFN variants were similar when compared with the episomal data, these results revealed that the requirements for ZFN activity change after integration of the target locus. In particular, gene targeting at a 16-bp spacer was much more efficient in a chromosomal context.
ZFN-induced gene targeting is cell-line dependent
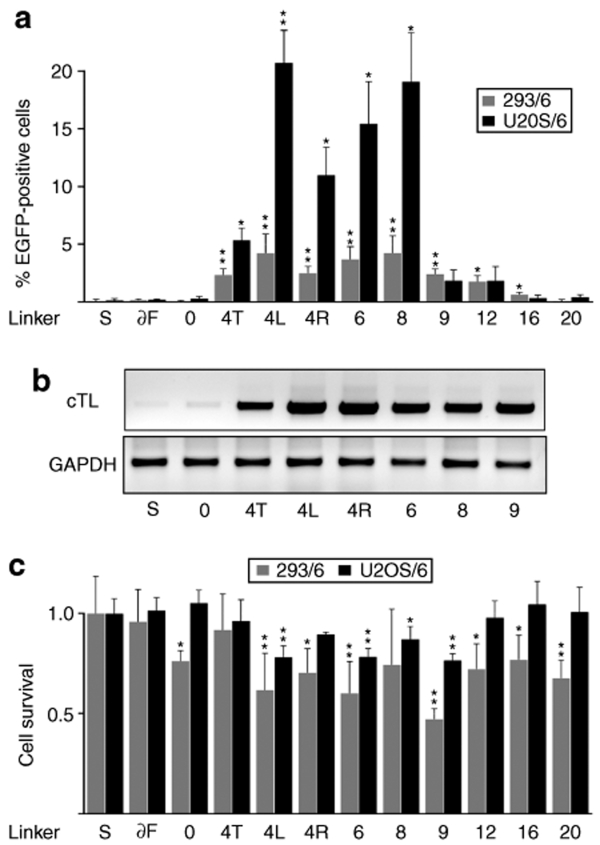
As noted in a previous study,29 the frequency of DSB-induced HR in the human osteosarcoma cell line U2OS is higher as compared to HEK293. To investigate whether the activity of the ZFN variants is cell-type-dependent, a polyclonal U2OS-based targeting cell line (which we termed as U2OS/6) was produced by lentiviral transduction. U2OS/6 harbors an integrated lacZ(fs)EGFP target locus with a 6-bp spacer identical to the one used in our HEK293 cell-based experiments (cell line 293/6). The efficiency of chromosomal gene targeting was determined 5 days after co-transfection of an HR donor plasmid with different ZFN expression vectors (Figure 5a). A comparison of the U2OS/6 results with those obtained with 293/6 cells indicated similar relative activities of all ZFN variants in the two cell lines but with up to fivefold higher gene targeting frequencies in the USOS cells. Linker variant 4L was again the most active ZFN, and induced gene editing in >20% of transfected cells.
Figure 5.
Cell line dependence of zinc-finger nuclease (ZFN)-mediated gene targeting and ZFN-associated toxicity. (a) Chromosomal recombination assay. U2OS/6 (black bars) and 293/6 (gray bars) target cell lines containing an integrated lacZfsEGFP locus with a 6-bp spacer were transfected with the various ZFN expression vectors, a donor plasmid, and a DsRed-Express (REx) expression vector. The REx marker was used to normalize for transfection efficiency and to follow the fate of transfected cells over time. The columns designate the percentage of EGFP-positive cells 5 days after transfection as determined by flow cytometry. I-SceI was included as a negative control to indicate the level of nonstimulated gene targeting. A statistically significant increase in gene targeting relative to nonstimulated HR is indicated by * (P < 0.05) or ** (P < 0.01). (b) Genotyping. U2OS/6 target cells were transfected as described in a and genomic DNA was isolated 5 days later. A nested PCR was performed with primers designed to amplify the corrected target locus (cTL). The positions of the primers used for the nested PCR protocol are indicated in Figure 2a. As an internal quality control, a fragment of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) locus was amplified. (c) Cell survival assay. U2OS/6 (black bars) and 293/6 (gray bars) target cells were transfected as described in a and the percentage of REx-positive cells was determined by flow cytometry 2 and 5 days after transfection. The columns represent the fraction of REx-positive cells at day 5 as compared to the fraction at day 2 after transfection, and are shown relative to transfection with an I-SceI expression plasmid. A statistically significant decrease in survival as compared to I-SceI is indicated by * (P < 0.05) or ** (P < 0.01).
To verify that EGFP expression in gene-corrected cells was indeed a result of a genuine HR event, PCR-based genotype analysis was performed on genomic DNA extracted from the transfected U2OS/6 target cell line. A nested PCR strategy was designed such that only sequences present in the corrected lacZ-EGFP target locus would be amplified (Figure 2a). In agreement with the flow cytometry data, only a few PCR products were detected from samples transfected with expression vectors for I-SceI or the ZFN variant 0, whereas an amplification product was readily detectable from cells transfected with the other ZFN linker variants analyzed (Figure 5b). As an internal quality control, a fragment of the glyceraldehyde 3-phosphate dehydrogenase locus was amplified. These results confirmed the data of the recombination assay and demonstrated that EGFP-positive cells arise as a result of genuine HR between the chromosomal target locus and the donor plasmid, and that the frequency of these events is stimulated by site-specific ZFNs.
ZFN-induced toxicity is cell line-dependent
ZFN-induced cytotoxicity has been hypothesized to result from extensive off-target activity of the nucleases. For assessing the toxicity of all ZFN linker variants, the fate of transfected 293/6 and U2OS/6 cells was followed over time by determining the percentage of REx-positive cells 2 and 5 days after transfection. Survival of transfected cells was calculated as the fraction of REx-positive cells at day 5 in relation to cells transfected with an I-SceI expression plasmid (Figure 5c). Although the same amounts of ZFN expression plasmids were transfected, the 293/6 cell line was more sensitive to overexpression of ZFNs than the U2OS/6 cell line was. At least in part, this can be explained by the slightly higher transfection efficiencies achieved in HEK293 cells. In both cell lines the ZFN variant with the 9-aa linker, which was the least selective with regard to spacer length (Figures 2c and 4b), revealed the highest toxicity; by contrast, variants with a more narrow activity spectrum, such as the 4-aa linker variants, induced less cytotoxicity. This suggests some correlation between target site selectivity, i.e., the activity spectrum of a specific ZFN linker variant, and toxicity.
Discussion
Despite much recent progress, the development of methods to better understand and quantify ZFN-associated genotoxicity remains an important priority for future research. Examination of the ZFN architecture has suggested three parameters that are likely to affect the frequency of off-target cleavage events: the DNA-binding specificity of each ZFN subunit, the stability of the ZFN dimerization interface, and the promiscuity of the ZFN dimer for spacers of different lengths. Recently published articles have indicated that ZFN-associated toxicity can be significantly reduced by improving the DNA-binding specificities of the zinc-finger domains23 and regulating the DNA-cleavage activity of the FokI nuclease domain.21,22 Although inter-domain ZFN linkers of various lengths have been tested in vitro and on plasmid DNA in Xenopus oocytes,7,24 a systematic analysis on chromosomal targets in human cells with regard to ZFN activity and toxicity has not been conducted to date. Our systematic analysis of 11 candidate-based linkers allowed us to define linker variants that either (i) have selective target-site activity or (ii) stimulate gene targeting at noncanonical 5- or 6-bp spacers.
Revisiting the parameters that determine the optimal linker–spacer combinations
Several predictions have been made on the basis of structural data with regard to the requirements of ZFN activity on a DNA target site. Although the structure of a ZFN–DNA complex has not been solved yet, structures of the FokI restriction enzyme and of zinc-finger proteins bound to DNA have been determined.9,30,31 Based on the helical structure of the DNA, molecular modeling predicted that a 6-bp spacer would locate the C-termini of the zinc-finger domains on either side of the DNA and hence close to the N-termini of the FokI cleavage domains.7,24 Moreover, since a B-form DNA helix features 10 bp per turn, a 16-bp spacer creates a structurally similar situation in which both termini to be connected are located on the same side of the DNA. Data generated by two laboratories largely confirmed these predictions. ZFNs with 18-aa linkers (22 aa as per our definition) were able to cleave target half-sites separated by 6–18 bp on episomal DNA in microinjected oocytes, while the use of a 0-aa linker (4 aa as per our definition) resulted in ZFN peak activity at a 6-bp spacer.7,24 A recently published protocol32 shows that the sequence between the last histidine in finger 3 and the start of the FokI cleavage domain of that “0-aa linker” was LRQK, which corresponds to the natural inter-domain linker of transcription factor Zif268. Two simple assumptions can be made from these earlier studies: (i) longer linkers bridge longer spacers while shorter linkers work on shorter spacers, and (ii) longer linkers are more promiscuous than shorter linkers are. In this study, we confirmed that these expectations also hold up for human cells, on both episomal and chromosomal targets. Short 4-aa linkers had an activity optimum at 5- and 6-bp spacers with the 6-aa linker being the most selective one with a peak activity at a 6-bp spacer. For longer linkers of 8 to 16 aa the activity spectrum became larger and shifted to up to 16 bp-spacers. Moreover, we demonstrated for the first time that, with the appropriate linkers, ZFNs can induce gene conversion almost as efficiently at a chromosomal target with a 16-bp spacer as at a 6-bp spacer. Importantly, however, we showed that not only does linker length matter but also that the sequence compositions of the ZFN inter-domain linker and—to a lesser extent—of the DNA spacer are important factors. For instance, single aa exchanges in the inter-domain linkers of the 4-aa variants led to up to fourfold differences in the ZFN activity on chromosomal targets in U2OS cells (P = 0.005 for 4L versus 4T).
As mentioned earlier in the text, the requirements for ZFN cleavage were more restrictive on episomal DNA than in vitro.7,24 When comparing our results obtained from the recombination assays on episomal targets with the chromosomal ZFN activity data, it became evident that the relative activities on the different spacer lengths also changed substantially. In the episomal assay, the best ZFN variants stimulated HR on a target with a 6-bp spacer about eightfold better than on targets with 5- or 16-bp spacers. However, when these targets were placed in a chromosomal context, the differences observed were less than twofold. Therefore, although transfected target plasmids will eventually be assembled into chromatin, the nucleosome structure on an integrated target locus may be considerably different and subject to dynamic changes, affecting both the accessibility and the topology of the ZFN target sites.
In conclusion, ideal linker–spacer combinations can be determined only on chromosomal target sites. Parameters that affect these combinations include not only the absolute length but also the sequence compositions, both of the ZFN linker and of the DNA spacer. In particular, the first two residues in the inter-domain linker affect ZFN activity and its preference for either AT- or GC-rich spacers.
Expanding or restricting the ZFN target site repertoire?
For therapeutic gene targeting, the DSB has to be introduced as close as possible to the mutation to be corrected. This is mainly because of the rather short DNA region that is exchanged between the target site and the donor DNA through HR. Increasing the distance of the DSB to the mutation leads to a rapid decrease in the frequency of gene conversion. For instance, the gene conversion frequency drops threefold if the DSB is inserted 50 bp away and about sixfold at a distance of 90 bp from the DSB.33 Given that highly specific zinc-finger arrays are difficult to engineer,3 and that for a significant fraction of potential 9-bp half-sites it might not be possible to generate them at all by using simple modular assembly-based zinc-finger engineering approaches26 or even by using more complex context-sensitive engineering platforms,28,34,35,36,37 it is important to expand the repertoire of putative ZFN target site beyond half-sites separated by the canonical 5- or 6-bp spacers.
Given the restraints imposed by the structures of the DNA, the FokI cleavage domains, and the zinc-finger DNA-binding domains, the composition and the length of the inter-domain linker are the only variables that can be adapted to fit the needs of a given target site. This study confirmed that ZFNs are most active on target sites with a 6-bp spacer. An ideal target site is composed of two target half-sites separated by an AT-rich 6-bp spacer. Surprisingly, the ZFN variant with an 8-aa linker was as active on a 7-bp spacer as the best 4-aa variants were on a 5-bp spacer. Moreover, the 9-aa linker variant stimulated gene targeting to a greater extent on 16-bp spacers than the best variants on 5- and 7-bp spacers. Our results therefore indicate that it is easy to expand or alter the target site repertoire for ZFNs by choosing an appropriate inter-domain linker.
A clear correlation between the linker length and ZFN-associated toxicity could not be established in this study. Rather, ZFNs that induced high-frequency gene conversion were also more toxic. It seems, however, that ZFN variants with a broad activity spectrum, such as the ZFN variant with a 9-aa linker, are more toxic than ZFN variants with a more narrow activity spectrum, such as variant 4R. Our results suggest, therefore, that if multiple target site options exist for a potential target locus, it will be advisable to choose a site which allows the use of a ZFN linker variant with the most narrowly defined target-site specificity to minimize ZFN-associated toxicity.
Cell-type dependence of ZFN performance
This study suggests that the overall efficiency of ZFN-induced genome editing may not only depend on ZFN parameters but also on the apoptotic threshold of a cell and its ability to activate the appropriate DNA repair pathways, both of which may vary significantly among different cell types. Here we compared the ZFN-stimulated gene conversion frequency and the ZFN-associated toxicity in two different human cell lines. Depending on the ZFN linker variant used, the frequency of gene targeting in U2OS cells was two- to sixfold higher than in HEK293 cells, reaching gene conversion frequencies of up to 20% in unselected cells. Moreover, U2OS cells were more resistant to overexpression of ZFNs, as demonstrated in our cell survival assays. Cell-type dependence of DSB-stimulated gene targeting has been observed before,17,29 and is an important parameter that should be factored in when planning therapeutic strategies using ZFNs.
In summary, our data suggest that the best first choice for ZFN-mediated genome modifications is the combination of the LGGS linker with a 6-bp spacer. However, the ideal combination of ZFN- and cell type-specific parameters can be determined only by performing cell type-specific assays, taking into account both ZFN activity on the chromosomal target site and ZFN-associated toxicity.
Materials and Methods
Plasmids. All expression plasmids containing the ZFN linker variants are based on plasmid pRK5.EB2-N23 and were assembled by overlap extension PCR cloning. The relevant sequences are shown in Figure 1a. Target plasmids bearing two EB half-sites (5′-gccgcagtg) as inverted repeats and separated by various spacers were created by oligonucleotide cloning into pCMV.LacZfsEGFP (Figure 2a). The resulting target loci LacZ(n)fsEGFP (n indicating the spacer length) were subsequently cloned in a lentiviral vector pLV-CMV.LacZs31∂GFPiNwpre29 carrying a neomycin cassette for the production of the target cell lines. The donor plasmids pUC.Zgfp/Rex for the episomal gene targeting assay and pUC.ZgfpiNwpre for the chromosomal gene targeting assay have been previously described.19,22
Cell lines. All cells were grown in DMEM (Gibco/Invitrogen, Kahlsruhe, Germany) supplemented with 10% fetal calf serum (Gibco/Invitrogen). Lentiviral vectors to establish the target cell lines were generated as previously described.29 To obtain cell lines with a single copy target locus, parental HEK293 or U2OS cells were infected with a small amount of lentivector that rendered <1% of cells LacZ-positive, as determined by X-Gal staining. Target cells were selected with 0.4 mg/ml geneticin (Gibco/Invitrogen) for at least 3 weeks. The sequence of the integrated target locus was verified by sequencing a PCR product amplified from genomic DNA, which encompassed the critical sequence around the ZFN target sites.
Quantitative recombination and toxicity assays. Assays were performed as previously described.29 Briefly, chromosomal gene targeting was carried out with 293/6 and U2OS/6 target cell lines in 12-well plates using 300 ng of the ZFN expression plasmid, 2 µg of the donor plasmid pUC.ZgfpiNwpre, and 10 ng of pDsRed-Express-N1 (REx; Clontech, Mountain View, CA). The cells were analyzed by flow cytometry 2 and 5 days after transfection. The number of REx-positive cells at day 2 was used for normalization of transfection efficiency. For the episomal gene targeting assay, HEK293T cells in 24-well plates were transfected with 100 ng of ZFN expression plasmid, 20 ng of target plasmid pCMV.LacZ(n)fsGFP, and 1 µg of donor plasmid pUC.Zgfp/REx. Two days later the percentage of EGFP- and REx-positive cells was determined by flow cytometry. For calculating the cell survival rate, the decrease in the number of REx-positive cells from day 2 to day 5 was determined and normalized to cells transfected with an I-SceI expression vector.
Immunoblotting. Immunoblotting was performed as previously described.19 Briefly, HEK293T cells in 12-well plates were transfected with 300 ng of expression plasmids harboring the ZFN linker variants, 100 ng of pEGFP-N1 (Clontech, Mountain View, CA), and pUC118 to 2 µg using the calcium phosphate precipitation method. The cells were harvested after 30 h, and 50 µg of lysate was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. After transfer to polyvinylidene difluoride membranes, ZFN and EGFP expression were detected simultaneously with antibodies directed against the HA tag (NB600-363; Novus Biologicals, Littleton, CO) and EGFP (MAB3580; Millipore, Billerica, MA), and visualized by Infrared Imaging after incubation with secondary antibodies conjugated with either IR-Dyes 680 or 800CW (LI-COR Biosciences, Bad Homburg, Germany).
Genotyping. Genomic DNA from cells was extracted using a Invisorb Spin Cell Mini Kit (Invitek, Berlin, Germany). A nested PCR was performed with 300 ng of genomic DNA and primers 5′-GCTGGCGTAATAGCGAAGAG and 5′-CATGGTGGCGATGGATCCTTTTT for 35 cycles in the first round and 5′-ACGGGTTGTTACTCGCTCAC and 5′-CAGCAGCAGAC CATTTTCAA for the second round, using SAWADY-Taq polymerase (PeqLab, Erlangen, Germany) for 27 cycles. PCR products were purified between the two rounds using Invisorb Spin PCRapaceKit (Invitek, Berlin, Germany). As an internal control, a fragment of the glyceraldehyde 3-phosphate dehydrogenase locus was amplified with primers 5′-TAGG CGAGATCCCTCCAAAA and 5′-AGTGATGGCATGGACTGTGG. PCR products were resolved by agarose gel electrophoresis.
Statistical analysis. All experiments were performed at least three times. Error bars represent SD. Statistical significance was determined using either a one-sided (Figure 5c) or a two-sided Student's t-test (Figures 2, 3, 4, and 5a) with unequal variance.
Supplementary MaterialTable S1. Candidate-based inter-domain linkers.Table S2. Relevant sequences of the target loci and the corrected target locus (cTL).
Supplementary Material
Candidate-based inter-domain linkers.
Relevant sequences of the target loci and the corrected target locus (cTL).
Acknowledgments
We thank Eva Guhl for technical assistance, and Tatjana I. Cornu and Katharina Gellhaus (all from Charité Medical School, Berlin) and J. Keith Joung (Harvard Medical School, Boston) for their valuable suggestions and critical reading of the manuscript. This work was supported by grant 01GU0618 (ITCF network) of the German Ministry of Education and Research (BMBF).
REFERENCES
- Rouet P, Smih F., and , Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulika A, Perrin A, Dujon B., and , Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathomen T., and , Keith Joung J. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- Kim YG, Cha J., and , Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitinaite J, Wah DA, Aggarwal AK., and , Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanamee ES, Santagata S., and , Aggarwal AK. FokI requires two specific DNA sites for cleavage. J Mol Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S., and , Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani M, Smith J, Kandavelou K, Berg JM., and , Chandrasegaran S. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem Biophys Res Commun. 2005;334:1191–1197. doi: 10.1016/j.bbrc.2005.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP., and , Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK., and , Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Beumer K, Bhattacharyya G, Bibikova M, Trautman JK., and , Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J, Davis MW, Jorgensen EM., and , Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci USA. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Plaisier CL, Carroll D., and , Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, et al. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 2005;44:693–705. doi: 10.1111/j.1365-313X.2005.02551.x. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Cathomen T, Weitzman MD., and , Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol Cell Biol. 2003;23:3558–3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwin S, Gere MB, Guhl E, Effertz K, Barbas CF, 3rd, Segal DJ, et al. Custom zinc-finger nucleases for use in human cells. Mol Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ., and , Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH., and , Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO., and , Joung JK. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc Natl Acad Sci USA. 2003;100:12271–12276. doi: 10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu TI., and , Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M, Rould MA, Nekludova L., and , Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- Wah DA, Hirsch JA, Dorner LF, Schildkraut I., and , Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- Carroll D, Morton JJ, Beumer KJ., and , Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A., and , Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greisman HA., and , Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- Isalan M, Klug A., and , Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH., and , Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Candidate-based inter-domain linkers.
Relevant sequences of the target loci and the corrected target locus (cTL).