Abstract
Osteopontin (OPN) is a secreted phosphoprotein which mediates tumorigenesis, local growth, and metastasis in a variety of cancers. It is a potential therapeutic target for the regulation of cancer metastasis. RNA aptamer technology targeting OPN may represent a clinically viable therapy. In this study, we characterize the critical sequence of an RNA aptamer, termed OPN-R3, directed against human OPN. It has a Kd of 18 nmol/l and binds specifically to human OPN as determined by RNA electrophoretic mobility assays. In MDA-MB231 human breast cancer cells examined under fluorescence microscopy, OPN-R3 ablates cell surface binding of OPN to its cell surface CD44 and αvβ3 integrin receptors. Critical enzymatic components of the OPN signal transduction pathways, PI3K, JNK1/2, Src and Akt, and mediators of extracellular matrix degradation, matrix metalloproteinase 2 (MMP2) and uroplasminogen activator (uPA), are significantly decreased following exposure to OPN-R3. OPN-R3 inhibits MDA-MB231 in vitro adhesion, migration, and invasion characteristics by 60, 50, and 65%, respectively. In an in vivo xenograft model of breast cancer, OPN-R3 significantly decreases local progression and distant metastases. On the basis of this “proof-of-concept” study, we conclude that RNA aptamer targeting of OPN has biologically relevance for modifying tumor growth and metastasis.
Introduction
Cancer progression depends on an accumulation of metastasis-supporting physiological changes which are regulated by cell signaling molecules. One such molecule, osteopontin (OPN), is a secreted phosphoprotein which functions as a cell attachment protein and cytokine that signals through two cell adhesion molecules: αvβ3-integrin and CD44.1,2,3 Initially discovered as an inducible, tumor-promoter gene, OPN is an acidic hydrophilic glycophosphoprotein which is overexpressed in human tumors and is the major phosphoprotein secreted by malignant cells in advanced metastatic cancer.4,5,6,7,8,9,10,11 Evidence has accumulated for involvement of OPN in increased cellular migratory and invasive behavior, increased metastasis, protection from apoptosis, promotion of colony formation and 3D growth ability, induction of tumor-associated inflammatory cells, and induction of expression of angiogenic factors.12 Gain- and loss-of function assays have demonstrated a critical role for OPN in tumor metastatic function in colon, liver, and breast cancers.13 Clinical studies certainly have linked serum OPN expression with increased metastatic tumor burden and poor patient outcomes.7,14 As a secreted phosphoprotein that is readily accessible in the extracellular milieu, OPN is an attractive therapeutic target for blockade of tumor growth and metastasis.
Recently, small structured single-stranded RNAs, also known as RNA aptamers, have emerged as viable alternatives to small-molecule and antibody-based therapy.15,16 RNA aptamers specifically bind target proteins with high affinity, are quite stable, lack immunogenicity, and elicit biological responses. Aptamers are evolved by means of an iterative selection method called SELEX (systematic evolution of ligands by exponential enrichment) to specifically recognize and tightly bind their targets by means of well-defined complementary three-dimensional structures. Typically, dissociation constants for these aptamer–target complexes are in the high pico-molar to low nano-molar range. As a secreted protein, OPN represents an ideal target for RNA aptamer mediated inhibition.
In this article, we utilize the MDA-MB231 human breast cancer cell line to demonstrate the effect of an OPN-directed RNA aptamer on in vitro and in vivo measures of growth and metastasis. This cell line expresses OPN and its cell surface target receptors, CD44 and αvβ3 integrin. Our data indicate that RNA aptamer binding of OPN blocks interaction with its cell surface receptors with significantly decreased: (i) expression of matrix metalloproteinase 2 (MMP2) and uroplasminogen activator (uPA), (ii) activation of CD44- and αvβ3-dependent signal transduction pathways, (iii) in vitro measures of adhesion, migration, and invasion, and (iv) in vivo local progression and distant metastases in a xenograft model. In this “proof-of-concept” study, our results suggest that RNA aptamer technology directed against OPN may be a viable therapeutic intervention to inhibit growth and metastasis in breast cancer.
Results
Characterization of an RNA aptamer directed against OPN
After successive rounds of SELEX, we characterized the Kd of the OPN aptamer, termed OPN-R3, using the double-filter nitrocellulose filter-binding method.17 SELEX is an iterative in vitro selection process consisting of sequential selection and amplification steps that can efficiently reduce a complex library of nucleic acids with randomized sequences (complexity of ~1014) to a minimized subset of one or more sequences that bind tightly to the target of choice. Fifty clones were sequenced following the eighth round of SELEX; 12 clones were the OPN-R3 aptamer. We compared OPN-R3 with the other 38 aptamer sequences, and there was no homology among them. The Kd value of OPN-R3 and 10 other clones were determined; the OPN-R3 Kd was the lowest at 18 ± 0.2 nmol/l.
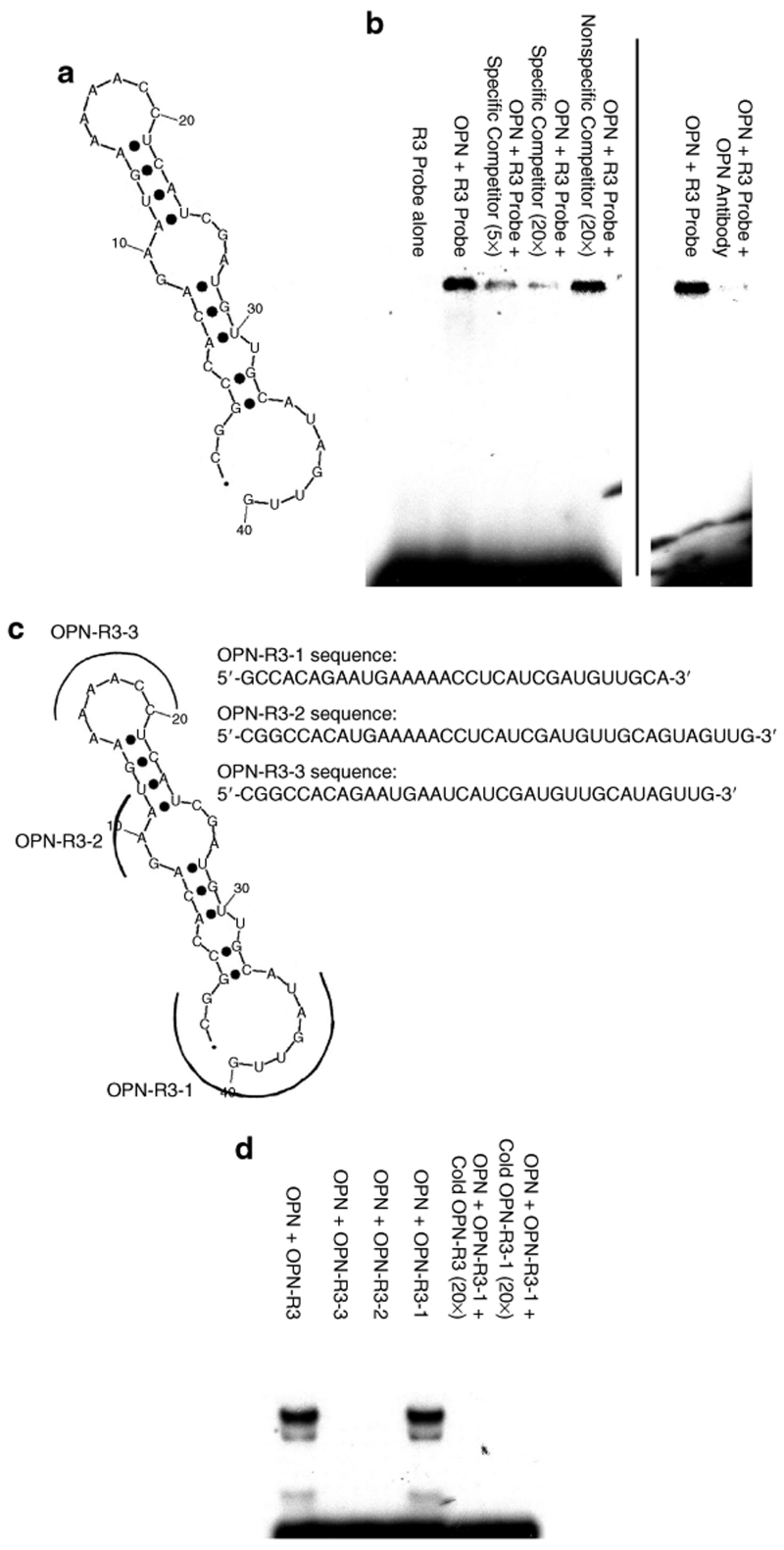
The predicted secondary structure of OPN-R3 contains the usual stem–loop structure of RNA aptamers and is shown in Figure 1a. To confirm in vitro binding of OPN-R3 to OPN, RNA electrophoretic mobility shift assays (REMSAs) were performed (Figure 1b). Human OPN bound to OPN-R3; increasing concentrations of unlabelled OPN-R3 probe effectively competed for OPN binding, whereas unlabelled nonspecific competitor RNA aptamer did not alter OPN binding to OPN-R3. Supershift assays using antibody to human OPN demonstrated decreased binding of OPN to OPN-R3. These data indicate that human OPN binds to OPN-R3 in a specific fashion.
Figure 1.
OPN-R3 structure and in vitro RNA binding. (a) OPN-R3 aptamer secondary structure model. (b) RNA electrophoretic mobility shift assays of OPN-R3. Blot is representative of four experiments. (c) Secondary structure models of mutant OPN RNA aptamers (OPN-R3-1, OPN-R3-2, OPN-R3-3). (d) RNA electrophoretic mobility shift assays of OPN-R3-1, OPN-R3-2, and OPN-R3-3. Blot is representative of four experiments.
Mutagenesis of OPN-R3 was then performed to determine the active binding site. Figure 1c depicts the regions of OPN-R3 that underwent deletion. Deletion constructs, OPN-R3-1, OPN-R3-2, and OPN-R3-3, were then tested in REMSA with human OPN (Figure 1d). Only OPN-R3-1 retained its OPN-binding abilities, suggesting that regions 2 and 3 are both required for in vitro binding to OPN. Binding affinity studies confirmed that OPN-R3-1 and OPN-R3 had identical Kd values.
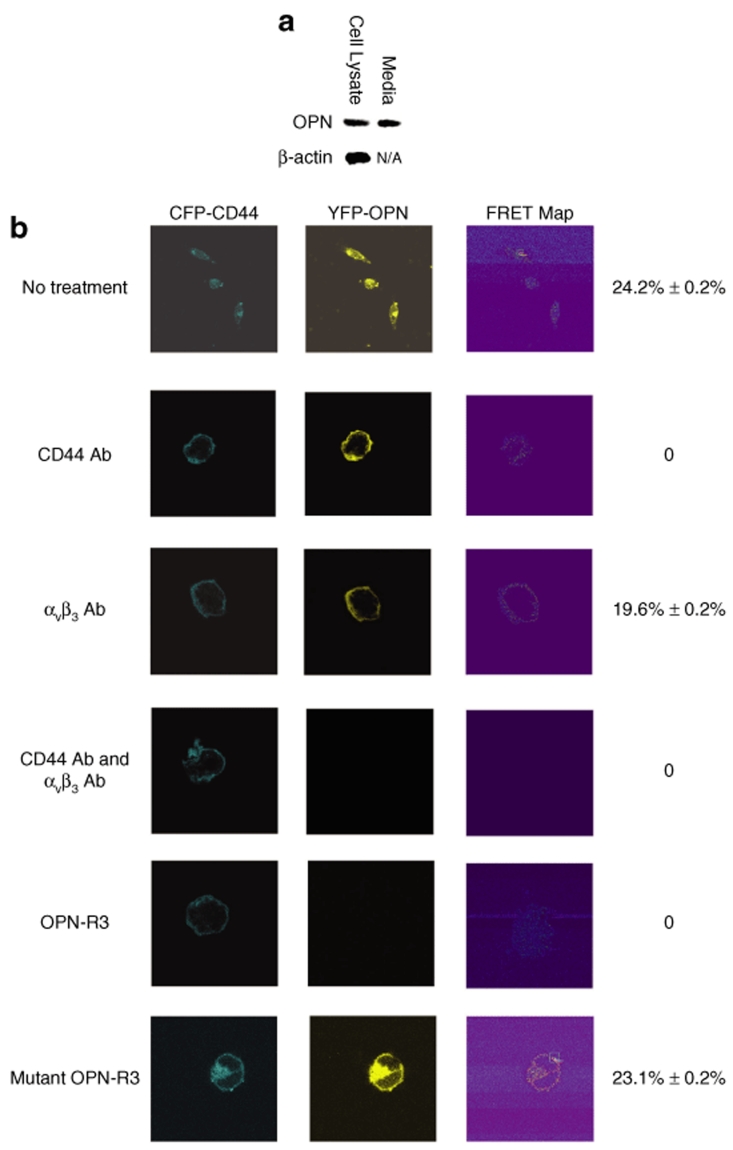
We then utilized Forster resonance energy transfer (FRET) confocal microscopy in MDA-MB231 human breast cancer cells to determine the efficacy of OPN-R3 for inhibition of OPN binding to its cell surface receptors, CD44 and αvβ3 integrin. Western blot analysis confirmed production of OPN in MDA cell lysates and media (Figure 2a). Human full-length CD44s cDNA and OPN cDNA were separately fused in frame into mammalian expression vector pECFP and pEYFP, respectively. MDA cells were then transfected with both plasmids. Confocal fluorescence microscopy showed overlapping localization of both CFP-tagged CD44 and YFP-tagged OPN to the MDA plasma membrane (Figure 2b). Acceptor photobleaching was then used to measure FRET between CFP-CD44 and YFP-OPN. If CFP-CD44 (donor) and YFP-OPN (acceptor) are within 10 nm of each other, and the fluorophore dipoles are aligned, resonance energy can be transferred from CFP to YFP.18 To perform acceptor photobleaching, a prebleach image was captured using the 458-nm laser line; a region of the plasma membrane was selectively irradiated using the 514-nm laser line. An increase in CFP fluorescence was observed following YFP photobleaching, and the mean FRET efficiency was 24.2 ± 0.2%. Acceptor photobleaching experiments were done on 50 MDA cells (three regions per cell) coexpressing CFP-CD44 and YFP-OPN as well as on a similar number of cells in which no FRET was expected. These additional controls included (i) cotransfection of CFP- and YFP-empty plasmids (FRET efficiency, 0.23 ± 0.2%) and (ii) transfection of a CFP-empty plasmid alone (FRET efficiency, 0.21 ± 0.1%). These data confirm interaction between OPN and CD44 on the MDA-MB231 cell surface.
Figure 2.
Characterization of OPN-R3 cell surface binding. (a) Western blot analysis of OPN expression in MDA-MB231 cell lysate and culture medium. Blot is representative of three experiments. N/A, not applicable. (b) FRET analysis of MDA-MB231 cells. FRET efficiency was calculated as 100 × [(CFP post-bleach ‐ CFP prebleach)/CFP post-bleach]; FRET was performed on 50 cells per treatment group with 3 regions per cell. Photos are representative of five experiments.
Subsequent FRET experiments were then performed in the presence of OPN-R3, blocking Ab to CD44, and/or blocking antibody to αvβ3 integrin. In the presence of CD44 Ab, cell surface binding of OPN was still present but no FRET was detected, indicating OPN binding to alternative αvβ3 integrin-binding sites (Figure 2b). In the presence of blocking Ab to αvβ3 integrin, FRET was detected (19.6 ± 0.2%; P = NS vs. CD44 Ab) and cell surface OPN was present suggesting that OPN was bound to native CD44 and/or CFP-CD44 receptors. However, in the presence of both CD44 Ab and αvβ3 integrin Ab, neither cell surface OPN nor FRET was detected. Finally, in the presence of OPN-R3 (100 nmol/l), FRET was totally ablated, and no cell surface OPN was found suggesting that the RNA aptamer blocked all interaction of OPN with its cell surface receptors, including CD44. As a control, mutated OPN-R3 aptamer was associated with FRET of 23.1 ± 0.2%.
OPN-R3 and OPN-dependent signaling pathways
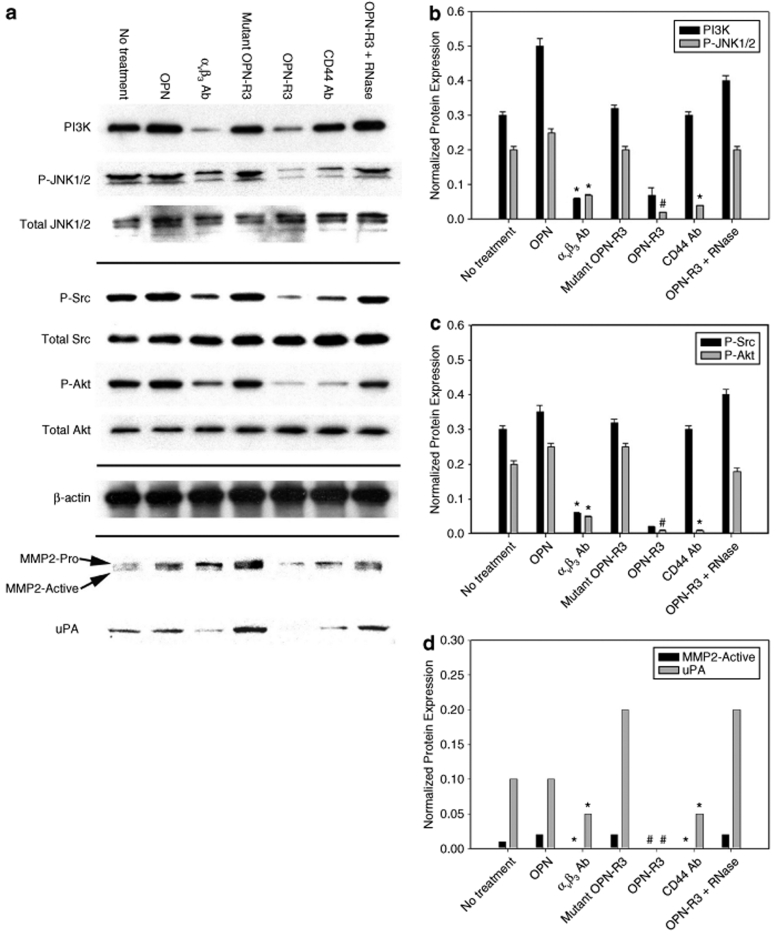
To determine the effect of OPN-R3 on OPN-dependent signal pathways, we performed western blots in MDA-MB231 for PI3K, JNK1/2, Src and Akt as constituents of the αvβ3 and/or CD44 pathways. The expression of these various markers were assessed in response to exposure to OPN-R3, exogenous OPN (20 nmol/l), αvβ3 Ab, CD44 Ab, mutant OPN-R3 and/or mutant OPN-R3 with RNase (Figure 3). Expression of phosphorylated JNK-1/2 (P-JNK1/2) and PI3k was detected in untreated MDA cells and was not altered in the presence of exogenous OPN. Exposure of the cells to αvβ3 Ab or OPN-R3 significantly decreased both P-JNK1/2 and PI3K expression. In contrast, mutant OPN-R3 and OPN-R3 + RNase did not alter levels of P-JNK1/2 and PI3K. Interestingly, exposure of the MDA cells to CD44 Ab did not alter PI3K, but did decrease P-JNK1/2 suggesting that crosstalk or overlap might exist between the CD44 and αvβ3 integrin signal transduction pathways. When phosphorylated-Src (P-Src) and -Akt (P-Akt) were addressed, expression of both proteins was detected in untreated MDA cells and was not altered in the presence of exogenous OPN (20 nmol/l). Exposure of the cells to CD44 Ab or OPN-R3 significantly decreased P-Src and P-Akt expression. In contrast, mutant OPN-R3 and OPN-R3 + RNase had no discernable effect. Ab to αvβ3 integrin decreased PI3k and P-Src expression also; this repeats the theme of overlapping signal transduction pathways between CD44 and αvβ3 integrin receptors.
Figure 3.
OPN-regulated signal transduction pathway. (a) Western blot of PI3K, JNK1/2, Src, Akt, MMP2, and uPA in MDA-MB231 cells. Blot is representative of four experiments. (b) Histogram of PI3K and P-JNK1/2 expression. PI3K and P-JNK1/2 are normalized to β-actin and total JNK expression, respectively, by laser densitometry. Data are presented as mean ± SEM of four experiments. *P < 0.01 vs. No treatment, OPN, Mutant OPN-R3, CD44 Ab, OPN-R3 + RNase; #P < 0.01 vs. No treatment, OPN, αvβ3 Ab, Mutant OPN-R3, CD44 Ab, OPN-R3 + RNase; **P < 0.01 vs. No treatment, OPN, Mutant OPN-R3, OPN-R3, OPN-R3 + RNase; @P < 0.01 vs. No treatment, OPN, Mutant OPN-R3, OPN-R3, OPN-R3 + RNase. (c) Histogram of P-Src and P-Akt expression. P-Src and P-Akt are normalized to total Src and total Akt expression, respectively, by laser densitometry. Data are presented as mean ± SEM of four experiments. *P < 0.01 vs. No treatment, OPN, Mutant OPN-R3, CD44 Ab, OPN-R3 + RNase; #P < 0.01 vs. No treatment, OPN, αvβ3 Ab, Mutant OPN-R3, CD44 Ab, OPN-R3 + RNase; **P < 0.01 vs. No treatment, OPN, Mutant OPN-R3, OPN-R3, OPN-R3 + RNase; @P < 0.01 vs. No treatment, OPN, Mutant OPN-R3, OPN-R3, OPN-R3 + RNase. (d) Histogram of MMP2 and uPA expression. MMP2 and uPA are normalized to β-actin expression by laser densitometry. Data are presented as mean ± SEM of four experiments.*P < 0.02 vs. No treatment, OPN, Mutant OPN-R3, OPN-R3 + RNase; #P < 0.01 vs. αvβ3 Ab, CD44 Ab.
OPN has previously been demonstrated to partially regulate expression of MMP2 and uPA as mediators of extracellular matrix degradation and facilitators of metastasis.19,20,21,22 In this setting, we examined pro- and active MMP2 and uPA expression in MDA-MB231 cells following exposure to OPN-R3 (Figure 3d). In a fashion similar to that seen for the previous proteins, pro-MMP2, active MMP2, and uPA were detected in untreated MDA-MB231 cells. Exogenous OPN did not significantly alter expression. OPN-R3 ablated uPA and active MMP2 levels, although pro-MMP2 was still readily detected. Ab to CD44 and αvβ3 integrin significantly decreased uPA, pro-MMP2, and active MMP2 levels. Mutant OPN-R3 and OPN-R3 + RNase had no effect. In total, these data indicate that OPN-R3 aptamer can significantly decrease activation and/or expression of various constituents of the CD44 and αvβ3 integrin signal transduction pathways and their downstream effector molecules in MDA-MB231 cells.
OPN-R3 and MD-MB231 adhesion, migration, and invasion
To assess the functional consequences of OPN-R3 ligation of OPN, in vitro adhesion, migration, and invasion assays were performed (Figure 4). When compared to untreated cells, adhesion, migration, and invasion in OPN-R3 treated cells were decreased by 60, 50, and 65%, respectively. In comparison, αvβ3 integrin Ab decreased adhesion, migration, and invasion by 30, 40, and 45%, respectively. Similarly, CD44 Ab decreased adhesion, migration, and invasion by 40, 30, and 48%, respectively. Exogenous OPN, mutant OPN-R3 and OPN-R3 + RNase had no effect on the three measures. These results indicate that OPN-R3 can effectively and significantly inhibit the in vitro correlates of adhesion, migration, and invasion in MDA-MB231 cells.
Figure 4.
OPN-R3 and adhesion, migration, and invasion of MDA-MB231 cells. In vitro adhesion, migration, and invasion assays were performed. Data are presented as mean ± SEM of four experiments. *P < 0.01 vs. No treatment, OPN, αvβ3 Ab, Mutant OPN-R3, CD44 Ab, OPN-R3 + RNase; #P < 0.01 vs. No treatment, OPN, Mutant OPN-R3, OPN-R3, OPN-R3 + RNase.
Functional in vivo activity of OPN-R3
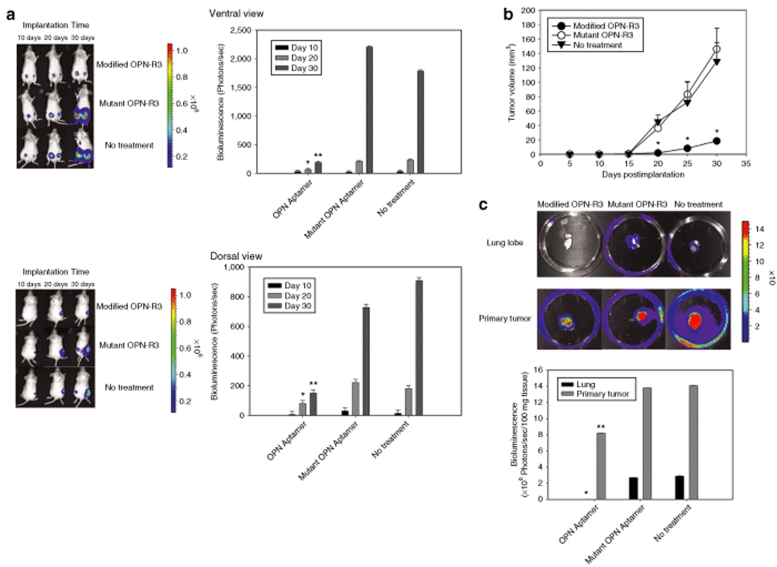
In the following in vivo studies, OPN-R3 (and mutant OPN-R3-2) was modified to increase its biologic half-life, incorporating 2′-O-methyl substituted nucleotides, 5′-cholesterol modification and 3′-inverted deoxythymidine. The sequence of OPN-R3 aptamer used in in vivo studies is same as OPN-R3-1. Based on the company's unpublished results (Dharmacon, Lafayette, CO), the half-life of the modified RNA APTAMER oligo is >24 hours in human serum at 37 °C. The half-life of both OPN-R3 and mutant OPN-R3-2 in Dulbecco's modified Eagle's medium with 10% normal mouse serum was 8 hours. The in vitro Kd of this Modified OPN-R3 was 18 nmol/l; in vitro specific binding of Modified OPN-R3 to OPN was again confirmed using REMSA (data not shown). We used a xenograft model of MDA-MB231 cell implantation into the mammary fat pads of female NOD scid mice; these MDA cells were previously engineered to express luciferase.23 Modified OPN-R3, Mutant OPN-R3 or vehicle were injected into the mouse tail veins every 2 days. Bioluminescence imaging data at days 10, 20, and 30 are displayed in Figure 5. Bioluminescence was significantly decreased in the Modified OPN-R3-treated animals by over 4- and 12-fold at 20 and 30 days after implantation, respectively, when compared to Mutant OPN-R3 or No treatment animals (P < 0.01 at 20 days and 30 days for OPN-R3 vs. Mutant OPN-R3 and No treatment). Tumor volumes were measured on a daily basis (Figure 5b). Similar to that seen with the bioluminescence data, tumor volume was significantly decreased in the Modified OPN-R3-treated animals. At day 20, tumor volume in the Modified OPN-R3 aptamer-treated group was 18–20-fold smaller than that noted in the Mutant OPN-R3 and No treatment groups (P < 0.01 vs. Mutant OPN-R3 and No treatment). At day 30, Modified OPN-R3 aptamer-treated group tumor was eightfold less than that of the Mutant OPN-R3 and the No treatment groups (P < 0.01 vs. Mutant OPN-R3 and No treatment). At 8 weeks, necropsy tissue from lung and primary tumor locations were examined for bioluminescence in a site for potential metastases and in the primary location (Figure 5c). In lung tissue, the measured bioluminescence in the Modified OPN-R3 group was <1% of that noted in the Mutant OPN-R3 and No treatment groups (P < 0.01 vs. Mutant OPN-R3 and No treatment). These data indicate that Modified OPN-R3 aptamer can significantly decrease both local tumor growth and distant metastases of MDA-MB231 cells in this xenograft model.
Figure 5.
In vivo OPN-R3 activity in a xenograft tumor model. (a) Mean bioluminescence of MDA-MB231 cells at the primary tumor site. Bioluminescence is reported as the sum of detected photons per second from a constant region of interest (photons/second/region of interest). Photos are representative of four animals in each group. *P < 0.01 day 20 Modified OPN-R3 vs. Mutant OPN-R3 and No treatment; **P < 0.01 day 30 Modified OPN-R3 vs. Mutant OPN-R3 and No treatment. (b) Volume of the primary tumors. Tumor volume (V) is calculated using the following formula: V = (1/2) S2 × L (S, the shortest dimension; L, the longest dimension). All data are presented as mean ± S.D (n = 4 per treatment group). *P < 0.01 Modified OPN-R3 vs. Mutant OPN-R3 and No treatment. (c) Mean bioluminescence of MDA-MB231 cells metastatic to the lung. Photos are representative of four animals in each group. *P < 0.01 Lung-Modified OPN-R3 vs. Mutant OPN-R3 and No treatment; **P < 0.01 Primary tumor–Modified OPN-R3 vs. Mutant OPN-R3 and No treatment.
Discussion
RNA aptamers represent a unique emerging class of therapeutic agents.15,16 They are relatively short (12–30 nt) ss RNA oligonucleotides that assume a stable three-dimensional shape to tightly and specifically bind selected protein targets to elicit a biological response. In contrast to antisense oligonucleotides, RNA aptamers can effectively target extracellular targets, such as OPN. Like antibodies, aptamers possess binding affinities in the low nanomolar to picomolar range. In addition, aptamers are heat stable, lack immunogenicity, and possess minimal interbatch variability. Chemical modifications, such as amino or fluoro substitutions at the 2′ position of pyrimidines, may reduce degradation by nucleases. The biodistribution and clearance of aptamers can also be altered by chemical addition of moieties such as polyethylene glycol and cholesterol. Furthermore, SELEX allows selection from libraries consisting of up to 1015 ligands to generate high-affinity oligonucleotide ligands to purified biochemical targets, such as OPN. Recently, the aptamer, pegaptanib, was approved for the treatment of age-related macular degeneration.24 With regard to the field of oncology, the DNA aptamer GBI-10, derived from a human glioblastoma cell line, was recently demonstrated to bind tenascin-C.25 Similarly, RNA aptamers have been demonstrated to target the Ku DNA repair proteins with resulting sensitization of breast cancer cells to etoposide.26 Our results suggest that RNA aptamer targeting of OPN may serve as an effective strategy for inhibiting OPN-dependent metastatic behavior.
OPN was initially characterized in 1979 as a phosphoprotein secreted by transformed, malignant epithelial cells.27 It is a member of the small integrin-binding ligand N-linked glycoprotein (SIBLING) family of proteins which include bone sialoprotein, dentin matrix protein 1, dentin sialoprotein, and matrix extracellular phosphoglycoprotein.28 The molecular structure of OPN is rich in aspartate and sialic acid residues and contains unique functional domains which mediate critical cell–matrix and cell–cell signaling through the αvβ3 integrin and CD44 receptors in a variety of normal and pathologic processes. Integrin αvβ3 is detected consistently in breast cancer bone metastases and αvβ3 contributes to metastatic behavior in several ways.29 OPN-integrin binding directly mediates migration and invasion of tumor cells, enhances endothelial cell migration, survival, and lumen formation during angiogenesis, represents a downstream target for vascular endothelial growth factor signaling results in microvascular endothelial cells with a direct role in angiogenesis, activates osteoclasts in lytic bone metastases, and alters host immunity by increasing IL-12 expression in murine macrophages and IFN expression in natural killer cells.30,31 CD44 variants, especially CD44v6, have been identified as protein markers for metastatic behavior in hepatocellular, breast, lung, pancreatic, colorectal, and gastric cancers and in lymphomas.32,33 OPN can interact specifically with CD44v6 and/or v7.32,33,34 CD44v7-10 ligation of OPN mediates chemotaxis and adhesion of fibroblasts, T cells, and bone marrow cells, downregulates the host-inflammatory response in a IL-10 mediated manner, and confers metastatic potential when overexpressed through plasmid vectors models of pancreatic cancer. Recently, we have shown that binding of OPN with αvβ3 integrin upregulates plasma membrane expression of CD44v6 and augments in vitro adhesion of HepG2 hepatocellular carcinoma cells.34 Lin et al. have demonstrated that increased survival and growth of IL-3-dependent mouse bone marrow cells is mediated by OPN, and CD44 antibody attenuates these effects.35 Studies also suggest that OPN and CD44 interact with the ezrin, radixin, and moesin proteins to alter cytoskeletal dynamics, cell adhesion, and motility through the cortical actin filaments.36,37 In this study using FRET-confocal microscopy, we demonstrate that OPN-R3 aptamer effectively eliminates OPN-cell surface binding to MDA-MB231 human breast cancer cells.
A critical component of tumorigenesis and metastasis is the degradation of the basement membrane and interstitial matrix by MMPs and uPA, as part of the plasminogen activator–plasmin system.19,20,21,22 MMPs are extracellular matrix-degrading enzymes that play a crucial role in embryogenesis, tissue remodeling, inflammation, and angiogenesis. MMP2 and MMP9 are important contributors to the process of invasion, tumor growth, and metastasis. uPA, its receptor uPAR and inhibitors PAI-1 and PAI-2, which together constitute the uPA system, play a vital role in not only cancer progression but also in several normal physiological process such as wound healing, liver regeneration, and homeostasis.21,22 High levels of uPA are associated with cancers of the lung, skin, breast, bladder, uterine cervix, and soft tissue sarcoma.22 Studies indicate that blocking of uPA activity or the uPA–uPAR interaction drastically downregulates tumor growth and metastasis.38,39 OPN appears to regulate the activity of at least two ECM-degrading proteins. Philip et al. demonstrate that OPN upregulates pro-MMP2 expression in a NF-kB-dependent fashion during extracellular matrix invasion.40 Transfection of IκBα abrogates OPN-induced MMP expression while MMP2 antisense olignonucleotides reduces OPN-mediated migration and ECM invasion in B16F10 murine melanoma cells. We have recently described a novel function of the thrombin-cleaved COOH-terminal fragment of OPN.18 This fragment binds cyclophilin C to the CD147 cell surface receptor to activate Akt1/2 and MMP2 to enhance matricellular proteolysis. OPN also increases cell invasiveness in human mammary carcinoma through stimulation of uPA.41 The uPA system is elevated in breast cancer patients with poor prognosis, in malignant cancers and in bone metastases.39,42 Das et al. have confirmed that OPN induction of uPA depends on PI 3′-kinase/Akt activity.8,21 We have demonstrated that OPN upregulates uPA and MMP2 activity through integrin-linked kinase and AP-1 signaling during tumor cell invasion.39
With regard to OPN signaling, our studies also address the effect of OPN-R3 ablation of OPN-cell surface binding on αvβ3 integrin and CD44 receptor–mediated signal transduction. We examine the effect of OPN-R3 on PI3K, JNK1/2, Src and Akt expression and/or activation. While clearly overlapping lines of activation exist between αvβ3 and CD44 pathways, OPN-R3 can significantly decrease activity and/or expression of all four proteins. Kundu's group has demonstrated that OPN induces PI3K activity and PI3K-dependent Akt phosphorylation through the αvβ3 integrin–mediated pathway in breast cancer cells.8,43 In addition, overexpression of PTEN, a phosphatase that can antagonize PI3K signaling, suppresses OPN-induced Akt activation during osteoclast differentiation and cell motility.31 OPN–CD44 interactions promote cell survival and motility through activation of PI3K-dependent pathways. OPN also stimulates Src-dependent AP-1 activation, regulates negative crosstalk between NIK/ERK and MEKK1/JNK1 pathways, and activates the mitogen-activated protein kinase pathway.31 All of these elements contribute to cancer cell motility, invasion, tumor growth, and metastasis.
We tested OPN-R3 on MDA-MB231 in vitro behavior in Matrigel-based adhesion, migration, and invasion assays. Matrigel is a solubilized tissue basement membrane matrix rich in extracellular matrix proteins.44 Although composed mainly of laminin, collagen IV, heparan sulfate, proteoglycans, and entactin, various growth factors such as transforming growth factor-β, fibroblast growth factor, and tissue plasminogen activator are also present. Under normal physiological conditions, Matrigel polymerizes to produce a reconstituted, biologically active stable matrix that is effective for the attachment of normal and transformed anchorage-dependent cell types. In this regard, OPN-R3 effectively inhibits MDA adhesion, migration, and invasion by 60, 50, and 65%, respectively. Finally, we utilized the xenograft model of MDA-MB231 breast cancer cells to demonstrate in vivo inhibition of tumor growth by Modified OPN-R3, as measured by bioluminescence and tumor volumes.
In conclusion, these results suggest that OPN-R3 aptamer may have potential as an agent to inhibit cancer growth and metastasis. Our studies indicate that RNA aptamer technology may have biologically relevant applicability in targeting OPN and modifying tumor growth and metastasis.
Materials and Methods
Materials. 2-F-RNA aptamers were used in SELEX, gel shift, and Kd measures. Synthesized 2-O-Me OPN-R3 and its mutant OPN-R3-2 with cholesterol and IDT modification were used in FRET, cell treatment, animal studies, and half-life measurements. Theoretical structure models were determined by the mFold program at http://www.idtdna.com/Scitools/Applications/mFold/.
RNA aptamer and SELEX. The SELEX selection procedure was described previously.45,46 Briefly, a random pool of RNA oligonucleotides was generated by in vitro transcription of synthetic DNA templates following the instructions in the DuraScribe T7 Transcription Kit (Epicentre Biotech, Madison, WI). 2-Fluorine-dCTP, 2-Fluorine-dUTP, normal GTP and ATP are efficiently incorporated into RNA transcripts through the DuraScribe T7 RNA polymerase. Nitrocellulose 0.45-µmol/l filter (Schleicher & Schuell, Keene N.H.) was used to perform a negative selection in phosphate buffered saline (PBS) buffer (137 nmol/l NaCl, 27 nmol/l KCl, 100 nmol/l Na2HPO4, 2 nmol/l K2HPO4, pH 7.4) at 37 °C to remove any filter-binding RNA aptamers from the aptamer library before SELEX selection. The binding reaction was setup by 5 µmol/l OPN protein incubated with 50 µmol/l RNA aptamer pool in PBS buffer at 37 °C for 4 hours rotation and was then loaded onto a Milliblot vacuum apparatus (Millipore, Billerica, MA) filled with 0.45-µmol/l nitrocellulose filter. After flow through of the binding reaction solution, the fragmented nitrocellulose filter was soaked in 1 ml PBS solution and extracted with phenol/chloroform (25:24). The aptamer RNA was precipitated at 15,000g for 30 min at 4 °C from the supernatant solution containing 5 mol/l ammonium acetate, 20 µg/ml linear acrylamide, and three volumes of 100% ethanol. This recovered RNA was followed by the RT-PCR and in vitro transcription for the next round of SELEX selection or binding affinity assays to measure its Kd value. After each round of SELEX, we performed a binding affinity assay to measure the aptamer pool's Kd value to ensure that the Kd values exhibited a decreasing trend.
We applied SELEX by alternating the bait protein between human OPN and mouse OPN in order to achieve RNA aptamer targeting to common features of both proteins. Selection after Round 8 through 11 was followed by ligation of 0.5 µg of the dsDNA pool into PGEM-T vector systems (Promega, Madison, WI) for sequencing. The DNA sequence used for in vitro transcription was 5′-GGGGGAATTCTAATACGACTCACTATAGGGAGGACGATGCGG- N40-CAGACGACTCGCTGAGGATCCGAGA-3′, where N40 represents the 40-nt RNA aptamer library sequence. The sequences for the aptamers are as follows: OPN-R3: 5′-CGGCCACAGAAUGAAAAACCUCAUCGAUGUUGCAUAGUUG-3′; Mutant OPN-R3: 5′-CGGCCACAGAA UGAAUCAUCGAUGUUGCAUAGUUG-3′, where C denotes 2-F-dCTP or 2-OMe-dCTP and U denotes 2-F-dUTP or 2-OMe-dUTP, as appropriate. This negative control mutant OPN-R3 is identical to OPN-R3-2. The randomized sequence length in our RNA aptamer pool was ~40 nt. The in vitro–transcribed RNA aptamer length was 80 nt. This 80-nt RNA aptamer was used for SELEX. We measured the binding affinity of this 80-nt OPN-R3 aptamer, commercially synthesized 40-nt OPN-R3, 32-nt OPN-R3, and OPN-R3 mutants (the mutation region was based on its mFOLD secondary structure). We found the Kd value to be the same among 80-nt OPN-R3, 40-nt OPN-R3, and 32-nt OPN-R3. The commercially synthesized OPN-R3 aptamers contain 2′-OMe C, 2′-OMe U, A, and G and were used for gel shift assay as well as in vitro cell treatment. Subsequent studies were performed using this commercially synthesized OPN-R3 (Dharmacon, Lafayette, IN) During SELEX, DuraScribe T7 RNA polymerase is preferred for incorporation of 2′-F-modified pyrimidine into nucleotide; it does not utilize 2′- OMe-modified pyrimidine for in vitro transcription reaction. Subsequent studies used the 2′-OMe modification instead of 2′-F modification.
Binding affinity assays. RNA–protein equilibrium dissociation constants (Kd) were determined by the double-filter nitrocellulose filter-binding method.17 For all binding assays, RNAs were dephosphorylated using bacterial alkaline phosphatase (Invitrogen, Carlsbad, CA) and 5′-end labeled using T4 polynucleotide kinase (New England Biolabs, Beverly, MA) and γ-32P-ATP (MP Biomedicals, Solon, OH). Direct binding assays were carried out by incubating 32P-labeled RNA at a concentration of <0.1 nmol/l and target protein at concentrations ranging from 300 to 10 nmol/l in selection buffer at 37 °C. The fraction of RNA bound was quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Raw binding data were corrected for nonspecific background binding of radiolabeled RNA to the nitrocellulose filter (see Supplementary Materials and Methods).
Transient transfection. Cells were transiently transfected using Lipofectamine 2000 according to manufacturer's instruction (Invitrogen, Frederick, MD). Briefly, 4 × 105 cells were seeded with antibiotic-free Dulbecco's modified Eagle's medium on each well of 12-well plates the day before transfection. Two micrograms of plasmid DNA and 4 µl Lipofectamine 2000, diluted with Opti-MEM medium, were mixed gently and incubated with cells. Culture medium was changed after 6-h transfection and incubated further at 37 °C for 24 h. The control cells received Lipofectamine 2000 alone.
REMSAs. REMSAs were conducted in buffers containing the protease inhibitors, and dithiothreitol (1 nmol/l) was freshly prepared. Recombinant human OPN (100 nmol/l) was dissolved in ice-cold buffer C containing 20 nmol/l HEPES, pH 7.9, 0.4 mol/l NaCl, 1.0 nmol/l EDTA, 1.0 nmol/l EGTA, 1.0 nmol/l dithiothreitol, pepstatin A (2 µg/ml), and 0.5 nmol/l phenylmethylsulfonyl fluoride, aliquoted and stored at −80 °C until use. OPN-R3 and mutant OPN-R3 RNA aptamers were synthesized and then end-labeled with [γ-32P] ATP (2,500 Ci/mmol) using T4 polynucleotide kinase (Promega), followed by G-50 column purification. Recombinant human OPN (1 nmol/l) and 10 nmol/l P32 end- labeled OPN-R3 or mutant OPN-R3 aptamer and/or 1 µg Ab were incubated in PBS (pH 7.4) solution at 37 °C for 30 min. The reactions were resolved on 6% native acrylamide gel in 0.5× Tris-borate/EDTA buffer and visualized by autoradiography. In specific competitive binding assays, unlabeled OPN-R3 type aptamers were added at a 20-fold molar excess. In nonspecific competitive binding assays, unlabeled mutant OPN-R3 aptamers were used. Antibody competition assays were performed by preincubating recombinant human OPN with rabbit anti-human OPN polyclonal Ab (Santa Cruz Biotechnology, Santa Cruz, CA).
Confocal microscopy and FRET acceptor bleaching assay. Cells were cultured on coverslips and then co-transfected with pECFP-CD44 FRET donor and pEYFP-OPN FRET acceptor plasmids. At 16 h after transfection, cells were exposed to 100 nmol/l aptamer and/or 2 µg Ab for 8 hours. After 24 h post transfection, the coverslips were rinsed three times with ice-cold PBS followed by fixation for 15 min with 1% (wt/vol) paraformaldehyde. Coverslips were rinsed three times with PBS and mounted onto a microscope slide using 50 µl mounting medium (Calbiochem, San Diego, CA). The coverslips were sealed using wax and kept at 4 °C until analysis. Leica TCS SP2 confocal microscope was used for image acquisition. CFP and YFP emission spectra were first optimized at 458 nm and 514 nm, respectively. FRET was measured by acceptor photobleaching using the FRET-AB wizard in the Leica TS software. A pair of prebleach images of CFP and YFP images was collected for the cells of interest. Randomly chosen regions of interest were irradiated (bleached) with the 514-nm laser line set at 100% intensity to photobleach YFP only for the minimum number of iterations of bleaching required. Post-bleach CFP and YFP images were collected following photobleaching. FRET was indicated by an increase in CFP donor fluorescence intensity following YFP photobleaching. FRET efficiency was calculated as 100 × [(Donor post-bleach ‐ Donor prebleach)/Donor post-bleach], taking into account CFP and YFP background noise in each channel; FRET efficiency was measured and calculated automatically by Leica LAS AF software.
In vivo OPN-R3 activity. Animal handling and procedures were approved by the Duke University Animal Care and Use Committee. 6-week-old female NOD scid mice were obtained from the Jackson Laboratory, Bar Harbor, Maine. 1 × 106 MDA-MB-231-luciferase-expressing cells (a gift of Mark Dewhirst, Duke University, NC) were suspended in 50% Matrigel-Hanks balanced salt solution and implanted into the R4 or L4 positions of the mice mammary fat pad (four per group). Modified OPN-R3 and Mutant OPN-R3 (500 µg/kg) were injected into the mouse tail vein every 2 days following cell implantation. Mice were anesthetized with intraperitoneal ketamine (75 mg/kg) and xylazine (10 mg/kg). For bioluminescent imaging, animals are placed in a light-tight chamber in which grayscale reference images were obtained under dim conditions. A pseudocolor image acquired in the dark will be superimposed on the grayscale image to represent photons emitted from tumors. Bioluminescence is reported as the sum of detected photons per second from a constant region of interest (photons/second/region of interest). Ten minutes after administration of luciferase substrates (D-luciferin, 150 mg/kg), anesthetized mice were imaged with the IVIS 100 Imaging System (Xenogen, Alameda, CA) following the company's manual. Initial in vivo images at day 2 were obtained to establish baseline tumor volume as measured by photon emission. For ex vivo imaging, after 8 weeks of Modified OPN R3 or Mutant OPN R3 treatment, D-luciferin (150 mg/kg) was injected into the mice before necropsy. Lung lobes were excised, weighed, placed into tissue culture plates with D-luciferin (300 µg/ml) in PBS, and imaged. The mean bioluminescence was quantified and analyzed using Living Image software (Xenogen). Bioluminescence from ROI was defined manually. Bioluminescence imaging was performed on all of the mice twice per week. The volume of the primary tumors will also be quantified with caliper measurements in two dimensions and tumor volume (V) calculated using the following formula: V = (1/2) S2 × L (S, the shortest dimension; L, the longest dimension).
Statistical analysis. All data are presented as mean ± SD. Analysis was performed using a Student's t-test. Values of P < 0.05 were considered significant.
Supplementary MaterialMaterials and Methods.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01AI44629 (P.C.K.), R21DK070642 (P.C.K.), R01GM65113 (P.C.K.), and HL65222 (B.A.S.)
REFERENCES
- Denhardt DT, Lopez CA, Rollo EE, Hwang SM, An XR., and , Walther SE. Osteopontin-induced modifications of cellular functions. Ann N Y Acad Sci. 1995;760:127–142. doi: 10.1111/j.1749-6632.1995.tb44625.x. [DOI] [PubMed] [Google Scholar]
- Denhardt DT, Giachelli CM., and , Rittling SR. Role of osteopontin in cellular signaling and toxicant injury. Annu Rev Pharmacol Toxicol. 2001;41:723–749. doi: 10.1146/annurev.pharmtox.41.1.723. [DOI] [PubMed] [Google Scholar]
- Weber GF, Ashkar S., and , Cantor H. Interaction between CD44 and osteopontin as a potential basis for metastasis formation. Proc Assoc Am Physicians. 1997;109:1–9. [PubMed] [Google Scholar]
- Denhardt DT., and , Chambers AF. Overcoming obstacles to metastasis-defenses against host defences: osteopontin as a chield against attack by sytotoxic host cells. J Cell Biochem. 1994;56:48–51. doi: 10.1002/jcb.240560109. [DOI] [PubMed] [Google Scholar]
- Brown LF, Papadopoulos-Sergiou A, Berse B, Manseau EJ, Tognazzi K, Perruzzi CA, et al. Osteopontin expression and distribution in human carcinomas. Am J Pathol. 1994;145:610–623. [PMC free article] [PubMed] [Google Scholar]
- Agrawal D, Chen T, Irby R, Quackenbush J, Chambers AF, Szabo M, et al. Osteopontin identified as lead marker of colon cancer progression, using pooled sample expression profiling. J Natl Cancer Inst. 2002;94:513–521. doi: 10.1093/jnci/94.7.513. [DOI] [PubMed] [Google Scholar]
- Coppola D, Szabo M, Boulware D, Muraca P, Alsarraj M, Chambers AF, et al. Correlation of osteopontin protein expression and pathological stage across a wide variety of tumor histologies. Clin Cancer Res. 2004;10:184–190. doi: 10.1158/1078-0432.ccr-1405-2. [DOI] [PubMed] [Google Scholar]
- Das R, Mahabeleshwar GH., and , Kundu GC. Osteopontin stimulates cell motility and nuclear factor kappaB-mediated secretion of urokinase type plasminogen activator through phosphatidylinositol 3-kinase/Akt signaling pathways in breast cancer cells. J Biol Chem. 2003;278:28593–28606. doi: 10.1074/jbc.M303445200. [DOI] [PubMed] [Google Scholar]
- Fedarko NS, Jain A, Karadag A, Van Eman MR., and , Fisher LW. Elevated serum bone sialoprotein and osteopontin in colon, breast, prostate, and lung cancer. Clin Cancer Res. 2001;7:4060–4066. [PubMed] [Google Scholar]
- Gotoh M, Sakamoto M, Kanetaka K, Chuuma M., and , Hirohashi S. Overexpression of osteopontin in hepatocellular carcinoma. Pathol Int. 2002;52:19–24. doi: 10.1046/j.1440-1827.2002.01316.x. [DOI] [PubMed] [Google Scholar]
- Grano M, Mori G, Minielli V, Colucci S, Vaira S, Giannelli G, et al. HGF and M-CSF modulate adhesion of MDA-231 breast cancer cell by increasing osteopontin secretion. J Biol Regul Homeost Agents. 2002;16:190–195. [PubMed] [Google Scholar]
- Tuck AB, Chambers AF., and , Allan AL. Osteopontin overexpression in breast cancer: knowledge gained and possible implications for clinical management. J Cell Biochem. 2007;102:859–868. doi: 10.1002/jcb.21520. [DOI] [PubMed] [Google Scholar]
- Wai PY., and , Kuo PC. The role of osteopontin in tumor metastasis. J Surg Res. 2004;121:228–241. doi: 10.1016/j.jss.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Mazar AP, Henkin J., and , Goldfarb RH. The urokinase plasminogen activator system in cancer: implications for tumor angiogenesis and metastasis. Angiogenesis. 1999;3:15–32. doi: 10.1023/a:1009095825561. [DOI] [PubMed] [Google Scholar]
- Que-Gewirth NS., and , Sullenger BA. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14:283–291. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- Ireson CR., and , Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- Gopinath SC. Methods developed for SELEX. Anal Bioanal Chem. 2007;387:171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- Mi Z, Oliver T, Guo H, Gao C., and , Kuo PC. Thrombin-cleaved COOH(-) terminal osteopontin peptide binds with cyclophilin C to CD147 in murine breast cancer cells. Cancer Res. 2007;67:4088–4097. doi: 10.1158/0008-5472.CAN-06-4066. [DOI] [PubMed] [Google Scholar]
- Deryugina EI., and , Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- Overall CM., and , Kleifeld O. Tumour microenvironment – opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- Das R, Philip S, Mahabeleshwar GH, Bulbule A., and , Kundu GC. Osteopontin: it's role in regulation of cell motility and nuclear factor kappa B-mediated urokinase type plasminogen activator expression. IUBMB Life. 2005;57:441–447. doi: 10.1080/15216540500159424. [DOI] [PubMed] [Google Scholar]
- Pillay V, Dass CR., and , Choong PF. The urokinase plasminogen activator receptor as a gene therapy target for cancer. Trends Biotechnol. 2007;25:33–39. doi: 10.1016/j.tibtech.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Kelly P, Moeller BJ, Juneja J, Booden MA, Der CJ, Daaka Y, et al. The G12 family of heterotrimeric G proteins promotes breast cancer invasion and metastasis. Proc Natl Acad Sci USA. 2006;103:8173–8178. doi: 10.1073/pnas.0510254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TY, Liew G., and , Mitchell P. Clinical update: new treatments for age-related macular degeneration. Lancet. 2007;370:204–206. doi: 10.1016/S0140-6736(07)61104-0. [DOI] [PubMed] [Google Scholar]
- Daniels DA, Chen H, Hicke BJ, Swiderek KM., and , Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc Natl Acad Sci USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yoo S, Dritschilo A, Belyaev I., and , Soldatenkov V. Targeting Ku protein for sensitizing of breast cancer cells to DNA-damage. Int J Mol Med. 2004;14:153–159. [PubMed] [Google Scholar]
- Senger DR, Wirth DF., and , Hynes RO. Transformed mammalian cells secrete specific proteins and phosphoproteins. Cell. 1979;16:885–893. doi: 10.1016/0092-8674(79)90103-x. [DOI] [PubMed] [Google Scholar]
- Fedarko NS, Jain A, Karadag A., and , Fisher LW. Three small integrin binding ligand N-linked glycoproteins (SIBLINGs) bind and activate specific matrix metalloproteinases. FASEB J. 2004;18:734–736. doi: 10.1096/fj.03-0966fje. [DOI] [PubMed] [Google Scholar]
- Liapis H, Flath A., and , Kitazawa S. Integrin alpha V beta 3 expression by bone-residing breast cancer metastases. Diagn Mol Pathol. 1996;5:127–135. doi: 10.1097/00019606-199606000-00008. [DOI] [PubMed] [Google Scholar]
- Wai PY, Mi Z, Guo H, Sarraf-Yazdi S, Gao C, Wei J, et al. Osteopontin silencing by small interfering RNA suppresses in vitro and in vivo CT26 murine colon adenocarcinoma metastasis. Carcinogenesis. 2005;26:741–751. doi: 10.1093/carcin/bgi027. [DOI] [PubMed] [Google Scholar]
- Rangaswami H, Bulbule A., and , Kundu GC. Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol. 2006;16:79–87. doi: 10.1016/j.tcb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Goodison S, Urquidi V., and , Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189–196. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponta H, Sherman L., and , Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- Gao C, Guo H, Downey L, Marroquin C, Wei J., and , Kuo PC. Osteopontin-dependent CD44v6 expression and cell adhesion in HepG2 cells. Carcinogenesis. 2003;24:1871–1878. doi: 10.1093/carcin/bgg139. [DOI] [PubMed] [Google Scholar]
- Lin YH, Huang CJ, Chao JR, Chen ST, Lee SF, Yen JJ, et al. Coupling of osteopontin and its cell surface receptor CD44 to the cell survival response elicited by interleukin-3 or granulocyte-macrophage colony-stimulating factor. Mol Cell Biol. 2000;20:2734–2742. doi: 10.1128/mcb.20.8.2734-2742.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar R, Suzuki N, Suzuki K, Arora P, Glogauer M, McCulloch CA, et al. Intracellular osteopontin is an integral component of the CD44-ERM complex involved in cell migration. J Cell Physiol. 2000;184:118–130. doi: 10.1002/(SICI)1097-4652(200007)184:1<118::AID-JCP13>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Zohar R, Cheifetz S, McCulloch CA., and , Sodek J.Analysis of intracellular osteopontin as a marker of osteoblastic cell differentiation and mesenchymal cell migration Eur J Oral Sci 1998106401–407.Suppl 1 [DOI] [PubMed] [Google Scholar]
- Bauer TW, Liu W, Fan F, Camp ER, Yang A, Somcio RJ, et al. Targeting of urokinase plasminogen activator receptor in human pancreatic carcinoma cells inhibits c-Met- and insulin-like growth factor-I receptor-mediated migration and invasion and orthotopic tumor growth in mice. Cancer Res. 2005;65:7775–7781. doi: 10.1158/0008-5472.CAN-05-0946. [DOI] [PubMed] [Google Scholar]
- Mi Z, Guo H, Wai PY, Gao C., and , Kuo PC. Integrin-linked kinase regulates osteopontin-dependent MMP-2 and uPA expression to convey metastatic function in murine mammary epithelial cancer cells. Carcinogenesis. 2006;27:1134–1145. doi: 10.1093/carcin/bgi352. [DOI] [PubMed] [Google Scholar]
- Philip S., and , Kundu GC. Osteopontin induces nuclear factor kappa B-mediated promatrix metalloproteinase-2 activation through I kappa B alpha /IKK signaling pathways, and curcumin (diferulolylmethane) down-regulates these pathways. J Biol Chem. 2003;278:14487–14497. doi: 10.1074/jbc.M207309200. [DOI] [PubMed] [Google Scholar]
- Tuck AB, Hota C., and , Chambers AF. Osteopontin(OPN)-induced increase in human mammary epithelial cell invasiveness is urokinase (uPA)-dependent. Breast Cancer Res Treat. 2001;70:197–204. doi: 10.1023/a:1013095329825. [DOI] [PubMed] [Google Scholar]
- Andreasen PA, Kjoller L, Christensen L., and , Duffy MJ. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72:1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Das R, Mahabeleshwar GH., and , Kundu GC. Osteopontin induces AP-1-mediated secretion of urokinase-type plasminogen activator through c-Src-dependent epidermal growth factor receptor transactivation in breast cancer cells. J Biol Chem. 2004;279:11051–11064. doi: 10.1074/jbc.M310256200. [DOI] [PubMed] [Google Scholar]
- Albini A. Tumor and endothelial cell invasion of basement membranes. The matrigel chemoinvasion assay as a tool for dissecting molecular mechanisms. Pathol Oncol Res. 1998;4:230–241. doi: 10.1007/BF02905254. [DOI] [PubMed] [Google Scholar]
- Lee SW., and , Sullenger BA. Isolation of a nuclease-resistant decoy RNA that selectively blocks autoantibody binding to insulin receptors on human lymphocytes. J Exp Med. 1996;184:315–324. doi: 10.1084/jem.184.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhaveri SD., and , Ellington AD. In vitro selection of RNA aptamers to a protein target by filter immobilization. Curr Protoc Mol Biol. 2001;Chapter 24:Unit 24.3. doi: 10.1002/0471142727.mb2403s52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.