Abstract
Pathways controlling cell proliferation and cell survival require flexible adaptation to environmental stresses. These mechanisms are frequently exploited in cancer, allowing tumor cells to thrive in unfavorable milieus. Here, we show that Hsp90, a molecular chaperone that is central to the cellular stress response, associates with survivin, an apoptosis inhibitor and essential regulator of mitosis. This interaction involves the ATPase domain of Hsp90 and the survivin baculovirus inhibitor of apoptosis repeat. Global suppression of the Hsp90 chaperone function or targeted Abmediated disruption of the survivin–Hsp90 complex results in proteasomal degradation of survivin, mitochondrial-dependent apoptosis, and cell cycle arrest with mitotic defects. These data link the cellular stress response to an antiapoptotic and mitotic checkpoint maintained by survivin. Targeting the survivin–Hsp90 complex may provide a rational approach for cancer therapy.
Tumor cells exhibit extraordinary plasticity to adapt to noxious stimuli and thrive in unfavorable environments. This typically involves increased resistance to apoptosis (1) by deregulated overexpression of cell death antagonists of the Bcl-2 (2) or inhibitor of apoptosis (IAP) gene family (3) or loss of cell death activators/effectors (4, 5). Another hallmark of cancer that promotes enhanced adaptation to environmental challenges is a constitutive up-regulation of the cellular stress response. This preserves folding of nascent polypeptides (6), prevents protein aggregation (7), and ensures specialized intracellular trafficking of client proteins (8). The protein folding quality control machinery is orchestrated by heat shock proteins (Hsps), a family of evolutionary conserved ATPase-directed molecular chaperones (9). In particular, Hsp90 (10, 11) controls the balance between folding/maturation (12) and proteasomal destruction (13) of a restricted number of client proteins (14) that are typically involved in signal transduction and cell proliferation (15). This pathway is exploited in cancer where Hsp90 is up-regulated (16) and may be linked to resistance to apoptosis (17) by inhibition of caspase-9 activation (18), induction of antiapoptotic Bcl-2 (19), or stabilization of survival kinases RIP-1 (20) or Akt (21). In this context, Hsp90 antagonists are being explored as novel cancer therapeutics (22).
Here, we show that Hsp90 associates with survivin (23), an IAP family protein (3) overexpressed in nearly every human tumor, and with essential roles in mitotic control and apoptosis inhibition (23). Disruption of the survivin–Hsp90 interaction destabilizes survivin, initiates mitochondrial apoptosis, and suppresses cell proliferation, suggesting its potential suitability for cancer therapies.
Materials and Methods
Abs. Abs to survivin were from Novus Biologicals (Littleton, CO) (24). Abs to Hsp90 or β-tubulin were from Transduction Laboratories (Lexington, KY) and Sigma, respectively. Abs to hemagglutinin or FLAG were from Roche and Sigma, respectively. An Ab to caspase-9 was from Cell Signaling Technology (Beverly, MA).
Cell Lines and Constructs. Cervical carcinoma HeLa or B lymphoblastoid Raji cells were from the American Type Culture Collection. A YUSAC-2 melanoma cell line stably transfected to express survivin on withdrawal of tetracycline (Tet) was described (25). WT or Apaf-1–/– mouse embryonic fibroblasts (MEF) were described (26). Hsp90α full-length (1–732) or the three fragments N (1–272), M (273–617), or C (629–732) were cloned by PCR in pGex-4T3 (Amersham Biosciences) and pFLAG-cytomegalovirus 6c (Sigma). A survivin Cys-84→Ala mutant [survivin(C84A)] was described (27). Recombinant proteins were expressed in Escherichia coli and purified as described (24). A CD11b integrin I domain was used as a control.
Affinity Chromatography, Coimmunoprecipitation, and Western Blotting. HeLa cell (1.5 × 108) extracts were applied to 0.5 ml of CNBr-activated Sepharose 4B (Amersham Biosciences) coupled to 5 mg of a polyclonal Ab to survivin. The bound material was eluted in 0.1 M glycine (pH 2.5) and neutralized in 1 M Tris (pH 8.0), and fractions were analyzed by Western blotting. Immunoprecipitation experiments from Raji (5 × 105) or HeLa (2 × 105) cell extracts followed by Western blotting of the immune complexes were carried out as described (24). HeLa cells transfected with GFP-survivin or GFP-survivin(C84A) were treated with cycloheximide (100 μg/ml), harvested after 0–60 min, and analyzed by Western blotting with an Ab to GFP. For heat shock, HeLa cells (2.5 × 105) were submerged in a preheated water bath for1hat 42°C and analyzed by Western blotting or DNA content during an 11-h recovery period at 37°C. To inhibit the chaperone function of Hsp90, HeLa, MEF, or YUSAC-2 cells (25) were treated with geldanamycin (GA, 0–10 μM, Sigma) with or without 5 μM of the proteasome inhibitor, lactacystin (Calbiochem), and analyzed after 24 h by Western blotting or cell cycle progression. HeLa cells treated with GA were simultaneously analyzed for in vivo caspase activity (CaspaTag, Intergen, Purchase, NY) and plasma membrane integrity by propidium iodide staining and multiparametric flow cytometry. For in vivo pull-downs, HeLa cells treated with or without microtubulestabilizing taxol (Sigma, 2 μM for 16 h) were harvested and incubated with survivin-Sepharose (3 mg/ml), and bound proteins were analyzed by Western blotting.
Structure/Function of Survivin–Hsp90 Interaction. For in vitro pull-downs, 10 μg of GST or GST fusion proteins bound to glutathione beads (20 μl) were incubated with recombinant survivin (30, 100, and 300 ng) and washed, and pellets or supernatants were analyzed by Western blotting. ELISA was carried out as described (24) by using survivin or Hsp90 (10 μg/ml) immobilized on microtiter wells (Dynatech). For peptide competition, Hsp90 (2.5 mg/ml) was incubated with overlapping synthetic peptides (24) duplicating the survivin sequence 1–101 (20 μg/ ml) for 16 h at 4°C, before determination of binding to survivin, by ELISA. For Ab competition, survivin (300 ng) was incubated with mAbs 58 or 8E2 (24) or IgG (3 μg) for 1 h at 22°C before addition to GST-Hsp90 or GST-N Hsp90 and analysis of bound proteins by Western blotting.
Intracellular Targeting of Survivin–Hsp90 Interaction. HeLa cells (2 × 105) were loaded with 5 μg of mAbs 8E2 or 58 to survivin (24) or mouse IgG in OptiMEM (Life Technologies, Grand Island, NY), in the presence of BioPorter protein transfection reagent (Gene Therapy Systems, San Diego). After 4 h, cells were supplemented with 100 μg/ml cycloheximide and analyzed (0–90 min) for survivin expression by Western blotting. The efficiency of Ab loading (90%) was determined by using FITC-conjugated IgG (5 μg) and fluorescence microscopy. Transduced HeLa or YUSAC-2 cells with or without Tet were analyzed for caspase-9 processing by Western blotting or in vivo caspase activity by flow cytometry (CaspaTag, Intergen).
Immunofluorescence Analysis. HeLa cells loaded with mAbs 58 or 8E2 to survivin or IgG were stained with a mAb to β-tubulin and analyzed by fluorescence microscopy. The mitotic index was calculated by counting mitotic cells at ×400 magnification in at least five fields containing an average of 100 cells per field.
Statistical Analysis. Data were analyzed by using the paired t test on a prism software package for Windows (GraphPad, San Diego). A P value of 0.05 was considered statistically significant.
Results and Discussion
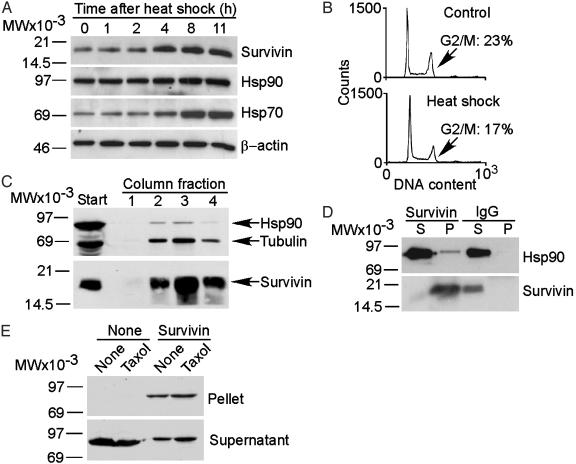
Identification of a Survivin–Hsp90 Interaction. We tested the effect of heat shock on expression levels of the IAP family (3) protein, survivin (23). A 42°C heat shock of HeLa cells caused time-dependent increase in survivin expression by Western blotting (Fig. 1A). This coincided with up-regulation of the inducible form of Hsp70, 4–11 h after heat shock, whereas Hsp90 only moderately increased, and β-actin levels remained unchanged (Fig. 1 A). Heat shock did not induce changes in cell cycle progression or G2/M arrest in HeLa cells (Fig. 1B), indicating that the induction of survivin under these conditions was independent of cell cycle transitions. To search for regulators of the survivin heat shock response, we coupled an Ab that recognizes the different subcellular pools of survivin (24) to Sepharose beads and fractionated HeLa cell extracts. A major band at 90 kDa coeluted with survivin and was identified as Hsp90 by Western blotting (Fig. 1C). Tubulin, a protein already known to interact with survivin (27), was eluted in the same fractions (Fig. 1C). Survivin immunoprecipitated from Raji cell extracts coassociated with Hsp90, by Western blotting, whereas immune complexes of a control IgG did not contain Hsp90 bands (Fig. 1D). In pull-down experiments, incubation of asynchronous or taxol-synchronized HeLa cell extracts with survivin-Sepharose resulted in coassociation of Hsp90 by Western blotting (Fig. 1E).
Fig. 1.
Survivin–Hsp90 interaction. (A) Heat shock. HeLa cells exposed to 42°C for 1 h were harvested at the indicated time intervals and analyzed by Western blotting. (B) Cell cycle analysis. HeLa cells were harvested 8 h after heat shock and analyzed for DNA content by flow cytometry. The percentage of G2/M cells is indicated. (C)Affinity purification. HeLa cell extracts were fractionated on a survivin-Sepharose column. Eluted proteins (arrows) were identified by Western blotting. (D) Coimmunoprecipitation. Raji cell extracts were immunoprecipitated with IgG or an Ab to survivin, and pellets (P) and supernatants (S) were analyzed by Western blotting. (E) In vivo pull-down. Asynchronous or taxol-treated HeLa cell extracts were incubated with Sepharose (None) or survivin-Sepharose (Survivin), and pellets or supernatants (25% of reaction) were analyzed for coassociated Hsp90 by Western blotting. MW, molecular weight.
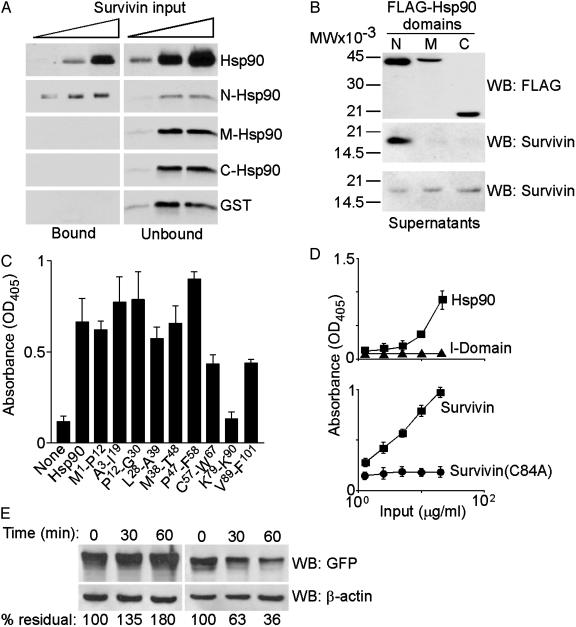
Structure/Function of Survivin–Hsp90 Interaction. To begin to identify the regions in Hsp90 that mediate binding to survivin, we expressed three regions of Hsp90 to encompass the NH2 terminus domain with the ATP-binding site (N) (12, 14), the middle domain (M), and the –COOH terminus domain (C) as GST fusion proteins and tested their association with survivin. Only full-length Hsp90 and its N domain bound recombinant survivin, whereas the M or C Hsp90 domains had no effect (Fig. 2A). Immunoprecipitation from HeLa cells transfected with the various FLAG-Hsp90 domains revealed that endogenous survivin coassociated with the N domain of Hsp90, but not with the M or C domains, in vivo (Fig. 2B). Comparable expression of the individual Hsp90 domains was confirmed by Western blotting of transfected HeLa cell extracts (Fig. 2B).
Fig. 2.
Structure/function of survivin–Hsp90 interaction. (A) In vitro pull-down. Survivin was incubated with Sepharose-GST, -GST-Hsp90, or the indicated -GST-Hsp90 N, M, or C domains, and protein binding was determined by Western blotting. (B) Immunoprecipitation. HeLa cells transfected with the indicated FLAG-Hsp90 domains were immunoprecipitated with a mAb to FLAG and analyzed by Western blotting. MW, molecular weight. (C) Peptide mapping. The indicated survivin peptides were used to compete the binding between Hsp90 and survivin by ELISA. (D) Folding requirement. Protein binding to immobilized survivin (Upper) or Hsp90 (Lower) was determined by ELISA. (E) In vivo turnover. HeLa cells transfected with GFP-survivin (Left) or GFP-survivin(C84A) (Right) were treated with cycloheximide, and protein turnover at the indicated time intervals was determined by Western blotting with an Ab to GFP.
To identify the complementary Hsp90-binding site(s) on survivin, we used partially overlapping synthetic peptides spanning the survivin sequence 1–101 (24) to compete the binding between survivin and Hsp90 in vitro. By ELISA, a Lys-79–Lys-90 survivin peptide completely abrogated the interaction between the two proteins, whereas none of the other survivin peptides was effective (Fig. 2C). The survivin Lys-79–Lys-90 peptide bound directly to Hsp90 with high affinity (Kd of 8.38 × 10–8 M), by surface plasmon resonance (data not shown). In the survivin crystal structure, the Lys-79–Lys-90 sequence connects the third core β strand of the conserved baculovirus IAP repeat to the α-4 and α-5 helices, and several of its residues (Lys-79, His-80, Lys-90, and Lys-91) form a surface-exposed “basic patch” in the survivin dimer (28).
Folding Requirement of Survivin–Hsp90 Interaction. Because of its role in protein folding quality control (12, 14), we asked whether Hsp90 bound mature or unfolded survivin. We expressed a survivin Cys-84→Ala mutant, which disrupts the Zn2+ coordination sphere in the baculovirus IAP repeat and generates an unfolded survivin molecule. By ELISA, WT survivin bound avidly to Hsp90 in a concentration-dependent manner, whereas control CD11b integrin I domain had no effect (Fig. 2D). In contrast, survivin(C84A) failed to associate with Hsp90 at all concentrations tested (Fig. 2D), suggesting that folding of survivin was required for Hsp90 binding. Next, we asked whether failure to bind Hsp90 correlated with reduced stability of survivin(C84A) in vivo. In cycloheximide block experiments, survivin(C84A) exhibited a 5-fold accelerated clearance compared with WT survivin (Fig. 2E), indicating that binding to Hsp90 may contribute to survivin stability in vivo.
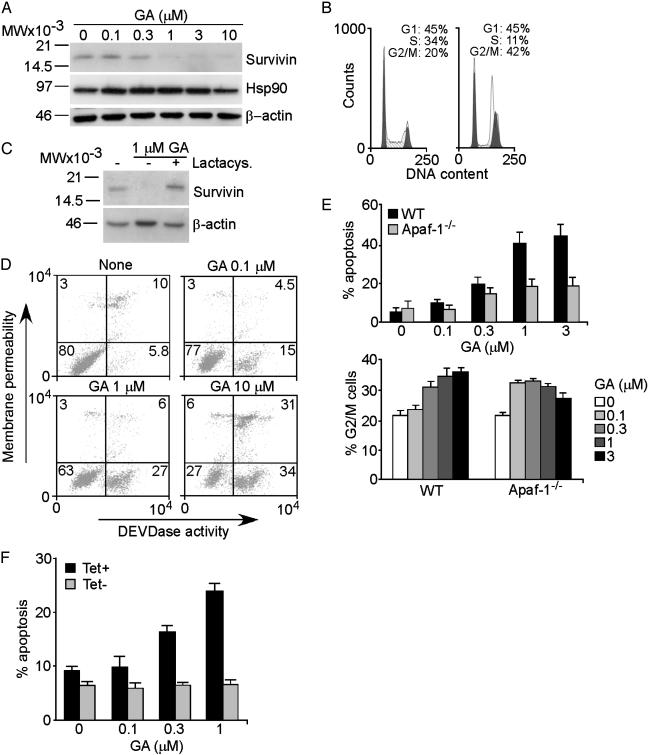
Regulation of Apoptosis and Cell Cycle by Survivin–Hsp90 Interaction. To test the chaperone function of Hsp90 in survivin binding, we used the Hsp90 inhibitor GA, which interferes with the ATPase cycles of the chaperone (29). Treatment of HeLa cells with GA resulted in concentration-dependent loss of survivin expression by Western blotting (Fig. 3A). In contrast, Hsp90 levels moderately increased in GA-treated cells, and β-actin was unchanged (Fig. 3A). The effect of GA on survivin levels was not due to cell cycle arrest before mitosis. Rather, GA induced a G2/M arrest in HeLa cells and no detectable G1 block (Fig. 3B), consistent with the inactivation of p53/Rb in these cells (30). Inhibition of the chaperone function of Hsp90 results in degradation of client proteins through the ubiquitin-proteasome pathway (13). Accordingly, addition of the proteasome inhibitor lactacystin prevented the loss of survivin induced by GA and resulted in increased survivin accumulation as compared with untreated cultures (Fig. 3C). Therefore, the chaperone function of Hsp90 is required for survivin stability in vivo and loss of survivin after Hsp90 inhibition involves proteasomal-dependent destruction.
Fig. 3.
Regulation of survivin stability by Hsp90. (A) Requirement for Hsp90 chaperone function. HeLa cells were treated with GA and analyzed after 24 h by Western blotting. MW, molecular weight. (B) Cell cycle analysis. Untreated (Left) or GA-treated (Right) HeLa cells were harvested after 24 h and analyzed for DNA content by flow cytometry. The percentage of cells at each cell cycle transition is indicated. Apoptotic cells were electronically excluded. (C) Proteasome inhibition. GA-treated HeLa cells with or without the proteasome inhibitor lactacystin were analyzed after 30 h by Western blotting. (D) GA-induced apoptosis. GA-treated HeLa cells were analyzed for plasma membrane integrity (red fluorescence) and caspase-3/7 activity (green fluorescence) by multiparametric flow cytometry. The percentage of cells in each quadrant is indicated. (E) Apoptosome requirement. WT or Apaf-1–/– MEF were treated with GA and analyzed for apoptosis (Upper) or G2/M arrest (Lower) by DNA content and flow cytometry. (F) Modulation of GA-induced apoptosis. YUSAC-2 cells conditionally expressing survivin after Tet withdrawal (25) were treated with GA with or without Tet and analyzed for apoptosis by DNA content and flow cytometry. Data in E and F are the means ± SD of three independent experiments.
Treatment of HeLa cells with GA resulted in concentration-dependent caspase-mediated apoptosis (Fig. 3D), in a reaction reversed by the broad-spectrum caspase inhibitor ZVAD-fmk (data not shown). To discriminate between the effect of GA on cell cycle progression and cell death, we used MEF-deficient in the upstream component of mitochondrial-dependent apoptosis, Apaf-1 (26). GA induced the dose-dependent apoptosis in WT MEF, but not in Apaf-1–/– MEF (Fig. 3E Upper). In contrast, GA-induced G2/M arrest was indistinguishable in WT or Apaf-1–/– MEF (Fig. 3E Lower). These data demonstrate that GA induces activation of mitochondrial apoptosis and that this occurs independently from dysregulation of cell cycle progression. To test whether survivin counteracted apoptosis induced by Hsp90 inhibition, we used stably transfected YUSAC-2 cells that conditionally express survivin after withdrawal of Tet (25). In the presence of Tet, GA-treated YUSAC-2 cells exhibited concentration-dependent apoptosis (Fig. 3F). In contrast, conditional expression of survivin in Tet-deprived YUSAC-2 cells (25), suppressed GA-induced apoptosis at all concentrations tested (Fig. 3F). Therefore, cytoprotection by the survivin–Hsp90 complex is centered on the mitochondrial pathway, in agreement with a role of survivin in regulating mitochondrial apoptosis (31) and caspase-9 recruitment to the Apaf-1 apoptosome (32).
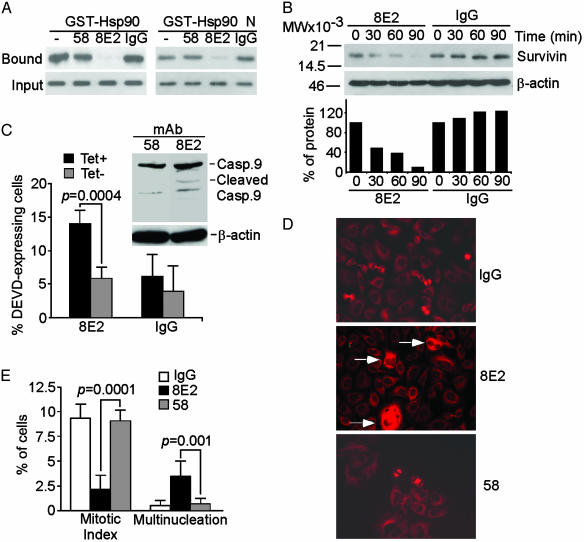
Selective Targeting of the Survivin–Hsp90 Interaction Causes Apoptosis and Mitotic Defects. To target more selectively the survivin–Hsp90 complex, we identified mAb 8E2 to survivin (24), which abolished the binding of survivin to Hsp90 or its N domain in vitro (Fig. 4A). In contrast, another Ab to survivin, mAb 58 (24), or nonimmune IgG, had no effect (Fig. 4A). Intracellular delivery of mAb 8E2 resulted in progressive loss of survivin expression in cycloheximide block experiments, whereas IgG did not affect survivin levels (Fig. 4B). Intracellular loading of mAb 8E2, but not IgG or mAb 58, resulted in increased caspase-3/7 activity in Tet-containing YUSAC-2 cultures (Fig. 4C) and proteolytic processing of caspase-9 in HeLa cells (Fig. 4C Inset). Conditional expression of survivin in Tet-deprived YUSAC-2 cells suppressed caspase-3/7 activity induced by mAb 8E2 (Fig. 4C). HeLa cells loaded with mAb 8E2 exhibited profound inhibition of cell proliferation and appearance of multinucleated cells as compared with IgG- or mAb 58-loaded cultures (Fig. 4 D and E). These data are consistent with a dual role of survivin in cytoprotection (32) and mitotic transition (23).
Fig. 4.
Ab disruption of survivin–Hsp90 complex induces apoptosis and cell cycle arrest. (A) In vitro targeting. Survivin was incubated with mAbs 58 or 8E2 to survivin (24) or IgG, and binding to GST-Hsp90 or GST-N-Hsp90 (GST-Hsp90 N) was determined by Western blotting. (B) In vivo targeting. HeLa cells were loaded intracellularly with mAb 8E2 or IgG, treated with cycloheximide, and analyzed by Western blotting (Upper). MW, molecular weight. Protein amounts were quantified by densitometry (Lower). (C) Caspase activity. YUSAC-2 cells loaded with mAb 8E2 or IgG were analyzed for caspase-3/7 activity by Asp-Glu-Val-Asp expression and flow cytometry in the absence (Tet–) or presence (Tet+) of Tet. Data are mean ± SD of three independent experiments. (Inset) Proteolytic processing of caspase-9 in HeLa cells transduced with mAbs 8E2 or 58 to survivin. The positions of proform (Casp.9) or cleaved (Cleaved) caspase-9 are indicated. (D) Mitotic defects. HeLa cells loaded with mAbs 8E2 or 58 to survivin or IgG were stained with an Ab to β-tubulin and analyzed by fluorescence microscopy. (Magnification, ×400.) Multinucleated cells in mAb 8E2-transduced cultures are indicated by arrows. (E) Summary of cell division defects. Mitotic or multinucleated cells in Ab-transduced HeLa cells were scored by fluorescence microscopy. Data are the means ± SEM of four independent determinations.
In summary, we identified the IAP family member (3) survivin as a new client protein for Hsp90 (14, 15), linking the cellular stress response to a dual cell viability/mitotic checkpoint. Because the conserved survivin baculovirus IAP repeat (3) contains the Hsp90-binding site, it is possible that Hsp90 recognition may involve other IAPs and thus affect their stability and maturation. This may have profound repercussions for cell viability because sudden losses in IAP expression trigger apoptosis (33), and mechanisms controlling IAP degradation regulate cell death (34). The association of survivin with Hsp90 may be required to accommodate its multiple molecular interactions (32) and dynamic subcellular localizations (24) required for cell death/mitotic regulation (23). In tumors, where both survivin and Hsp90 are up-regulated (16, 23), this interaction may provide a broad permissive environment (18, 21), inhibiting apoptosis (32) and obliterating mitotic checkpoints (24). Conversely, molecular antagonists of the survivin–Hsp90 interaction may provide a rational approach to critically lower this antiapoptotic threshold and promote targeted elimination of cancer cells.
Acknowledgments
This work was supported by National Institutes of Health Grants CA78810, HL54131, and CA90917. P.F. is a fellow of the Lymphoma Research Foundation of America.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IAP, inhibitor of apoptosis; GA, geldanamycin; MEF, mouse embryonic fibroblast; Tet, tetracycline.
References
- 1.Hanahan, D. & Weinberg, R. A. (2000) Cell 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 2.Cory, S. & Adams, J. M. (2002) Nat. Rev. Cancer 2, 647–656. [DOI] [PubMed] [Google Scholar]
- 3.Salvesen, G. S. & Duckett, C. S. (2002) Nat. Rev. Mol. Cell. Biol. 3, 401–410. [DOI] [PubMed] [Google Scholar]
- 4.Soengas, M. S., Capodieci, P., Polsky, D., Mora, J., Esteller, M., Opitz-Araya, X., McCombie, R., Herman, J. G., Gerald, W. L., Lazebnik, Y. A., et al. (2001) Nature 409, 207–211. [DOI] [PubMed] [Google Scholar]
- 5.Shin, M. S., Kim, H. S., Kang, C. S., Park, W. S., Kim, S. Y., Lee, S. N., Lee, J. H., Park, J. Y., Jang, J. J., Kim, C. W., et al. (2002) Blood 99, 4094–4099. [DOI] [PubMed] [Google Scholar]
- 6.Hartl, F. U. & Hayer-Hartl, M. (2002) Science 295, 1852–1858. [DOI] [PubMed] [Google Scholar]
- 7.Schiene, C. & Fischer, G. (2000) Curr. Opin. Struct. Biol. 10, 40–45. [DOI] [PubMed] [Google Scholar]
- 8.Matouschek, A., Pfanner, N. & Voos, W. (2000) EMBO Rep. 1, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindquist, S. & Craig, E. A. (1988) Annu. Rev. Genet. 22, 631–677. [DOI] [PubMed] [Google Scholar]
- 10.Pratt, W. B. & Toft, D. (1997) Endocr. Rev. 18, 306–360. [DOI] [PubMed] [Google Scholar]
- 11.Prodromou, C., Panaretou, B., Chohan, S., Siligardi, G., O'Brien, R., Ladbury, J. E., Roe, S. M., Piper, P. W. & Pearl, L. H. (2000) EMBO J. 19, 4383–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchner, J. (1999) Trends Biochem. Sci. 24, 136–141. [DOI] [PubMed] [Google Scholar]
- 13.Hohfeld, J., Cyr, D. M. & Patterson, C. (2001) EMBO Rep. 2, 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearl, L. H. & Prodromou, C. (2000) Curr. Opin. Struct. Biol. 10, 46–51. [DOI] [PubMed] [Google Scholar]
- 15.Young, J. C., Moarefi, I. & Hartl, F. U. (2001) J. Cell Biol. 154, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jolly, C. & Morimoto, R. I. (2000) J. Natl. Cancer Inst. 92, 1564–1572. [DOI] [PubMed] [Google Scholar]
- 17.Nollen, E. A. & Morimoto, R. I. (2002) J. Cell Sci. 115, 2809–2816. [DOI] [PubMed] [Google Scholar]
- 18.Pandey, P., Saleh, A., Nakazawa, A., Kumar, S., Srinivasula, S. M., Kumar, V., Weichselbaum, R., Nalin, C., Alnemri, E. S., Kufe, D., et al. (2000) EMBO J. 19, 4310–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dias, S., Shmelkov, S. V., Lam, G. & Rafii, S. (2002) Blood 99, 2532–3540. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, J., Devin, A., Miller, A., Lin, Y., Rodriguez, Y., Neckers, L. & Liu, Z. G. (2000) J. Biol. Chem. 275, 10519–10526. [DOI] [PubMed] [Google Scholar]
- 21.Sato, S., Fujita, N. & Tsuruo, T. (2000) Proc. Natl. Acad. Sci. USA 97, 10832–10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isaacs, J. S., Xu, W. & Neckers, L. (2003) Cancer Cells 3, 213–217. [DOI] [PubMed] [Google Scholar]
- 23.Altieri, D. C. (2003) Nat. Rev. Cancer 3, 46–54. [DOI] [PubMed] [Google Scholar]
- 24.Fortugno, P., Wall, N. R., Giodini, A., O'Connor, D. S., Plescia, J., Padgett, K. M., Tognin, S., Marchisio, P. C. & Altieri, D. C. (2002) J. Cell Sci. 115, 575–585. [DOI] [PubMed] [Google Scholar]
- 25.Grossman, D., Kim, P. J., Schechner, J. S. & Altieri, D. C. (2001) Proc. Natl. Acad. Sci. USA 98, 635–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida, H., Kong, Y.-Y., Yoshida, R., Elia, A. J., Hakem, A., Hakem, R., Penninger, J. M. & Mak, T. W. (1998) Cell 94, 739–750. [DOI] [PubMed] [Google Scholar]
- 27.Li, F., Ambrosini, G., Chu, E. Y., Plescia, J., Tognin, S., Marchisio, P. C. & Altieri, D. C. (1998) Nature 396, 580–584. [DOI] [PubMed] [Google Scholar]
- 28.Verdecia, M. A., Huang, H., Dutil, E., Kaiser, D. A., Hunter, T. & Noel, J. P. (2000) Nat. Struct. Biol. 7, 602–608. [DOI] [PubMed] [Google Scholar]
- 29.Prodromou, C., Roe, S. M., O'Brien, R., Ladbury, J. E., Piper, P. W. & Pearl, L. H. (1997) Cell 90, 65–75. [DOI] [PubMed] [Google Scholar]
- 30.Srethapakdi, M., Liu, F., Tavorath, R. & Rosen, N. (2000) Cancer Res. 60, 3940–3946. [PubMed] [Google Scholar]
- 31.Blanc-Brude, O. P., Mesri, M., Wall, N. R., Plescia, J., Dohi, T. & Altieri, D. C. (2003) Clin. Cancer Res. 9, 2683–2692. [PubMed] [Google Scholar]
- 32.Marusawa, H., Matsuzawa, S., Welsh, K., Zou, H., Armstrong, R., Tamm, I. & Reed, J. C. (2003) EMBO J. 22, 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, Y., Fang, S., Jensen, J. P., Weissman, A. M. & Ashwell, J. D. (2000) Science 288, 874–877. [DOI] [PubMed] [Google Scholar]
- 34.Martin, S. J. (2002) Cell 109, 807–809.12110178 [Google Scholar]