Ongoing investigations into the fundamental aspects of virus biology continue to provide crucial insights for the development of more effective virus-based therapeutics. Within the field of gene therapy, these discoveries have translated into virus vectors with improved tissue tropism, prolonged transgene expression, and enhanced safety. However, radical modifications to the viral genome or capsid constituents can have marked effects on virion stability. In particular, recent insights into the role of viral genomes in contributing to the physical stability of the virion highlight the need to reconsider this heretofore underappreciated aspect of vector design. Here we discuss the impact of the genome on virion stability, with particular emphasis on the development of adenovirus (Ad)-based vectors.
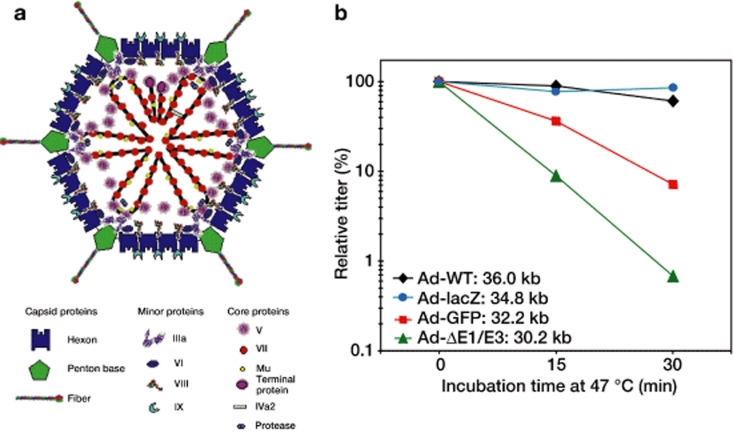
Ad virions are characterized by a nucleoprotein core containing a linear double-stranded genome (~30–40 kb) surrounded by an icosahedral, nonenveloped capsid (~70 to 100 nm in diameter) (Figure 1a).1 Interestingly, despite differences in host and tissue tropism, little variation exists among Ads with regard to the genomic and structural parameters. Human Ad serotype 5, the most extensively characterized Ad, contains an ~36 kb genome that encodes genes that are divided into early (E1–E4) and late (L1–L5) transcripts depending on whether they are expressed before or after DNA replication (refer to ref. 1 for an in-depth review). In addition to encoding the necessary elements for virion production, the Ad genome contains inverted terminal repeats and a packaging sequence, which are required for the replication and encapsidation of the viral DNA, respectively.
Figure 1.
Adenoviral genome size affects virion stability. (a) A schematic representation of adenovirus (Ad) virion structure based on cryoelectron microscopy and crystallography. (Reprinted from ref. 1 with permission of W.C. Russell and the Society for General Microbiology. (b) Heat stability of Ad virions is significantly impacted by the size of the packaged genome. Ad virions ranging in size from 30.2 to 36 kb were incubated at 47 °C for 0, 15, or 30 minutes and the residual virus titer was determined by standard plaque assay. (Adapted from ref. 22.) GFP, green fluorescent protein; WT, wild type.
Our current understanding of the interactions between the encapsidated Ad DNA and the major capsid proteins is incomplete.2 The association of the viral DNA with three highly basic proteins—proteins VII (pVII), V (pV), and μ (mu)—is believed to play an essential role.3,4,5 pVII, a protamine-like protein, is responsible for wrapping and condensing the viral DNA and thereby facilitating packaging within the physical constraints of the capsid.6 Early studies examining the organization of the nucleoprotein complex formed by the interaction of pVII with the viral DNA have revealed a high level of organization within the Ad virion. In particular, 12 large spherical units, termed adenosomes, extend from a central dense core and are directed toward each vertex of the icosahedral capsid.7,8 pV is believed to form a shell around the pVII–DNA complex7,9 and acts to tether the pVII-wrapped DNA to the inner capsid at the vertices.10,11 The indirect interaction of pV with the capsid is mediated by the minor capsid protein VI, which is directly connected to each peripentonal hexon.12 A recent study by Silvestry et al.13 suggests that protein IIIa may mediate an interaction between core and penton protein in the immature virion (i.e., before proteolytic cleavage of six structural proteins, including pre-protein IIIa, by the Ad-encoded protease), but this interaction seems significantly reduced in the mature virion. The remaining constituents of the Ad5 capsid, which include three major (II, III, and IV) and three other minor (IVa2, VIII, and IX) polypeptides (Figure 1a), do not appear to play a critical role in mediating the interaction of the viral DNA with the capsid in the mature virion. Thus, the only discernible contact between the viral DNA and the capsid exists at the vertices, bridged through proteins V and VI. These data suggest a role for an indirect interaction between the adenosome and capsid constituents in maintaining capsid stability.
An understanding of the interaction(s) between the viral genome and capsid proteins has a direct relevance in the continuing development of Ad as an effective and safe gene-delivery tool. Within the context of current therapeutic strategies, E1-deleted vectors or first-generation Ad (fgAd) are the most commonly used vehicles for delivery of foreign genes into mammalian cells. Although Ad vectors have been generated with approximately 108% of the wild-type genome length, these vectors grow poorly, and the genome tends to be unstable (spontaneously rearranging), with a resulting outgrowth of vectors with smaller genomes.14 Vectors with genomes of approximately 105% of the wild-type genome length are relatively stable. Thus, deletion of the E1 and E3 regions permits the inclusion of ~8 kb of foreign DNA. However, the full-vector genome size can be significantly smaller than this; for example, fgAds containing expression cassettes for short-hairpin RNA may have a genome size of only ~30 kb.15
Another class of Ad vector that has shown great promise in numerous preclinical applications consists of helper-dependent Ad vectors (hdAd), which are devoid of all viral-protein coding sequences.16 Currently, methods for hdAd production make use of a helper virus to permit the replication and packaging of the hdAd genome, and Cre or Flp recombinase–mediated excision of the helper virus DNA packaging element prevents packaging of helper virus genomes.17 However, contamination of hdAd stock with residual helper virus does occur despite removal of the DNA packaging element. Therefore, hdAd vectors are typically designed to be ~27–30 kb to allow maximal separation of the hdAd from the residual helper virus on a cesium chloride gradient.18
Thus, pushing the requirements of the Ad vector system has caused a general reduction in the size of the packaged genome to ~75–83% of the natural genome length. The impact of reducing the genome size on virion function or stability has only recently become clearer.
As part of our continuing efforts to improve the production of hdAds, we developed a method for generating hdAd that was dependent on DNA size restrictions imposed by a protein IX (pIX)–deficient capsid.19 The resulting hdAds, deficient in pIX, were predicted to display reduced heat stability compared with hdAds propagated with a pIX-expressing helper virus, in that pIX is involved in stabilizing the Ad capsid.20, 21 Surprisingly, the heat stability of hdAd was identical whether or not pIX was present in the hdAd capsid. Furthermore, normal and pIX-deficient hdAds were significantly less stable than wild-type Ad, which showed no drop in titer after incubation at 47 °C for 30 minutes. 22 The hdAd generated with the pIX+ helper virus had virion protein constituents that were essentially identical to those of the parental helper virus and wild-type Ad, suggesting that the defect underlying the capsid instability was not due to an overt deficiency in a capsid protein, as was previously observed in virions with very small genomes (<12 kb; refs. 23, 24). Interestingly, increasing the genome size of the hdAd from 30 to 34 kb resulted in a significant increase in heat stability, indicating that for hdAd the size of the genome can significantly influence overall virion stability. A similar phenomenon was observed for fgAd; vectors constructed to be larger than ~33 kb showed heat stability identical to that of wild-type Ad, whereas smaller vectors showed a significant decrease in heat stability that correlated with the size of the genome (Figure 1b). For the smallest vector, an E1/E3-deleted vector with no transgene (30.2 kb), the time required to decrease the titer by half was only 4 minutes, whereas the titer of wild-type Ad was unaffected by heating. Further analysis demonstrated that heating of vectors with small genomes resulted in a rapid loss of capsid integrity. Specifically, heating resulted in a release of the vertex proteins, fiber, and penton (and probably the peripentonal hexon), followed by complete disintegration of the capsid structure. Whether similar virion instability occurs for vectors with genomes larger than wild-type size is unknown. Taken together, these results indicate that the Ad DNA genome contributes to the physical stability of the Ad capsid, and that reduction of the genome size in fgAd and hdAd vectors can reduce stability of the vector.
The relationship between genome size and capsid stability has been extensively investigated in other viruses as well. Spooling of full-length viral DNA into preformed capsids by bacteriophage lambda, another icosahedral virus, results in significant internal pressure that is subsequently used to propel the phage DNA into the bacteria during the infection process.25 Using atomic-force microscopy, Ivanovska et al. demonstrated that capsids with DNA of wild-type length can withstand twice the force of capsids containing small genomes (~78% of wild-type length), indicating a direct relationship between viral genome size and capsid stability.26 Similarly, the parvovirus minute virus of mice (MVM) is encoded by a single-stranded DNA genome that also acts to stabilize the capsid. Atomic-force microscopy measurements of empty and DNA-containing MVM capsids yielded up to a 140% increase in capsid stiffness when DNA was present.27 Structural studies have further demonstrated that the MVM genome is bound within 60 concavities on the inner surface of the capsid. This direct interaction of the genome with the capsid acts to effectively increase the thickness of the capsid wall and provide structural stiffness and stability.27 Further observations in cowpea chlorotic mottle virus and bean pod mottle virus, both multipartite single-stranded RNA viruses, is suggestive of a conserved mechanism involving genome-mediated capsid stabilization through direct physical interactions between the viral genome and a capsid protein.28,29,30
The genome-mediated capsid stabilization in Ad is unlikely to occur in a manner analogous to any of the aforementioned viruses. Although both bacteriophage lambda and Ad use a proteinaceous icosahedral capsid to package double-stranded DNA, the mechanism used by bacteriophage is distinct, in that the internal pressure used to stabilize the bacteriophage capsid is caused by electrostatic repulsion of the packaged DNA31 and such charges are neutralized in the Ad virion. Similarly, because Ad DNA does not make direct contact with any capsid constituent, it is unlikely that the Ad genome stabilizes the capsid structure in a manner similar to that observed for MVM, cowpea chlorotic mottle virus, and bean pod mottle virus. The mechanism of nucleic acid–mediated mechanical stabilization of the capsid probably depends on the virus and nucleic acid involved. For Ad the mechanism is currently unclear, but one can be envisaged where the tight packaging of DNA of wild-type length into the capsid may be required to force the adenosome DNA in the vertex regions into the proper position/orientation to achieve an indirect linkage between the Ad DNA and hexon. Reducing the size of the genome could, within this model, misalign or even prevent this indirect interaction and ultimately destabilize the capsid. Although Ad vectors with very small genomes (>12 kb) show a marked difference in protein content in the virion,23, 24 the small-genome vectors we tested (~30 kb) did not show any overt difference in protein content relative to wild-type Ad. It is likely that there are small differences in the two virions (for example, pVII should be reduced by ~17% compared with wild-type Ad), but it is not clear whether this is the cause of the reduced stability or simply a symptom of the reduced genome size.
The impact of genome size on Ad virion stability has important implications for the design of Ad-based vectors. For example, an ~32-kb fgAd vector encoding green fluorescent protein has inherently reduced stability as compared to an ~35-kb vector encoding lacZ (Figure 1b). However, most applications of Ad vectors generally do not require the use of elevated temperatures, and we did not observe a difference in virion stability under other more physiologically relevant conditions such as extended heating at 37 °C, repeated freeze–thaw cycles, or altered pH (unpublished data). Nevertheless, we have tested only a small fraction of possible conditions under which Ad vectors may be used in experimental and clinical settings, and other conditions may exist in which genome-dependent changes in Ad vector stability manifest. For example, whether the effect of genome-dependent differences in virion stability translates into altered transduction efficiencies in vivo has yet to be addressed. Furthermore, evidence suggests that Ad-induced activation of innate immune signaling pathways is enhanced by delaying the kinetics of Ad endocytic escape, a process that is dependent on capsid destabilization.32 In light of these findings, the role of genome-dependent capsid stability in activation of the host immune response provides an interesting area for further research.
Whether these observations with Ad-based vectors can be extended to other vector systems also remains an important question. Adeno-associated virus (AAV), a parvovirus being developed for use in a variety of therapeutic applications, could also exhibit genome-dependent capsid stability similar to that observed for MVM.27 Indeed, structural analysis of AAV serotypes 4 and 8 has revealed the presence of a single nucleotide within a binding pocket of the capsid architecture.33,34 This strongly suggests that an interaction between the genome and capsid exists and may even serve to provide stability or rigidity to the AAV virion as seen with MVM.
As with all viruses, wild-type Ad has evolved to optimize both its genome and capsid to provide maximal fitness. Gene therapy vectors based on Ad frequently compromise this relationship through deletion of viral genes and insertion of therapeutic genes, which alters the interaction between the genome and physical structure of the virion. Detailed analysis of the mechanism(s) by which the genome of Ad, and other viral vector systems, promotes capsid stability may lead to currently unappreciated opportunities to improve gene delivery.
REFERENCES
- Russell WC. Adenoviruses: update on structure and function. J Gen Virol. 2009;90:1–20. doi: 10.1099/vir.0.003087-0. [DOI] [PubMed] [Google Scholar]
- Fabry CM, Rosa-Calatrava M, Conway JF, Zubieta C, Cusack S, Ruigrok RW, et al. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 2005;24:1645–1654. doi: 10.1038/sj.emboj.7600653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel JV, Jr, White DO., and , Scharff MD. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology. 1968;36:126–136. doi: 10.1016/0042-6822(68)90122-0. [DOI] [PubMed] [Google Scholar]
- Russell WC, Laver WG., and , Sanderson PJ. Internal components of adenovirus. Nature. 1968;219:1127–1130. doi: 10.1038/2191127a0. [DOI] [PubMed] [Google Scholar]
- Chatterjee PK, Vayda ME., and , Flint SJ. Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J Mol Biol. 1986;188:23–37. doi: 10.1016/0022-2836(86)90477-8. [DOI] [PubMed] [Google Scholar]
- Mirza MA., and , Weber J. Structure of adenovirus chromatin. Biochim Biophys Acta. 1982;696:76–86. doi: 10.1016/0167-4781(82)90012-4. [DOI] [PubMed] [Google Scholar]
- Brown DT, Westphal M, Burlingham BT, Winterhoff U., and , Doerfler W. Structure and composition of the adenovirus type 2 core. J Virol. 1975;16:366–387. doi: 10.1128/jvi.16.2.366-387.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Boring JW., and , Brown JC. Ion etching of human adenovirus 2: structure of the core. J Virol. 1984;51:52–56. doi: 10.1128/jvi.51.1.52-56.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt E, Sundquist B, Pettersson U., and , Philipson L. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology. 1973;52:130–147. doi: 10.1016/0042-6822(73)90404-2. [DOI] [PubMed] [Google Scholar]
- Everitt E, Lutter L., and , Philipson L. Structural proteins of adenoviruses. XII. Location and neighbor relationship among proteins of adenovirion type 2 as revealed by enzymatic iodination, immunoprecipitation and chemical cross-linking. Virology. 1975;67:197–208. doi: 10.1016/0042-6822(75)90417-1. [DOI] [PubMed] [Google Scholar]
- Chatterjee PK, Vayda ME., and , Flint SJ. Interactions among the three adenovirus core proteins. J Virol. 1985;55:379–386. doi: 10.1128/jvi.55.2.379-386.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PL, Fuller SD., and , Burnett RM. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestry M, Lindert S, Smith JG, Maier O, Wiethoff CM, Nemerow GR, et al. Cryo-electron microscopy structure of adenovirus type 2 temperature-sensitive mutant 1 reveals insight into the cell entry defect. J Virol. 2009;83:7375–7383. doi: 10.1128/JVI.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bett AJ, Prevec L., and , Graham FL. Packaging capacity and stability of human adenovirus type 5 vectors. J Virol. 1993;67:5911–5921. doi: 10.1128/jvi.67.10.5911-5921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi H, Funakoshi N, Hosono T, Sakurai F, Kawabata K, Yamaguchi T.Rapid construction of small interfering RNA-expressing adenoviral vectors on the basis of direct cloning of short hairpin RNA-coding DNAs Hum Gene Ther 20071874–80.et al [DOI] [PubMed] [Google Scholar]
- Palmer DJ., and , Ng P. Helper-dependent adenoviral vectors for gene therapy. Hum Gene Ther. 2005;16:1–16. doi: 10.1089/hum.2005.16.1. [DOI] [PubMed] [Google Scholar]
- Parks RJ, Chen L, Anton M, Sankar U, Rudnicki MA., and , Graham FL. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci U S A. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks RJ., and , Graham FL. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J Virol. 1997;71:3293–3298. doi: 10.1128/jvi.71.4.3293-3298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent K, Ng P, Evelegh C, Graham FL., and , Parks RJ. Development of a size restricted pIX deleted helper virus for amplification of helper dependent adenovirus vectors. Gene Ther. 2004;11:504–511. doi: 10.1038/sj.gt.3302107. [DOI] [PubMed] [Google Scholar]
- Colby WW., and , Shenk T. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J Virol. 1981;39:977–980. doi: 10.1128/jvi.39.3.977-980.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks RJ. Adenovirus protein IX: a new look at an old protein. Mol Ther. 2005;11:19–25. doi: 10.1016/j.ymthe.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Smith AC, Poulin KL., and , Parks RJ. DNA genome size affects the stability of the adenovirus virion. J Virol. 2009;83:2025–2028. doi: 10.1128/JVI.01644-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber A, He CY, Kirillova I., and , Kay MA. Recombinant adenoviruses with large deletions generated by Cre-mediated excision exhibit different biological properties compared with first-generation vectors in vitro and in vivo. J Virol. 1996;70:8944–8960. doi: 10.1128/jvi.70.12.8944-8960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayakhmetov DM, Li ZY, Gaggar A, Gharwan H, Ternovoi V, Sandig V, et al. Genome size and structure determine efficiency of postinternalization steps and gene transfer of capsid-modified adenovirus vectors in a cell-type-specific manner. J Virol. 2004;78:10009–10022. doi: 10.1128/JVI.78.18.10009-10022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurmemmedov E, Castelnovo M, Catalano CE., and , Evilevitch A. Biophysics of viral infectivity: matching genome length with capsid size. Q Rev Biophys. 2007;40:327–356. doi: 10.1017/S0033583508004666. [DOI] [PubMed] [Google Scholar]
- Ivanovska I, Wuite G, Jonsson B., and , Evilevitch A. Internal DNA pressure modifies stability of WT phage. Proc Natl Acad Sci U S A. 2007;104:9603–9608. doi: 10.1073/pnas.0703166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco C, Carreira A, Schaap IA, Serena PA, Gómez-Herrero J, Mateu MG, et al. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc Natl Acad Sci U S A. 2006;103:13706–13711. doi: 10.1073/pnas.0601881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chen Z, Johnson JE., and , Thomas GJ., Jr Conformations, interactions, and thermostabilities of RNA and proteins in bean pod mottle virus: investigation of solution and crystal structures by laser Raman spectroscopy. Biochemistry. 1992;31:6673–6682. doi: 10.1021/bi00144a006. [DOI] [PubMed] [Google Scholar]
- Da Poian AT, Johnson JE., and , Silva JL. Protein-RNA interactions and virus stability as probed by the dynamics of tryptophan side chains. J Biol Chem. 2002;277:47596–47602. doi: 10.1074/jbc.M209174200. [DOI] [PubMed] [Google Scholar]
- Michel JP, Ivanovka IL, Gibbons MM, Klug WS, Knobler CM, Wuite GJ, et al. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc Natl Acad Sci U S A. 2006;103:6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzlil S, Kindt JT, Gelbart WM., and , Ben-Shaul A. Forces and pressures in DNA packaging and release from viral capsids. Biophys J. 2003;84:1616–1627. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejer G, et al. Key role of splenic myeloid DCs in the IFN-alphabeta response to adenoviruses in vivo. PLoS Pathog. 2008;4:e1000208. doi: 10.1371/journal.ppat.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HJ, Lane MD, Padron E, Gurda B, McKenna R, Kohlbrenner E, et al. Structure of adeno-associated virus serotype 8, a gene therapy vector. J Virol. 2007;81:12260–12271. doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini JA, et al. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol. 2006;80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]