Abstract
Tumor necrosis factor-α (TNF-α) is upregulated in psoriatic skin and represents a prominent target in psoriasis treatment. The level of TNF-α-encoding mRNA, however, is not increased in psoriatic skin, and it remains unclear whether intervention strategies based on RNA interference (RNAi) are therapeutically relevant. To test this hypothesis the present study describes first the in vitro functional screening of a panel of short hairpin RNAs (shRNAs) targeting human TNF-α mRNA and, next, the transfer of the most potent TNF-α shRNA variant, as assessed in vitro, to human skin in the psoriasis xenograft transplantation model by the use of lentiviral vectors. TNF-α shRNA treatment leads to amelioration of the psoriasis phentotype in the model, as documented by reduced epidermal thickness, normalization of the skin morphology, and reduced levels of TNF-α mRNA as detected in skin biopsies 3 weeks after a single vector injection of lentiviral vectors encoding TNF-α shRNA. Our data show efficient lentiviral gene delivery to psoriatic skin and therapeutic applicability of anti-TNF-α shRNAs in human skin. These findings validate TNF-α mRNA as a target molecule for a potential persistent RNA-based treatment of psoriasis and establish the use of small RNA effectors as a novel platform for target validation in psoriasis and other skin disorders.
Introduction
Psoriasis is a chronic inflammatory skin disorder affecting 1–3% of the world population. The disease is characterized by demarcated erythematous scaly plaques in which keratinocytes exhibit hyperproliferation and abnormal differentiation leading to epidermal hyperplasia. Infiltration by lymphocytes as well as dilation and extension of superficial blood vessels are additional hallmarks of the disease.1 Psoriasis is generally believed to be driven by a complex network of immune cells and inflammatory cytokines. Within psoriatic lesional skin, expression of cytokines belonging to the T helper 1 and 17 subtypes is predominating,2,3 and increased levels of proinflammatory cytokines such as tumor necrosis factor-α (TNF-α), interferon-γ, interleukin-1, interleukin-15, and interleukin-20 have attracted attention. Although these proinflammatory cytokines may not be the initial cause of psoriasis, they are likely to play key roles in the disease by maintaining skin inflammation. Thus, reduction or inhibition of one or several of these proinflammatory cytokines is an attractive strategy for psoriasis treatment.
In particular, inhibition of TNF-α by systemically administered TNF-α-specific antibodies (infliximab and adalimumab) or soluble receptor constructs (etanercept) has been successful and is now widely employed clinically in the treatment of psoriasis and psoriasis arthritis.4 The increased level of TNF-α in psoriasis skin may be caused by a variety of cell types in the skin, including keratinocytes, activated T cells, macrophages, and dendritic cells.4 The ability of TNF-α to target various cells in the skin is thought to affect the pathogenesis in several ways including induction of other inflammatory cytokines, chemokines, and growth factors as well as endothelial adhesion molecules.5,6,7 Production of TNF-α is increased also in other chronic inflammatory diseases such as psoriatic arthritis,8 rheumatoid arthritis,9 and inflammatory bowel disease.10
TNF-α-specific antagonists that are currently used in the clinique inhibit the effects of TNF-α by binding directly to TNF-α protein. In lesional psoriatic skin, TNF-α is upregulated at the post-transcriptional level by mechanisms controlled by increased levels of activated MAPK-activated protein kinase 2 (MK2).11 Hence, induced translation, and not upregulated transcription, seems to account for an increased TNF-α level in psoriasis skin. It remains unclear, therefore, whether intervention strategies that target the TNF-α mRNA rather than TNF-α protein show relevance in psoriasis treatment. In the present report, we examined the potential use of DNA-encoded small RNA effectors to target TNF-α mRNA by RNA interference (RNAi).
RNAi is a naturally occurring mechanism of gene regulation that induces sequence-specific knockdown of gene expression at the post-transcriptional level12 with importance for both regulation of cellular genes and the defence against foreign genetic elements,13 Regulation of gene expression by RNAi uses endogenous cellular pathways in which double-stranded RNA molecules produced from endogenous or foreign DNA are processed into short double-stranded RNA molecules of 21–23 nucleotides.14 These small interfering RNA (siRNA) molecules are incorporated into an RNA-induced silencing complex which facilitates sequence-specific cleavage and degradation of the target mRNA guided by perfect complementarity between one of the two siRNA strands and the target sequence.15 Both synthetic siRNA duplexes or DNA-encoded short hairpin RNAs (shRNAs) are efficiently processed by the cellular RNAi machinery16,17 and can target predetermined mRNA targets in laboratory animals.18,19,20,21 RNAi-based tools are already widely used in animals, and small RNA effectors have the potential to become therapeutics in humans. Synthetic siRNA molecules typically trigger a transient RNAi response, whereas expression of shRNAs from viral vectors has been found to induce a robust and persistent gene-regulating effect.22,23
In the present study, we used lentiviral vectors as potent carriers of shRNA-encoding gene cassettes to human skin. Lentiviral vectors are attractive gene vehicles primarily due to their ability to transduce both dividing and nondividing cells and to establish persistent expression owing to genomic integration of the vector DNA reverse-transcribed from virally delivered single-stranded RNA. Efficient lentiviral transduction of xenografted human skin on immunodeficient mice has been demonstrated previously.24,25 Furthermore, shRNAs are efficiently produced from lentiviral templates.23,26
To examine the use of RNAi-based therapeutics in psoriasis skin we selected, in this study, several potentially favourable target sequences within the TNF-α mRNA sequence. In vitro screening assays enabled us to identify a single shRNA variant with consistent and reproducible ability to inhibit expression of TNF-α when expressed from transfected DNA vectors as well as from transduced lentiviral vectors. Moreover, by employing a lentiviral vector expressing green fluorescent protein (GFP), efficient lentiviral gene transfer to lesional human psoriatic skin transplanted onto immunodeficient mice was observed. Resolution of the psoriasis phenotype, as determined by improved clinical scores, reduced epidermal thickness, and reduced levels of TNF-α mRNA in skin treated in vivo by a single intradermal injection of shRNA-encoding lentiviral vectors in the psoriasis xenograft transplantation model27 led to the demonstration of TNF-α mRNA as a useful target for treatment of psoriasis. Our study provides proof-of-concept that targeting cytokines with RNAi is therapeutically applicable in treatment of psoriasis and proposes a new shRNA-based strategy for evaluating potential targets for treatment of psoriasis.
Results
Identification of effective shRNAs targeting transiently expressed human TNF-α
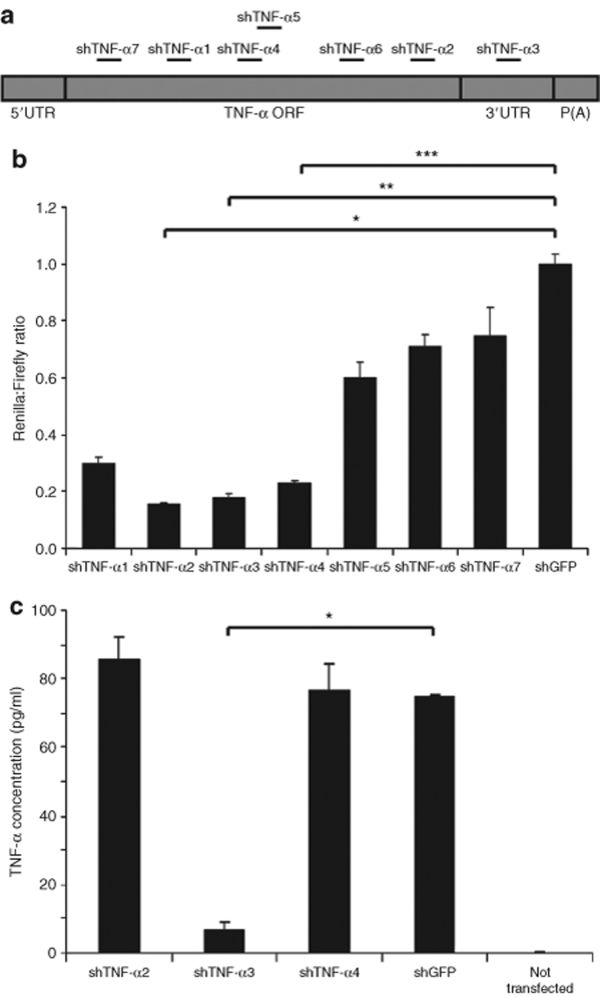
Designed shRNA variants directed against different target sites in a defined mRNA species may diverge greatly in their potential to trigger mRNA degradation. Therefore, seven shRNAs targeting different sites in the human TNF-α mRNA were designed in this study. Six target sequences were located within the coding region of the TNF-α gene and one target was located within the 3′-untranslated region (3′-UTR) of the gene (Figure 1a). To examine shRNA efficiency, we transiently transfected HEK293 cells with plasmid DNA (pBC.H1-shTNF-α1 through pBC.H1-shTNF-α7) expressing each of the shRNAs from the H1 promoter. By using a dual-luciferase assay in which the TNF-α complementary DNA (cDNA) was transiently expressed as a fusion RNA with the coding sequence of the Renilla luciferase (R-luc) gene, we measured the ability of each of the shRNA variants to downregulate expression of the R-luc reporter. Transfections of pBC.H1-shTNF-α2, pBC.H1-shTNF-α3, and pBC.H1-shTNF-α4 reduced R-luc expression with >75% compared to plasmid DNA encoding an irrelevant shRNA (pBC.H1-shGFP), whereas the remaining shRNA variants caused smaller reductions of R-luc expression (Figure 1b).
Figure 1.
Functional screening of potential shRNA variants targeting human TNF-α. (a) Schematic representation of the TNF-α mRNA sequence and the distribution of shRNA target sequences. 5′ UTR and 3′ UTR indicate the 5′- and 3′-untranslated regions, respectively. ORF represents the open reading frame and P(A) the polyadenylation sequence. (b) Targeting the TNF-α sequence as detected by the dual-luciferase reporter assay. TNF-α cDNA was fused to the Renilla luciferase gene in the psiCHECK-2 vector and the plasmid co-transfected together with pBC.H1-shTNF-α1 through pBC.H1-shTNF-α7 and pBC.H1-shGFP (negative control) into HEK293 cells. Luciferase activities were determined 48 hours after transfection and the normalized values calculated relative to the negative shRNA control (pBC.H1-shGFP). P-values for the comparisons indicated by brackets were as follows: *P < 0.0001; **P = 0.0001; ***P = 0.0004. (c) Downregulation of transiently expressed TNF-α by transfection with lentiviral vector constructs encoding shRNAs. HEK293 cells were transfected with pSBT/CMV-TNF-α.EF1α-zeo and lentiviral vector constructs encoding shRNAs as indicated. TNF-α expression was determined by measuring TNF-α concentration in the medium by enzyme-linked immunosorbent assay. The asterisk indicates significant difference between the two groups indicated by the bracket (P = 0.0004). All experiments were performed in triplicates. Data are presented as mean ± SD. shRNA, short hairpin RNA; TNF-α, tumor necrosis factor-α.
Based on the initial efficiency screening assay, lentiviral vector constructs (pLV) encoding shTNF-α2, shTNF-α3, shTNF-α4, and shGFP were generated. TNF-α knockdown was verified by co-transfecting these plasmid vectors into HEK293 cells with plasmid DNA transiently expressing TNF-α from a cytomegalovirus (CMV) promoter. As shown in Figure 1c, transient expression of shTNF-α3 from the lentiviral vector construct resulted in a 92% reduction of the TNF-α protein expression compared to the level of TNF-α protein in the negative control (P = 0.0004). Notably in this assay, expression of neither shTNF-α2 nor shTNF-α4 did succeed to downregulate the production of TNF-α protein (Figure 1c), likely reflecting the importance of the context of the mRNA target sequence. In summary, of the seven shRNA variants targeting TNF-α mRNA, one shRNA, shTNF-α3, was found to efficiently reduce TNF-α gene expression.
Downregulation of stable TNF-α expression following lentiviral transfer of shTNF-α3
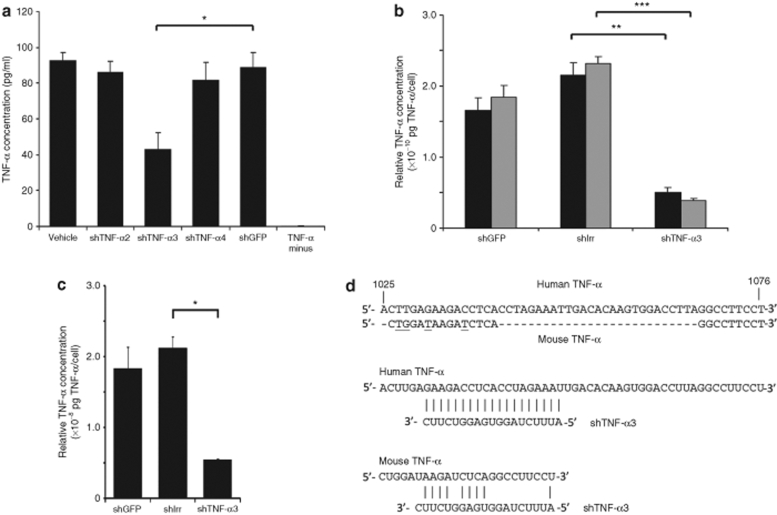
The ability of a single or a few copies of lentiviral shRNA templates to facilitate sufficient production of functional shRNAs is a crucial achievement toward obtaining functionality of RNAi effectors upon lentiviral delivery in an experimental or therapeutic scenario. To examine the potential of TNF-α-directed shRNAs to inhibit TNF-α expression when expressed from lentivirally delivered templates, we produced vesicular stomatitis virus G–pseudotyped lentiviral vectors, LV/shTNF-α2, LV/shTNF-α3, and LV/shTNF-α4, expressing shRNAs with variable functional efficacies according to our initial screen. We also generated a HEK293-derived cell line, designated 293-TNF-α, which stably expressed TNF-α in the presence of zeocin. These cells were treated with lentiviral vectors at an approximate multiplicity of infection (MOI) of 40, and the concentration of secreted TNF-α was subsequently determined by enzyme-linked immunosorbent assay (ELISA). LV/shTNF-α2 and LV/shTNF-α4 did not have any significant impact on the production of TNF-α as compared to a control vector expressing an irrelevant shRNA (LV/shGFP) (Figure 2a). In contrast, lentivirally delivered shTNF-α3 led to a clear reduction in the level of secreted TNF-α protein 2 days after vector treatment resulting in a TNF-α concentration in the medium that was 48% of the level measured with LV/shGFP (P = 0.003) (Figure 2a). As shTNF-α3 was consistently found to be the most potent RNAi effector, we focused on this particular shRNA in subsequent experiments.
Figure 2.
In vitro knockdown of stable TNF-α expression by shTNF-α3. (a,b) Knockdown of stable TNF-α expression following transduction with lentiviral vector-encoded shTNF-α3. 293-TNF-α cells were transduced with LV/shTNF-α2, LV/shTNF-α3, LV/shTNF-α4, LV/shGFP, or LV/shIrr, as indicated below each column, at an estimated MOI of ~40. The column labeled “Vehicle” (a) represents cells that were transduced with LV/PGK-puro and therefore did not receive any shRNA. “TNF-α minus” refers to HEK293 cells that did not express TNF-α. The concentration of TNF-α in the medium of the transduced cells was determined (a) after 2 days or (b) after 2 and 4 days. Black and gray columns represent concentrations measured after 2 and 4 days, respectively. Relative TNF-α concentrations (normalized for cell number and amount of viral particles transferred to the cells) are presented in b. Significant differences between the most relevant groups (indicated by brackets) are indicated by asterisks representing the following P values; *P = 0.003; **P = 0.002; ***P = 0.0007. (c) Stable downregulation of TNF-α expression after vector treatment. 293-TNF-α cells were transduced with lentiviral vectors at an estimated MOI of ~2 followed by selection of transduced cells with puromycin. TNF-α expression was determined after 11 days by measuring TNF-α concentration in the medium by enzyme-linked immunosorbent assay. The relative amounts of TNF-α were determined by correlation of TNF-α concentration to the number of cells and to the amount of lentiviral vectors used for transduction (pg p24 Gag) (TNF-α concentration/cell number/pg p24 Gag). Significant difference (P = 0.0002) between cells treated with shIrr and shTNF-α3 is indicated by an asterisk. All experiments were done in triplicates. Data are presented as mean ± SD. (d) shTNF-α3 guide strand sequence complementarity with human and mouse TNF-α mRNA. Alignment at top shows the degree of sequence similiarity between the human (NM_000594) and mouse (NM_013693) TNF-α genes in the segment of the gene containing the shTNF-α3 target sequence. Numbers refer to distance from first nucleotide in start codon of the human TNF-α sequence. Nucleotides that differ between the two sequences are underlined in the mouse TNF-α sequence. Nucleotides that are not present in mouse sequence are indicated with hyphens. The two alignments below demonstrate that shTNF-α3 has perfect match with human TNF-α mRNA but matches only 9 out of 19 positions within this region of mouse TNF-α mRNA. TNF-α, tumor necrosis factor-α; shIrr, irrelevant shRNA; MOI, multiplicity of infection; shRNA, short hairpin RNA.
One of the advantages of a lentiviral delivery approach is the integration of vector DNA into the genome of treated cells resulting in persistence of gene expression. To confirm the consistency of the effects caused by shRNA expression, the persistence of TNF-α knockdown in 293-TNF-α cells following lentiviral transfer was investigated. We included in these experiments an additional negative control vector encoding a previously used irrelevant shRNA (shIrr) which does not match any known sequence in the human genome.28 First, the TNF-α concentration in the medium was determined 2 and 4 days after transductions performed at an approximate MOI of 40 and normalized for the number of cells and the amount of viral vector (as measured by p24 Gag protein) added in each experiment (Figure 2b). Two days after lentiviral transduction, the relative amount of TNF-α was significantly reduced with 77% in samples treated with the shTNF-α3-encoding vector compared to samples treated with shIrr (P = 0.002). In accordance, the relative amount of TNF-α was reduced with 83% after 4 days in samples treated with shTNF-α3 compared to samples treated with lentivirally delivered shIrr (P = 0.0007). The reduction in relative amount of TNF-α from day 2 to 4 in samples treated with shTNF-α3 was moderate (P = 0.11), indicating that persistent knockdown had been obtained after 2 days.
To verify that a single or only a few inserted copies of the shTNF-α3-expressing vector had a persistent effect in TNF-α-expressing cells, we transduced 293-TNF-α cells with LV/shTNF- α3 at an MOI of 2. After transduction, the cells were selected by drug resistance for presence of the vector and the relative amount of TNF-α produced by a pool of cells at day 11 after transduction was determined (Figure 2c). Compared to both negative controls, cells treated with shTNF-α3 secreted a significantly reduced amount of TNF-α. Hence, the shTNF-α3-encoding lentiviral vector led to a 75% reduction in the relative amount of TNF-α compared to shIrr (P = 0.0002), providing proof of a persistent regulatory effect of lentivirally encoded shTNF-α3 effectors.
As our main objective was to investigate the use of TNF-α-directed shRNAs in human skin transplanted onto mice (see below), potential regulation by shTNF-α3 of mouse TNF-α would be of concern in this model. However, processed shTNF-α3 targets a 19-nucleotide target site within the 3′ untranslated region of human TNF-α mRNA. As this target site is not intact in murine TNF-α mRNA (allowing basepairing at only 9 out of the 19 positions) (Figure 2d) shTNF-α3 is not likely to target mouse TNF-α mRNA. Therefore, adding to the potency by which shTNF-α3 targets human TNF-α, this particular RNA effector is perfectly suited for studies in the psoriasis xenograft transplantation model providing means of downregulating human TNF-α locally in transplanted human skin without posing the risk of affecting the TNF-α level systemically in the animal.
Lentiviral penetration and transduction of skin in vivo leading to epidermal and dermal expression of eGFP
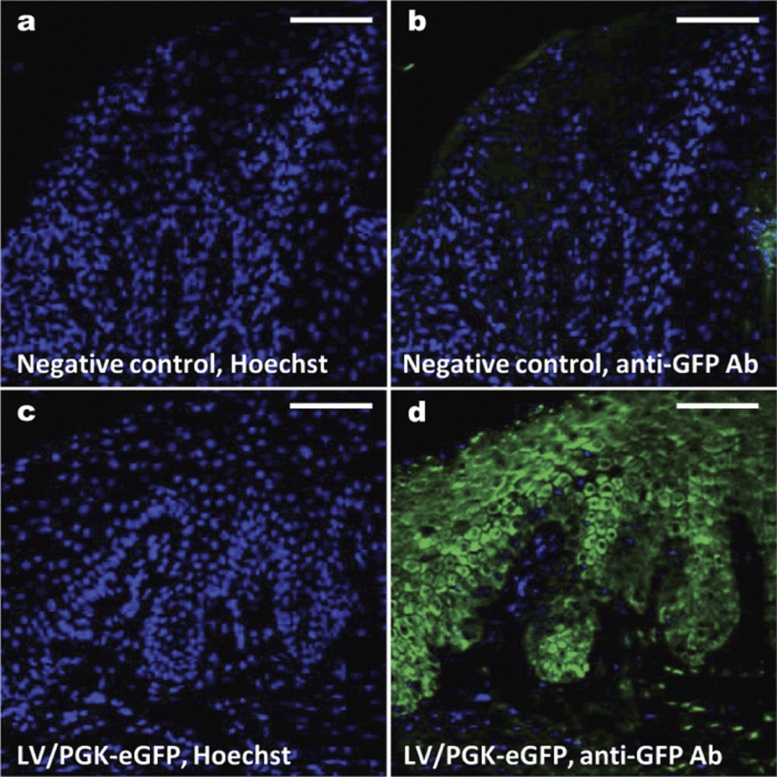
To investigate the efficacy of lentiviral vectors to penetrate and transduce human psoriasis skin, we injected lentiviral vectors encoding enhanced GFP (eGFP) intradermally into human psoriatic skin grafted onto the back of severe combined immunodeficiency mice in the xenograft transplantation model. Two transplanted mice received a single dose (150 µl each) of vesicular stomatitis virus G–pseudotyped LV/PGK-eGFP vector corresponding to 5.3 µg p24 Gag per graft. The mice were killed 3 days after transduction and eGFP expression in the psoriatic skin grafts was examined (Figure 3). For both mice, fluorescence microscopy of the biopsies illustrated eGFP expression in all cell layers of the epidermis and to some extent in the dermis (Figure 3d), whereas skin from untreated negative control mice was found to be negative as expected (Figure 3b). In conclusion, gene transfer to lesional human psoriatic skin grafted onto mice was efficiently accomplished by a single intradermal administration of lentiviral vectors. Our findings demonstrate that vector penetration of the skin following intradermal injection causes an initial boost of transgene expression. Characterization of potential transduction of stem cells and long-term effects in psoriatic skin, in which a high turnover rate of transduced keratinocytes could result in loss of transgene expression, is currently being investigated.
Figure 3.
In vivo transduction of human psoriasis skin with eGFP-encoding lentiviral vectors. Psoriatic plaque skin biopsies were transplanted onto severe combined immunodeficiency mice in the psoriasis xenograft transplantation model. Following healing, eGFP-encoding lentiviral vectors were injected intradermally into the human psoriasis skin grafts in an amount corresponding to 5.3 µg p24 Gag/mouse. Three days after transduction skin biopsies were taken, fixed, embedded in paraffin, and stained with anti-eGFP antibody. eGFP expression was analyzed by fluorescence microscopy. (a) Negative control (untreated transplanted grafts) stained with Hoechst to visualize cell nuclei. (b) Negative control stained with anti-GFP antibody. (c) Skin biopsy treated with lentiviral-mediated transfer of eGFP (LV/PGK-eGFP) and stained with Hoechst. (d) Skin biopsy treated with LV/PGK-eGFP and stained with anti-GFP antibody. Bar = 100 µm. eGFP, enhanced GFP.
Amelioration of psoriasis by TNF-α knockdown in shTNF-α3-treated lesional skin
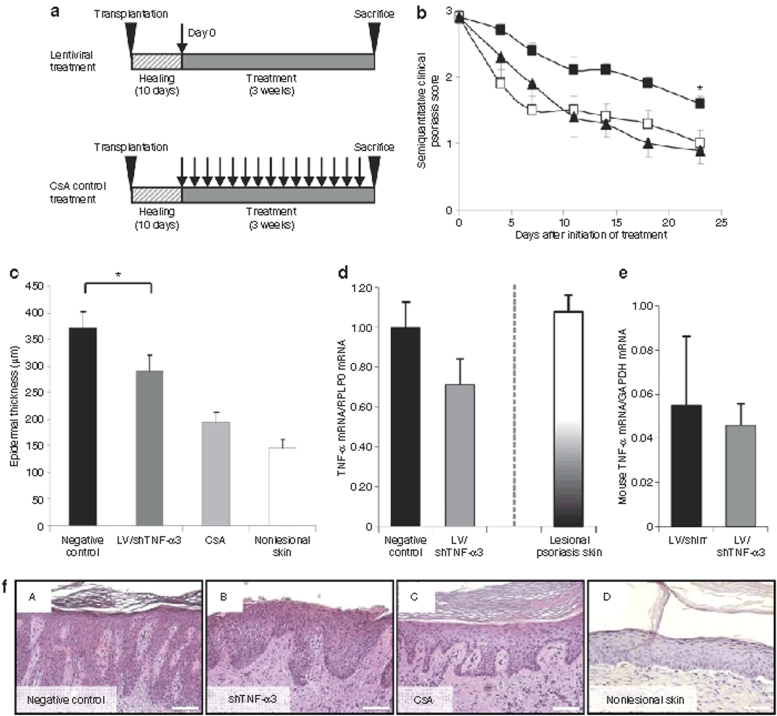
To investigate the effects of lentivirally delivered shTNF-α3 in human psoriatic skin in vivo, we used the psoriasis xenograft transplantation model. Upon transplantation of psoriatic keratome skin biopsies, the mice were divided into four groups. The first group (n = 10) was treated with a single intradermal dose of LV/shTNF-α3 (51.5 ± 11.0 µg p24 Gag/ml in 150 µl, corresponding to ~7.7 × 108 infectious particles), whereas the second group (n = 12) was treated with five weekly intraperitoneal injections of cyclosporin A (CsA), which has previously been shown to efficiently treat psoriasis in the xenotransplantation model and therefore has become the standard positive control in this particular model.29 The third group (n = 7) was treated with a single intradermal dose of LV/shIrr (same dose as group one) as negative control. As an additional negative control group a fourth group of mice (n = 10) was treated with an irrelevant monoclonal antibody as previously described.29 The mice were assigned a baseline semiquantitative clinical psoriasis score based on the average erythema, thickness and scaliness of the psoriatic plaques. The effects of treatment were evaluated in a blinded fashion by semiquantitative clinical scores given twice weekly for 3 weeks until killing. The final endpoint in evaluating the effect on psoriasis was determined histologically by measuring epidermis thickness and by quantification of the amount of TNF-α mRNA in the biopsies obtained from the psoriatic skin grafts.
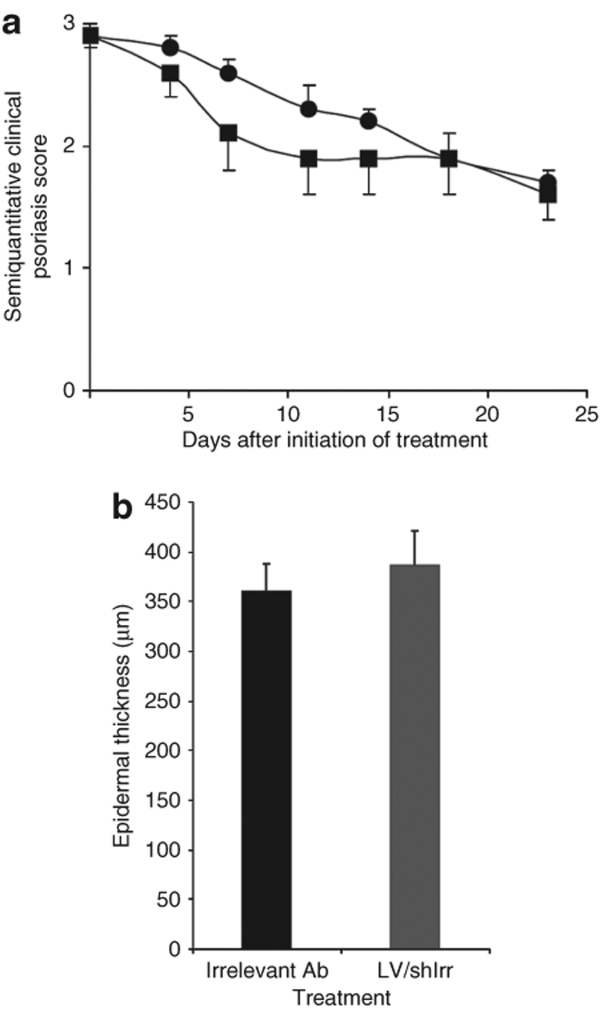
Mice treated with the lentiviral shIrr-encoding vector and the irrelevant antibody obtained similar semiquantitative clinical scores for the duration of the experiment (Figure 4a). Moreover, the epidermal thickness was indistinguishable between mice receiving the lentiviral control vector and the irrelevant antibody (385.9 ± 35.4 µm versus 360.7 ± 26.6 µm; P = 0.538) (Figure 4b), demonstrating a lack of phenotypical difference between these two groups. For the subsequent analyses we therefore collected all mice of groups 3 and 4 in a single negative control group (n = 17). It has to be mentioned that all mice showed normal well-being and weight gain throughout the study, suggesting that the treatment had no systemic influence on the physiology of the mice.
Figure 4.
Comparison of control treatments of psoriatic plaques in the xenograft transplantation model. (a) Semiquantitative clinical psoriasis scores were given twice weekly for 3 weeks to mice treated with the irrelevant monoclonal antibody (filled circles, n = 10) or LV/shIrr (filled squares, n = 7). The two control treatments showed no significant difference in semiquantitative clinical psoriasis scores. (b) Epidermal thickness of skin biopsies after treatment with the irrelevant monoclonal antibody and LV/shIrr. Upon sacrifice 3 weeks after treatment biopsies were taken, fixed and paraffin-embedded, and stained. Epidermal thickness was measured in irrelevant antibody-treated (black column, n = 10) or LV/shIrr-treated (gray column, n = 7) mice. The two control treatments showed no significant difference in the epidermal thickness. Data are presented as mean + SEM. shIrr, irrelevant shRNA.
To compare the effect of treatment with a single injection of shTNF-α3-encoding lentiviral vectors with five weekly injections of CsA (Figure 5a), the semiquantitative clinical psoriasis scores assigned to shTNF-α3- and CsA-treated mice were compared to those of the negative control group. During the 3 weeks of incubation, the scores of shTNF-α3-treated grafts reduced gradually in a pattern similar to the scores for CsA-treated mice (Figure 5b). shTNF-α3 significantly reduced the semiquantitative clinical psoriasis scores of the treated grafts compared to grafts treated with negative controls (P = 0.03). Histological examination of skin biopsies from psoriasis skin grafts revealed a significant decrease in epidermal thickness corresponding to a 22% reduction (P = 0.03) of the thickness in shTNF-α3-treated grafts relative to negative control-treated grafts (Figure 5c). The effect of repeated administration of CsA was confirmed in this assay (48% reduction of epidermal thickness in mice treated with CsA compared to the negative control group). Additionally, we investigated whether TNF-α mRNA was downregulated in psoriasis skin grafts treated with LV/shTNF-α3 and, therefore, performed quantitative reverse transcription (RT)-PCR on RNA extracted from treated skin grafts. The TNF-α mRNA level in the grafts treated with lentiviral shTNF-α3 vector was reduced with 29% relative to grafts isolated from mice in the negative control groups (Figure 5d). Moreover, the presence of lentiviral DNA in the treated psoriatic skin grafts after 3 weeks was confirmed by PCR analysis (data not shown). To verify that the level of TNF-α mRNA in the untreated lesional skin grafts was not artificially increased in the transplanted skin relative to psoriatic skin biopsies from patients, we also quantified TNF-α mRNA in lesional skin biopsies derived from seven patients. Notably, comparable levels of TNF-α mRNA were detected in lesional skin biopsies and untreated grafted lesional skin (Figure 5d), supporting the notion that shTNF-α3 in the xenotransplantation model did in fact reduce the level of TNF-α mRNA below the range observed in patients. Histological assessment of skin grafts revealed amelioration of the psoriasis phenotype by the injected lentiviral vectors. Negative control grafts had retained a characteristic psoriatic phenotype with elongated rete pegs (Figure 5f, panel A). In contrast, xenografts from both shTNF-α3- and CsA-treated animals (Figure 5f, panels B and C) demonstrated a psoriatic skin phenotype with 20–30% reduction of the epidermal thickness along with histological characteristics of prominent rete ridges, acanthosis and parakeratosis and thus partial correction of psoriasis with indications of progression toward skin normalization (as demonstrated by a comparison with a nonlesional skin sample, shown in Figure 5f, panel D).
Figure 5.
In vivo knockdown of TNF-α in human psoriasis skin by shTNF-α3. (a) The schedule for treatment of psoriasis plaques in vivo. Psoriatic plaque skin biopsies were transplanted onto SCID mice in the psoriasis xenograft transplantation model. All mice were allowed to heal for 10 days before onset of treatment. Grafts were treated either by a single injection with LV/shTNF-α3 or LV/shIrr, or five times weekly with CsA injections (positive control) and killed 3 weeks after treatment. Arrows indicate injection time points. (b) Semiquantitative clinical psoriasis scores were given twice weekly for 3 weeks to mice treated with negative control (filled squares, n = 17), LV/shTNF-α3 (open squares, n = 10), or CsA (positive control) (filled triangles, n = 12). shTNF-α3 significantly decreased the semiquantitative clinical psoriasis score (*P = 0.03). Intradermal injections were performed at day 0. (c) LV/shTNF-α3 treatment decreased the epidermal thickness in psoriasis skin grafts. Mice were killed after treatment and biopsies were taken, fixed, paraffin-embedded and stained with H&E. Epidermal thickness was measured in negative control-treated (black column, n = 17), LV/shTNF-α3-treated (dark gray column, n = 10), and CsA-treated (light gray column, n = 10) grafts. shTNF-α3 significantly decreased the epidermal thickness as compared to negative control (*P = 0.03). A measurement of nonlesional skin (n = 12) is included for comparison (white column). (d) Reduction of TNF-α gene expression in psoriasis skin in LV/shTNF-α3-treated skin. Biopsies were taken 3 weeks after treatment and TNF-α mRNA levels were measured by qPCR in negative control-treated (black column, n = 17) and in LV/shTNF-α3-treated (dark gray column, n = 9) grafts (P = 0.10). The black/white-graduated column (n = 7) represents the mean level of TNF-α mRNA detected in psoriatic plaque skin biopsies obtained from seven skin donors and which were not grafted onto mice. (e) The level of mouse TNF-α mRNA in the spleen is unaffected by shTNF-α3 treatment. Quantitative reverse transcription-PCR was performed on total RNA derived from spleens of mice treated with LV/shIrr (black column; n = 7) and LV/shTNF-α3 (dark gray column; n = 10), respectively (P = 0.77). (f) Histological assessment of grafted skin samples. Representative tissue samples of each treatment group are shown. Bar = 100 µm. All data are presented as mean + SEM. TNF-α, tumor necrosis factor-α; SCID, severe combined immunodeficiency; CsA, cyclosporin A; shIrr, irrelevant shRNA; H&E, hematoxylin and eosin.
shTNF-α3 does not target murine TNF-α (Figure 2d) and therefore is unlikely to affect the expression of murine TNF-α in the psoriasis xenograft transplantation model. To verify that the expression level of murine TNF-α was not affected in LV/shTNF-α3-treated animals, we performed quantitative RT-PCR on total RNA derived from the spleen of LV/shIrr- and LV/shTNF-α3-treated animals. Among the two groups (7 mice treated with LV/shIrr and 10 treated with LV/shTNF-α3), we did not detect any measurable difference in the RNA level (Figure 5e). As an additional control of potential systemic effects of the lentiviral vector, we analyzed DNA samples from spleens of LV/shIrr- and LV/shTNF-α3-treated animals for the presence of lentiviral DNA. Using different primer sets and a range of different PCR conditions, we were not able to detect DNA remnants of the shRNA-encoding vector (data not shown). Taken together, these findings suggest that putative long-term systemic effects affecting expression of endogenous TNF-α in the LV-treated psoriasis xenograft transplantation model are limited under the conditions used.
In summary, our findings demonstrate an efficient RNAi response in human psoriatic skin triggered by lentivirally delivered small RNAs targeting the 3′ UTR of the TNF-α gene followed by alleviation of the psoriasis phenotype in the xenograft transplantation model.
Discussion
TNF-α is upregulated in lesional psoriatic skin and plays a key role in psoriasis development.3,11,30 Treatment with specific inhibitors of TNF-α-protein, including antibodies and soluble receptors, causes resolution of psoriasis providing proof that TNF-α is indeed a crucial therapeutic target.31,32,33 Positive clinical effects of TNF-α antagonists, such as infliximab, adalimumab, and etanercept, require repeated systemic administration with potential negative and long-term side effects including increased risk of cancer and tuberculosis.34 Interests have been given therefore to alternative methods of intervention that may offer both a local and target-specific therapy. The ability to control disease-associated genes makes RNAi one of these potential alternatives for treatment of psoriasis. RNA effectors can be designed to target any gene involved in a disease and may potentially be administered locally to minimize side effects due to systemic administration of TNF-α antagonists in the patient. A recent report demonstrated that the increased TNF-α protein expression in lesional psoriatic skin is not the result of abnormal high levels of TNF-α mRNA but is caused by upregulation of gene expression at the post-transcriptional level triggered by an increased level of activated MK2.11 This could argue that targeting TNF-α mRNA molecules by RNAi-based intervention would have a limited, if any, effect. However, we hypothesized that reducing the pool of TNF-α mRNA would lead to a reduction of TNF-α protein, provided that regulation at the post-transcriptional level was not affected. Hence, targeting of TNF-α mRNA by local RNAi would lead to a reduction in the level of TNF-α protein in the skin and serve as a potential alternative strategy toward treatment of psoriasis.
To test this hypothesis we generated a panel of TNF-α-targeted shRNAs and demonstrated that the naturally occurring endogenous RNAi pathway can be harnessed in human skin to control production of cytokines involved in development of psoriasis. Based on functional evaluations of a panel of shRNAs directed against TNF-α-encoding mRNA, we identified one shRNA variant, designated shTNF-α3, which led to consistent downregulation of TNF-α expression. Among seven tested shRNA targets, shTNF-α3 was the only shRNA designed to target the 3′ untranslated region of the gene. Importantly, the murine TNF-α gene does not contain the shTNF-α3 target sequence, and the shRNA therefore should not potentially target mouse TNF-α mRNA but only human TNF-α mRNA in the psoriasis xenograft transplantation model. As lentiviral vectors pseudotyped with vesicular stomatitis virus G envelope protein facilitated efficient gene transfer to both epidermis and dermis, we employed lentiviral vectors for transfer of shTNF-α3-encoding cassettes to xenografted psoriatic skin. Following a single, intradermally administered dose of lentiviral vector in human skin, we detected a reduction in the level of TNF-α mRNA in shTNF-α3-treated skin samples as well as clinical and histological improvements of the psoriasis phenotype. The severity of the psoriasis phenotype, as evaluated by the semiquantitative clinical psoriasis score, improved following lentiviral treatment to an extent that was comparable with that observed in mice receiving systemically administered CsA five times weekly for the duration of the experiment. In addition, treatment with the shTNF-α3-encoding vector reduced epidermal thickness significantly in the skin xenografts. Together, our data showed that persistent TNF-α knockdown mediated by RNAi leads to reduced epidermal thickness and reduced clinical severity.
Recent in situ investigations have demonstrated that the TNF-α mRNA levels are not increased in lesional relative to nonlesional skin.11 Our results support a model by which a reduction of the TNF-α mRNA level in xenografts treated with potent TNF-α-directed shRNAs leads to an overall decrease of the production of TNF-α protein and amelioration of the psoriasis phenotype. These findings demonstrate the therapeutic potential of RNAi-based intervention in the xenotransplantation model and lend hope for the future use of RNA-based drugs in human skin. In further support of a claim of therapeutic utility of a TNF-α RNAi treatment in patients, we showed comparable levels of TNF-α mRNA in lesional skin biopsies and untreated lesional skin grafts. These findings suggest that TNF-α mRNA production in the lesional grafts was not artificially increased due to surgical stress or the process of wound healing and, hence, that a therapeutic effect of shTNF-α3 in the model was biologically relevant. To our knowledge, this is the first study employing siRNA technology in the xenograft transplantation model; however, at present we cannot exclude that specific properties of the model may influence the RNAi response either positively or negatively. Furthermore, it has to be mentioned that the level of TNF-α-encoding mRNA is variable among psoriatic skin from distinct patients which may to some degree influence the response to standard anti-TNF-α biological treatments. In accordance, it is not possible at this preliminary stage to predict whether an RNAi-based intervention has the full potential to manage the variable levels of TNF-α mRNA in lesional skin from different patients.
Our findings reveal the potential of small RNA effectors in treatment of skin disorders and establish a new platform for carrying out systematic validations of therapeutic targets among numerous potential target molecules involved in psoriasis development. Previous such in vivo validations have relied on systemic administration of antibodies with specificity for relevant proteins. Importantly, shRNAs can be designed to target any known gene based only on the primary gene sequence. Hence, by using lentiviral vectors for shRNA delivery it may be possible to target any predetermined gene in transplanted psoriasis skin and evaluate and compare degrees of psoriasis resolution. Furthermore, by construction of lentiviral vectors expressing two (or potentially more) different shRNAs, it should be possible to predict potential beneficial effects of combinatorial treatments targeting several key components in disease development.
In vivo lentiviral gene transfer to human skin xenografts has been carried out previously with results ranging from transduction of <1% of the human keratinocytes35 or just around the needle tract site of injection24 to efficient transduction resulting in transgene expression in all layers of the epidermis.36 Moreover, based on a single injection of lentiviral vectors encoding luciferase or human erythropoietin, Siprashvilli and Khavari documented efficient gene transfer to human skin transplanted onto severe combined immunodeficiency mice.25 The latter is consistent with our findings showing eGFP expression in all layers of the epidermis as well as in dermis, although eGFP detected as early as 3 days after injection most likely is derived from episomal vector DNA intermediates as well as integrated vectors. Furthermore, lentiviral vector DNA was easily detected in genomic DNA isolated from biopsies of treated skin 3 weeks after injection. Recent findings have demonstrated efficient and persistent gene expression from episomal lentiviral vector forms in quiescent cells in vivo.37 In the case of hyperproliferative psoriatic skin, unintegrated vector DNA will be diluted upon cell growth, and persistent shRNA expression leading to TNF-α downregulation is most likely entirely derived from integrated vectors. However, we cannot exclude that the beneficial effects of TNF-α-directed shRNAs are supported by an initial boost of shRNA production encoded by episomal templates shortly after the injection of vector particles.
As an easily accessible organ, skin is an attractive target for RNAi-based therapeutics. Recent findings have documented targeting of chimeric luciferase reporter RNA molecules by synthetic siRNAs injected into the paws of mice.38,39 Such data have led to a proposed clinical trial using siRNA directed against keratin genes in patients with dominant-negative keratin disorders.40 Other groups have reported effects of siRNAs delivered to skin by electroporation41,42 or by cream-emulsified preparations of siRNA43 as well as in human organotypic skin models,44 supporting potential future use of synthetic siRNAs in skin. However, recent striking reports exploring gene-regulatory efficacy of synthetic siRNAs directed against vascular endothelial growth factor in the eye have demonstrated that therapeutic effects of sequence-specific RNA agents administered as ex vivo synthesized RNAs are not necessarily triggered by sequence-specific degradation of mRNA. Notably, the positive effects were caused rather by the interaction of siRNAs with Toll-like receptors on the cell surface and induction of a cellular signalling cascade leading to inhibition of angiogenesis.45 Such findings call for extra caution when considering the beneficial effects of synthetic siRNAs. The present report provides evidence of in vivo knockdown of an endogenous gene in human skin facilitated by DNA-encoded shRNA molecules produced inside the treated cells. By exploring the potential of shRNA expression in skin our proof-of-concept study documents the potential of using RNAi for treatment of psoriasis, a disease that widely affects the human population worldwide and, moreover, provide a new strategy for validating psoriasis targets. We anticipate that RNAi in such studies of target validation will impact future regiments for treatment of psoriasis.
Materials and Methods
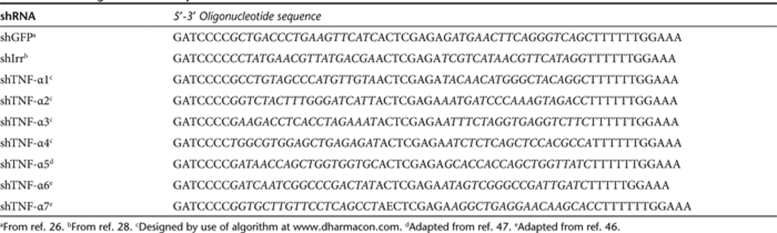
Plasmid construction. The sense strand of all shRNA-encoding DNA oligonucleotides designed for this study is presented in Table 1 with target sequences underlined. shTNF-α1 through shTNF-α4 were designed by using an algorithm provided by Dharmacon (Dharmacon siDESIGN http://www.dharmacon.com/ ). shTNF-α5 through shTNF-α7 were designed on the basis of murine TNF-α targets which have previously been found to be efficiently targeted by siRNAs.46,47 These sequences were modified to target the corresponding sequences in human TNF-α mRNA. All DNA oligonucleotides were obtained from DNA Technology (Risskov, Denmark). Complementary sense and antisense DNA oligonucleotides were annealed by incubation for 5 minutes at 100 °C followed by slow cooling. Annealed oligonucleotides carrying terminal BglII and XbaI overhangs were inserted downstream of the H1 promoter in BglII/XbaI-digested pBC.H1,23 generating pBC.H1-shTNF-α1 through pBC.H1-shTNF-α7. Similarly, shRNA-encoding cassettes, one with specificity for eGFP (shGFP) and one without any known target sequence in the human genome (shIrr, referred to as “scrRNA” in ref. 28), were inserted into pBC.H1 to provide appropriate negative controls. H1-shRNA expression cassettes were isolated from pBC.H1-shTNF-α1 through pBC.H1-shTNF-α7, pBC.H1-shGFP, and pBC.H1-shIrr by digestion with ClaI and inserted into the self-inactivating 3′ long terminal repeat (LTR) region of BbsI-treated pBSK-3′ LTR, as previously described,23 generating pBSK-3′LTR.H1-shTNF-α1 through pBSK-3′LTR.H1-shTNF-α7, pBSK-3′LTR.H1-shGFP, and pBSK-3′LTR.H1-shIrr, respectively. To generate the lentiviral vector plasmid pLV/PGK-puro, the puromycin resistance gene (puro) was amplified by PCR from pSBT/PGK-puro, previously described and referred to as pTpuro,48 digested with BamHI/XhoI, and inserted into the BamHI/XhoI-digested pCCL-WPS-PGK-eGFP-WHV (here referred to as pLV/PGK-eGFP).23 The original 3′ LTR of pLV/PGK-puro was replaced with 3′ LTRs containing the shRNA expression cassettes by digesting pBSK-3′LTR.H1-shTNF-α2 through pBSK-3′LTR.H1-shTNF-α4, pBSK-3′LTR.H1-shGFP, and pBSK-3′LTR.H1-shIrr with ScaI/KpnI and inserting the resulting fragments into ScaI/KpnI-digested pLV/PGK-puro, generating pLV/shTNF-α2 through pLV/shTNF-α4, pLV/shGFP, and pLV/shIrr. To generate psiCHECK-2-TNF-α, mRNA was isolated from human Psor T cells49 using Tri Reagent (Sigma, St Louis, MO) according to manufacturer's instructions. The mRNA was reverse-transcribed into cDNA using Cloned AMV First-Strand cDNA synthesis kit (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. TNF-α cDNA was amplified by PCR using TNF-α-specific primers 5′-AAAGCGGCCGCGAGCAGAG GCTCAGCAATGA-3′ and 5′-AAACTCGAGCCCTGACAAGCTGCCAG GC-3′. The TNF-α fragment was digested with XhoI/NotI and inserted into XhoI/NotI-digested psi-CHECK-2 (Promega, Madison, WI). A Sleeping Beauty DNA transposon vector encoding the TNF-α cDNA was created by amplification of TNF-α cDNA from psi-CHECK-2-TNF-α using primers 5′-AAAGCTAGCCCTGACAAGCTGCCAGGCA-3′ and 5′-AAAGCT AGCCTCATTTAGATCCTCACAC-3′. The PCR fragment was digested with NheI and inserted after the CMV promoter into NheI-digested pSBT/CMV.EF1α-zeo, creating a transposon, pSBT/CMV-TNF-α.EF1α-zeo, with two expression cassettes; TNF-α driven by the CMV promoter and the zeocin resistance gene driven by the EF1α promoter.
Table 1.
shRNA oligonucleotide sequences
Cell lines. HEK293, 293-TNF-α (see below), and 293T cells were cultured at 37 °C in 5% (vol/vol) CO2 and maintained in Dulbecco's modified Eagle's medium (Cambrex, Verviers, Belgium) supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (0.1 mg/ml), and L-glutamine (265 mg/l). Psor T cells49 were likewise cultured at 37 °C in 5% (vol/vol) CO2 and maintained in RPMI 1640 medium (Cambrex, Verviers, Belgium) supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (0.1 mg/ml), L-glutamine (265 mg/l), and recombinant human interleukin-2 (50 ng/ml; Chiron, Emeryville, CA). For generation of the 293-TNF-α cell line, an approach based on the Sleeping Beauty DNA transposon system was employed. HEK293 cells were seeded at a density of 2 × 104 cells/cm2 in 6-well dishes 1 day prior to transfection. Cotransfections were performed with a total of 2 µg DNA (1 µg pSBT/CMV-TNF-α.EF1α-zeo and 1 µg pCMV-SB50 the latter encoding the SB10 transposase) using FuGENE-6 (Roche, Basel, Switzerland) according to manufacturer's instructions. Two days after transfection cells were trypsinized and reseeded in appropriate dilutions. Diluted cells were grown in medium containing zeocin (200 µg/ml; Invitrogen) until single clones could be isolated and subcloned. Subsequently, 293-TNF-α cells were continuously grown in medium containing zeocin (200 µg/ml). TNF-α expression was verified by ELISA using a Human TNF-α Ready-SET-Go! kit (eBioscience, San Diego, CA).
In vitro analysis of shRNA efficiency. To perform the dual-luciferase reporter assay, HEK293 cells were seeded with a density of 104 cells/cm2 in 24-well dishes the day before transfection. Cells were transfected with a total of 0.4 µg DNA (0.04 µg psi-CHECK-2-TNF-α and 0.36 µg pBC.H1-shTNF-α1 through –shTNF-α7 and pBC.H1-shGFP) using FuGENE-6. Forty-eight hours after transfection, Renilla and Firefly luciferase activity was analyzed by using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to manufacturer's instructions. Reactions were performed in 96-well arrays and reading was performed in a multi-sample plate-reading luminometer (Berthold, Bad Wildbad, Germany). R-luc activity was normalized to Firefly luciferase activity and is presented relative to the negative shRNA control (pBC.H1-shGFP). In assays based on transient expression of TNF-α, HEK293 cells were seeded with a density of 5 × 104 cells/cm2 in 6-well dishes 1 day prior to transfection. Transfections were performed with a total of 2 µg DNA (0.2 µg pSBT/CMV-TNF-α.EF1α-zeo and 1.8 µg of pLV/shTNF-α2, pLV/shTNF-α3, pLV/shTNF-α3, or pLV/shGFP) using FuGENE-6. The medium was changed 24 hours after transfection and 48 hours after transfection the medium was collected and analyzed for TNF-α expression by ELISA, as described above. In transduction studies of shRNA-encoding lentiviral vectors, 293-TNF-α cells were seeded with a density of 2.5 × 104 cells/cm2 in 6-well dishes. On the following day, lentiviral supernatant corresponding to an MOI of ~40 was added to the cells and allowed to incubate overnight. Vector-containing medium was collected 48 hours after transduction and analyzed for TNF-α expression by ELISA. To follow TNF-α expression for 4 days after transduction, cells transduced with crude viral supernatants were trypsinized and reseeded at 5 × 104 cells/cm2 in 6-well dishes. After 48 hours, the medium was harvested and analyzed for TNF-α expression by ELISA. For analysis of TNF-α expression following lentiviral transduction at a lower MOI (MOI of ~2), 293-TNF-α cells were seeded at 5 × 103 cells/cm2 in 6-well dishes. On the following day the cells were transduced overnight with serially diluted lentiviral vectors and selected with puromycin (3 µg/ml; Sigma) for 8 days. Subsequently, the resistant population was seeded in normal medium which was collected after 24 hours for determination of the TNF-α concentration by ELISA. Whenever TNF-α expression was correlated to the number of cells and the lentiviral load, the cells were counted upon collection of medium for TNF-α analysis and the concentration of p24 Gag was determined by ELISA (see below).
Lentiviral vector production. For production of lentiviral vectors, 293T cells were seeded at a density of 5 × 104 cells/cm2 in 10-cm dishes 1 day before transfection. Cells were transfected by CaPO4 treatment with 3.75 µg pMD.2G, 3 µg pRSV-Rev, 13 µg pMDGP-Lg/RRE (constructs as in ref. 23), and 13 µg pLV/shTNF-α2, pLV/shTNF-α3, pLV/shTNF-α4, pLV/shGFP, pLV/shIrr, or pLV/PGK-eGFP. Forty-eight hours after transfection the viral supernatant was harvested and passed through 0.45 µm filters to remove cellular debris (Sarstedt, Nümbrecht, Germany). The resulting lentiviral vectors were designated LV/shTNF-α2, LV/shTNF-α3, LV/shTNF-α4, LV/shGFP, LV/shIrr, and pLV/PGK-eGFP. Colony-forming titer assays were performed on HEK293 cells, allowing us to determine the MOI for subsequent experiments. Briefly, HEK293 cells were seeded at a density of 5 × 103 cells/cm2 in 6-well dishes 1 day before transduction. Lentiviral supernatants were serially diluted and supplemented with polybrene (8 µg/ml; Sigma-Aldrich, Milwaukee, WI) before addition to the cells. Transduced cells were grown in medium containing puromycin (3 µg/ml; Sigma) for ~10 days. For in vivo transductions, the lentiviral supernatants were ultracentrifuged for 2 hours (4 °C at 25.000 r.p.m.) in a SW28 rotor (Beckman Coulter, Fullerton, CA). Virus pellets were resuspended overnight in PBS−/− at 4 °C in a volume of 1/300 of the original volume. The lentiviral vector yield was determined by measuring the amount of p24 Gag protein using a HIV-1 p24 Antigen ELISA Kit (ZeptoMetrix, Buffalo, NY) according to manufacturer's instructions.
Psoriasis xenograft transplantation model. Four psoriatic plaque skin biopsies were obtained from donors with moderate to severe plaque psoriasis aged 39–67 years (mean, 54 + 12 years). The psoriasis of the participants was untreated for at least 1 month prior to the time of biopsy. Informed consent was obtained and the study was approved by the Central Ethical Committee and conducted according to the Declaration of Helsinki protocols. Animal studies were carried out with permission from the Danish Experimental Animal Inspectorate. Each keratome skin biopsy, containing both epidermis and dermis, was split into several grafts (each 1.5 × 1.5 × 0.05 cm) and transplanted onto C.B-17 severe combined immunodeficiency mice, 6–8 weeks old (Taconic M & B, Silkeborg, Denmark), as described.27 Shortly, the mice were anesthetized prior to surgery by a subcutaneous injection of Ketaminol (ketamine, 100 mg/kg; Intervet, Skovlunde, Denmark) and Narcoxyl (xylazine, 10 mg/kg; Intervet, Skovlunde, Denmark). The back was shaved and part of the exposed skin removed. The grafts were sutured with absorbable 6–0 suture (Caprosyn, Tyco, Copenhagen, Denmark) and covered with Xeroform dressings (Sherwood Medical Company; Markham, Ontario, Canada) for 1 week. The mice were kept under pathogen-free conditions throughout the study. The grafts healed for 10 days before the mice were randomized and subjected to treatment as indicated.
Lentiviral vector penetration in human psoriatic skin. To examine the ability of lentiviral vectors to penetrate psoriatic skin, 150 µl of the LV/PGK-eGFP vector preparation was injected intradermally into grafted skin at a dose of 35 µg p24 Gag/ml. The mice were killed after 3 days and biopsies from the center of the graft were isolated, fixed, and embedded in paraffin. Sections were incubated overnight with mouse anti-GFP antibody (5 µg/ml; Clontech, Mountain View, CA, USA) and 30 minutes with the secondary antibody Alexa Fluor 488 conjugated with goat-anti-mouse IgG (5 µg/ml; Invitrogen) and finally embedded with antifade containing Hoechst 33258 (1 µg/ml; Molecular Probes, Eugene, OR). All sections were treated identically during the staining process and subsequently analyzed by fluorescence microscopy.
In vivo administration of shRNA-encoding lentiviral vectors. LV/shTNF-α3 and LV/shIrr were administered intradermally into the graft (at a dose of 51 ± 11.0 µg p24 Gag/ml in 150 µl) as a single treatment. As an additional negative control we injected mice IP with an irrelevant mAb previously used as a negative control treatment in this model.29 As positive control, CsA (20 mg/kg; CsA; Sandimmun, Novartis, Copenhagen, Denmark) was administered IP five times weekly (Figure 5a).
Xenograft evaluation following treatment. The severity of psoriatic lesions in the grafts was assessed blinded twice weekly and scored semiquantitatively according to the clinical signs: scaliness, induration, and erythema. On a scale from 0 to 3 a maximal score of 3 represents severe scale, induration, and erythema of the psoriatic xenografts, as described.29 After treatment, biopsies from the center of the graft were obtained, fixed, embedded in paraffin. The remaining grafted skin was snap-frozen in liquid nitrogen and stored at −80 °C for further analyses. Employing standard methods sections were stained histochemically with hematoxylin and eosin. Epidermal thickness was measured on three equally distantly cut hematoxylin and eosin-stained sections. All sections were blinded prior to evaluation and evaluated randomly. Mean epidermal thickness values for each graft in each treatment group were calculated, and the data summarized as mean ± SEM.
RNA isolation and quantitative RT-PCR. Skin biopsies, samples of grafted skin, or spleen were incubated in RNAlater-ICE (Ambion, Austin, TX) for 24 hours prior to RNA isolation, and RNA was purified employing the SV Total RNA Isolation System (Promega, Madison, WI) according to the manufacturer's instructions. In the lysis buffer the biopsies were homogenized 2 × 2 minutes at 25 Hz using a TissueLyser (Qiagen, Ballerup, Denmark). Isolated RNA was dissolved in RNase-/DNase-free water and stored until further use at −80 °C. For RT Taqman reagents (Applied Biosystems, Foster City, CA) were used according to the manufacturer's instructions. RT thermal cycling was performed using a Peltier Thermal Cycler-200 (MJ Research, Waltham, MA); 25 °C for 10 minutes, 48 °C for 30 minutes, and 95 °C for 5 minutes. Complementary DNA was stored at −80 °C. Human TNF-α mRNA expression was determined employing TNF-α (Hs00174128_m1, Applied Biosystems) and RPLP0 (Hs99999902_ml, Applied Biosystems) primers and probes (FAM-labeled MGB-probes) using a Taqman 20× Assay-By-Design gene expression assay mix (Applied Biosystems). Mouse TNF-α mRNA was measured using TNF-α (Mm00443258_m1, Applied Biosystems) and GAPDH (Mm99999915_g1, Applied Biosystems) primers. Expression of each gene was analyzed in triplicate. PCR conditions were 10 minutes at 95 °C, 50 cycles of 15 seconds at 95 °C and 60 seconds at 60 °C. A Rotorgene-3000 (Corbett Research, Sydney, Australia) real-time PCR machine was used. Relative gene expression levels were determined by using the relative standard curve method as outlined in User Bulletin no. 2 (ABI Prism 7700 sequencing detection system, Applied Biosystems). Briefly, a standard curve for each gene was made from fivefold serial dilutions of total RNA isolated from normal human epidermal keratinocytes. The curve was then used to calculate relative amounts of target mRNA in the biopsies. Mean mRNA values for all grafts in each treatment group were calculated, and the data summarized as mean ± SEM.
DNA isolation and PCR. Genomic DNA was isolated from another sample of the grafted skin by salt precipitation. Samples were lyzed by addition of lysisbuffer (0.5 ml; 10 mmol/l Tris, 1 mmol/l EDTA, 150 mmol/l NaCl, 0.5% SDS; pH 10.5) and Proteinase K was added (100 µg; Invitrogen) followed by incubation for 2 hours at 55 °C. 6 mol/l NaCl (165 µl) and samples were mixed by shaking followed by centrifugation at 4 °C, 13,000 r.p.m. for 10 minutes. Two volumes ice-cold 99% ethanol was added to the supernatant and DNA was precipitated by centrifugation 4 °C, 13,000 r.p.m. for 10 minutes. DNA pellets were resuspended in TE-buffer. The presence of lentiviral DNA was confirmed by PCR (using sense primer 5′-AATCAACCTCTGGATTAC-3′ and antisense primer 5′-GACTCTAGAACCACGGAATTGTCAGTGCC-3′) and sequencing of PCR amplicons.
Statistical analyses. The two-tailed Student's t-test was used to test the null hypothesis of no difference between treatment groups studied in tissue culture experiments. The assumption of equal variances was tested by the F-test. The nonparametric Mann–Whitney test was used to test for differences between treatment groups in semiquantitative clinical psoriasis scores and the t-test was used to test the null hypothesis of no difference between treatment groups for epidermal thickness and TNF-α gene expression. Each patient donated skin to several mice; however, observations made for different mice were assumed to be independent of each other. In all statistical analyses, P-values <0.05 were considered significant.
Acknowledgments
We thank Winnie Heidelmann for technical assistance and Frederik Dagnaes-Hansen for technical advice related to the animal work. This study was made possible through support by the Danish Medical Research Council, the Novo Nordisk Foundation, the Carlsberg Foundation, the Danish Cancer Society, Kgl. Hofbuntmager Aage Bangs Foundation, the Psoriasis Foundation, the Augustinus Foundation, and the EU (EU-FP6-STREP, contract number 018961). B.M. was funded by a grant from the Danish Cancer Society.
REFERENCES
- Krueger JG., and , Bowcock A.Psoriasis pathophysiology: current concepts of pathogenesis Ann Rheum Dis 200564ii30–ii36.Suppl 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nograles KE, Zaba LC, Guttman-Yassky E, Fuentes-Duculan J, Suárez-Fariñas M, Cardinale I, et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol. 2008;159:1092–1102. doi: 10.1111/j.1365-2133.2008.08769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyemura K, Yamamura M, Fivenson DF, Modlin RL., and , Nickoloff BJ. The cytokine network in lesional and lesion-free psoriatic skin is characterized by a T-helper type 1 cell-mediated response. J Invest Dermatol. 1993;101:701–705. doi: 10.1111/1523-1747.ep12371679. [DOI] [PubMed] [Google Scholar]
- Chong BF., and , Wong HK. Immunobiologics in the treatment of psoriasis. Clin Immunol. 2007;123:129–138. doi: 10.1016/j.clim.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borish LC., and , Steinke JW.2. Cytokines and chemokines J Allergy Clin Immunol 2003111S460–S475.2 Suppl [DOI] [PubMed] [Google Scholar]
- Ozawa M., and , Aiba S. Immunopathogenesis of psoriasis. Curr Drug Targets Inflamm Allergy. 2004;3:137–144. doi: 10.2174/1568010043343868. [DOI] [PubMed] [Google Scholar]
- Markham T, Mullan R, Golden-Mason L, Rogers S, Bresnihan B, Fitzgerald O, et al. Resolution of endothelial activation and down-regulation of Tie2 receptor in psoriatic skin after infliximab therapy. J Am Acad Dermatol. 2006;54:1003–1012. doi: 10.1016/j.jaad.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Partsch G, Steiner G, Leeb BF, Dunky A, Bröll H., and , Smolen JS. Highly increased levels of tumor necrosis factor-alpha and other proinflammatory cytokines in psoriatic arthritis synovial fluid. J Rheumatol. 1997;24:518–523. [PubMed] [Google Scholar]
- Tetta C, Camussi G, Modena V, Di Vittorio C., and , Baglioni C. Tumour necrosis factor in serum and synovial fluid of patients with active and severe rheumatoid arthritis. Ann Rheum Dis. 1990;49:665–667. doi: 10.1136/ard.49.9.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimund JM, Wittersheim C, Dumont S, Muller CD, Baumann R, Poindron P, et al. Mucosal inflammatory cytokine production by intestinal biopsies in patients with ulcerative colitis and Crohn's disease. J Clin Immunol. 1996;16:144–150. doi: 10.1007/BF01540912. [DOI] [PubMed] [Google Scholar]
- Johansen C, Funding AT, Otkjaer K, Kragballe K, Jensen UB, Madsen M, et al. Protein expression of TNF-alpha in psoriatic skin is regulated at a posttranscriptional level by MAPK-activated protein kinase 2. J Immunol. 2006;176:1431–1438. doi: 10.4049/jimmunol.176.3.1431. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE., and , Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Bagasra O., and , Prilliman KR. RNA interference: the molecular immune system. J Mol Histol. 2004;35:545–553. doi: 10.1007/s10735-004-2192-8. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W., and , Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N., and , Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R., and , Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ., and , Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplen NJ, Parrish S, Imani F, Fire A., and , Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K., and , Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ., and , Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Raoul C, Abbas-Terki T, Bensadoun JC, Guillot S, Haase G, Szulc J, et al. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- Baek SC, Lin Q, Robbins PB, Fan H., and , Khavari PA. Sustainable systemic delivery via a single injection of lentivirus into human skin tissue. Hum Gene Ther. 2001;12:1551–1558. doi: 10.1089/10430340152480276. [DOI] [PubMed] [Google Scholar]
- Siprashvili Z., and , Khavari PA. Lentivectors for regulated and reversible cutaneous gene delivery. Mol Ther. 2004;9:93–100. doi: 10.1016/j.ymthe.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Abbas-Terki T, Blanco-Bose W, Déglon N, Pralong W., and , Aebischer P. Lentiviral-mediated RNA interference. Hum Gene Ther. 2002;13:2197–2201. doi: 10.1089/104303402320987888. [DOI] [PubMed] [Google Scholar]
- Dam TN, Kang S, Nickoloff BJ., and , Voorhees JJ. 1alpha,25-dihydroxycholecalciferol and cyclosporine suppress induction and promote resolution of psoriasis in human skin grafts transplanted on to SCID mice. J Invest Dermatol. 1999;113:1082–1089. doi: 10.1046/j.1523-1747.1999.00811.x. [DOI] [PubMed] [Google Scholar]
- Thakker DR, Natt F, Hüsken D, Maier R, Müller M, van der Putten H, et al. Neurochemical and behavioral consequences of widespread gene knockdown in the adult mouse brain by using nonviral RNA interference. Proc Natl Acad Sci USA. 2004;101:17270–17275. doi: 10.1073/pnas.0406214101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenderup K, Rosada C, Worsaae A, Dagnaes-Hansen F, Steiniche T, Hasselager E, et al. Interleukin-20 plays a critical role in maintenance and development of psoriasis in the human xenograft transplantation model. Br J Dermatol. 2009;160:284–296. doi: 10.1111/j.1365-2133.2008.08890.x. [DOI] [PubMed] [Google Scholar]
- Ettehadi P, Greaves MW, Wallach D, Aderka D., and , Camp RD. Elevated tumour necrosis factor-alpha (TNF-alpha) biological activity in psoriatic skin lesions. Clin Exp Immunol. 1994;96:146–151. doi: 10.1111/j.1365-2249.1994.tb06244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew AL, Bennett A, Smith CH, Barker J., and , Kirkham B. Successful treatment of severe psoriasis and psoriatic arthritis with adalimumab. Br J Dermatol. 2004;151:492–496. doi: 10.1111/j.1365-2133.2004.06105.x. [DOI] [PubMed] [Google Scholar]
- Gisondi P, Gubinelli E, Cocuroccia B., and , Girolomoni G. Targeting tumor necrosis factor-alpha in the therapy of psoriasis. Curr Drug Targets Inflamm Allergy. 2004;3:175–183. doi: 10.2174/1568010043343903. [DOI] [PubMed] [Google Scholar]
- Gottlieb AB, Evans R, Li S, Dooley LT, Guzzo CA, Baker D, et al. Infliximab induction therapy for patients with severe plaque-type psoriasis: a randomized, double-blind, placebo-controlled trial. J Am Acad Dermatol. 2004;51:534–542. doi: 10.1016/j.jaad.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Lin J, Ziring D, Desai S, Kim S, Wong M, Korin Y, et al. TNFalpha blockade in human diseases: an overview of efficacy and safety. Clin Immunol. 2008;126:13–30. doi: 10.1016/j.clim.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Terunuma A, Pfutzner W, Foster RA., and , Vogel JC. In vivo assessment of gene delivery to keratinocytes by lentiviral vectors. J Virol. 2002;76:1496–1504. doi: 10.1128/JVI.76.3.1496-1504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya A, Sriwiriyanont P, Patel A, Saito N, Ohuchi A, Kitahara T, et al. Gene transfer in human skin with different pseudotyped HIV-based vectors. Gene Ther. 2007;14:648–656. doi: 10.1038/sj.gt.3302915. [DOI] [PubMed] [Google Scholar]
- Yáñez-Muñoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- Hickerson RP, Smith FJ, Reeves RE, Contag CH, Leake D, Leachman SA, et al. Single-nucleotide-specific siRNA targeting in a dominant-negative skin model. J Invest Dermatol. 2008;128:594–605. doi: 10.1038/sj.jid.5701060. [DOI] [PubMed] [Google Scholar]
- Smith FJ, Hickerson RP, Sayers JM, Reeves RE, Contag CH, Leake D, et al. Development of therapeutic siRNAs for pachyonychia congenita. J Invest Dermatol. 2008;128:50–58. doi: 10.1038/sj.jid.5701040. [DOI] [PubMed] [Google Scholar]
- Leachman SA, Hickerson RP, Hull PR, Smith FJ, Milstone LM, Lane EB, et al. Therapeutic siRNAs for dominant genetic skin disorders including pachyonychia congenita. J Dermatol Sci. 2008;51:151–157. doi: 10.1016/j.jdermsci.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Sugimoto M, Sakurai T, Saito R, Futaki N, Hashimoto Y, et al. Modulation of scratching behavior by silencing an endogenous cyclooxygenase-1 gene in the skin through the administration of siRNA. J Gene Med. 2007;9:994–1001. doi: 10.1002/jgm.1091. [DOI] [PubMed] [Google Scholar]
- Nakai N, Kishida T, Shin-Ya M, Imanishi J, Ueda Y, Kishimoto S, et al. Therapeutic RNA interference of malignant melanoma by electrotransfer of small interfering RNA targeting Mitf. Gene Ther. 2007;14:357–365. doi: 10.1038/sj.gt.3302868. [DOI] [PubMed] [Google Scholar]
- Ritprajak P, Hashiguchi M., and , Azuma M. Topical application of cream-emulsified CD86 siRNA ameliorates allergic skin disease by targeting cutaneous dendritic cells. Mol Ther. 2008;16:1323–1330. doi: 10.1038/mt.2008.91. [DOI] [PubMed] [Google Scholar]
- Mildner M, Ballaun C, Stichenwirth M, Bauer R, Gmeiner R, Buchberger M, et al. Gene silencing in a human organotypic skin model. Biochem Biophys Res Commun. 2006;348:76–82. doi: 10.1016/j.bbrc.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyhan AA, Vlassov AV, Ilves H, Egry L, Kaspar RL, Kazakov SA, et al. Complete, gene-specific siRNA libraries: production and expression in mammalian cells. RNA. 2005;11:837–846. doi: 10.1261/rna.7285805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen DR, Leirdal M., and , Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J Mol Biol. 2003;327:761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- Yant SR., and , Kay MA. Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol Cell Biol. 2003;23:8505–8518. doi: 10.1128/MCB.23.23.8505-8518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltoft K, Hansen BH, Pedersen CB, Pedersen S., and , Thestrup-Pedersen K. Common clonal chromosome aberrations in cytokine-dependent continuous human T-lymphocyte cell lines. Cancer Genet Cytogenet. 1995;85:68–71. doi: 10.1016/0165-4608(95)00118-2. [DOI] [PubMed] [Google Scholar]
- Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z., and , Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]