Abstract
RNA interference (RNAi) is a widely used gene suppression tool that holds great promise as a novel antiviral approach. However, for error-prone viruses including human immunodeficiency virus type 1(HIV-1), a combinatorial approach against multiple conserved sequences is required to prevent the emergence of RNAi-resistant escape viruses. Previously, we constructed extended short hairpin RNAs (e-shRNAs) that encode two potent small interfering RNAs (siRNAs) (e2-shRNAs). We showed that a minimal hairpin stem length of 43 base pairs (bp) is needed to obtain two functional siRNAs. In this study, we elaborated on the e2-shRNA design to make e-shRNAs encoding three or four antiviral siRNAs. We demonstrate that siRNA production and the antiviral effect is optimal for e3-shRNA of 66 bp. Further extension of the hairpin stem results in a loss of RNAi activity. The same was observed for long hairpin RNAs (lhRNAs) that target consecutive HIV-1 sequences. Importantly, we show that HIV-1 replication is durably inhibited in T cells stably transduced with a lentiviral vector containing the e3-shRNA expression cassette. These results show that e-shRNAs can be used as a combinatorial RNAi approach to target error-prone viruses.
Introduction
RNA interference (RNAi) is an evolutionarily conserved gene silencing mechanism that is induced by double-stranded RNA. RNAi plays an important role in the regulation of cellular gene expression as well as in innate antiviral immune responses.1,2,3 Besides its natural functions, RNAi is widely used as a tool to silence specific genes, with an associated array of therapeutic possibilities. Transfection of plasmids that express short hairpin RNAs (shRNAs) is commonly used to induce RNAi in mammalian cells.4,5 Like double-stranded RNA, these shRNAs are processed by the cellular Dicer endonuclease into ~22 base pairs (bp) small interfering RNA duplexes (siRNAs).2 One strand of the siRNA, the so-called “guide strand”, is incorporated into the RNA-induced silencing complex and programs this complex to cleave the perfectly complementary mRNA target.6,7 The other strand of the siRNA, the passenger strand, is degraded.8,9
RNAi targeted toward the human immunodeficiency virus type 1 (HIV-1) RNA genome via stable intracellular shRNA expression is highly effective in suppressing viral replication.10,11,12 However, the therapeutic use of a single shRNA is limited because of the rapid emergence of RNAi-resistant virus variants.13,14 These variants contain a deletion or point mutation within the target sequence that abolish the antiviral effect.15,16 To reduce the chance of escape from RNAi attack, the virus should be targeted simultaneously with multiple shRNAs. There are several combinatorial RNAi strategies to express multiple effective siRNAs.17,18 One can combine multiple shRNA-expression cassettes in a single vector.19,20,21 Alternatively, one can construct a microRNA-like polycistronic transcript that encodes multiple antiviral siRNAs.22 Another possibility is to express long hairpin RNAs (lhRNAs), from which multiple siRNAs can be processed.23 Several reports described virus inhibition using lhRNAs against HIV-1,24,25,26,27 hepatitis C virus,28 and hepatitis B virus.29 In contrast to transfection of double-stranded RNA molecules larger than 30 bp, the intracellular expression of lhRNA at an effective dose does not readily induce the interferon (IFN) response.24 However, it is important to note that even smaller RNA duplexes can activate the IFN response in a dose-dependent manner.30,31
We previously described a set of shRNAs with potent anti-HIV activity.19 Based on these shRNAs, we constructed extended shRNAs (e-shRNAs) that encode two siRNAs by stacking of the shRNA units on top of each other (e2-shRNAs).32 We showed that the siRNA derived from the base of the e2-shRNA is efficiently produced and fully active. However, the top siRNA was only produced when the hairpin stem reached a length of 43 bp.32 In this study, we designed and tested antiviral e-shRNAs that encode three or four siRNAs (e3 and e4-shRNAs). We show that intracellular expression of three appropriately stacked inhibitors as part of the 66 bp e3-shRNA is possible without triggering the IFN response. Expression of longer e-shRNAs resulted in an overall reduced RNAi activity. We show that the RNAi activity of the e-shRNAs correlates with the efficiency of expression and proper intracellular processing of these transcripts into functional siRNAs. Finally, we show that HIV-1 replication is durably inhibited in T cells expressing a stably integrated e3-shRNA expression cassette. These results provide important insight for the design of multi-shRNA hairpin constructs.
Results
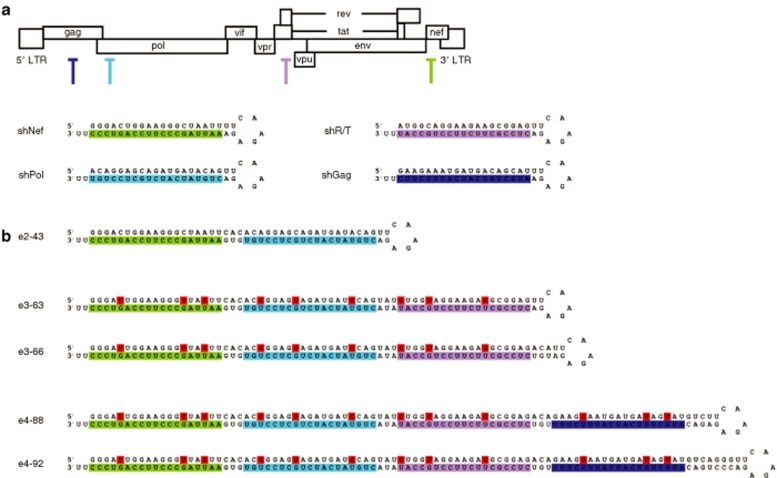
Design of e-shRNAs encoding three and four siRNAs against HIV-1
Previously, we demonstrated that a minimal hairpin stem length of 43 bp is required to generate two functional siRNAs from an e2-shRNA.32 In an attempt to construct e-shRNAs that can prevent the onset of HIV-1 escape, we designed and constructed e-shRNAs encoding three or four highly potent anti-HIV-1 siRNAs. We selected four potent antiviral shRNAs against different HIV-1 regions: nef, pol, rev/tat (r/t), and gag (Figure 1a).19 We build further on the successful e2-43 hairpin design (Figure 1b), with shNef at the base and shPol at the top of the hairpin stem and we used the same expression cassette driven by the H1 polymerase III promoter.32 The e-shRNAs have identical 5 nucleotide (nt) loop sequences and the top 2 bp of the pSUPER system4 and end with a 3′ UU overhang derived from the polymerase III termination signal. We made e-shRNA constructs that encode three siRNAs (e3-shRNAs) of 63 and 66 bp or four siRNAs (e4-shRNAs) of 88 and 92 bp (Figure 1b). These length variations were tested because the exact cleavage sites of Dicer on these extended stems are not known and inappropriate processing will affect the RNAi activity. The additional bp between the siRNAs were obtained by extending the passenger strand of the siRNA at its 3′ end with flanking HIV-1 derived sequences. We introduced multiple mutations in the passenger strand of the e-shRNA to generate relatively weak G-U bp in the hairpin stem (marked red in Figure 1b). We introduced 3 G-U bp/shRNA unit, thus yielding a total of 9 G-U bp in e3-shRNA and 12 G-U bp in e4-shRNA. This G-U bp modification has been shown to avoid IFN induction, to improve the stability of the plasmid in Escherichia coli and to allow sequencing of stem-loop structures.23,33 Importantly, introduction of G-U bp does not negatively affect the RNAi efficacy.33
Figure 1.
Design of e-shRNAs encoding 3 and 4 siRNAs. (a) The HIV-1 genome and the position of target sequences for the original 21 bp shRNAs (shNef, shPol, shR/T, and shGag). (b) Structure of the e2-shRNA and the e-shRNAs encoding 3 or 4 siRNAs of 63, 66, 88, or 92 bp. The e3 and e4 constructs were made with mutations in the passenger strand of the hairpins (indicated in red), which results in destabilizing G-U bp in the hairpin stem. The guide strand sequences are marked in colors. bp, base pair; HIV-1, human immunodeficiency virus type 1; shRNA, short hairpin RNA; siRNA, small interfering RNA.
RNAi activity of the antiviral e-shRNAs
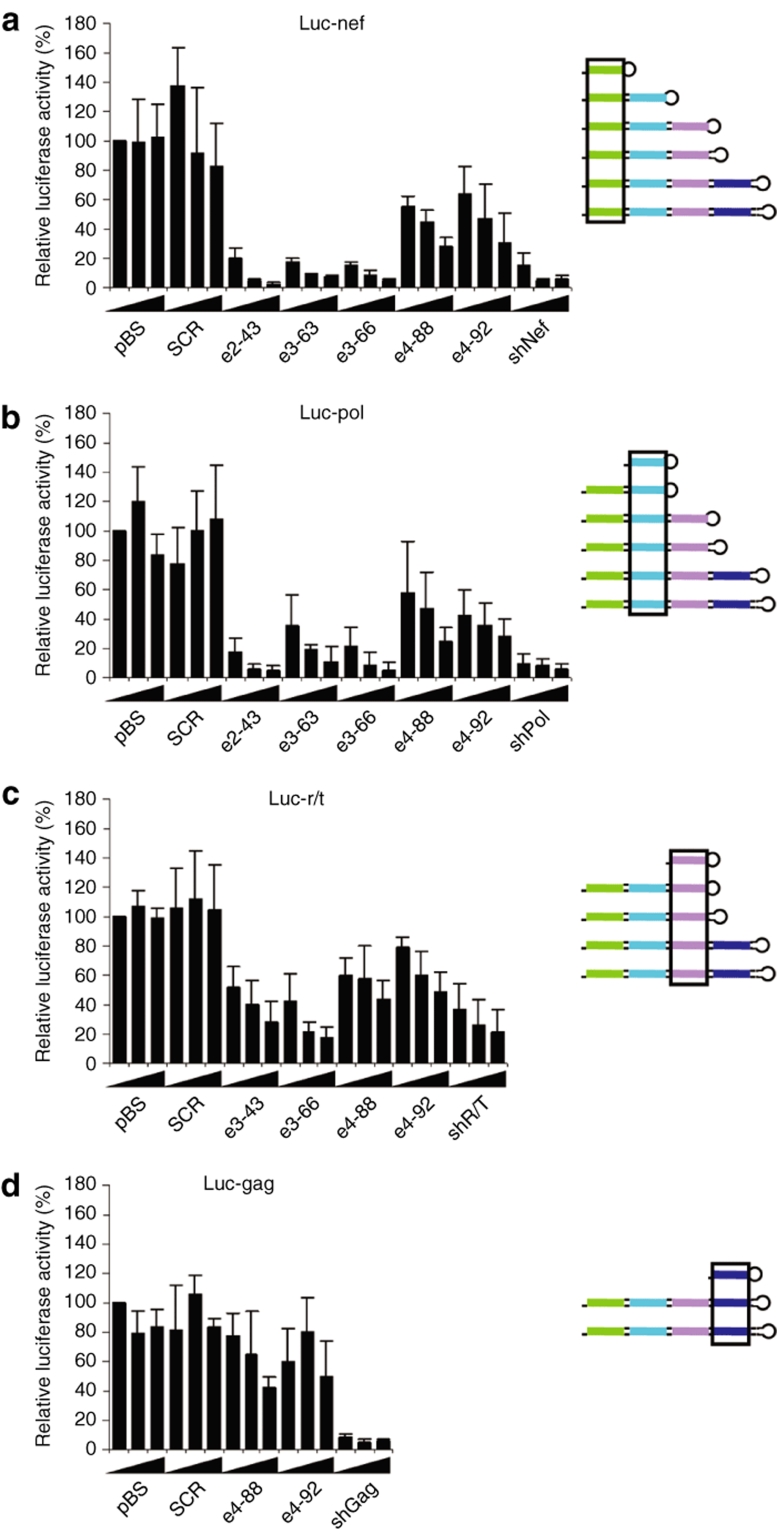
To evaluate the suppressive activities of the novel e3 and e4-shRNA constructs, we co-transfected human embryonic kidney (HEK) 293T cells with an increasing amount of these inhibitors together with a fixed amount of a luciferase reporter construct with the respective target. As positive controls, we transfected the original shRNAs: shNef, shPol, shR/T, shGag, and the e2-shRNA inhibitor e2-43. As negative controls, we included the unrelated plasmid pBluescript and the control scrambled hairpin construct that harbors the scrambled sequence of e2-43. The renilla luciferase expression plasmid was included to control for transfection efficiency. Firefly and renilla luciferase expression was measured 2 days post-transfection and the ratio was used to calculate the relative luciferase activity. The firefly/renilla ratio in the presence of pBluescript was set at 100%.
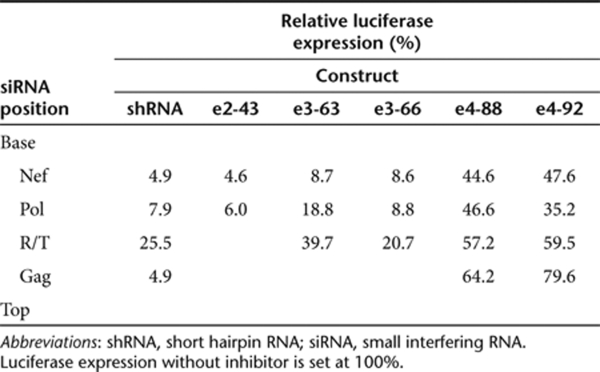
We first focus on the activity of the bottom shNef unit that is present in all e-shRNA designs. The e-shRNAs encoding two and three siRNAs (e2-43, e3-63, and e3-66) were equally active as the original shNef inhibitor on the Luc-nef reporter (Figure 2a). These results indicate that the first siRNA from the base of the e-shRNAs encoding two and three siRNAs is produced and functional. In contrast, the e4-88 and e4-92 constructs showed a significantly reduced inhibition of Luc-nef expression. Similar results were obtained when the e-shRNA constructs were tested on the Luc-pol reporter construct for the second shRNA unit (Figure 2b). Equally efficient luciferase knockdown was observed for e2-43 and the original shPol inhibitor, indicating that the production of the second siRNA in this e-shRNA is equally efficient as in the original shRNA construct. However, the production of the second siRNA targeting the pol gene seems slightly impaired for the e3-shRNAs, e3-63 and e3-66, and even more for the e4 constructs e4-88 and e4-92. On the Luc-r/t reporter, which was used to determine the activity of the third siRNA in the novel e-shRNAs, we observed good inhibition of luciferase expression by e3-63 and e3-66 that is comparable to the activity of shR/T (Figure 2c). In fact, e3-66 showed an increased knockdown of luciferase expression compared to e3-63, indicating that the addition of 3 bp improved the processing and/or activity of the top siRNA. Minimal knockdown of Luc-r/t expression was observed for the e4-88 and e4-92 constructs. We also tested the activity of the fourth siRNA encoded by e4-88 and e4-92 on the Luc-gag reporter construct (Figure 2d). Consistent with the previous results, hardly any RNAi activity was measured. These combined results indicate that effective e3-shRNAs can be build with a stem length of up to 66 bp. The addition of a fourth shRNA unit as in e4-88 and e4-92 has a general negative impact on all four units, including the shRNA at the base of the hairpin.
Figure 2.
RNAi activity of the different siRNAs produced by e-shRNAs. Luciferase reporter constructs encoding 50-nt HIV-1 sequences (a) Luc-nef, (b) Luc-pol, (c) Luc-r/t, and (d) Luc-gag, the e-shRNA variants and the renilla luciferase expression plasmid (pRL) were co-transfected into HEK 293T cells. Two days post-transfection, firefly and renilla luciferase expression levels were determined and the ratio was plotted as relative luciferase activity. Luciferase expression in the presence of the pBS control was set at 100%. A scrambled hairpin (SCR) was used as a negative control and the original shRNAs (shNef, shPol, shR/T, and shGag) were included as positive controls. Bars represent the average values from five independent transfections and error bars show the SD. HEK, human embryonic kidney; HIV, human immunodeficiency virus; nt, nucleotide; RNAi, RNA interference; shRNA, short hairpin RNA; siRNA, small interfering RNA.
RNAi activity of lhRNAs with different hairpin stem length targeting a consecutive target sequence
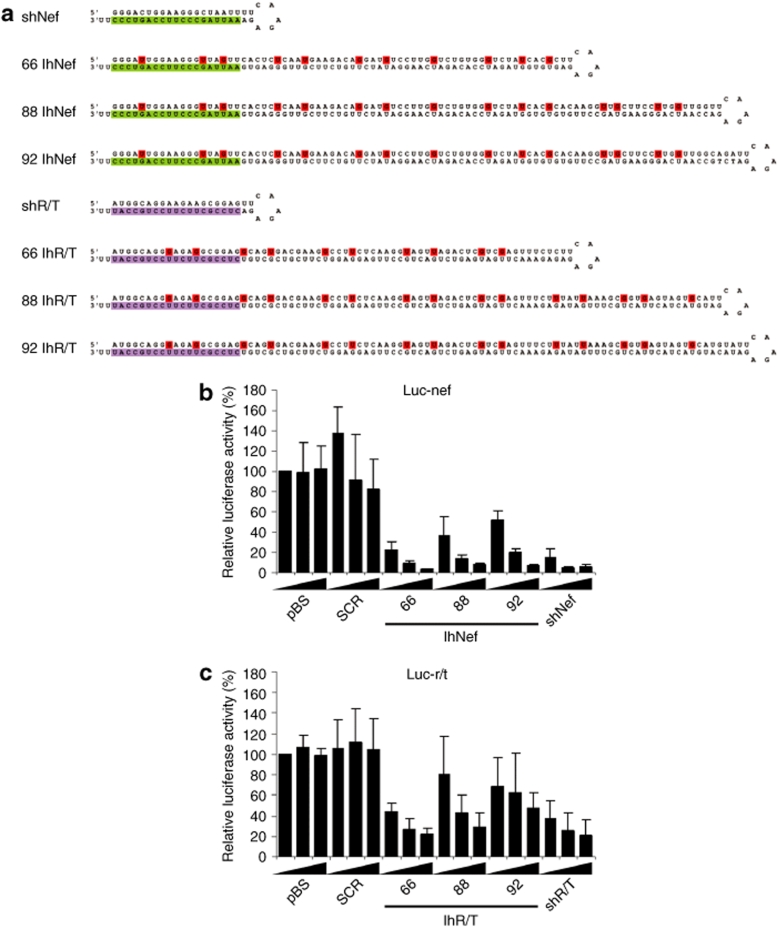
To test whether the length of the hairpin stem is the cause of the reduced RNAi activity, we also constructed lhRNA constructs of 66, 88, and 92 bp in size (Figure 3a). These hairpins were designed to contain an active siRNA sequence at the base of the hairpin, which is extended by adding consecutive HIV-1 sequences. We made two lhRNA sets with the siRNA sequence of shNef or shR/T at the base of the hairpin (lhNef and lhR/T). Theoretically, these constructs can produce three (66 bp) and four (88 and 92 bp) siRNAs that target adjacent regions of the HIV-1 RNA genome. In contrast to the e-shRNA design, we only know up front that the siRNA at the base of the hairpin represents a potent inhibitor. We again introduced point mutations in the passenger strand to generate G-U bp in the hairpin stem that facilitate cloning and sequencing (Figure 3a).
Figure 3.
RNAi activity of lhRNAs of different hairpin stem length. (a) Design of two sets of lhRNAs targeting a consecutive sequence of the HIV-1 nef and r/t gene of 66, 88, and 92 bp in length (lhNef and lhR/T). The shRNA targeting nef and r/t is placed at the base of the hairpin and is marked in green or purple, respectively. Mutations were introduced in the passenger strand of the hairpin (indicated in red) to create G-U bp in the hairpin stem (b) HEK 293T cells were co-transfected with the lhRNA constructs, the corresponding Luc-nef or Luc-r/t reporter and pRL. Two days post-transfection luciferase activities were measured and used to calculate the relative luciferase expression (firefly/renilla ratio). Luciferase expression in the presence of pBS was set at 100%. A scrambled hairpin (SCR) was used as negative control and the original shRNA, shNef, or shR/T, as positive control. Bars represent the average values from five independent transfections and error bars show the SD. bp, base pair; HEK, human embryonic kidney; lhRNA, long hairpin RNA; RNAi, RNA interference; shRNA, short hairpin RNA.
To quantify the RNAi activity of these hairpins, we co-transfected HEK 293T cells with an increasing amount of lhRNA constructs and a fixed amount of the corresponding luciferase reporter. Two days post-transfection, firefly and renilla luciferase expression was measured and the ratio was used to calculate the relative luciferase activity. The RNAi activity of lhNef showed a slight decrease when the hairpin stem length was extended from 66 bp to 88 or 92 bp (Figure 3b). The lhR/T showed a stronger reduction in luciferase knockdown with increasing stem length. In general, only the lhRNAs of 66 bp showed an RNAi activity that is comparable to that of the original shNef and shR/T inhibitors (Figure 3b).
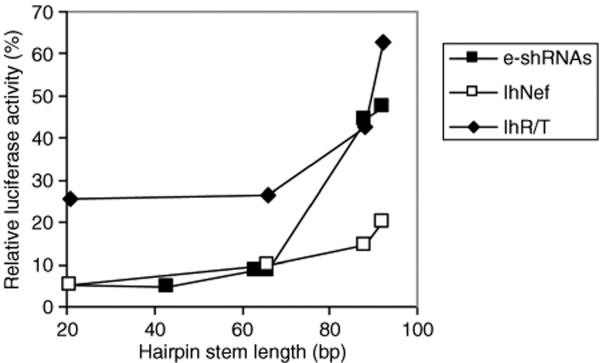
To summarize the data obtained for the e-shRNA and lhRNA constructs, we plotted the relative luciferase activity of the base shRNA against the hairpin stem length (Figure 4). A general trend is that a reduction in RNAi activity is observed when the hairpin stem length exceeds 66 bp. These results suggest that there are also strict limitations to the length of active e-shRNAs and lhRNAs.
Figure 4.
RNAi activity of hairpins with increasing hairpin stem length. The relative luciferase activity of the base shRNA of the e-shRNA constructs and the two lhRNA series were plotted against the hairpin stem length. Luciferase expression in the absence of inhibitor was set at 100%. lhRNA, long hairpin RNA; RNAi, RNA interference; shRNA, short hairpin RNA.
Inhibitory effect of each siRNA derived from the e-shRNAs
Several recent reports indicate that Dicer-mediated processing of lhRNAs may not be equal along the hairpin stem and that Dicer favors the production of siRNAs from the base of the hairpin stem.27,29,34,35 This may create a gradient of siRNA activity from the base (very active) to the top (poorly active). To see whether this also holds true for the e-shRNA design, we summarized the data obtained in the luciferase assays (Table 1). We observed similar knockdown efficiencies for the two siRNAs in the e2-43 hairpin when compared to the relative activities of the original shRNAs. For e3-63 and e3-66, shNef activity from the base of the hairpin was equal (8.7 and 8.6%, Table 1). For e3-63, we measured a gradient of siRNA production with the highest RNAi activity for the siRNA at the base and a gradual reduction in RNAi activity for the second and third siRNA toward the loop of the hairpin when compared to the shRNA controls. The optimized e3-66 construct resulted in enhanced activity of the second and third siRNA, resulting in a comparable RNAi activity as measured for the individual shRNAs. As already mentioned, further extension of the hairpins to 88 or 92 bp resulted in reduced knockdown of luciferase expression. We observed modest RNAi activity for the first three siRNAs and hardly any knockdown for the fourth siRNA. Thus, we observed a decrease in RNAi activity from the base to the top of the hairpin, which is consistent with previous studies.27,29,34
Table 1.
Differential siRNA activity along the e-shRNA stem
Processing of e-shRNAs and lhRNAs into siRNAs
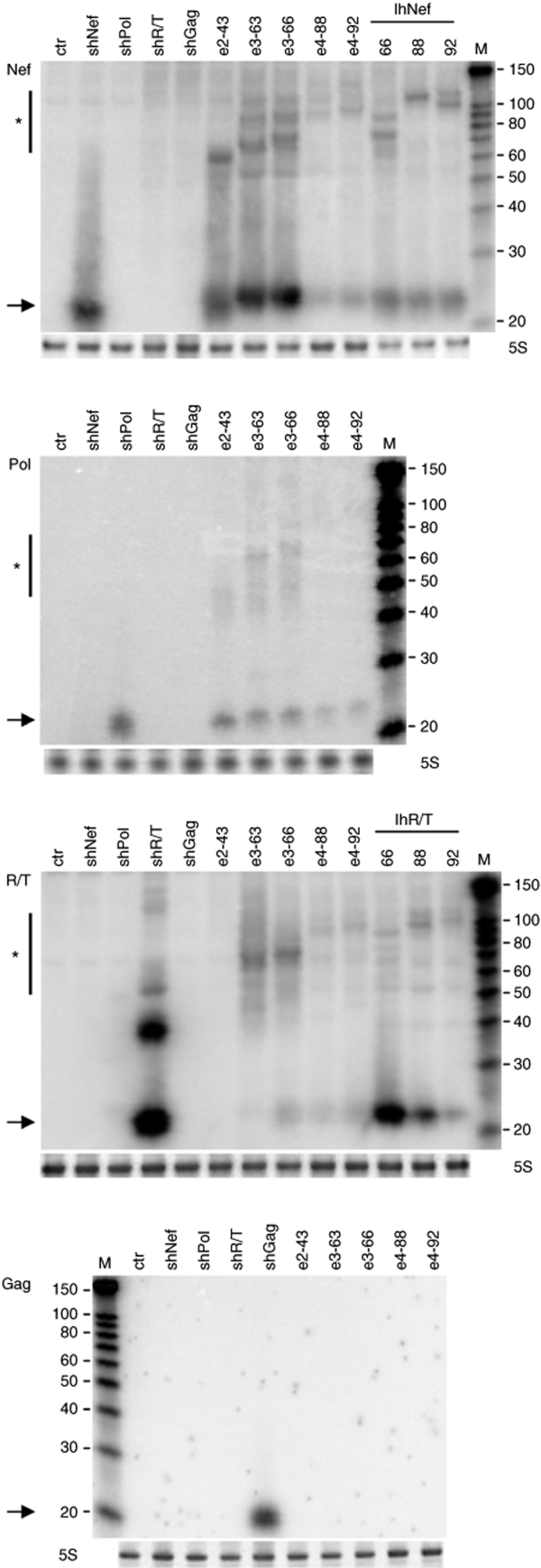
We next studied whether the differences in silencing activity of the units within the e-shRNA constructs correlated with their ability to be processed into functional siRNAs. For comparison, we also included the lhRNAs against nef and r/t sequences. We transfected HEK 293T cells with the hairpin RNA constructs and examined siRNA production by northern blotting. To detect siRNAs, we used 5′ end-labeled 19-nt locked nucleic acid probes that are complementary to the guide strand of the siRNA nef, pol, r/t, and gag.
A fair amount of fully processed siRNAs against nef, which is derived from the base of the e-shRNAs, was detected for e2-43, e3-63, and e3-66 (Figure 5, top panel). The original shNef was used as a positive control. A remarkable decrease in siRNA production was detected for the e-shRNAs encoding four siRNAs (e4-88 and e4-92). This result correlates well with the reduced RNAi activity of these constructs. A similar picture was obtained using the probe that detects the second siRNA against HIV-1 pol. Slightly less siRNAs were produced by e2-43, e3-63, e3-66 compared to the shPol control, but an obvious reduction in siRNA production was apparent for e4-88 and e4-92. A small reduction in siRNA production was apparent for the e3-shRNAs compared to the e2 constructs. For the third siRNA R/T, a more complex picture was observed. The amount of siRNA produced by all e-shRNAs is much reduced compared to the shR/T control. Nevertheless, it is clear that e3-66 is more efficiently processed than e3-63, e4-88, and e4-92, which is fully consistent with the measured RNAi activities. The gag probe that should detect the top siRNA of the e4-shRNAs constructs did not yield any signal, whereas a fair siRNA signal was detected for the shGag control (Figure 5, lower panel).
Figure 5.
Northern blot analysis of the e-shRNAs derived siRNAs. Total RNA was purified from HEK 293T cells transfected with the indicated e-shRNAs and lhRNAs and 10 µg was used to detect siRNAs against nef, pol, r/t, and gag with 19-nt complementary LNA oligonucleotide probes. As negative controls, the pSUPER vector (ctr) and unrelated shRNAs were used. The original shRNAs were used as positive controls, showing the processed ~22 nt siRNA. Precursor hairpin RNAs are indicated by asterisks. Bands that correspond to siRNAs are indicated by arrows. Ethidium bromide staining of the 5S rRNA band is shown below each panel as a control for equal sample loading. M represents the RNA size marker (in nt) and the probe name is indicated next to the blot. lhRNA, long hairpin RNA; LNA, locked nucleic acid; nt, nucleotide; shRNA, short hairpin RNA; siRNA, small interfering RNA.
For the lhRNA designs targeting Nef of 66, 88, and 92 bp, we detected an approximate steady level of siRNA from the base of the hairpin (Figure 5, top panel). These results are consistent with the luciferase knockdown experiments in which we measured only a slight decrease in RNAi activity when the hairpin stem length is extended from 66 bp to 88 or 92 bp. In contrast, the lhRNAs against the R/T region showed a strong reduction in siRNA production and RNAi activity with increasing stem length, suggesting that the magnitude of activity loss due to increased stem length may vary for different hairpins.
The e-shRNAs and lhRNAs do not induce the IFN response
To test whether the e-shRNAs induce the IFN response, we performed reverse transcriptase-PCR analyses for markers of the IFN pathway on transfected HEK 293T cells. Expression of the e3 and e4-shRNA constructs at the efficacious dose did not result in an upregulation of IFN-β, 2′-5′ oligoadenylate synthase, retinoic acid-inducible gene-I, ISG56, signal transduction and activator of transcription-1, and myxovirus resistant gene A mRNAs in HEK 293T cells (Figure 6a). We measured the β-actin mRNA levels as a control. As a positive control for IFN induction, HEK 293T cells were transfected with poly I:C, a synthetic double-stranded RNA that simulates a viral infection. Cells transfected with poly I:C caused an upregulation of IFN-related mRNAs, indicating that these cells are able to activate an innate IFN response. We performed the same analysis for the lhNef and lhR/T constructs, which also did not trigger the IFN response (Figure 6b).
Figure 6.
The e-shRNA and lhRNA transcripts do not induce the interferon response. Total RNA was purified from HEK 293T cells transfected with (a) e-shRNAs and (b) lhRNAs at an efficacious dose. The IFN-β, OAS, RIG-I, ISG56, STAT-1, and MxA expression levels were determined by RT-PCR. β-actin mRNA expression serves as an internal control. We included poly I:C transfected cells as positive control. As negative controls, cells were Mock transfected or with the pSUPER vector (ctr). We also performed a control PCR on RNA extracts derived from poly I:C transfected cells that were not subjected to reverse transcription. Either the 200 or 400 bp lane of the SmartLadder (M) (Eurogentec) is shown as size reference. HEK, human embryonic kidney; IFN-β, interferon-β; lhRNA, long hairpin RNA; MxA, myxovirus resistant gene A; OAS, 2′-5′ oligoadenylate synthase; RIG-I, retinoic acid-inducible gene-I; shRNA, short hairpin RNA; STAT-1, signal transduction and activator of transcription-1.
Efficient inhibition of HIV-1 production by e3-shRNAs
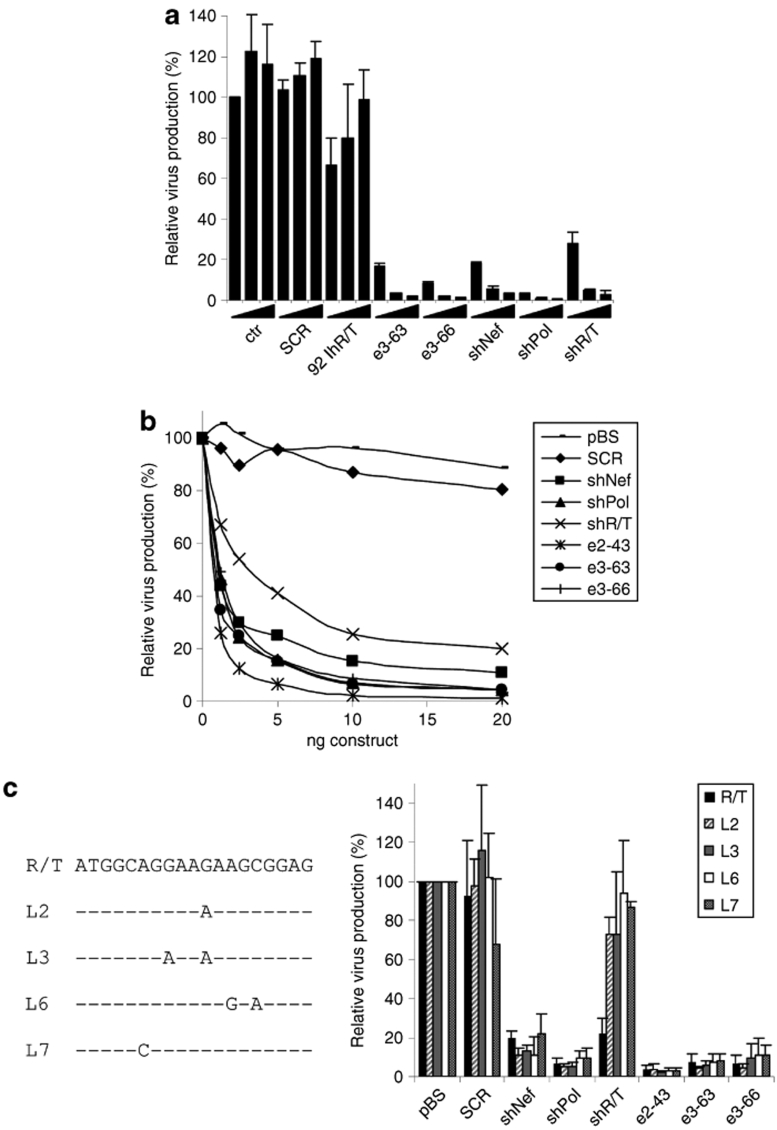
Among the novel e-shRNA designs, the e3-shRNAs e3-63 and e3-66 showed potent RNAi activity against all three HIV-1 target sites. We next tested whether these hairpins are indeed potent HIV-1 inhibitors. To quantify the antiviral effects of these hairpins we co-transfected the HIV-1 molecular clone LAI with an increasing amount of e3-63 and e3-66 into HEK 293T cells. Two days post-transfection, capsid p24 (CA-p24) levels in the culture supernatant were measured as an indicator of virus production. CA-p24 levels were normalized to the renilla luciferase expression of the co-transfected control plasmid. Virus production in the presence of the empty vector was set at 100%. We measured a potent knockdown of HIV-1 production with e3-63 and e3-66 (Figure 7a). At the lowest concentration, it is apparent that e3-66 is more efficient than e3-63. These e3-shRNAs show similar or better inhibition of virus production than the original shRNAs. The scrambled hairpin that was used as a negative control was not able to inhibit HIV-1 production, indicating that virus suppression by the e-shRNAs is sequence-specific. For the lhR/T of 92 bp hardly any knockdown of HIV-1 production was measured, providing further evidence that hairpin RNAs exceeding 66 bp are not efficient RNAi inducers.
Figure 7.
Inhibition of wild-type and mutant HIV-1 variants by e-shRNAs. (a) HEK 293T cells were co-transfected with the HIV-1 molecular clone pLAI, pRL, and increasing amounts of e3-shRNA constructs. Two days post-transfection, HIV-1 production was measured by CA-p24 quantification in the culture supernatant. CA-p24 levels were normalized for renilla expression; virus production with the pSUPER vector (ctr) was set at 100%. The original shRNAs were used as positive controls. The SCR served as negative control. (b) Inhibition of HIV-1 production by e-shRNAs at low concentrations. Transfections were performed as described for a, but now including the e2 design and we used reduced shRNA concentrations to carefully examine their potency. The SCR and pBS served as negative control. (c) Inhibition of RNAi-escape viruses by e-shRNAs. Sequence of the L2, L3, L6, and L7 HIV-1 variants that contain mutations in the R/T siRNA target region. Inhibition of CA-p24 expression was determined as described above. The mean values and standard deviations are based on three independent experiments. HEK, human embryonic kidney; HIV, human immunodeficiency virus; RNAi, RNA interference; SCR, scrambled hairpin; shRNA, short hairpin RNA.
To examine the potency of the e-shRNAs compared to shRNAs, we performed co-transfection experiments in which we carefully titrated the amount of hairpin RNA constructs (Figure 7b). In general, more potent inhibition was observed for the e2 and e3-shRNAs compared to the individual shRNAs. In fact, the highest knockdown efficiency was observed for the e2-shRNA. This may relate to the optimal expression and silencing activity of the second siRNA against Pol of e2-shRNA versus both e3 constructs (Figures 5 and 2, respectively). However, the e3-shRNA seems more attractive as anti-escape therapeutic because it targets three different sites in the HIV-1 genome.
To test the importance of targeting multiple sites as anti-escape approach, we tested the ability of the e-shRNAs to inhibit RNAi-escape mutants L2, L3, L6, and L7 with single or double point mutations in the viral target sequence (Figure 7c). Such mutations were previously observed in a large scale HIV-1 escape study.16 The three shRNA controls show the expected pattern. The shNef and shPol inhibitors do potently suppress production of the R/T-escape viruses, but the shR/T inhibitor looses its activity on all R/T-escape variants. The e2-shRNA does not encode siRNA against R/T and is thus also not sensitive to mutations in the R/T target. The siRNA against R/T is encoded at the top of both e3 constructs, but we nevertheless measured potent inhibition of all R/T-escape viruses. This inhibition is obviously due to the activity of the other two siRNA units of the e3 molecules. These results show the importance of targeting multiple sites to avoid the emergence of viral escape.
HIV-1 replication is suppressed in cells expressing e3-66
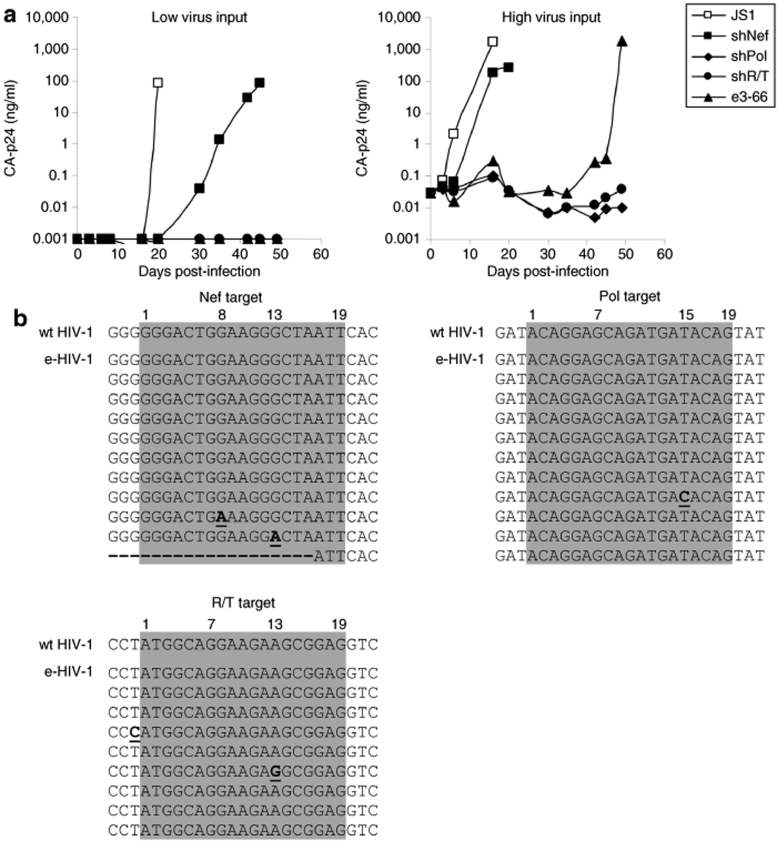
To follow long-term inhibition of virus replication by the e3-66 transcript, we transduced SupT1 T cells with a lentiviral vector (JS1) expressing the e3-66 hairpin or the control shNef. The titer of the e3-66 lentiviral vector was very poor, which is likely caused by the presence of Nef target sequences in the vector backbone.36 We were able to overcome this problem by saturation of the RNAi machinery in the producer cell with an excess of unrelated shRNAs.37 Transduced SupT1 cells were selected by fluorescence-activated cell sorting for green fluorescent protein expression. The cells were subsequently expanded and infected with HIV-1. Cells transduced with the empty JS1 vector were used as a negative control. Virus replication was followed for 49 days by monitoring the CA-p24 level in the culture supernatant. Virus replication and virus-induced cytopathic effects were observed in cells expressing the empty JS1 lentiviral vector at day 20 (Figure 8a, left panel). At day 35, virus replication was also observed in the shNef cells, however virus replication was not observed for the cells expressing the potent inhibitors shPol, shR/T, and e3-66. When we used a high amount of challenge virus, we observed virus replication and virus-induced cytopathic effects in cells expressing the empty JS1 lentiviral vector already at day 6 (Figure 8a, right panel). At day 16, virus replication was observed in the cells expressing shNef, whereas virus replication is inhibited for 49 days in the cells expressing e3-66. Eventually viral breakthrough occurred in these cells. The finding that HIV-1 is controlled significantly longer in cells expressing the e3-66 hairpin compared to those expressing shNef suggests that the second and third siRNA contribute to the virus inhibition by the e3-66 construct. We performed clonal sequencing of the viruses that appeared in the e3-66 expressing cells to determine if these represent true escape variants. The three target sequences were analyzed. We detected G-to-A mutations at position 8 and 13 of the Nef target (Figure 8b). In addition, one clone contained a big deletion of 312 nt in the Nef region, which includes 17 nt of the Nef target. One clone contained a T-to-C point mutation at position 15 in the Pol target. For the R/T target region we detected a single clone with an A-to-G point mutation at position 13 of the target. We also detected a T-to-C point mutation 1 nt upstream of the target sequence.
Figure 8.
Suppression of HIV-1 replication in e3-66 expressing T cells. SupT1 cells were transduced with the lentiviral vector encoding e3-66 shNef, shPol, or shR/T. The empty JS1 vector was used as a negative control. (a) Transduced GFP-positive cells were selected by sorting and infected with low (left panel) and high (right panel) virus input. Virus replication was monitored for 49 days by measuring CA-p24 in the culture supernatant. (b) Clonal sequencing reveals the genetic alterations in the three target regions in different viral clones. GFP, green fluorescent protein; HIV, human immunodeficiency virus.
Discussion
Previous studies showed that RNAi induced against HIV-1 via stable shRNA expression can efficiently suppress viral replication.10,11,12 However, the impact of a single shRNA inhibitor is not sufficient, because of the rapid emergence of RNAi-resistant virus variants with a mutation in the target sequence.13,14,15 To prevent the onset of HIV-1 escape, a combinatorial RNAi approach is required in which the virus is targeted simultaneously with multiple siRNAs. We previously generated e2-shRNAs that encode two potent siRNAs against different HIV-1 sequences.32 Based on this e2-shRNA design we now build e3 and e4-shRNAs that encode three and four siRNAs, respectively. We included mutations in the passenger strand of the e3 and e4-shRNAs to obtain relatively unstable G-U bp in the hairpin. This G-U design facilitates cloning and sequencing of the hairpin encoding inverted repeats, without negatively affecting the hairpin's RNAi activity.23,33
We showed that the e4-shRNAs have reduced RNAi activity compared to the e3-shRNAs. The same length effect was observed for lhRNAs that target a consecutive viral target sequence. In agreement with our results, previous reports also suggested that hairpin transcripts have an upper size limit for effective production of multiple, functional siRNAs.27,34 In general, the hairpins seem to lose activity when they get larger than ~66 bp. Northern blot analyses showed a coinciding decrease in siRNA production from the e4-shRNAs. This decrease in siRNA production could be due to reduced e4-shRNA expression, diminished stability, hampered nuclear export, or poor processing into functional siRNAs. In the latter case, more precursor transcripts or intermediately processed products should have been observed on northern blot, which was not the case. These combined results indicate that the reduced RNAi activity of the e4-shRNAs is due to diminished expression and/or stability. Diminished expression of the lhRNA transcripts could be related to the use of the H1 pol III promoter. It could be that the e4-88 (~187 nt) and e4-92 (~195 nt) hairpin transcripts are less efficiently transcribed by this promoter. In this case, other promoters could be tested, for example: the strong and ubiquitous pol II promoter of the human U1 small nuclear RNA.20,38
Another possibility for the inactivity of the e4-transcripts is that the wrong strand of the siRNA, the passenger strand, is selected for incorporation into the RNA-induced silencing complex. To test this, we performed northern blot analyses with probes that detect the passenger strand, but did not detect increased passenger strand expression for the e4-shRNAs compared to the e2 and e3-shRNAs (results not shown).
We succeeded to generate an e3-66 molecule that produces three active siRNAs, where our previous hairpin stacking attempts stopped at two siRNAs.32 By size optimization, more specifically the addition of 3 bp to e3-63, the processing of the top siRNA was activated in e3-66. We observed a gradient of siRNA production from the base toward the top of the hairpin. We compared the potency of the e-shRNAs compared to the shRNAs to inhibit HIV-1 production. In general, we observed more potent inhibition for the e2 and e3-shRNAs than the individual shRNAs. Overall, we measured optimal HIV-1 inhibitory activity of the e2 design, but the advantage of e3 molecules against an evolving viral target is that the selection of escape-variants is more difficult. For RNAi inhibition of static cellular mRNAs, the e2 design seems optimal. In case an even more complete gene knockdown is required, it would seem preferential to use two e2 molecules in a combinatorial RNAi approach.17,18
We previously showed that combinatorial RNAi using four separate shRNA expression cassettes efficiently prevents the chance of HIV-1 escape from RNAi.20 This approach requires the use of different pol III promoters to prevent the loss of individual shRNA expression cassettes from the lentiviral vector via homologous recombination. The e-shRNA design aimed to provide combinatorial RNAi from a single expression cassette. HIV-1 replication is suppressed for >7 weeks in cells that stably express the e3-66 hairpin, whereas viral breakthrough is readily apparent for the single shNef control. Nevertheless, virus replication was observed in the cells expressing e3-66 when a high amount of challenge virus was used. These results illustrate the effectiveness of the e3-shRNA design in providing durable HIV-1 inhibition compared to the shNef. These data indicate that the expression of the second and third siRNA from the e3-66 hairpin does significantly contribute to the observed virus inhibition. To further optimize the e3-66 hairpin design, we should place a more potent shRNA at the base of the hairpin since shNef is at most a modest inhibitor. An important advantage of the e3-shRNA design is that it causes only a modest reduction in the lentiviral vector titer, whereas the titer of the four shRNA vector is much reduced.20 The ability to obtain high lentiviral vector titers is critical for the clinical development of a gene therapy.
We performed clonal sequencing of the viruses that appeared in the e3-66 expressing cells to determine whether genetic alterations were selected in the target sequences. We found point mutations and a gross deletion in the Nef target sequence. These genetic alterations likely do not impact on virus replication because the Nef protein is not required for viral replication in vitro. In addition, we found a silent point mutation in the Pol target that has also been detected as a popular escape route in a large-scale evolution study.16 We also detected point mutations in the R/T target. One mutation is in fact positioned 1 nt upstream of the R/T target, suggesting that siRNAs of variable lengths are generated. Another mutation was located in the target region and was also previously described as escape route, although this mutation causes an amino acid change in both the Tat and Rev proteins.16 It is important to notice that we only observed scattered escape mutations, as none of them became dominant in the viral quasispecies. These results indicate that it is difficult for the virus to develop full resistance to the e3-shRNA.
Besides escape, another major challenge toward an effective anti-HIV therapy is the need to target the vast number of different virus variants that are present in the epidemic and in an infected individual. Multiple e2 or e3-shRNA expression cassettes may provide the profound and broad inhibition that is required to simultaneously block different HIV-1 variants. Another possibility is to use multiple e3-shRNA expression cassettes to target HIV-1 and cellular cofactors, which may result in an even more potent and durable inhibition. An adverse effect of this approach is the increased chance of off-target effects or saturation of the RNAi machinery. Future experiments in an appropriate in vivo model should reveal whether multiple e-shRNAs can provide durable HIV inhibition without toxicity.39
In conclusion, we showed that stacked e-shRNA constructs have potential for long-term inhibition of HIV-1 replication. However, there is an upper size limit such that maximally three siRNAs can be generated from a single e-shRNA. Moreover, we showed that a gradient of siRNA activity is observed along the hairpin stem, with the most efficiently expressed siRNA from the base of the hairpin. Importantly, we demonstrated that HIV-1 replication is durably inhibited in T cells expressing a stably integrated e3-shRNA expression cassette. These results provide important insights for the use of e-shRNAs for a durable inhibition of escape-prone viral pathogens. In addition, the e-shRNA design may present a useful new research tool for the knockdown of multiple genes.
Materials and Methods
DNA constructs. The e-shRNA constructs were made by annealing of complementary oligonucleotides (containing the BamHI and HindIII sites) and inserting them into the BglII and HindIII sites of the pSUPER vector.4 The lhRNAs targeting the nef and r/t region were made by PCR amplification of the guide strand of the hairpin RNA with a primer including the 5-nt loop sequences and top 2 bp of the pSUPER system. The passenger strand of the hairpin RNA was made by annealing oligonucleotides containing the loop and some additional nucleotides that are complementary to the guide strand of the hairpin. The passenger and guide strand of the hairpin was combined by a fusion PCR with primers containing the BamHI and HindIII sites. The PCR products were purified from agarose gels and digested with BamHI and HindIII and inserted into the BglII and HindIII sites of the pSUPER vector. The 43 bp control scrambled hairpin construct encodes the scrambled sequence of e2-43 and has been described previously.32 The construct expressing the 5xshRNA Gag5, Pol1, Pol6, Pol9, Pol47 from repeated H1 promoters was constructed as described previously.40
The pLAI plasmid encoding the HIV-1 isolate LAI41 was used to study inhibition of HIV-1 production. For the R/T mutants, a single mutation was introduced at position 11 for L2 and position 6 for L7 and double mutations were introduced at position 8/11 and 13/15 in the target sequence for L3 and L6, respectively. Such mutations have been previously observed in a large scale HIV-1 RNAi-escape study.16 Mutations were introduced by fusion PCR using oligonucleotides with the desired mutations.
Firefly luciferase reporter constructs (pGL3; Promega, Madison, WI): Luc-nef, Luc-pol, Luc-r/t, and Luc-gag were made by insertion of a 50–70-nt HIV-1 sequence, with the 19-nt target region in the centre, in the EcoRI and PstI sites of pGL3-Nef.15,19,20
Lentiviral vector plasmids are derived from the construct (pRRLcpptpgkgfppreSsin),42 which we renamed as JS1. The plasmids JS1-shNef, JS1-shPol, JS1-shR/T and JS1-e3-66 were obtained by digestion of the pSUPER construct with XhoI and PstI and inserting this fragment into the corresponding sites of JS1.
All DNA constructs were sequence verified using the BigDye terminator cycle sequencing kit (ABI, Foster City, CA). Hairpin RNA constructs were sequenced using a sample denaturation temperature of 98 °C and upon addition of 1 mol/l betaine. We used the Mfold program to calculate the thermodynamic stability of the RNA secondary structure of the antiviral transcripts.43
Cell culture, transfections, and virus infection. HEK 293T cell line was cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (FCS) (Hybond, Escondido, CA), penicillin (100 U/ml), streptomycin (100 µg/ml), and minimal essential medium nonessential amino acids (DMEM/10% FCS) at 37 °C and 5% CO2. For luciferase assays, HIV-1 inhibition assays and the IFN assay, HEK 293T cells were plated 1 day before transfection in 24-well plates at a density of 1.4 × 105 cells/well in 1 ml DMEM/10% FCS without antibiotics. Cells were transfected with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Transfection experiments were corrected for between-session variation as described previously.44
For siRNA analysis, HEK 293T cells were plated 1 day before transfection in 6-well plates at a density of 6.0 × 105 cells/well in 2 ml DMEM/10% FCS without antibiotics. The next day, cells were transfected with 5 µg of the hairpin RNA constructs with Lipofectamine 2000 reagent (Invitrogen) as suggested by the manufacturer.
The human T-cell line SupT1 was cultured in Advanced RPMI (Gibco BRL, Carlsbad, CA) supplemented with L-glutamine, 1% FCS, penicillin (30 U/ml), and streptomycin (30 µg/ml) at 37 °C and 5% CO2. SupT1 cells (200.000) were infected with HIV-1 (0.03 or 0.001 ng CA-p24) produced in HEK 293T cells. When HIV-induced cytopathic effects were observed, cell and supernatant samples were stored at −80 °C. Virus spread was followed by measuring the CA-p24 levels in the culture supernatant by enzyme-linked immunosorbent assay.
Luciferase assays. HEK 293T cells were co-transfected with 100 ng of the firefly luciferase expression plasmid, 1 ng of renilla luciferase expression plasmid (pRL-CMV) and 5, 25, or 100 ng of hairpin RNA expression constructs. We added pBluescript SK-(pBS) (Promega) to all transfections to obtain equal DNA concentrations. Two days post-transfection, firefly and renilla luciferase activities were assessed using the Dual-luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Relative luciferase activities were calculated from the ratio between firefly and renilla luciferase activities. Transfection experiments were corrected for between-session variation as described previously.44
HIV-1 inhibition assay. HEK 293T cells were co-transfected with 250 ng of the HIV-1 infectious molecular clone pLAI, 1 ng of pRL-CMV and 1.25, 2.5, 5, 10, 20, or 25 ng of the hairpin RNA constructs. We added pBluescript to obtain an equal DNA concentration in each transfection. To determine inhibition of the RNAi-escape variants L2, L3, L6, and L7, 250 ng of the molecular clones were co-transfected with 1 ng of pRL-CMV and 10 ng of the e-shRNA constructs. Virus production was determined at 2 days post-transfection by measuring the CA-p24 level in the culture supernatant by enzyme-linked immunosorbent assay as described previously.45 Cells were lysed and 5 µl of the lysate was used to measure the renilla luciferase activities using the Renilla Luciferase Assay System (Promega). The relative CA-p24 production was calculated as the quotient between the CA-p24 level and the renilla luciferase activity, which was corrected for between-session variation.44
Northern blot analyses. Total cellular RNA was extracted from transfected HEK 293T cells at 2 days post-transfection using the mirVana microRNA isolation kit (Invitrogen) according to the manufacturer's protocol. For northern blot analysis, 10 µg of total RNA per lane was resolved on urea denaturing 15% polyacrylamide gel (Invitrogen). RNA molecular weight markers (Ambion, Foster City, CA) were prepared as suggested by the manufacturer's protocol and run alongside the cellular RNA. To check for equal sample loading, ribosomal RNA in the gel was stained with 2 µg/ml ethidium bromide for 20 minutes. Destaining was performed by rinsing the gel three times with milliQ water for 10 minutes. The ribosomal RNA bands were visualized under ultraviolet light. The RNA samples in the gel were electrotransferred to a positively charged nylon membrane (Boehringer Mannheim, Mannheim, Germany) and crosslinked to the membrane using ultraviolet light at a wavelength of 254 nm (1,200 µJ × 100). Locked nucleic acid oligonucleotide probes were 5′ end labeled with the kinaseMax kit (Ambion) in the presence of 1 µl of [γ-32P]adenosine triphosphate (0.37 MBq/µl; Perkin Elmer, Waltham, MA). The probes were purified on Sephadex G-25 spin columns (Amersham Biosciences, Piscataway, NJ) to remove unincorporated nucleotides. Hybridizations were performed with labeled locked nucleic acid oligonucleotides in 10 ml ULTRAhyb hybridization buffer (Ambion) at 42 °C according to the manufacturer's instructions. We used oligonucleotide probes (locked nucleic acid-positions underlined): nef probe: 5′ GGGACTGGAAGGGCTAATT 3′, pol probe: 5′ ACAGGAGCAGATGATACAG 3′, r/t probe: 5′ ATGGCAGGAAGAAGC GGAG 3′, gag probe: 5′ GAAGAAATGATGACAGCAT 3′. After overnight hybridization, the membranes were washed twice for 5 minutes at 42 °C in 2 × SSC/0.1% sodium dodecyl sulfate and twice for 15 minutes at 42 °C in 0.1 × SSC/0.1% sodium dodecyl sulfate. Signals were detected using a phosphorimager (Amersham Biosciences).
IFN assay. To determine whether the extended hairpin RNA constructs induce the IFN pathway at their efficacious dose, HEK 293T cells were transfected with 25 ng of the hairpin encoding constructs using Lipofectamine 2000 reagent (Invitrogen). Transfection of 2 µg poly I:C was used as a positive control for IFN-β induction as described previously.24 Total RNA was isolated from cells 1 day post-transfection using the RNeasy mini kit (Invitrogen) according to the manufacturer's protocol. Genomic DNA was removed by DNase treatment using the TURBO DNA-free kit (Ambion). One microgram of total RNA, Thermoscript RT (Invitrogen) and random hexamer primers (Invitrogen) were used to reverse transcribe the RNA into complementary DNA according to the manufacturer's instructions (Invitrogen).
PCR amplification was performed on 2 µl of this complementary DNA-containing mixture with IFN-β, 2′-5′ oligoadenylate synthases, retinoic acid-inducible gene-I, ISG56, signal transduction and activator of transcription-1, myxovirus resistant gene A, and β-actin specific primers. The following primer combinations were used:
IFN-β, f: 5′-GCCGCATTGACCATCTATGAGA-′3; r: 5′-GAGATC TTCAGTTTCGGAGGTAAC-′3 (345 bp product), 2′-5′ oligoadenylate synthase, f: 5′-TCAGAAGAGAAGCCAACGTGA-′3; r: 5′-CGGAGAC AGCGAGGGTAAAT-′3 (399 bp product),46 retinoic acid-inducible gene-I, f: 5′-GAGTGTCTTTTCTTATGTGATTTT-′3; r: 5′-GCAGGCAA GTCTTACATGGCAGCA-′3 (268 bp product), ISG56, f: 5′-CTTGAGCC TCCTTGGGTTCG-′3; r: 5′-GCTGATATCTGGGTGCCTAAGG ′3 (137 bp product),47 signal transduction and activator of transcription-1, f: 5′-TTCTGTGTCTGAAGTGTAAGTGAA-′3; r: 5′-TAACACGGGGAT CTCAACAAGTTC-′3 (159 bp product), myxovirus resistant gene A, f: 5′-AGTATGGTGTCGACATACCGGA-′3; r: 5′-GAGTCTGGTAAACAGCC GAATG-′3 (145 bp product),47 β-actin f: 5′-GACTACCTCATGAAGATCC TCAC-3′; r: 5′-ATTGCCAATGGTGATGACCTG-3′ (197 bp product).
For PCR amplification, the Reddymix Master Mix (Abgene, Epsom, UK) was used in a 50 µl reaction using the following PCR program: 3 minutes at 95 °C, 30 cycles of 30 seconds at 95 °C, 30 seconds at 57 °C, 45 seconds at 72 °C and a final extension for 8 minutes at 72 °C. The PCR products were analyzed on a 1.5% agarose gel with the SmartLadder (Eurogentec, San Diego, CA) as a size reference.
Lentiviral vector production and transduction. Lentiviral vector plasmids are derived from the construct JS1 (pRRLcpptpgkgfppreSsin).42 For production of lentiviral vector, 6.0 × 105 HEK 293T cells were seeded per well in 6-well plates in 4 ml of DMEM/10% FCS without antibiotics. The next day, medium was replaced with 0.4 ml medium without antibiotics. Subsequently, the empty JS1 vector and JS1 vectors expressing shNef, shPol, shR/T and e3-66 (0.95 µg) were co-transfected with packaging plasmids pSYNGP (0.6 µg),48 RSV-rev (0.25 µg), pVSVg (0.33 µg),49 and a 5 shRNA-expressing construct, p5xshRNA (2.9 µg) with Lipofectamine 2000 reagent in accordance with the manufacturer's recommendations (Invitrogen). The second day, medium was replaced with 1 ml optimem. On the third day, medium containing lentiviral vector was harvested. Cellular debris was removed by centrifugation for 5 minutes at 1,200 r.p.m. Lentiviral stocks were titrated on SupT1 cells to determine the vector titer. SupT1 cells were transduced with the lentiviral vectors at a multiplicity of infection of 0.15 as described previously.19 Four days post-transduction, live transduced cells were sorted with fluorescence-activated cell sorting and single green fluorescent protein-positive cells were selected. Sorted cells were expanded and resorted at 30 days post-transduction. These cells were used to determine long-term inhibition of HIV-1 replication.
Sequencing proviral target regions. Cellular DNA of infected SupT1 cells with the integrated provirus was isolated as described previously.50 Integrated proviral DNA sequences were PCR amplified with the following primer pairs (indicated 5′-3′). Nef region, forward: ATTCG CCACATACCTAGAAG, reverse: GACTCTGGTAACTAGAGATCCC TCAGACC; Pol region, forward: AGGCTAATTTTTTAGGGAAGA TCTGGCCTTCC, reverse: GCAAATACTGGAGTATTGTATGGATT TTCAGG and R/T region, forward: ACCTTGTCTAGAATGGAGCCAG TAGATCCTAGACTAGAGCCCTG, reverse: TTCCACACAGGTA CCCCATAATAGACTGTGACCCACA. The PCR products were cloned into the pCR2.1 TOPO vector and subsequently clones were sequenced with the T7 or M13 reverse primers.
Acknowledgments
We thank Stephan Heynen for performing the CA-p24 ELISA. We thank Mustafa Ceylan for the generous gift of the 5 shRNA-expressing construct p5xshRNA. RNAi research in the Berkhout laboratory is sponsored by NWO-CW (TOP grant).
REFERENCES
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE., and , Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Meister G., and , Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Haasnoot J, Westerhout EM., and , Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R., and , Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ., and , Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D., and , Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA., and , Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP., and , Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S., and , Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Jacque JM, Triques K., and , Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Miyoshi H, Yamamoto N, Yamamoto N, Inoue J., and , Tsunetsugu-Yokota Y. Lentivirus vectors expressing short hairpin RNAs against the U3-overlapping region of HIV nef inhibit HIV replication and infectivity in primary macrophages. Blood. 2006;108:3305–3312. doi: 10.1182/blood-2006-04-014829. [DOI] [PubMed] [Google Scholar]
- Boden D, Pusch O, Lee F, Tucker L., and , Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, et al. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhout EM, Ooms M, Vink M, Das AT., and , Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eije KJ, ter Brake O., and , Berkhout B. Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J Virol. 2008;82:2895–2903. doi: 10.1128/JVI.02035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., and , Kay MA. Combinatorial RNAi: a winning strategy for the race against evolving targets. Mol Ther. 2007;15:878–888. doi: 10.1038/sj.mt.6300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP., and , Berkhout B. Combinatorial RNAi strategies against HIV-1 and other escape-prone viruses. Int J BioSci Technol. 2008;1:1–10. [Google Scholar]
- ter Brake O, Konstantinova P, Ceylan M., and , Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- ter Brake O, ‘t Hooft K, Liu YP, Centlivre M, von Eije KJ., and , Berkhout B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- Henry SD, van der Wegen P, Metselaar HJ, Tilanus HW, Scholte BJ., and , van der Laan LJ. Simultaneous targeting of HCV replication and viral binding with a single lentiviral vector containing multiple RNA interference expression cassettes. Mol Ther. 2006;14:485–493. doi: 10.1016/j.ymthe.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Liu YP, Haasnoot J, ter Brake O, Berkhout B., and , Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Miyagishi M, Yokota T, Watanabe T, Hino T, Nishina K, et al. Escape from the interferon response associated with RNA interference using vectors that encode long modified hairpin-RNA. Mol Biosyst. 2005;1:382–390. doi: 10.1039/b510159j. [DOI] [PubMed] [Google Scholar]
- Konstantinova P, de Vries W, Haasnoot J, ter Brake O, de Haan P., and , Berkhout B. Inhibition of human immunodeficiency virus type 1 by RNA interference using long-hairpin RNA. Gene Ther. 2006;13:1403–1413. doi: 10.1038/sj.gt.3302786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitsuji H, Kohara M, Kannagi M., and , Masuda T. Effective suppression of human immunodeficiency virus type 1 through a combination of short- or long-hairpin RNAs targeting essential sequences for retroviral integration. J Virol. 2006;80:7658–7666. doi: 10.1128/JVI.00078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barichievy S, Saayman S, von Eije KJ, Morris KV, Arbuthnot P., and , Weinberg MS. The inhibitory efficacy of RNA POL III-expressed long hairpin RNAs targeted to untranslated regions of the HIV-1 5' long terminal repeat. Oligonucleotides. 2007;17:419–431. doi: 10.1089/oli.2007.0095. [DOI] [PubMed] [Google Scholar]
- Sano M, Li H, Nakanishi M., and , Rossi JJ. Expression of long anti-HIV-1 hairpin RNAs for the generation of multiple siRNAs: advantages and limitations. Mol Ther. 2008;16:170–177. doi: 10.1038/sj.mt.6300298. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sudoh M, Miyagishi M, Akashi H, Arai M, Inoue K, et al. Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype. Gene Ther. 2006;13:883–892. doi: 10.1038/sj.gt.3302734. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Ely A, Barichievy S, Crowther C, Mufamadi S, Carmona S, et al. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol Ther. 2007;15:534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL., and , Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Sledz CA, Holko M, de Veer MJ, Silverman RH., and , Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Liu YP, Haasnoot J., and , Berkhout B. Design of extended short hairpin RNAs for HIV-1 inhibition. Nucleic Acids Res. 2007;35:5683–5693. doi: 10.1093/nar/gkm596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M, Sumimoto H, Miyoshi H, Kawakami Y., and , Taira K. Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J Gene Med. 2004;6:715–723. doi: 10.1002/jgm.556. [DOI] [PubMed] [Google Scholar]
- Saayman S, Barichievy S, Capovilla A, Morris KV, Arbuthnot P., and , Weinberg MS. The efficacy of generating three independent anti-HIV-1 siRNAs from a single U6 RNA Pol III-expressed long hairpin RNA. PLoS ONE. 2008;3:e2602. doi: 10.1371/journal.pone.0002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soifer HS, Sano M, Sakurai K, Chomchan P, Saetrom P, Sherman MA, et al. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 2008;36:6511–6522. doi: 10.1093/nar/gkn687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Brake O., and , Berkhout B. Lentiviral vectors that carry anti-HIV shRNAs: problems and solutions. J Gene Med. 2007;9:743–750. doi: 10.1002/jgm.1078. [DOI] [PubMed] [Google Scholar]
- Poluri A., and , Sutton RE. Titers of HIV-based vectors encoding shRNAs are reduced by a dicer-dependent mechanism. Mol Ther. 2008;16:378–386. doi: 10.1038/sj.mt.6300370. [DOI] [PubMed] [Google Scholar]
- Denti MA, Rosa A, Sthandier O, De Angelis FG., and , Bozzoni I. A new vector, based on the PolII promoter of the U1 snRNA gene, for the expression of siRNAs in mammalian cells. Mol Ther. 2004;10:191–199. doi: 10.1016/j.ymthe.2004.04.008. [DOI] [PubMed] [Google Scholar]
- ter Brake O, Legrand N, von Eije KJ, Centlivre M, Spits H, Weijer K, et al. Evaluation of safety and efficacy of RNAi against HIV-1 in the human immune system (Rag-2(−/−)(c)(−/−)) mouse model Gene Ther 2008. epub ahead of print [DOI] [PubMed]
- ter Brake O., and , Berkhout B. Therapeutic Oligonucleotides. RSC Publishing: Cambridge; 2008. Development of an RNAi-based gene therapy against HIV-1. In: Kurreck, J (ed.) pp. 296–311. [Google Scholar]
- Peden K, Emerman M., and , Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- Seppen J, Rijnberg M, Cooreman MP., and , Oude Elferink RP. Lentiviral vectors for efficient transduction of isolated primary quiescent hepatocytes. J Hepatol. 2002;36:459–465. doi: 10.1016/s0168-8278(01)00308-7. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Thygesen HH, Schoneveld OJ, Das AT, Berkhout B., and , Lamers WH. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology. 2006;3:2. doi: 10.1186/1742-4690-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K., and , Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehaud C, Megret F, Lafage M., and , Lafon M. Virus infection switches TLR-3-positive human neurons to become strong producers of beta interferon. J Virol. 2005;79:12893–12904. doi: 10.1128/JVI.79.20.12893-12904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Ziegler K, Ananthula P, Co JK, Frisque RJ, Yanagihara R, et al. JC virus induces altered patterns of cellular gene expression: interferon-inducible genes as major transcriptional targets. Virology. 2006;345:457–467. doi: 10.1016/j.virol.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Kotsopoulou E, Kim VN, Kingsman AJ, Kingsman SM., and , Mitrophanous KA. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J Virol. 2000;74:4839–4852. doi: 10.1128/jvi.74.10.4839-4852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinova P, de Haan P, Das AT., and , Berkhout B. Hairpin-induced tRNA-mediated (HITME) recombination in HIV-1. Nucleic Acids Res. 2006;34:2206–2218. doi: 10.1093/nar/gkl226. [DOI] [PMC free article] [PubMed] [Google Scholar]