Abstract
The utilization of organized supramolecular assemblies to exploit the synergistic interactions afforded by close proximity, both for enzymatic synthesis and for the degradation of recalcitrant substrates, is an emerging theme in cellular biology. Anaerobic bacteria harness a multiprotein complex, termed the “cellulosome,” for efficient degradation of the plant cell wall. This megadalton catalytic machine organizes an enzymatic consortium on a multifaceted molecular scaffold whose “cohesin” domains interact with corresponding “dockerin” domains of the enzymes. Here we report the structure of the cohesin–dockerin complex from Clostridium thermocellum at 2.2-Å resolution. The data show that the β-sheet cohesin domain interacts predominantly with one of the helices of the dockerin. Whereas the structure of the cohesin remains essentially unchanged, the loop–helix–helix–loop–helix motif of the dockerin undergoes conformational change and ordering compared with its solution structure, although the classical 12-residue EF-hand coordination to two calcium ions is maintained. Significantly, internal sequence duplication within the dockerin is manifested in near-perfect internal twofold symmetry, suggesting that both “halves” of the dockerin may interact with cohesins in a similar manner, thus providing a higher level of structure to the cellulosome and possibly explaining the presence of “polycellulosomes.” The structure provides an explanation for the lack of cross-species recognition between cohesin–dockerin pairs and thus provides a blueprint for the rational design, construction, and exploitation of these catalytic assemblies.
Protein–protein recognition plays a pivotal role in an array of biological processes. One fundamental example is the degradation of the most abundant reservoir of organic carbon in the biosphere, the plant cell wall, by anaerobic organisms. These organisms utilize a high molecular mass (megadalton) cellulase–hemicellulase complex termed the “cellulosome,” in which an extensive repertoire of glycoside hydrolases are grafted on a macromolecular scaffold (1, 2). It is generally believed that assembly of the catalytic components into a complex enhances the synergistic interactions between enzymes with complementary activities, leading to more efficient plant cell wall degradation (3, 4). The cellulosome of the anaerobic bacterium Clostridium thermocellum has been extensively studied (1, 4, 5). In this complex, the enzymes are bound to a noncatalytic protein termed the “scaffoldin” (CipA), which, in turn, binds to cell-surface anchoring proteins (4). CipA contains nine reiterated sequences referred to as “cohesin” domains (6) that interact with the catalytic subunits (7). Enzymes that are components of the C. thermocellum cellulosome contain a noncatalytic domain, referred to as the “dockerin,” which comprises a 23-residue tandemly repeated sequence (8). Cellulosome assembly is mediated by the interaction of the dockerin domains of each enzyme with one of the complementary cohesin domains of CipA (7). In C. thermocellum, the nine cohesin domains of CipA are unable to discriminate between the individual dockerins present in the various enzymes, thus any individual cellulosome complex may comprise a different ensemble of catalytic subunits appended to CipA (9, 10).
Recent structural studies in conjunction with mutagenesis approaches have started to dissect the molecular determinants that underpin cohesin–dockerin (Coh-Doc) recognition. The 3D structure of cohesin domains from CipA of C. thermocellum (11, 12) and CipC of C. cellulolyticum (13) have both been solved and reveal similar elongated β-barrel “jelly roll” topologies. The solution structure of the dockerin domain from the C. thermocellum enzyme CelS revealed a flexible protein that contains two calcium-binding loop–helix motifs connected by a linker (14). Mutagenesis studies, informed by sequence conservation and structural data, have probed the apparent lack of cross-specificity between C. thermocellum and C. cellulolyticum dockerin–cohesin pairs (15, 16) but have been restricted by the absence of 3D information on the Coh-Doc complex itself.
Here we report the 3D structure of the Coh-Doc complex. The structure of the complex shows that protein–protein recognition is mediated mainly by hydrophobic interactions between one of the faces of the cohesin and α-helices 1 and 3 of the dockerin; there are relatively few direct hydrogen bonds between the two protein molecules. Ser-45 and Thr-46 dominate the hydrogen-bonding network between the dockerin and cohesin. Although Ser-10 and Thr-11 do not play a direct role in protein–protein recognition in this complex, given that the tandem repeats of the dockerin are also manifested in structural similarity (the dockerin possesses near perfect internal twofold symmetry), the “symmetric” binding mode featuring Ser-10 and Thr-11 is possible. The structure of the Coh-Doc complex sheds light on the lack of cross-species recognition between Coh-Doc pairs and should direct and inform future strategies designed to introduce novel specificities into a multifaceted supramolecular assembly.
Methods
Cloning and Expression. DNA encoding the dockerin domain from xylanase 10B (Xyn-10B) (17) (residues 733–791) and the cohesin 2 from CipA (6) (residues 182–328) were amplified by PCR from C. thermocellum genomic DNA, and the products were ligated into NdeI/BamHI-digested pET3a and NheI/XhoI-restricted pET21a, respectively (Novagen). Recombinant cohesin contained a C-terminal His-6 tag. To express the dockerin and the cohesin genes in the same plasmid, the recombinant pET3a derivative was digested with BglII and BamHI, to excise the dockerin gene under the control of the T7 promoter, which was subcloned into the BglII site of recombinant pET21a, so that both genes were organized in tandem. The region of the Xyn-10B gene encoding the dockerin and the C-terminal family 22 carbohydrate-binding module was amplified by PCR and cloned into NheI/XhoI-restricted pET21a. BL21 cells, transformed with pET21a derivatives, were grown at 37°C to OD600 0.5, and recombinant protein expression was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside and incubation for a further 3 h at 37°C.
Protein Purification. The dockerin domain fused to the C-terminal family 22 carbohydrate-binding module, the recombinant cohesin and the Coh-Doc complex were purified by ion metal affinity chromatography. Fractions containing the purified complex were buffer exchanged, in PD-10 Sephadex G-25M gel filtration columns (Amersham Pharmacia Biosciences), into 20 mM Tris·HCl, pH 8.0 containing 2 mM CaCl2. A further purification step by anionic exchange chromatography was performed by using a column loaded with Source 30Q media and a gradient elution of 0–1 M NaCl (Amersham Pharmacia Biosciences). Fractions containing the complex were buffer exchanged, in the same PD-10 columns, into 20 mM NaHepes, pH 7.5, containing 200 mM NaCl and 2 mM CaCl2. The purified complex was then concentrated with Amicon 10-kDa molecular-mass centrifugal membranes and washed three times with 2 mM CaCl2. The final protein concentration was adjusted to 12 g/liter in 2 mM CaCl2.
Isothermal Calorimetry of Coh-Doc Binding. Isothermal titration calorimetry was carried essentially as described (18, 19), except that measurements were made at 65°C, and proteins were dialyzed into 50 mM NaHepes, pH 7.5, containing 2 mM CaCl2. During titration, the dockerin (20 μM), which is fused to C-terminal family 22 carbohydrate-binding module, was stirred at 300 rpm in the reaction cell, which was injected with 25 successive 10-μl aliquots of ligand comprising cohesin (350 μM) at 200-s intervals. Integrated heat effects, after correction for heats of dilution, were analyzed by nonlinear regression by using a single site-binding model (Microcal origin, Ver. 5.0, Microcal Software, Northampton, MA). The fitted data yield the association constant (KA) and the enthalpy of binding (ΔH). Other thermodynamic parameters were calculated by using the standard thermodynamic equation: –RTlnKA = ΔG = ΔH–TΔS. The c values (product of the molar concentration of binding sites × the association constant) were ≈100.
Complex Crystallization. Crystals were grown in two different conditions, the first consisting of 2 mM CaCl2, 0.2 M KNO3, and 20% (wt/vol) polyethylene glycol 3350 (protein concentration of 6g/liter); and the second using 0.2 M Na2HPO4 instead of KNO3 (protein concentration of 12 g/liter). Crystals grew over a period of 5–6 days and were cryoprotected with 40% to 20% (vol/vol) of glycerol. Preliminary x-ray diffraction analyses revealed that both conditions produce crystals belonging to cubic space group P213. However, the unit cell dimensions differed by 1 Å (see Tables 1 and 2, which are published as supporting information on the PNAS web site, for details). Crystals grown with Na2HPO4 had cell dimensions a = b = c = 98.9 Å (data set Coh-Doc1), whereas crystals grown with KNO3 had cell dimensions a = b = c = 97.9 Å (data set Coh-Doc2).
X-Ray Data Collection and Processing. A complete diffraction data set from the Coh-Doc1 (Table 1) crystals, to 2.50-Å resolution, was collected on a MAR-Research (Hamburg, Germany) imaging plate system by using graphite monochromated CuKα radiation from an Enraf–Nonius rotating anode generator operated at 4.5 kW. The Coh-Doc2 (Table 1) diffraction data, to 2.20-Å resolution, were collected at BM30 (European Synchrotron Radiation Facility, Grenoble, France). Both data sets were processed and scaled with programs mosflm and scala from the ccp4 suite of programs (20). The Matthews parameter of the Coh-Doc crystals is 3.0 Å3·Da–1 for one Coh-Doc heterodimer in the asymmetric unit, with a solvent content of 60%.
Structure Determination, Refinement, and Model Building. The structure was solved by Patterson search methods with cns (21) by using, as search model, the known structure for the cohesin 2 domain of the cellulosome from C. thermocellum (PDB ID code 1ANU) and, as observed structure factors, the Coh-Doc1 data set (Table 1), to 2.5-Å resolution. Solvent flipping, with CNS, improved the phases and allowed model building of the dockerin domain, comprising 56-aa residues. Subsequent structure refinement was performed with refmac5 (22), applying bulk solvent and isotropic B factor corrections. The final model (Rcryst = 22.2%; Rfree = 25.9%) was refined as two polypeptide chains, one for the cohesin domain, with 143 residues and a second polypeptide chain of 56 residues belonging to the dockerin domain. Eighty-eight water molecules were included, as well as two Ca2+, one Na+, one Cl –, and two glycerol molecules. Eight residues had one or more side chain atoms with no occupancy, all located in the molecular surface. The Coh-Doc1 model was used as phases for the Coh-Doc2 data set (Table 1), to 2.20-Å resolution. Model refinement was performed with refmac5 (22), as described above. The final model has Rcryst = 21.0% and Rfree = 24.2% and includes 115 water molecules, as well as two Ca2+, one Cl–, two NO–3, and one ethyleneglycol molecule. Both Coh-Doc models have all residues in the allowed regions of the Ramachandran plot. The refinement statistics are summarized in Table 1.
Results and Discussion
Affinity of the Xyn-10B Dockerin with Cohesin 2 from CipA. To study the affinity and thermodynamics of Coh-Doc interactions, the second cohesin domain (cohesin 2) of C. thermocellum CipA and a fusion protein comprising the dockerin domain and the C-terminal family 22 carbohydrate-binding module of C. thermocellum Xyn-10B (17, 23) were expressed as discrete entities. The interaction between the two proteins was evaluated by using isothermal titration calorimetry at 65°C, the temperature of the microbial niche occupied by C. thermocellum. The data (not shown) reveal a macromolecular association with a Ka of 6.2 × 106 M–1, a stoichiometry of 1:1 with ΔH and TΔS of –36.1 kcal·mol–1 and –25.7 kcal·mol–1, respectively. Although a previous study showed that the Coh-Doc interaction, between 25 and 50°C, was both enthalpically and entropically favorable, there was a linear relationship between the increase in temperature and a decrease in both enthalpy and entropy. Extrapolation of the data of Schaeffer et al. (24) to 65°C would also have generated negative entropic and enthalpic values and a Ka of 2.5 × 107, ≈40× greater than for the Xyn-10B dockerin–cohesin 2 interaction. Recent data by other groups have also indicated that the affinity of different Type I Coh-Doc interactions can vary between pairs.
Production of the Coh-Doc Complex for Structural Determination. The dockerin could not be produced as a discrete entity due to degradation in Escherichia coli. Based on the assumption that the dockerin was more stable when bound to the cohesin, both proteins were expressed in E. coli from a single plasmid with the cohesin containing a His tag (C terminal). Ion metal affinity chromatography was used to purify the cohesin protein both as a discrete entity and in complex with the dockerin. The protein complex was then purified from unbound cohesin by anion exchange chromatography.
Overall Architecture. The polypeptide chain of the cohesin domain is well defined in the electron density, with a mean B value of 34.5 Å2. However, the first five N-terminal residues and the last three C-terminal residues (Met-1A, Ala-2A, Ser-3A, Asp-4A, Gly-5A and Asn-145A, Ala-146A, Thr-147A, respectively) were disordered. The structure of this protein as a discrete entity (12) or in complex with its target dockerin is extremely similar, reflected in an rms deviation of 0.43 Å for 138 Cα atoms, indicating that the cohesin does not undergo a significant conformation change when binding to its ligand. The cohesin domain of the Coh-Doc complex consists of a nine-stranded flattened β-barrel with jelly-roll topology, defined by two β-sheets. The first β-sheet comprises β-strands 5, 6, 3, and 8, whereas β-strands 4, 7, 2, 1, and 9 define the second β-sheet, with β-strands 9 and 1, the C and N termini, respectively, running parallel. The core of the nine β-strands assembly is extensively aromatic.
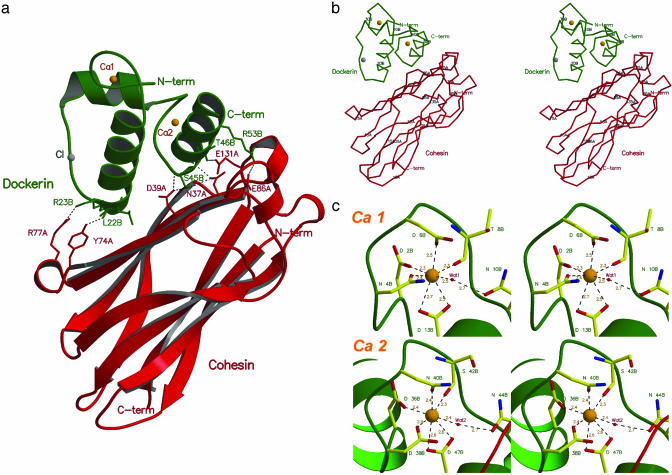
The dockerin subunit was modeled as a single polypeptide chain (chain B) of 57 residues that form 3 α-helices (segments Ser-11B—Leu-22B, Thr-28B—Asp-36B, and Ser-45B—Leu-55B), in a conformation defined by a loop–helix motif followed by a helix–loop–helix motif, connected by a six-residue segment. The electron density is absent for the three residues at the C terminus, as well as for the N-terminal methionine, as observed for the dockerin solution structure. Two Ca2+ ions were identified in the electron density maps, coordinated by several amino acid residues, in a 12-residue EF-hand loop motif (25). One of the Ca2+ ions of the Coh-Doc complex, Ca 1, is located close to the N terminus end of the dockerin domain and is coordinated by the side chains of residues Asp-2B, Asn-4B, Asp-6B, and Asp-13B (both the OD1 and OD2), the latter belonging to the first α-helix segment of this domain. The octahedral geometry of the coordination of Ca 1 is fulfilled by the main chain oxygen atom of residue Thr-8B and by a water molecule (Wat-1), which bridges to Asn-10B (Fig. 1c). The second Ca2+ site, Ca 2, stabilizes the loop connecting α-helices 2 and 3 of the dockerin domain. This Ca2+ ion is coordinated by the side chains of residues Asp-36B, Asp-38B, Asn-40B, and Asp-47B (both the OD1 and OD2), as well as by the main chain oxygen atom of residue Ser-42B and by water molecule Wat-2, which is H bonded to Asn-44B. Both Ca2+ sites show coordination to residues n, n + 2, n + 4 and n + 6 (main-chain O atom) and n + 11, with a water molecule bridging to residue n + 8 (Fig. 1c). A similar calcium coordination is observed in calcyclin (1K96), calbindin D9k (1HT9), calmodulin (1CLL), and troponin C (1NCX), where the metal ion contributes to the stabilization of the 3D structure of this domain. Ca2+ is also essential for dockerin stability (26). Fig. 2 shows the secondary structure of the Coh-Doc complex along its amino acid sequence.
Fig. 1.
Structure of the type I Coh-Doc complex. (a) The complex is formed between a cohesin 2 molecule (red) and a Ca2+-bound dockerin (green). The residues involved in domain contacts are shown as stick models. The two Ca2+-binding sites of the dockerin domain are represented as orange spheres. (b) Cα representation of the Coh-Doc structure with every 10th residue labeled. (c) Ca2+ coordination in the dockerin domain. The Ca2+-bound residues are shown as stick models with green labels.
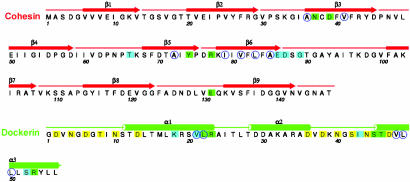
Fig. 2.
Amino acid sequence of Coh-Doc with secondary structure. Residues involved in direct contacts between the two subunits are marked with green squares. Blue squares represent contacts mediated by bridging water molecules. The blue open circles indicate hydrophobic residues in the Coh-Doc contact surface. The residues involved in Ca2+ coordination are marked with yellow squares.
The NMR structure of the dockerin domain shows deviation from the canonical EF-hand Ca2+-binding motif (14). Furthermore, the crystal structure of this protein in complex with the cohesin domain is in a “tighter” conformation, which brings the Cα atoms of α-helices 1 and 3 to within a distance of 6–7 Å compared with 9–11 Å for the solution structure. It is likely that the dockerin has a flexible conformation in solution, consistent with its rapid degradation in E. coli, but that, on binding the cohesin domain, the protein–protein interactions impose structural constraints on the dockerin fold such that it adopts a single conformation. Indeed, overlaying the solution structure of the dockerin with the Coh-Doc complex shows that α-helix 3 (from Ser-45B to Leu-56B) of the solution structure is too distant to interact with the cohesin domain without conformational change (Fig. 3).
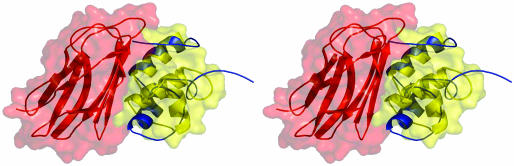
Fig. 3.
Stereo picture of the cohesin (red)–dockerin (yellow) complex. The dockerin solution structure (1DAQ) is overlaid in blue and reveals the movement of α-helix 3 away from the cohesin.
The Coh-Doc Complex. The Coh-Doc contacts are located mainly on one face of the cohesin β-barrel. Several hydrophobic residues participate in complex formation: Ala-36A, Val-41A, Ala-72A, Ile-79A, Val-81A, Leu-83A, and Ala-85A from β-strands 3, 5, and 6 of the cohesin domain, and residues Val-21B, Leu-22B, Val-48B, Leu-49B, and Leu-50B, from α-helices 1 and 3 of the dockerin domain. The importance of hydrophobic interactions in Coh-Doc association is consistent with the negative heat capacity of the binding event of –306 cal–1·mol–1·K–124. The proteins also interact via a series of hydrogen bonds (see Table 2 for details). The electrostatic surface potential calculated for the cohesin domain reveals that the face that interacts with the dockerin is predominantly negatively charged, Fig. 4a, as proposed by Bayer et al. (18) based on sequence comparisons. This same region of the cohesin is also responsible for the cohesin dimer formation, a process inhibited by dockerin–cohesin interaction (11, 13).
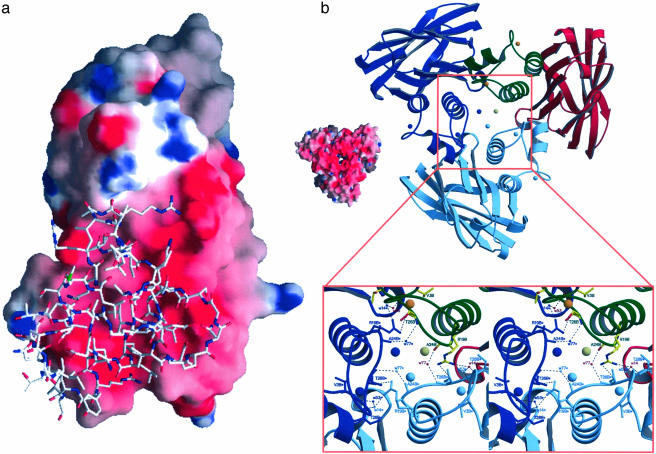
Fig. 4.
Electrostatic surface potential of cohesin 2 and structure of the Coh-Doc crystallographic trimer. (a) 3D structure of the dockerin domain superimposed on the electrostatic surface potential of the cohesin 2 domain. (b) Structure of the Coh-Doc crystallographic “trimer.” The cohesin 2 model is in red; the dockerin, green; the Ca2+ ions, orange; and the Cl– ion, gray. The symmetry-related molecules are shown in dark and light blue. A closer view along the crystallographic threefold axis shows the contacts among symmetry-related molecules.
Recent site-directed mutagenesis studies on the seventh cohesin domain of C. thermocellum CipA (24, 27) propose that residues Asp-39, Tyr-74, and Glu-86 and Gly-89 of this domain play a key role in the formation of Coh-Doc complexes and cohesin–cohesin dimers (27). The crystal structure of the Coh-Doc complex reveals that the equivalent residues in cohesin 2, Asp-39A, Tyr-74A, and Glu-86A, respectively, do indeed interact with the dockerin. Although Gly-89A (equivalent to Gly-89 in the seventh cohesin domain of CipA) does not play a direct role in complex formation, this residue does, however, make a water-mediated link from its main-chain amide to the Nε of dockerin Arg-53B. Gly-89A also displays ϕ/ψ angles of 94.5°, –10.6° that lie in a forbidden region of the Ramachandran plot; mutations at this position would both destabilize the structure and most likely lose the solvent-mediated interaction. Interestingly, both deletion of Gly-89 and substitution of Ala-94 for leucine seem to have little effect on overall affinity but instead increase the enthalpy of the binding event, suggesting that these changes affect the cohesin structure such that the hydrophilic contact between the two proteins is increased (27).
In the dockerin domain, the residues that make direct hydrogen bond to the cohesin (Table 2) are Leu-22B (main chain O), Arg-23B (main chain O), Ser-45B (Oγ and main chain N), Thr-46B (Oγ1 and main chain N), and Arg-53B (Nε and Nη2). All of these residues are strictly conserved in the internal sequence duplication of the dockerin, the implications of which are discussed below. Mutagenesis studies have also suggested that the equivalent residue to Arg-57B plays a role in cohesin recognition (15). In the Coh-Doc complex reported here, Arg-57B is disordered and thus not interacting with the cohesin. Given the internal sequence duplication and the internal structural symmetry, described below (Fig. 5), it is likely that Arg-57B plays the same role in binding in the “symmetry-related” orientation as Arg-23B does in the observed model.
Fig. 5.
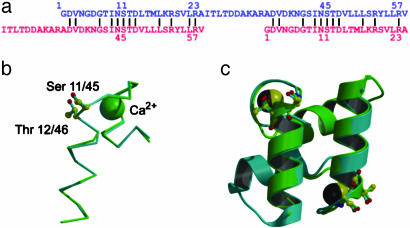
Internal symmetry of the dockerin. (a) Sequence alignment showing the tandem repeat within the dockerin sequence (residues mentioned in the text are indicated). (b) 3D overlap of residues 1–22 with 35–56 showing the near-perfect coincidence of the structure, the Ca2+ loop, and Ser-11 with Ser-45 and Thr-12 with Thr-46. Other residues at the Coh-Doc interface are also conserved (see text). (c) The internal symmetry of the dockerin. Not only do residues 1–22 overlap with 35–56, but the reverse is also true, because the dockerin shows internal twofold symmetry.
The residues in the dockerin domain of Xyn-10B that interact with the cohesin are highly conserved in the dockerins located in other C. thermocellum cellulosomal enzymes, although Arg-23B is often substituted for a lysine, and in two of the dockerin domains Leu-22B is replaced with an isoleucine. Similarly the residues in cohesin 2 of CipA, which the Coh-Doc structure show make direct hydrogen bonds with the dockerin domain, are completely conserved in the other cohesin modules in the C. thermocellum scaffoldin protein. This conservation in the residues involved in cellulosome assembly in both the dockerin and cohesin domains is completely consistent with the inability of the cohesin domains of C. thermocellum CipA to discriminate between the different dockerins appended to cellulosomal enzymes (9, 10). When we compare different species, however, the lack of conservation of Ser-45B and Thr-46B and the replacement of Arg-53B with a lysine in the dockerin domains of C. cellulolyticum cellulosome enzymes provide a partial explanation for why cohesins or dockerin domains from one Clostridium do not recognize the complementary protein partner from a different clostridial species.
Oligomeric State. Although the Coh-Doc complex behaves as a monomer in solution (no oligomerization occurs, as judged by gel filtration), the crystal structure presents a crystallographic trimer formed around the threefold axis (Fig. 4b), with approximate dimensions 68×60×52 Å. Each dockerin contacts its complementary cohesin molecule, as described above, and two symmetry-related dockerins through the hydrogen bond Arg-19B—Thr-26B. Each cohesin domain has a total surface area of 6,812 Å2, 24% of which (1,640 Å2) contacts the dockerin domain of the complex (calculations performed with areaimol from the ccp4 program suite (20). On the other hand, the dockerin domain has 12% (472 Å2) of its surface area in close contact with a symmetry-related dockerin, along the threefold axis. Besides the complementary cohesin and the two symmetry-related dockerins, each dockerin also makes hydrogen bonds with a second symmetry cohesin, through residues Thr-26B, Thr-28B, Asp-29B, and Asp-30B. Because trimer assembly does not occur in solution, and in nature the dockerin domains are appended to large catalytic domains, we conclude that in the absence of further evidence, the trimeric structure of the Coh-Doc complex seems unlikely to have any significant biological relevance.
Structural Basis for Cross-Species Specificity of Coh-Doc Pairs. The capacity of cohesins and dockerins to interact with their respective complementary protein partners from different bacteria has been extensively studied. In particular, it has been proposed that two serine/threonine pairs, (equivalent to positions 11–12 and 45–46, respectively, in the Xyn-10B dockerin), which are highly conserved in C. thermocellum dockerins but not in the equivalent domains from other bacteria, play a key role in defining the specificity of these protein domains (28). Consequently, these residues have been targeted for mutagenesis (15, 16, 24). The T11L mutation of the C. thermocellum dockerin enabled the protein to recognize both the C. cellulolyticum and C. thermocellum cohesins, while introducing serine (at positions 10 and 44) and threonine (at positions 11 and 45) into the C. cellulolyticum dockerin conferred significant recognition of the C. thermocellum cohesin. Based on the structure of the C. thermocellum Coh-Doc complex, although it is apparent why the introduction of hydroxy amino acids into the C. cellulolyticum dockerin conferred recognition of the C. thermocellum cohesin, the structural basis for the change in specificity mediated by the reciprocal mutations (introduction of hydrophobic residues in place of hydroxy amino acids into the C. thermocellum dockerin) is less apparent. The corresponding residues in the C. cellulolyticum cohesin to Asn-37, Asp-39, and Glu-131 (the residues in the C. thermocellum cohesin that form H bonds with Ser-45 and Thr-46) are also polar and thus would not form hydrophobic interactions with the C. cellulolyticum alanine, leucine, and phenylalanine residues that are equivalent to Ser-11, Thr-12, Ser-45, and Thr-46 in the C. thermocellum dockerin. The basis for lack of cross-species specificity will be fully understood only when the structure of the C. cellulolyticum Coh-Doc complex is known.
Plasticity in Coh-Doc Recognition. A well-documented feature of the dockerin sequence is the presence of a tandem sequence duplication equating to residues 1–23 with 35–57 (Fig. 5a). A structural overlap of these regions (Fig. 5b) reveals that the 3D structures are also extremely similar with a rms deviation of just 0.36 Å for all main-chain atoms. Furthermore, there is both sequence and structural conservation of the whole EF hand and of all of the residues that interact with the dockerin, described above. Even more striking is that the dockerin possesses near-perfect internal twofold symmetry such that residues 1–22 overlay onto 35–56, and at the same time residues 35–56 overlay onto 1–22 (Fig. 5c). This is reflected in a rms main-chain deviation of just 0.6 Å for the 44 equivalent residues.
The implications of this internal symmetry are profound. First, it would seem likely that both halves of the dockerin could interact with the cohesin in almost identical manners, only one of which is revealed here. The extent and significance of the “symmetry-related” binding mode would most likely also depend on the particular Coh-Doc pair. Indeed, this proposal is entirely consistent with site-directed mutagenesis data reported previously. For example, whereas introduction of either of the double mutations S10L/T11L (equivalent to Ser-11 and Thr-12 in the Xyn-10B dockerin) and S46M/S47Q (equivalent to Ser-45 and Thr-46), respectively, into a C. thermocellum dockerin derived from CelD did not influence affinity for C. thermocellum cohesins, combining both these mutant pairs to generate S10L/T11L/S46M/S47Q caused a 1,000-fold reduction in affinity (24). Likewise, mutation of the residues equivalent to Arg-23 and Arg-57 disrupted binding, yet in the model presented here, Arg-57 was disordered. If one considers the internal symmetry-related binding mode, Arg-57B would play the same role in binding as Arg-23B does in the observed model.
These observations imply that there is plasticity in cohesin recognition by the dockerin with either the N- or C-terminal helix, containing the conserved Ser-Thr motif, capable of interacting with its protein ligand. Furthermore, there would appear to be little or no steric barrier to the simultaneous binding of two cohesins by a single dockerin species. In the case of the Xyn-10B dockerin and cohesin 2 of CipA described here, utilization of both putative binding faces simultaneously would cause a clash involving residues 64–67 of the cohesin at the newly generated twofold axis. Small conformational changes in the cohesin or in the binding to two different cohesins could result in the binding of two cohesins simultaneously by a single dockerin. Such an interaction, which would necessarily vary between different Coh-Doc pairs (and might also reflect the position of the dockerin in a multimodular protein) would not only provide a higher level of structure to the cellulosome but might also permit the “cross-linking” of two scaffoldins through a single dockerin and thus provide an explanation for the presence of “polycellulosomes.”
Conclusion
The Coh-Doc interaction is crucial for biomass conversion by anaerobic organisms, because the enzyme complexes synthesized by these organisms are among the most potent hydrolytic enzyme systems known. The resolution of the 3D structure of a dockerin–cohesin complex reveals the mechanism of macromolecular association leading to cellulosome assembly, both explaining mutagenesis studies on interspecies chimeras and also informing mutagenesis strategies for the design of specific Coh-Doc pairs. The C. thermocellum CipA provides a macromolecular scaffold with nine potential cohesin “landing platforms” for appropriate dockerins. Engineering of their surfaces to interact with tailored dockerins will allow the construction of designer macromolecular assemblies not merely to generate highly ordered enzyme complexes in the sphere of plant cell wall degradation but also to orchestrate any enzyme-catalyzed reactions that might benefit from component proximity, such as electron or glycosyl transfer and metabolite channeling. The Coh-Doc structure will thus form a blueprint for tailored multicomponent catalytic machines across a range of biological processes.
Supplementary Material
Acknowledgments
We thank the beamline scientists at BM30, European Synchrotron Radiation Facility, and are grateful for insightful discussions with Andy Karplus and Ed Bayer at the 2003 Gordon Research Conference on “Cellulases and Cellulosomes.” This work was supported by Grants SFRH/BPD/9446/2002 (to A.L.C.) and PraxisXXI/BD/21250 (to F.M.V.D.) from the Fundação para a Ciência e a Tecnologia–Ministério da Ciência e Ensino Superior, Portugal. G.J.D. is a Royal Society University Research Fellow. G.J.D. and H.J.G. are supported by the Biotechnology and Biological Sciences Research Council.
Abbreviation: Coh-Doc, cohesin–dockerin.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1ohz).
References
- 1.Shoham, Y., Lamed, R. & Bayer, E. A. (1999) Trends Microbiol. 7, 275–281. [DOI] [PubMed] [Google Scholar]
- 2.Béguin, P. & Aubert, J. P. (1994) FEMS Microbiol. Rev. 13, 25–58. [DOI] [PubMed] [Google Scholar]
- 3.Fierobe, H. P., Bayer, E. A., Tardif, C., Czjzek, M., Mechaly, A., Belaich, A., Lamed, R., Shoham, Y. & Belaich, J. P. (2002) J. Biol. Chem. 277, 49621–49630. [DOI] [PubMed] [Google Scholar]
- 4.Béguin, P. & Lemaire, M. (1996) Crit. Rev. Biochem. Mol. Biol. 31, 201–236. [DOI] [PubMed] [Google Scholar]
- 5.Bayer, E. A., Shimon, L. J., Shoham, Y. & Lamed, R. (1998) J. Struct. Biol. 124, 221–234. [DOI] [PubMed] [Google Scholar]
- 6.Gerngross, U. T., Romaniec, M. P., Kobayashi, T., Huskisson, N. S. & Demain, A. L. (1993) Mol. Microbiol. 8, 325–334. [DOI] [PubMed] [Google Scholar]
- 7.Salamitou, S., Raynaud, O., Lemaire, M., Coughlan, M., Beguin, P. & Aubert, J. P. (1994) J. Bacteriol. 176, 2822–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayer, E. A., Morag, E. & Lamed, R. (1994) Trends Biotechnol. 12, 379–386. [DOI] [PubMed] [Google Scholar]
- 9.Lytle, B., Myers, C., Kruus, K. & Wu, J. H. (1996) J. Bacteriol. 178, 1200–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaron, S., Morag, E., Bayer, E. A., Lamed, R. & Shoham, Y. (1995) FEBS Lett. 360, 121–124. [DOI] [PubMed] [Google Scholar]
- 11.Tavares, G. A., Béguin, P. & Alzari, P. M. (1997) J. Mol. Biol. 273, 701–713. [DOI] [PubMed] [Google Scholar]
- 12.Shimon, L. J., Bayer, E. A., Morag, E., Lamed, R., Yaron, S., Shoham, Y. & Frolow, F. (1997) Structure (Cambridge, U.K.) 5, 381–390. [DOI] [PubMed] [Google Scholar]
- 13.Spinelli, S., Fierobe, H. P., Belaich, A., Belaich, J. P., Henrissat, B. & Cambillau, C.. (2000) J. Mol. Biol. 304, 189–200. [DOI] [PubMed] [Google Scholar]
- 14.Lytle, B. L., Volkman, B. F., Westler, W. M., Heckman, M. P. & Wu, J. H. (2001) J. Mol. Biol. 307, 745–753. [DOI] [PubMed] [Google Scholar]
- 15.Mechaly, A., Fierobe, H. P., Belaich, A., Belaich, J. P., Lamed, R., Shoham, Y. & Bayer, E. A. (2001) J. Biol. Chem. 276, 9883–9888. [DOI] [PubMed] [Google Scholar]
- 16.Mechaly, A., Yaron, S., Lamed, R., Fierobe, H. P., Belaich, A., Belaich, J. P., Shoham, Y. & Bayer, E. A. (2000) Proteins 39, 170–177. [DOI] [PubMed] [Google Scholar]
- 17.Fontes, C. M., Hazlewood, G. P., Morag, E., Hall, J., Hirst, B. H. & Gilbert, H. J. (1995) Biochem. J. 307, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayer, E. A., Chanzy, H., Lamed, R. & Shoham, Y. (1998) Curr. Opin. Struct. Biol. 8, 548–557. [DOI] [PubMed] [Google Scholar]
- 19.Charnock, S. J., Bolam, D. N., Nurizzo, D., Szabo, L., McKie, V. A., Gilbert, H. J. & Davies, G. J. (2002) Proc. Natl. Acad. Sci. USA 99, 14077–14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 21.Brünger, A. T., Adams, P. D., Clore, G. M., Delano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 23.Charnock, S. J., Bolam, D. N., Turkenburg, J. P., Gilbert, H. J., Ferreira, L. M., Davies, G. J. & Fontes, C. M. (2000) Biochemistry 39, 5013–5021. [DOI] [PubMed] [Google Scholar]
- 24.Schaeffer, F., Matuschek, M., Guglielmi, G., Miras, I., Alzari, P. M. & Beguin, P. (2002) Biochemistry 41, 2106–2114. [DOI] [PubMed] [Google Scholar]
- 25.Kretsinger, R. H. & Nockolds, C. E. (1973) J. Biol. Chem. 248, 3313–3326. [PubMed] [Google Scholar]
- 26.Lytle, B. L., Volkman, B. F., Westler, W. M. & Wu, J. H. (2000) Arch. Biochem. Biophys. 379, 237–244. [DOI] [PubMed] [Google Scholar]
- 27.Miras, I., Schaeffer, F., Béguin, P. & Alzari, P. M. (2002) Biochemistry 41, 2115–2119. [DOI] [PubMed] [Google Scholar]
- 28.Pagès, S., Belaich, A., Belaich, J. P., Morag, E., Lamed, R., Shoham, Y. & Bayer, E. A. (1997) Proteins 29, 517–527. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.