Abstract
Bone marrow transplantation can provide an effective cell-based strategy to enhance bone repair. However, the fate of implanted cells and the extent of their contribution to bone osteoinduction remain uncertain. To define the fate of bone marrow–derived cells and their contribution in vivo, we used a bone-specific collagen I promoter (2.3Col) driving green fluorescent protein (GFP) (2.3ColGFP) within a lentiviral vector. Prior to in vivo cell fate determination, we verified a high efficiency of lentiviral transduction in human bone marrow stromal cells (hBMSCs), without altering the proliferation or differentiation potential of these cells. We showed that the 2.3ColGFP marker responded to endogenous transcriptional regulation signals. In a mouse ossicle model, we demonstrated that the 2.3ColGFP marker is able to specifically define human bone marrow–derived stem cells that enter the osteoblast lineage in vivo. In addition, cells labeled with 2.3ColGFP with the donor origin, directly make a major contribution to bone formation. Furthermore, we also demonstrated in a calvarial defect model that a mixture of human bone marrow–derived populations, have stronger bone regenerative potential than that of hBMSCs, and an optimal dose is required for bone regeneration by the mixed populations.
Introduction
Given the severe tissue loss associated with traumatic events such as auto accidents, industrial accidents, and war wounds, there is an immediate need for effective cell therapy to regenerate bone and other lost tissues. One therapy to regenerate tissue lost to injury is administration of human bone marrow–derived stem cells, which constitute an exciting therapeutic possibility, because they give rise to a number of cell types, including osteoblasts (bone), chondrocytes (cartilage), adipocytes (fat), and cells needed to reconstruct vascular beds.1 Indeed, the successful use of freshly isolated autologous bone marrow cells, or bone marrow–derived osteoprogenitors has been reported in several retrospective case series of nonunion fractures,2,3 osteonecrosis,4 and spinal fusion.5
In regenerative medicine, knowledge of the mechanisms by which cell grafts contribute to bone repair and regeneration is limited. Furthermore, the fate of transplanted stem cells and the extent of their direct contribution to tissue regeneration remain controversial. Techniques which have been used with limited success to define the fate of cells in new tissue formation included: (i) in situ hybridization to detect donor-specific chromosome,6,7 (ii) species-specific gene sequences,8 (iii) the β-gal transgene reporter or PCR for detecting neomycin or other reporter genes,6 and (iv) immunocytochemistry.9 Green fluorescent protein (GFP) reporters, including tissue-specific promoter-directed GFP reporters, are widely used to track cell lineage progression.10,11,12,13,14 Several studies employing ubiquitously expressed GFP or lacZ to investigate the fate of engrafted mouse or rat cells during bone formation in vivo were frustrated by the technical difficulties due to loss of GFP expression during tissue processing.15,16,17 It has been assumed that contribution to bone formation should be originated from both donor and host sides. However, the exact contribution of implanted human cells in in vivo bone formation models also remains uncertain.
To address this issue, we used a rat bone–specific promoter (collagen type I 2.3 kb) driving GFP (2.3ColGFP) within a lentiviral vector for defining the fate of these human stem and progenitor cells and their contribution to bone generation in vivo, as this 2.3ColGFP reporter has previously been demonstrated to distinguish specific stages of osteoblast differentiation in murine stem and progenitor cells.18
Two ex vivo expanded cell populations were examined in this study to assess their contribution to bone regeneration in vivo. One is the population of human bone marrow stromal cells (hBMSCs) expanded with a traditional cell culture method,19 which have previously been reported to contribute to bone formation in vivo.20 The other is an ex vivo expanded mixture of human bone marrow–derived stem and progenitor cells including cells from the mesenchymal, hematopoietic, and endothelial lineages. This mixed population, termed tissue repair cells (TRCs), is generated in a 12-day culture of bone marrow mononuclear cells (BMMNCs), without the need for passage-purification.21 A proprietary cell production system has been developed for the ex vivo production of TRCs,22 and cells generated in this system have been used in the US Food and Drug Administration–approved clinical trials for several indications, including bone regeneration.23,24,25 To determine whether the mixed population (e.g., TRCs) is more suitable for osteoinduction than that of relatively pure population (hBMSCs), we also compare the potential of bone formation between these ex vivo expanded populations using total bone marrow cells from the same human donors.
Results
Lentiviral transduction produces GFP-labeled human cells
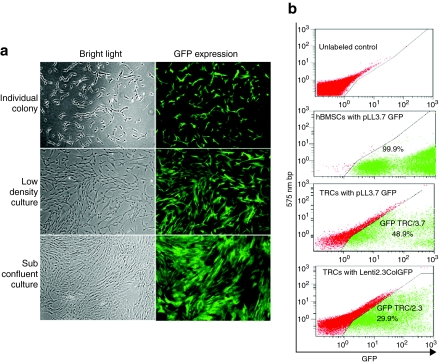
To evaluate transduction efficiencies of hBMSCs or TRCs with a lentivector containing a bone-specific promoter–driven GFP, we firstly used pLL3.7 with a titer of 5 × 105 viral particles/ml, which have the same lentiviral vector with a nonspecific promoter cytomegalovirus (CMV) driving GFP expression, to reflect its transduction efficiency. Two days after lentiviral transduction, microscopic evaluation revealed GFP expression in colony-forming fibroblasts in the hBMSCs culture, and all cells were visually GFP+ with a fibroblast-like morphology by 7 days after transduction (Figure 1a). Fluorescence-activated cell-sorting (FACS) analysis confirmed that 99.9% of transduced cells were GFP+ (Figure 1b), indicating high-transduction efficiency by the lentivirus in hBMSCs. We noticed the stable GFP expression over 40 passages or in liquid nitrogen storage for >4 months (data not shown). The transduced cells maintained 99.9% of GFP+ just prior to transplantation.
Figure 1.
Transduction efficiency of hBMSCs and TRCs with a CMVGFP lentivirus. (a) GFP expression in transduced hBMSCs visualized by fluorescent microscopy. The left panel is a bright field image and right panel is fluorescence to visualize GFP+ cells. Original magnification: ×40. (b) Quantitative analysis of transduction efficiency of hBMSCs and TRC with pLL3.7 or Lenti2.3ColGFP vector by flow cytometry as shown in a two parameter plot. GFP, green fluorescent protein; hBMSCs, human bone marrow stromal cells; TRCs, tissue repair cells.
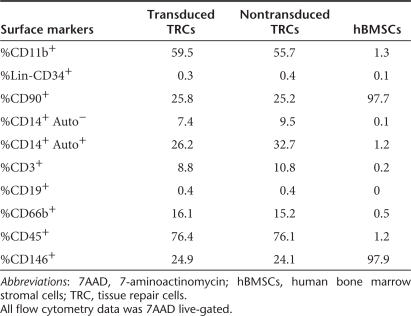
Because a high titer of a lentivirus was required to transduce total bone marrow cells based on a previous report,26 we performed the transduction of human BMMNCs with a concentrated lentivirus at 108 infectious units/ml twice within 3 days prior to 12 days of TRCs culture.21 Typically, transduction of human BMMNCs with pLL3.7 resulted in 16% at TRCs harvest, 49% of GFP expression at 2 weeks of T-flask culture post-TRC harvest, and 29.9% in TRCs with Lenti2.3ColGFP transduction after 2 weeks culture in T-flask post-TRC harvest (Figure 1b). Because the transduction with pLL3.7 vector caused two GFP+ populations (GFPlow and GFPhigh) (Figure 1b), we carried out a flow cytometric analysis to exclude a possibility that might cause any changes in immunophenotypes of TRCs. As shown in Table 1, the lentiviral transduction did not affect immunophenotypes and compositions of TRCs. As the majority of hBMSCs are CD90+ or CD146+ (97%), and no changes were seen in these cell population in TRCs post-transduction (shown in Table 1), we did not perform a flow cytometric analysis of immunophenotype in transduced hBMSCs.
Table 1.
Flow cytometric analysis of transduced, nontransduced TRCs, and hBMSCs
Lentiviral transduction does not affect proliferation or differentiation of hBMSCs
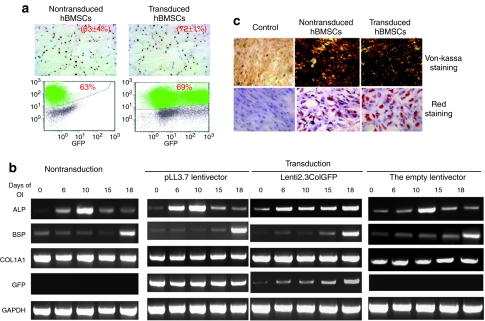
Prior to in vivo defining the fate of implanted stem and progenitor cells, it was critical to ensure that the lentiviral transduction protocol did not alter the ability of cells to proliferate or differentiate. We selected hBMSCs for cell proliferation and differentiation assessment as a representative human cell population because TRCs have shown to contain all osteopotential of hBMSCs.21 The effect of lentiviral transduction on cell proliferation in hBMSCs was examined by 5′-bromo-2-deoxyuridine (BrdU) incorporation. As shown in Figure 2a, the percentage of BrdU+ cells in transduced hBMSCs did not differ significantly from that of nontransduced hBMSCs by both immunohistochemistry (transduced hBMSCs 72 ± 1% versus nontransduced hBMSCs 63 ± 4%) and flow cytometry (transduced hBMSCs 69% versus nontransduced hBMSCs 62%).
Figure 2.
Characterization of transduced hBMSCs in vitro. (a) Influence of lentiviral transduction on hBMSCs. BrdU incorporation assay was used to assess the proliferation of both transduced and nontransduced cells by immunocytochemistry (top panel) and FACS analysis (bottom panel). The numbers indicate the percentage of BrdU+ cells. Original magnification: ×40. (b). Analysis of the differentiation potential of transduced hMSCs by RT-PCR. GAPDH was used as a house-keeping gene. (c) von Kossa staining and Oil Red O staining of nontransduced and transduced cells after 4-week induction of osteogenic differentiation or adipogenic differentiation. ALP, alkaline phosphatase; BrdU, 5′-bromo-2-deoxyuridine; BSP, bone sialoprotein; Col1A1, collagen 1A1; FACS, fluorescence-activated cell-sorting; GADPH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; hBMSCs, human bone marrow stromal cells; OI, osteogenic induction.
Next, to confirm that lentiviral transduction did not modify the differentiation potential of hBMSCs, nontransduced, and transduced hBMSCs with three lentiviral vectors (an empty lentiviral vector, pLL3.7, or Lenti2.3colGFP vector) were induced to differentiate into either osteoblast or adipocyte lineages. GFP expression, controlled by the CMV promoter, could be observed throughout the culture period (days 0–18) in the transduced cells; 2.3ColGFP expression increased with induction of osteoblast differentiation (Figure 2b). Reverse transcription–PCR was used to compare the expression of several osteoblast differentiation markers including alkaline phosphatase, bone sialoprotein, and collagen 1A1. After cells were cultured in osteogenic media, the timing of expression of these genes in transduced hBMSCs with the empty lentiviral vector, pLL3.7, or Lenti2.3ColGFP vectors was similar to nontransduced hBMSCs (Figure 2b), and mineralization based on von Kossa staining, occurred in both cell groups (Figure 2c). When hBMSCs were grown in adipogenic media, both transduced and nontransduced cells differentiated into adipocytes as determined by Oil Red O staining (Figure 2d). Collectively, these data show that lentiviral transduction does not appear to alter the proliferative or differentiate potential of hBMSCs, which are 97.7% CD90+ (Table 1). Because previous studies have shown that the in vitro osteogenic potential of TRCs is contained entirely within the CD90+ fraction, e.g., hBMSCs,21 therefore, separate studies were not performed to examine the osteogenic potential of TRCs after lentiviral transduction, but a similar result to transduced hBMSCs is expected for transduced TRCs.
2.3ColGFP expression is associated with osteoblast differentiation in hBMSCs
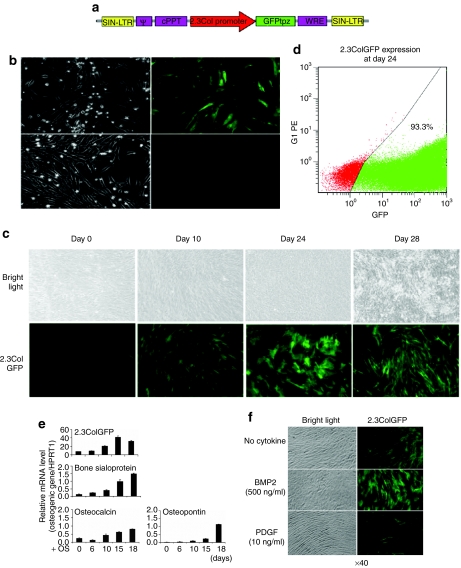
We next verified that the bone-specific promoter 2.3ColGFP would be able to define the fate of cells differentiating into the osteoblast lineage in vitro. Human BMSCs were transduced with either the nonspecific pLL3.7 vector or the Lenti2.3ColGFP. Expression of GFP was visualized using fluorescence microscopy in culture. 2.3ColGFP was very weakly expressed and only in a few cells at early stages of hBMSC culture, compared to the nonspecific CMV promoter vigorously driving GFP expression in vitro (Figure 3b). As time in culture increased, and cells adopted an osteogenic phenotype, both the intensity of GFP expression and the number of GFP+ osteogenic cells increased (Figure 3c) in cells containing 2.3ColGFP. Expression of 2.3ColGFP peaked at day 24, prior to initiation of mineralization, and then decreased after mineralization (Figure 3c). For analysis of GFP expression, cells were harvested from six-well plates with trypsin at day 24 and then subjected to FACS analysis. The percentage of GFP+ was 93.3% at this time point in osteoblast differentiation culture. Transduction with the Lenti2.3ColGFP vector did not produce two separated GFP+ populations (lower or high) (Figure 3d). This bone-specific GFP expression also indicates a high efficiency of the lentiviral transduction as reflected by the CMVGFP lentivector as seen in Figure 1.
Figure 3.
The specificity of 2.3ColGFP expression in transduced hMSCs. (a) A map of Lenti2.3ColGFP vector. (b) Comparison of GFP expression in cells transduced with the nonspecific pLL3.7 CMVGFP (top panel) and the osteo-specific Lenti2.3ColGFP (bottom panel) at an early stage of culture (1-week post-transduction using fluorescence microscopy. Original magnification: ×100). (c) Induction of 2.3ColGFP expression with time during in vitro osteogenic differentiation. Top and bottom panels show photomicrographs under bright field and fluorescence, respectively. Original magnification: ×40. (d) FACS analysis of GFP expression in hBMSCs transduced Lenti2.3ColGFP vector at day 24. (e) Association of 2.3ColGFP expression with other typical osteoblast differentiation markers. qRT-PCR was carried out to evaluate the pattern of expression of BSP, osteocalcin, and osteopontin. Total RNA was extracted from 2.3ColGFP-transduced hBMSCs cultured in osteogenic medium at 0, 6, 10, 15, and 18 days of culture. (f) Regulation of the 2.3ColGFP expression by osteoblast stimulation and inhibition factors. Human BMSCs transduced with Lenti2.3ColGFP cells were treated with either BMP-2 at 500 ng/ml or PDGF at 10 ng/ml for 2 days when culture became subconfluent. BMP-2, bone morphogenetic protein-2; BSP, bone sialoprotein; FACS, fluorescence-activated cell-sorting; GFP, green fluorescent protein; PDGF, platelet-derived growth factor; qRT-PCR, quantitative reverse-transcription–PCR.
To determine whether there was an association between 2.3ColGFP and osteoblast differentiation markers, the temporal pattern for 2.3ColGFP expression was evaluated by quantitative reverse-transcription–PCR using total RNA extraction from cultured hBMSCs transduced with the 2.3ColGFP vector. As expected, at the transcriptional level, this transgene marker increased 20–40-fold with induction of osteoblast differentiation as shown from days 15–18, which is supported by results obtained by fluorescence microscopy as shown in Figure 3c. Interestingly, this sequential expression paralleled other typical osteoblast differentiation markers, such as osteocalcin (OC) and osteopontin, and was particularly closely associated with bone sialoprotein (Figure 3e). We also found that 2.3ColGFP expression responded to osteoblast regulatory factors. GFP expression in these cells was increased remarkably when stimulated by bone morphogenetic protein-2 at a concentration of 500 ng/ml, with a corresponding change in cell morphology to that of osteoblast-like cells (Figure 3f). In contrast, GFP expression decreased, and cells had fibroblast-like morphology when treated with 10 ng/ml platelet-derived growth factor (Figure 3f).
These results demonstrate that 2.3ColGFP expression, which responds to endogenous transcriptional regulation, is strongly correlated with typical osteoblast differentiation markers. The tempo of this transgenic marker reflects the progression of hBMSCs differentiation into the osteoblast lineage. Therefore, it provides us with a powerful tool for visually determining the fate of hBMSCs or TRCs into the osteoblast lineage in vivo during bone formation in a mouse ectopic model.
In vivo bone formation by human cells in a mouse ectopic model
To evaluate the osteoinductive potential of cells transduced with a lentivirus containing either CMVGFP (control), or the bone-specific promoter 2.3ColGFP (test), transduced hBMSCs or TRCs were loaded onto the gelatin sponge (Gelfoam) and placed subcutaneously in immunodeficient (severe combined immunodeficiency) mice. GFP expression was evaluated in the resulting ossicles.
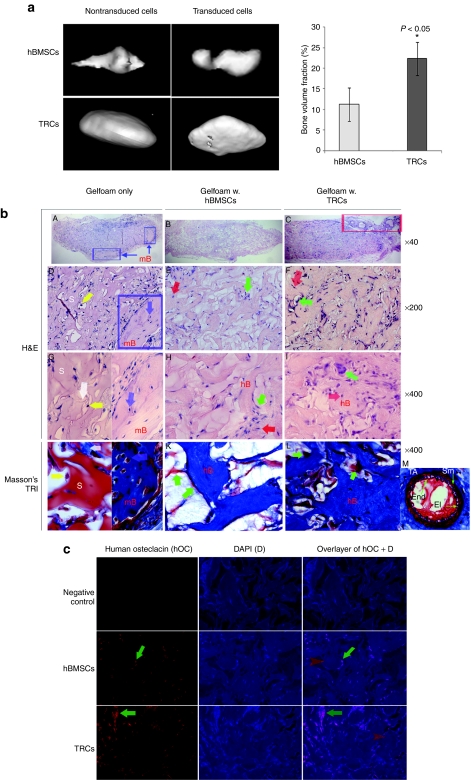
Both nontransduced and transduced hBMSCs and TRCs induced mineralized bone ossicles after 6 weeks of implantation (Figure 4a). Interestingly, micro-computed tomography (µ-CT) data indicated TRCs generated significantly more bone volume (12,912 ± 4,878 µm3, n = 8) than that of hBMSCs (6,657 ± 1,277 µm3, n = 6) (P < 0.05; Figure 4a, right panel). In addition, we did not observe any differences in bone formation potential between transduced cells and nontransduced cells [transduced hBMSCs (n = 6) versus nontransduced hBMSCs (n = 6), P > 0.05; transduced TRCs (n = 8) versus nontransduced TRCs (n = 8), P > 0.05] after analysis of bone volume and bone mineral density measured by µ-CT, indicating that transduction did not affect in vivo bone formation potential of both hBMSCs and TRCs.
Figure 4.
Detection of bone formation by implanted hBMSCs or TRCs in the mouse ectopic model. Human BMSCs or TRCs were implanted at 3 × 106 cells per implant for 6 weeks in the mouse ectopic model. (a) µ-CT detection of new bone formation in ossicles after 6 weeks of implantation. GEMS MicroView software was used to make a 3D reconstruction (left panel) and to calculate bone volume (right panel) from the set of scans with a fixed thresholds of 900 (n = 5). (b) The histological examination of newly formed bone by H&E and Masson's Trichrome stainings. H&E staining: from A to I. Cell-free Gelfoam: A, D, and G; Gelfoam loaded with hBMSCs: B, E, and H; Gelfoam loaded with TRCs: E, F, and I. For G, a representative of scaffold areas is shown in the left and a representative of mouse laminar bone is shown in the right. Blue squares indicate mouse laminar bone; a blue arrow indicates a mouse osteocyte in lacunae; and a white arrow points to a fibrous tissue inside scaffold. A red square indicates an area of vascular formation. Red arrows indicate osteocytes and green arrow indicate osteoblasts on the surface of bony structure. S, scaffold, e.g., Gelfoam; mB, laminar bone–derived host mouse cells; hB, woven bone derived from human donor cells. Masson's Trichrome staining from J to M at a original magnification of ×400. J: scaffold only, a representative of scaffold areas is shown in the left and a representative of mouse laminar bone is shown in the right; K: Gelfoam loaded with hBMSCs; L: Gelfoam loaded with TRCs; M: a representative of vessels (an artery) in C. End, endothelial cells in tunica intima; El, elastic lamellae; TA, tunica adventia; Sm, smooth muscle. (c) Immunofluorescent detection of hOC expression in the sections of implants from either hBMSCs or TRCs. A mouse isoform matched IgG was used as a negative control. Green arrows indicate cells expressing hOC and red arrows point to matrix areas containing hOC. H&E, hematoxylin and eosin; hOC, human osteocalcin; hBMSCs, human bone marrow stromal cells; IgG, immunoglobin G; TRCs, tissue repair cells.
Followed by µ-CT detection of mineralization, implanted samples derived either from cell-free scaffolds (Gelfoam only), or scaffold loaded with hBMSCs or TRCs were processed for hematoxylin and eosin (H&E) staining. Among six samples of the cell-free scaffolds, we observed that there was no bone formation in three samples, in which a few cells with lymphocyte-like morphology were present in the empty spaces of scaffold (data not shown); whereas in another three samples, we observed that a large number of lymphocyte-like cells were infiltered into the scaffold structure and a few fibroblast-like tissues with minimal bone formation (indicated in blue square) were located at the edge of scaffold (Figure 4b-A, at a magnification of ×40). The scaffold structure (Gelfoam) was seen in the majority area of the implants. At a higher magnifications (×400), the minimal bone formation in the scaffold only was seen to be lamellar form with a few osteocytes in lacunae; lymphocyte-like cells attached to the edge of scaffold with smooth surface (Figure 4b-D,G).
In the implants of hBMSCs or TRCs loaded into the scaffold, woven bone structure with disorganized/randomly-oriented collagen fibers were observed, in which some cells were present on the surface of newly formed bone and a few cells were embedded in bone matrix as osteocytes (Figure 4b-B,E,G for hBMSCs and Figure 4b-E,F,I for TRCs at magnifications of ×40 and ×400, respectively). Interestingly, we observed that TRCs formed denser woven bone structure with more cells (Figure 4b-F,I), and small vascular vessels were associated at fibrous tissues surrounding the ossicles (indicated in red square) (Figure 4b-E).
To confirm the results of H&E staining, we carried out a Masson's Trichrome staining to demonstrate the bony structure of the implants. As shown in Figure 4b-J, the scaffold was stained in red with smooth surface and a few fibrous attached at the edges stained in blue. In contrast, the structure of the implant sections from either hBMSCs or TRCs was stained in blue, indicating that these were bony structure (Figure 4b-K,L). A representative of vessels in Figure 4b-C was shown in Figure 4b-M by Masson's Trichrome staining.
Furthermore, we also performed immunofluorescent staining to detect human OC (hOC) expression in these bony structures, using a mouse-derived monoclonal antibody against hOC, a noncollagenous protein found in bone and dentin.27 It is secreted by osteoblasts and thought to play a role in mineralization and calcium ion homeostasis. This protein has been recognized as a specific marker for osteoblasts and bone tissues. As shown in Figure 4c, we found hOC expressed in cells on the surface of the bony structure or cells embedded in the structure. In addition, this protein was also found in the tissue matrix indicated by red arrows. Therefore, the immunodetection of hOC in the structure of implants clarifies woven bone formation inside ossicles.
Determining the fate of GFP-labeled cells for bone formation in a mouse ectopic model
The location of GFP+ cells adjacent to ossicle bone surfaces was examined at 4 weeks of implantation using fluorescence microscopy of 5-µm frozen sections of the ossicles. In contrast to the low intensity of the GFP expression observed in ossicles induced by hBMSCs or TRCs transduced with pLL3.7, GFP expression was bright (high intensity) in ossicles induced by bone-specific 2.3ColGFP-grafted hBMSCs or TRCs (data not shown). It indicates that 2.3ColGFP is more suitable for mapping the osteoblast lineage fate in vivo, which led us to utilize 2.3ColGFP for further tracking the fate of TRC for bone formation in the following experiments.
We noticed that at 4 weeks after cell engraftment, ossicles consisted primarily of cells with minimal bone formation; after 6–8 weeks, bone-like tissue dominated the structure of the ossicle and the number of cells in the ossicle greatly decreased. Due to lack of cellular content in older ossicles, we elected to analyze only 4-week ossicles for in vivo cell fate determination with a GFP technology.
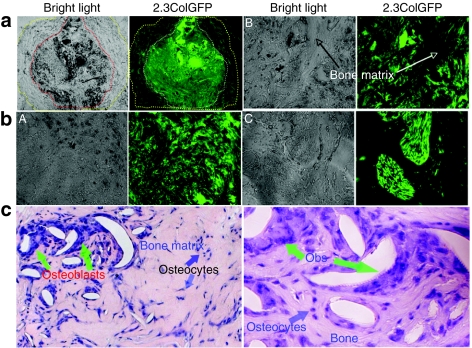
As shown in Figure 5a, the bone-specific 2.3ColGFP was strongly expressed in each individual cells within the TRCs-derived ossicle and the layer of fibroblasts surrounding the ossicle exhibited much weaker GFP expression, which may represent populations of cells at early stages of osteoblast lineage. Three representative areas are shown at a high magnification to demonstrate GFP+ cells within the ossicle (Figure 5b). GFP+ cells were embedded within bone of the ossicle, as osteocytes, as well as being on the surface of the trabecular bone-like structures, as osteoblasts. We observed segregated clusters of cells with intense GFP expression and speculate that these clusters may represent clonal expansion of isolated stem or progenitor cells in TRCs, which then adopted an osteogenic fate. By matching and overlaying bright field and GFP expression images, we confirmed that GFP− areas represented areas of bone matrix. Followed by GFP imaging, H&E staining confirmed the newly formed bone and distribution of cells (nuclei stained in blue color) on surface or embedded in bone matrix within the ossicle at a magnification of ×200 (left) and ×400 (right), respectively (Figure 5c).
Figure 5.
The specificity of 2.3ColGFP expression in transduced TRCs in vivo. (a). GFP expression in a representative area of a whole TRCs ossicle frozen-section from a 4-week 2.3ColGFP implant. The left panel is a bright field in which a dotted line in red outlines a bone formation area inside the ossicle and the area between the red line and yellow line is a fibrous tissue. The right panel is a fluorescence phase, in which a dotted white line outlines the corresponding bone formation area inside the ossicle; the area between the white line and yellow line is the corresponding fibrous tissue. Original magnification: ×40. (b). Three representative areas from panel b were examined at a higher magnification (×200). A, an area full of cells with strong GFP expression. B, a field containing a bony structure in the middle along with GFP+ cells on both sides. C, a display of two clusters of cells with extremely high expression of 2.3ColGFP. (c). H&E staining of the ossicle section at a magnification of ×200 (left) and ×400 (right). GFP, green fluorescent protein; hBMSCs, human bone marrow stromal cells.
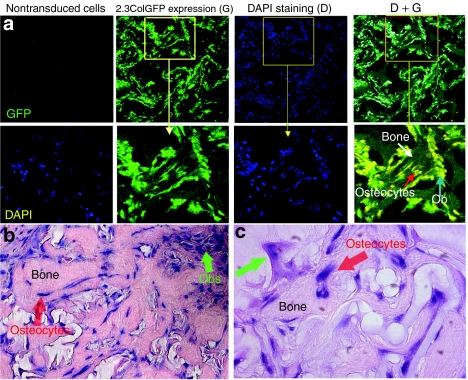
We further examined GFP expression in TRC-derived ossicles by confocal microscopy (Figure 6a). We used 4′6-diamidino-2-phenylindole nuclei staining to identify the distribution of all cells. Most cells located over bone surfaces or within the bone matrix strongly expressed GFP. A representative area was examined at higher magnification as shown in the lower panels. All 4′6-diamidino-2-phenylindole positive cells were also 2.3ColGFPGFP+ and layered over the surfaces of bone matrix with cuboidal and/or flattened osteoblast-like morphology. Some 2.3ColGFP+ cells were embedded in the bone-like structure, indicating that these TRCs fully differentiated and became osteocytes. H&E demonstrated newly formed woven bone and cell distribution at a high magnification of ×200 in Figure 6b and at ×400 in Figure 6c. These observations were similar with 2.3ColGFP-labeled bone of transgenic mice.18
Figure 6.
The confocal imaging of 2.3ColGFP expression in transduced TRCs in vivo. (a) Confocal imaging of 2.3ColGFP expression in ossicles generated by 4-week implantation of TRCs. Nontransduced cells served as a negative control and DAPI was used for staining cellular nuclei (left panel). DAPI staining indicates cellular distribution or existence inside the ossicle from either donor or host origin, whereas GFP indicates the donor origin of cells. Both GFP expression (G) and DAPI staining (D) were examined under Zeiss confocal microscopy at original magnification ×200 (middle two panels). The right panel is an overlay of GFP expression (G) and DAPI staining (D). A representative area with bone structure is amplified to exhibit the distribution of GFP+ cells (lower panels). (b) H&E of the ossicle section at a original magnification ×200. (c) H&E staining of the ossicle section at a original magnification ×400. DAPI, 4,6′-diamidino-2-phenylindole; GFP, green fluorescent protein.
Assessment of bone regeneration potential of TRCs in a mouse calvarial critical-sized defect model
To further quantitatively evaluate the bone regeneration potential of TRCs and address whether an optimal dose is required for bone formation by TRCs or this bone regeneration process is in a dose-dependent manner, we employed a standardized mouse calvarial critical-sized defect model to answer the questions, as this calvarial defect model is believed as a reproducible model for fracture repair. Therefore, we took this model to further examine the fracture repair/regeneration ability of implanted cells. Designed doses of TRCs (0, 0.1, 0.5, 1, or 3 million cells/scaffold) were implanted onto standard sized, mouse critical defect calvarial defects (drilled through the calvaria of immunodeficient mice, 5 mm in diameter). After 7 weeks of implantation, the mice were killed and the calvaria were resected and evaluated by µ-CT. The experiment was replicated twice and each time it showed equivalent outcomes.
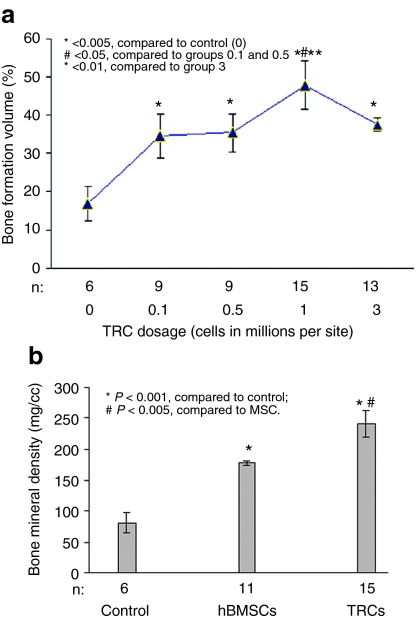
By assessing bone volume fraction transplantation of 0.1, 1, or 3 × 106 of TRCs resulted in significantly more bone formation than that by the cell-free group (Gelfoam only) in the standardized defect area in healing process (P < 0.05, P < 0.001, and P < 0.01, respectively) (Figure 7a). (µ-CT scans and histological images of these samples are shown in Supplementary Figure S1.) The highest bone volume values were associated with a given dose of 1 × 106 cells (Figure 7a). Therefore, 1 million TRCs appeared to be an optimal dose required for bone formation in the mouse calvarial defect model. There were no statistically significant differences in bone volume fraction of a standardized area in the healing calvaria between mice given 105–0.5 × 106 cells and those given 3 × 106 cells (Figure 7a). Given a dose >1 million cells/scaffold did not produced more bone formation as expected; instead, increasing doses above the optimal one appears to cause a reduction in bone formation. It supports our hypothesis that bone regeneration requires a threshold dosage, but does not appear to be in a cell-dose dependent manner.
Figure 7.
Bone regeneration potential in a mouse calvarial critical-sized defect model. (a) µ-CT images evaluation of calvarial defect repair by implantation of different doses of TRCs. Data are presented as the mean ± SEM of three individual experiments while n is the sample size of each group. (b) The comparison of bone regenerative ability between hBMSCs and TRCs in the calvarial defect model by implantation of 1 million cells of hBMSCs or TRCs (n indicated the sample numbers). Human BMSCs, human bone marrow stromal cells; TRC, tissue repair cells.
Because we found that transplantation of 1 million TRCs generated the maximum generated bone formation for repairing the calvarial defect as described above, we further compared the bone regenerative ability of TRCs with hBMSCs by implanting 1 million cells of either hBMSCs or TRCs onto the defect sites in the calvarial defect model. As shown in Figure 7b, implantation of 1 million TRCs resulted in significantly higher bone volume than that of hBMSCs at the same dose (P < 0.005), indicating the higher regenerative capability of TRCs than that of hBMSCs at this same dose. This is consistent with the outcome from the ectopic model (Figure 4a), suggesting that a similar trend was observed from both models. This suggests that the TRC ex vivo expansion process, most notably the presence of multiple cell types, might enhance the osteogenic potential of human bone marrow stem and progenitor cells.
Discussion
The current study demonstrates that the 2.3ColGFP lentivector can specifically and selectively define the fate of hBMSCs or TRCs when they commit to osteoblast lineage in vitro or in vivo; the implanted stem and progenitor cells directly make a major contribution to bone formation in the ectopic model. TRCs, a mixed population of bone marrow–derived cells which contain ~20% mesenchymal progenitors (Table 1),21 exhibited higher bone osteoinductive potential than that of conventionally cultured, passaged hBMSCs, and an optimal dose is required for this process by TRCs (Figures 4 and 5).
The rationale to take hBMSCs in vitro study was to demonstrate the efficiency of the transduction protocol reflected by CVM-driven GFP reporter, and to ensure that this protocol would not affect the potential of proliferation and differentiation in the tested cells. In addition, it is necessary to reveal the responsiveness of the 2.3ColGFP reporter to the endogenous transcriptional regulation in human cells. Compared to the culture system of TRCs which is required to be carried out in a closed-automated AastromReplicell Cell Production System (Aastrom Biosciences, Ann Arbor, MI) for a 12-day period, the procedure for culturing hBMSCs is more easily manipulated, which enables us to daily monitor the GFP expression in a culture flask. Because previous studies have shown that the in vitro osteogenic potential of TRCs is entirely contained within hBMSCs fraction,21 therefore, a separate study was not performed to examine the TRCs in vitro after lentiviral transduction. However, we demonstrated that the lentiviral transduction did not affect the in vivo osteogenic potential of both hBMSCs and TRC in the ectopic model (Figure 4a). After we demonstrated that a highly efficient transduction protocol did not alter the proliferation and differentiation potential of hBMSCs, and because the tempo of 2.3ColGFP expression represents the osteoblast differentiation progression when hBMSCs enter the osteoblast lineage, we moved forward to define the fate of implanted TRCs by this powerful 2.3ColGFP in vivo.
The power of 2.3ColGFP strategy is that we can directly visualize the implanted cells in real-time when they enter the osteoblast lineage and clearly determine their donor origin and contribution. We have performed different time points for implantation, for example, 4-, 6-, 8-, and 16 weeks. We found that after 6 weeks, bony structure dominated the ossicles, relatively few GFP+ cells left on the surface of trabecular-bone or embedded in mineralized matrix and the majority of engrafted cells died as reported in previous publications. We believe that choosing a timing window is critical for visualizing cellular events and defining the fate of these cells during the bone generation process. Based on our preliminary study, we chose a 4-week point as a critical timing window at the time we can visualize all 2.3ColGFP+ cells functioning for bone formation. After 6 weeks, we observed much less 2.3ColGFP+ cells on surface or embedded in mineralized bone matrix, as the majority of grafted cells have completed their jobs and cannot be detected (data not shown). Therefore, it will be tricky to make a conclusion on the contribution of the implanted cells. This GFP marker has been demonstrated to function in a murine model for >6 months in the surviving cells in the calvarial defect model.
A self-inactivated lentiviral vector, in which low level of long-term repeat transcription is completely terminated, is ideal for lineage-specific gene expression to track the fate of stem cells. Previous studies demonstrated a lineage-specific expression driven by a tissue-specific promoter in hematopoietic cells.13,28,29,30 In this study, we used a bone-specific promoter, 2.3Col, which is a rat 2.3 kb type I collagen 1A1 promoter. The 2.3ColGFP reporter has been well documented in a transgenic mouse model as GFP-labeled osteoprogenitor cells differentiate to osteoblasts and osteocytes.18 Our data show that expression of 2.3ColGFP is very low at the beginning of culture in the osteogenic medium and that it increases steadily over time as the number of osteoblasts increases (Figure 3). The GFP-labeled cells differentiated at the same tempo as unmodified hBMSCs, and responded appropriately to transcriptional regulation (Figure 3). Taken together, these data demonstrates the specificity of the rat-derived 2.3Col promoter–driven GFP for osteogenic cells originating from human bone marrow. Therefore, the 2.3ColGFP reporter can be utilized as a valuable tool for determining the fate of human bone marrow stem and progenitor cells into osteoblast lineage.
As mentioned previously, technical difficulties have limited knowledge of the mechanisms in which cell grafts contribute to the repair or regeneration of bone tissue. To overcome loss of GFP signal during tissue processing for paraffin embedding in this study, we used a frozen-section approach to preserve consistent, robust GFP expression. With the osteoblast-specific 2.3ColGFP reporter, we successfully demonstrated that the implanted TRCs entered into the osteoblast lineage (Figure 5). In addition, we clearly demonstrated that almost every individual cell inside the generated ossicles was GFP+ (Figure 6). This suggests that the bone forming cell populations are of donor origin, and that these cells directly make a major contribution to bone formation. In contrast, cells of host origin are not detectable because cells with 4′6-diamidino-2-phenylindole staining inside ossicles are all GFP+ at the time examined (4 weeks of implantation). It remains to be determined whether the host-origin cells participate in the modeling of bone-generating structures at late stages.
For the repair of complex tissues containing many cell types, a cell product with multi-lineage potential is highly desired. For example, in bone generation, the development of vascular structures is required prior to new bone formation.31 We have observed that TRCs had higher bone osteoinductive potential than conventional cultured hBMSCs in the mouse ectopic ossicle model at a dose of 3 million cells/scaffold (Figure 4) and in the mouse calvarial defect repair model at a dose of 1 million cells/scaffold (Figure 7). One of the key features of TRCs is the presence of ex vivo expanded mixed population cells from multiple lineages in a single pass perfusion bioreactor, including hematopoietic (nonadherent cells) and mesenchymal (adherent cells). During culture, the CD90+ cells in TRCs, which represent mesenchymal stem and progenitor cells, are enriched >135-fold compared to the starting BMMNC population. CD90+ TRCs have been found to strongly correlate with colony-forming unit in fibroblastic colony-forming ability and osteogenic potential.21 In addition to CD90+ cells, TRCs are enriched for cell populations expressing endothelial lineage markers, including von Willebrand factor and vascular endothelial-cadherin and can form tube-like structures in a collagen matrix.22 Therefore, unlike conventional hBMSCs, TRCs may provide both osteogenic and vascular components. As demonstrated by histological staining (Figures 4 and 5), in comparison to ossicles formed by hBMSCs, there were more bone forming cells present in the surrounding layer of fibroblasts with weak expression of 2.3ColGFP in TRCs-derived ossicles, demonstrating that TRCs are able to provide more cells for bone formation. This may be one of mechanisms responsible for TRCs' stronger bone osteoinductive potential.
From a clinical aspect, it is essential to consider a dosage for implanted cells for meeting mass transport processes and metabolic demand (reviewed by Muschler et al.).32 To regenerate bone, a sufficient number of stem cells must be available for regeneration.33 Limited clinical investigations indicate a positive correlation between bone marrow osteogenic capacity and cell concentration.5,34 A balance between consumption clearance and local delivery of substrate molecules (oxygen, glucose, and amino acids) is critical for cell survival.32 We used a mouse calvarial defect model to examine whether the potential for bone regeneration depends on dosage or it requires a threshold dosage. As shown in Figure 7, we observed that maximum bone regeneration is produced by an optimal dose of implanted TRCs, indicating that a threshold is required. Given the space within the scaffold for hosting the grafted cells and local vascular penetration are limited, and optimal dose is required to keep in balance of consumption clearance and local delivery of nutrition (oxygen, glucose, and amino acids) for cell survival, proliferation, and function. Cells less than the optimal dose provided to the defect site will not reach to the maximum of regenerative function; while increasing the cells over the threshold will lead to interrupt the balance, leading to cell death and delaying the regeneration process. Therefore, a cell-dose threshold is required for bone regeneration by TRCs. Although we did not perform an optimal dosing experiment for hBMSCs, this general principal may be applied to all cells for tissue regeneration and their optimal doses may be different. This observation may provide an insight for designing clinical practice of stem cell therapy–based bone regeneration.
In conclusion, we used a lentivirus to efficiently deliver GFP reporter driving by a bone-specific promoter into bone marrow–derived stem cells populations. This in vivo cell fate determination method enabled us to define the fate and contribution of TRCs and hBMSCs to bone formation. We also found that TRCs, as an innovative mixture of stem and progenitor cells, had stronger in vivo osteogenic potential than traditionally cultured hBMSCs. Finally, we determined that an optimal cell dose is required for bone regeneration by TRCs.
Materials and Methods
Cell culture. Human BMMNCs were purchased from Poietics (Walkersville, MD) for producing hBMSCs and TRCs from the same donors (n = 8). Human BMSCs were cultured as previously described;19 in brief, a single cell suspension of freshly obtained BMMNCs was plated in T-75 flasks at a density of 22.5 million cells per flask (0.3 million cells/cm2) and cultured in growth medium consisting of α-minimum essential medium (Invitrogen, Grand Island, NY), 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 2 mmol/l L-glutamine, 10−8 mmol dexamethasone (Sigma, St Louis, MO), 100 U/ml penicillin, 100 µg/ml streptomycin sulfate (Biofluids, Rockville, MD). Nonadherent cells were removed on day 3 at the first medium change and fresh medium was replaced twice per week thereafter. The cells were passaged two times prior to transplantation. For TRC cell production, human BMMNCS were cultured in a single perfusion bioreactor system in long-term bone marrow culture medium consisting of Iscove's modified Dulbecco's medium, 10% fetal bovine serum, 10% horse serum, 5 µmol/l hydrocortisone.21 After inoculation of cells, the bioreactor was placed into the automated AastromReplicell Cell Production System (Aastrom Biosciences) and medium was exchanged at a controlled ramped perfusion schedule during a 12-day culture process.35 Cultures were maintained at 37 °C with 5% CO2 and 20% O2. The TRC product was then harvested with 0.025% trypsin in 0.9% sodium chloride and used for these studies. Neither hBMSCs nor TRCs were cultured in any osteoinduction medium prior to implantation in vivo. In addition, a similar TRC product could be achieved with T-flask based small-scale culture at a seeding density of 0.3 million BMMNCs/cm2 as described previously.36
Lentiviral constructs. A CMV promoter driving GFP lentiviral vector, pLL3.737 was purchased from the American Type Culture Collection (Manassas, VA). A lentiviral vector containing a bone-specific 2.3ColGFP was constructed to track osteoblast lineage determination. The 2.3 kb collagen 1A1 promoter was derived from an original plasmid, Col3.6Puc12 (ref. 38) by restriction enzyme digestion with HindIII and ligated with a PacI linker. The 2.3-kb fragment was further cut with PacI and BamHI, and subcloned into a lentiviral vector FUGW13 at corresponding sites. The resulting vector was designated as Lenti2.3ColGFP (Figure 3a).
Lentiviral production. Third generation packaging systems for lentiviral production were used as described previously.13 The isolated media was then passed through a 0.4-µm filter, after which the virus was concentrated by ultracentrifugation for 2 hours at 25,000 r.p.m. in a Beckman SW28 rotor. Titers were determined by infecting 293 FT cells with serial dilutions of concentrated lentivirus. GFP expression of infected cells was determined by FACS analysis (Epics XL-MCL; Beckman Coulter, Fullerton, CA) 3–4 days after transduction. Typically, the titer was expected to be ~5 × 106 infectious units/ml and 107–108 infectious units/ml after concentration.
Lentiviral transduction. We conducted lentiviral transductions of primary hBMSCs with a lentivirus (pLL3.7 or Lenti2.3ColGFP, MOI: 5 × 105 viral particles/ml) when individual colonies began to appear in primary culture of hBMSCs, typically from days 4–7 of culture. Transductions were repeated twice before examination for GFP expression under a fluorescent microscope or by flow cytometry. For lentiviral transduction of TRCs, we performed transduction of BMMNCs prior to initiating the 12-day culture process to generate TRCs. Briefly, human BMMNCS were seeded in T-75 flasks at a density of 45 × 106 cells/flask in 5 ml of long-term bone marrow culture medium and 5 ml of a concentrated lentivirus with a titer of ~8 × 107 viral particles/ml in the presence of protamine sulfate (8 µg/ml; Sigma) for overnight. Next day, cells were rinsed with 10-ml phosphate buffered saline and cultured in 20 ml long-term bone marrow culture medium for a day before repeating the second viral transduction as described above. On day 3, the transduced BMMNCs were harvested by trypsinization and cultured for TRC production as described earlier and previously.39 The efficiency of transduction was determined by flow cytometric analysis of GFP expression.
Flow cytometric analysis of cell immunophenotypes. We performed flow cytometry to compare immunophenotypes of TRCs and hBMSCs and assess whether lentiviral transduction affected cell immunophenotypes by an Epics XL-MCL flow cytometer (Beckman Coulter) as previously described.21 All PE- or FITC-conjugated antibodies were purchased from Bechman Coulter and all flow cytometry data was 7-aminoactinomycin live-gated.
Cell proliferation and differentiation assay. To monitor cell proliferation, we used a BrdU incorporation assay according to the manufacturer's instructions (FITC BrdU Flow Kit; BD Pharmingen, San Diego, CA) in conjunction with immunocytochemistry or FACS. Briefly, passaged nontransduced hBMSCs, and hBMSCs transduced with the pLL3.7 lentivirus, were cultured on four-well chamber slides (2 × 105 cells/well) for 5 days, and exposed overnight to 10 µmol/l BrdU before fixation with 4% paraformaldehyde. Immunohistochemical staining for BrdU was visualized by microscopy. For FACS analysis of BrdU+ cells, 2 × 106 cells were cultured in a 10 cm2 dish for 24 hours and then labeled with BrdU at a final concentration of 10 µmol/l for 17 hours. Flow cytometric analysis of stained cells was conducted using an APC BrdU flow kit (BD Biosciences, San Diego, CA) per the manufacturer's instructions and a flow cytometer (Epics Altra; Beckman Coulter).
To induce osteogenic differentiation, passaged hBMSCs, with or without lentiviral transduction were treated with osteogenic medium consisting of α-minimum essential medium supplemented with 10% fetal bovine serum, 50 µg/ml ascorbic acid (Sigma), 10 nmol/l dexamethasone (Sigma), and 5 mmol/l β-glycerophosphate (Sigma) for 4 weeks as described previously.19 To identify newly deposited mineral in the cell culture, the cells were fixed with 10% normal buffered formalin for 20 minutes at room temperature, and stained with 1% silver nitrate (Sigma; von Kossa staining) for 20 minutes under ultra violet light to identify mineralization. To test for potential of adipogenic differentiation, hBMSCs, or lentiviral transduced hBMSCs were treated with adipogenic medium consisting of α-minimum essential medium supplemented with 10% fetal bovine serum, 1 µmol/l hydrocortisone (Sigma), 0.1 mmol/l indomethacin (Sigma), and 0.5 mmol/l 3-isobutyl-1-methylxanthine (Sigma) for 3 weeks. Then cells were fixed with 4% formaldehyde, stained with Oil Red O (Sigma) for 10 minutes, and then counterstained with hematoxylin (Sigma) for 1 minute.
Reverse transcription–PCR analysis of osteoblast differentiation markers. Human BMSCs were cultured in 10-cm dishes in the osteogenic medium for 0, 6, 10, 15, or 18 days. Total RNA extraction from cultured hBMSCs at designated time points was done with Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA was further purified by PureLink Micro-to-Midi total RNA Purification System (Invitrogen, Grand Island, NY) according to the manufacturer's instructions. The mRNA from 1 µg purified total RNA was reverse transcribed to complementary DNA using SuperScript III-First-Strand Synthesis SuperMix (Invitrogen, Carlsbad, CA) and complementary DNA was amplified using Platinum PCR SuperMix (Invitrogen, Grand Island, NY) and an ABI GeneAmp PCR System 97 °C (Applied Biosystems, Boston, MA) at 94 °C for 30 seconds, 55 °C for 30 seconds, and 72 °C for 60 seconds for 30 cycles, after initial denaturation at 94 °C for 2 minutes. Primers used for amplification were described previously.19 PCR products were separated on a 2% agarose gel by electrophoresis. Quantitative real-time PCR was carried out using TaqMan Universal PCR Master Mix and TaqMan premade primers (Applied Biosystems). Primers for bone morphogenetic protein-2 are: forward 5′ GGTGGAATGACTGGATTG 3′; reverse 5′ GCATCGAGATAGCACTG 3′. All experiments were done with two biological and two technical replicates. Values were calculated using the comparative C(t) method and normalized to hypoxanthine phosphoribosyltransferase expression.
Mouse ossicle and calvarial critical-sized defect models. All experimental protocols were approved in accordance with the University Committee on Use and Care of Animals at the University of Michigan. Animals were housed in a light- and temperature-controlled environment and given food and water ad libitum. The ossicle bone formation model has been described previously.40,41 Briefly, up to 3 million fresh TRCs or passaged hBMSCs (passages 2–3) were collected in 1.5-ml microcentrifuge tubes by centrifuging for 5 minutes at a speed of 1,500 r.p.m. and resuspending in 40 µl Dulbecco's modified Eagle's medium (Invitrogen, Grand Island, NY). Concentrated cells were loaded into 3.5-mm Gelfoam (a gelatin scaffold; Pharmacia & Upjohn, Kalamazoo, MI) cubes for the ossicle model and Gelfoam discs with a diameter of 5 mm and thickness of 3.5 mm for a mouse calvarial critical-sized defect model. Five-week-old female severe combined immunodeficiency mice (NIH-bg-nuxid BR; Harlan Sprague Dawley, Indianapolis, IN) were anesthetized by peritoneal injection of an anesthetic cocktail (75 mg/kg ketamine, 10 mg/kg xylazine of body weight). An incision was made on the dorsal surface of skin in each mouse. Two or four subcutaneous pockets per mouse were created by blunt dissection and three severe combined immunodeficiency mice per cell group were used. A single implant was placed into each pocket and incisions were closed with surgical staples. Mice were killed at 4, 6, or 8 weeks after surgery with inhaled CO2 and their ossicles were harvested.
For quantitative analysis of bone formation by TRCs or hBMSCs, we utilized a critical-sized calvarial defect model in 5-week-old female severe combined immunodeficiency mice as previously described.8,40 Briefly, a linear scalp incision was made from the nasal bone to the occiput and full-thickness flaps were elevated. The periosteum overlying the calvarial bone was completely resected. A trephine was used to create a craniotomy defect (5 mm in diameter) centered on the sagittal sinus and the wounds were copiously irrigated with Hanks' balanced salt solution (Invitrogen, Grand Island, NY) while drilling. The calvarial disc was removed carefully in order to avoid injury to the underlying dura or brain. After careful hemostasis, gelatin sponges previously loaded with cells were placed into the defects. The cells were concentrated by centrifugation in 1.5-ml tubes before being loaded onto gelatin sponges. The cells loaded onto the gelatin sponge were incubated for 30 minutes before implantation. The sponges were placed to fill the entire defect and allowed to attach to the bone edges around the entire periphery. The incision was closed with 4-0 Chromic Gut suture (Ethicon/Johnson & Johnson, Sommerville, NJ). Mice were killed 7 weeks after surgery with inhaled CO2 and their calvaria were harvested for µ-CT analysis of calvarial defect repair, followed by histological examination.
µ-CT analysis. Implants from the ossicle and calvarial model were harvested at 6 or 7 weeks after surgery, respectively. After fixation in aqueous buffered zinc formalin for 24 hours at 4 °C, we conducted µ-CT to assess bone regeneration such as bone mineral density, bone volume, and bone volume fraction as previously described.41 In general, specimens were scanned at an 8.93-µm voxel resolution on an EVS Corporation µ-CT scanner (London, Ontario, Canada), with a total of 667 slices per scan. GEMS MicroView software (GE, Medical Systems, Toronto, Ontario, Canada) was used to make a 3D reconstruction from the set of scans. Fixed thresholds of 1,500 and 500 were used to extract the mineralized bone phase of ossicles and calvaria, respectively. Actual bone volumes were then calculated.
Histological examination. For histology and detection of GFP expression in tissues, fixed implants were decalcified in 10% EDTA for 3 days. For H&E staining, decalcified implants were embedded in paraffin and sectioned (5-µm thick) at the Histology Core Facility, University of Michigan School of Dentistry. For detection of GFP expression, a CryoJane Tape-Transfer System (Instrumedics, Hackensack, NJ) was utilized to section frozen decalcified samples (5-µm thick) as previously described.42 GFP expression was assessed using a Zeiss Axioplan 200 inverted microscope equipped with epifluoresence and a Zeiss AxioCam color digital camera. The filter combinations that were used to distinguish the GFP signal from the autofluorescent background of bone marrow were: Excitation 475/25; Emission 495; Dichroic 525/45. Confocal imaging of GFP expression was done at the Imaging Core of the University of Michigan Medical School (Zeiss LSM 510 meta, excitation 488 and emission 505/550). Documentation of H&E staining for implant sections was recorded by Zeiss Observer Z1 with AxioCam HRC camera (Carl Zeiss MicroImaging GmbH, Göttingen, Germany). Immunofluorescent staining of hOC was performed following the manufacturer's instruction (R&D Systems, Minneapolis, MN). A mouse-derived monoclonal anti-hOC antibody was purchased from R&D Systems and it was diluted by 1:200 to 10 µg/ml for immunofluorescent detection of human osteocalcin expression in the ossicle sections. A mouse isotype prematched immunoglobulin, immunoglobulin G, of irrelevant antispecificity at the same concentration was used as a negative control. Human OC expression was visualized by rhodamine red coupled anti-mouse immunoglobulin G under fluorescent microscopy and cell nuclei were stained with 4′6-diamidino-2-phenylindole. Masson's Trichrome staining was carried out according to Sigma–Aldrich's instruction for Trichrome Stain (Masson) kit (Sigma; cat. no. HT15), which stains nuclei in black; cytoplasm, muscle and erythrocytes in red, and bone/collagen in blue.
Statistics. Data are presented as the mean ± SD or the mean ± SEM, dependent on sample numbers. For ossicle samples, Student's t-test for independent groups was used for statistical analysis. P value <0.05 was considered statistically significant. Significant differences between mean values in calvarial samples were evaluated by Dunnett's method with either hBMSC or acellular group as a control using JMP 6.0.3 software (SAS Institute, Cary, NC).
SUPPLEMENTARY MATERIALFigure S1. µ-CT images and H&E staining of calvarial defect repair by implantation of different doses of TRCs (a magnification of ×200).
Supplementary Material
µ-CT images and H&E staining of calvarial defect repair by implantation of different doses of TRCs (a magnification of ×200).
Acknowledgments
This work was supported by Aastrom Biosciences Inc. The scientific data presented in the manuscript was approved by the company, but we did not select results preferable to the company. We thank Ronnda L. Bartel and Kristin L. Goltry for reading the manuscript.
REFERENCES
- Caplan AI. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol. 2007;213:341–347. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
- Connolly JF. Injectable bone marrow preparations to stimulate osteogenic repair. Clin Orthop Relat Res. 1995. pp. 8–18. [PubMed]
- Connolly JF, Guse R, Tiedeman J., and , Dehne R. Autologous marrow injection as a substitute for operative grafting of tibial nonunions. Clin Orthop Relat Res. 1991. pp. 259–270. [PubMed]
- Gangji V., and , Hauzeur JP.2005Treatment of osteonecrosis of the femoral head with implantation of autologous bone-marrow cells. Surgical technique J Bone Joint Surg Am 87suppl. 1) (Pt 1): 106–112. [DOI] [PubMed] [Google Scholar]
- Muschler GF, Nitto H, Matsukura Y, Boehm C, Valdevit A, Kambic H, et al. Spine fusion using cell matrix composites enriched in bone marrow-derived cells. Clin Orthop Relat Res. 2003. pp. 102–118. [DOI] [PMC free article] [PubMed]
- Dominici M, Pritchard C, Garlits JE, Hofmann TJ, Persons DA., and , Horwitz EM. Hematopoietic cells and osteoblasts are derived from a common marrow progenitor after bone marrow transplantation. Proc Natl Acad Sci USA. 2004;101:11761–11766. doi: 10.1073/pnas.0404626101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson SK, Hulspas R, Weier HU., and , Quesenberry PJ. In situ detection of individual transplanted bone marrow cells using FISH on sections of paraffin-embedded whole murine femurs. J Histochem Cytochem. 1996;44:1069–1074. doi: 10.1177/44.9.8773573. [DOI] [PubMed] [Google Scholar]
- Mankani MH, Kuznetsov SA, Wolfe RM, Marshall GW., and , Robey PG. In vivo bone formation by human bone marrow stromal cells: reconstruction of the mouse calvarium and mandible. Stem Cells. 2006;24:2140–2149. doi: 10.1634/stemcells.2005-0567. [DOI] [PubMed] [Google Scholar]
- Goshima J, Goldberg VM., and , Caplan AI. The origin of bone formed in composite grafts of porous calcium phosphate ceramic loaded with marrow cells. Clin Orthop Relat Res. 1991. pp. 274–283. [PubMed]
- Sena G, Jung JW., and , Benfey PN. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development. 2004;131:2817–2826. doi: 10.1242/dev.01144. [DOI] [PubMed] [Google Scholar]
- Balsam LB, Wagers AJ, Christensen JL, Kofidis T, Weissman IL., and , Robbins RC. Haematopoietic stem cells adopt mature haematopoietic fates in ischaemic myocardium. Nature. 2004;428:668–673. doi: 10.1038/nature02460. [DOI] [PubMed] [Google Scholar]
- Landmann F, Quintin S., and , Labouesse M. Multiple regulatory elements with spatially and temporally distinct activities control the expression of the epithelial differentiation gene lin-26 in C. elegans. Dev Biol. 2004;265:478–490. doi: 10.1016/j.ydbio.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ., and , Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Bilic-Curcic I, Kalajzic Z, Wang L., and , Rowe DW. Origins of endothelial and osteogenic cells in the subcutaneous collagen gel implant. Bone. 2005;37:678–687. doi: 10.1016/j.bone.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen X, Armstrong MA., and , Li G. Survival of bone marrow-derived mesenchymal stem cells in a xenotransplantation model. J Orthop Res. 2007;25:926–932. doi: 10.1002/jor.20385. [DOI] [PubMed] [Google Scholar]
- Hannouche D, Raould A, Nizard RS, Sedel L., and , Petite H. Embedding of bone samples in methylmethacrylate: a suitable method for tracking LacZ mesenchymal stem cells in skeletal tissues. J Histochem Cytochem. 2007;55:255–262. doi: 10.1369/jhc.6A7063.2006. [DOI] [PubMed] [Google Scholar]
- Tu Q, Valverde P, Li S, Zhang J, Yang P., and , Chen J. Osterix overexpression in mesenchymal stem cells stimulates healing of critical-sized defects in murine calvarial bone. Tissue Eng. 2007;13:2431–2440. doi: 10.1089/ten.2006.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, et al. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- Liu P, Kalajzic I, Stover ML, Rowe DW., and , Lichtler AC. Human bone marrow stromal cells are efficiently transduced by vesicular stomatitis virus-pseudotyped retrovectors without affecting subsequent osteoblastic differentiation. Bone. 2001;29:331–335. doi: 10.1016/s8756-3282(01)00590-7. [DOI] [PubMed] [Google Scholar]
- Bruder SP, Kurth AA, Shea M, Hayes WC, Jaiswal N., and , Kadiyala S. Bone regeneration by implantation of purified, culture-expanded human mesenchymal stem cells. J Orthop Res. 1998;16:155–162. doi: 10.1002/jor.1100160202. [DOI] [PubMed] [Google Scholar]
- Dennis JE, Esterly K, Awadallah A, Parrish CR, Poynter GM., and , Goltry KL. Clinical-scale expansion of a mixed population of bone-marrow-derived stem and progenitor cells for potential use in bone-tissue regeneration. Stem Cells. 2007;25:2575–2582. doi: 10.1634/stemcells.2007-0204. [DOI] [PubMed] [Google Scholar]
- Gastens MH, Goltry K, Prohaska W, Tschöpe D, Stratmann B, Lammers D, et al. Good manufacturing practice-compliant expansion of marrow-derived stem and progenitor cells for cell therapy. Cell Transplant. 2007;16:685–696. doi: 10.3727/000000007783465172. [DOI] [PubMed] [Google Scholar]
- Stiff P, Chen B, Franklin W, Oldenberg D, Hsi E, Bayer R, et al. Autologous transplantation of ex vivo expanded bone marrow cells grown from small aliquots after high-dose chemotherapy for breast cancer. Blood. 2000;95:2169–2174. [PubMed] [Google Scholar]
- Pecora AL, Stiff P, LeMaistre CF, Bayer R, Bachier C, Goldberg SL, et al. A phase II trial evaluating the safety and effectiveness of the AastromReplicell system for augmentation of low-dose blood stem cell transplantation. Bone Marrow Transplant. 2001;28:295–303. doi: 10.1038/sj.bmt.1703137. [DOI] [PubMed] [Google Scholar]
- Jimenez ML, Lyon T, Nowinski G, Balazsy J, Goulet JA, Wolff S, et al. Stem and progenitor cell therapy for management of refractory long bone nonunions: a multicenter clinical feasibility study. American Academy of Orthopaedic Surgeons Annual Meeting (abstract) 2007.
- Chen W, Wu X, Levasseur DN, Liu H, Lai L, Kappes JC, et al. Lentiviral vector transduction of hematopoietic stem cells that mediate long-term reconstitution of lethally irradiated mice. Stem Cells. 2000;18:352–359. doi: 10.1634/stemcells.18-5-352. [DOI] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ., and , Stein JL. The osteocalcin gene: a model for multiple parameters of skeletal-specific transcriptional control. Mol Biol Rep. 1997;24:185–196. doi: 10.1023/a:1006803615430. [DOI] [PubMed] [Google Scholar]
- Moreau-Gaudry F, Xia P, Jiang G, Perelman NP, Bauer G, Ellis J, et al. High-level erythroid-specific gene expression in primary human and murine hematopoietic cells with self-inactivating lentiviral vectors. Blood. 2001;98:2664–2672. doi: 10.1182/blood.v98.9.2664. [DOI] [PubMed] [Google Scholar]
- Scott BB., and , Lois C. Generation of tissue-specific transgenic birds with lentiviral vectors. Proc Natl Acad Sci USA. 2005;102:16443–16447. doi: 10.1073/pnas.0508437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui K, Furuta RA, Matsumoto K, Tani Y., and , Fujisawa J. HIV-1-derived self-inactivating lentivirus vector induces megakaryocyte lineage-specific gene expression. Microbes Infect. 2005;7:240–247. doi: 10.1016/j.micinf.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Carano RA., and , Filvaroff EH. Angiogenesis and bone repair. Drug Discov Today. 2003;8:980–989. doi: 10.1016/s1359-6446(03)02866-6. [DOI] [PubMed] [Google Scholar]
- Muschler GF, Nakamoto C., and , Griffith LG. Engineering principles of clinical cell-based tissue engineering. J Bone Joint Surg Am. 2004;86-A:1541–1558. doi: 10.2106/00004623-200407000-00029. [DOI] [PubMed] [Google Scholar]
- Triffitt JT. Osteogenic stem cells and orthopedic engineering: summary and update. J Biomed Mater Res. 2002;63:384–389. doi: 10.1002/jbm.10260. [DOI] [PubMed] [Google Scholar]
- Hernigou P, Poignard A, Beaujean F., and , Rouard H. Percutaneous autologous bone-marrow grafting for nonunions. Influence of the number and concentration of progenitor cells. J Bone Joint Surg Am. 2005;87:1430–1437. doi: 10.2106/JBJS.D.02215. [DOI] [PubMed] [Google Scholar]
- Mandalam RK., and , Smith AK.2002Ex vivo expansion of bone marrow and cord blood cells to produce stem and progenitor cells for hematopoietic reconstitution Mil Med 1672 suppl.): 78–81. [PubMed] [Google Scholar]
- Koller MR, Manchel I., and , Palsson BO. Importance of parenchymal:stromal cell ratio for the ex vivo reconstitution of human hematopoiesis. Stem Cells. 1997;15:305–313. doi: 10.1002/stem.150305. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Pavlin D, Lichtler AC, Bedalov A, Kream BE, Harrison JR, Thomas HF, et al. Differential utilization of regulatory domains within the alpha 1(I) collagen promoter in osseous and fibroblastic cells. J Cell Biol. 1992;116:227–236. doi: 10.1083/jcb.116.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundell BI, Mandalam RK., and , Smith AK. Clinical scale expansion of cryopreserved small volume whole bone marrow aspirates produces sufficient cells for clinical use. J Hematother. 1999;8:115–127. doi: 10.1089/106161299320398. [DOI] [PubMed] [Google Scholar]
- Krebsbach PH, Kuznetsov SA, Satomura K, Emmons RV, Rowe DW., and , Robey PG. Bone formation in vivo: comparison of osteogenesis by transplanted mouse and human marrow stromal fibroblasts. Transplantation. 1997;63:1059–1069. doi: 10.1097/00007890-199704270-00003. [DOI] [PubMed] [Google Scholar]
- Wang Z, Song J, Taichman RS., and , Krebsbach PH. Ablation of proliferating marrow with 5-fluorouracil allows partial purification of mesenchymal stem cells. Stem Cells. 2006;24:1573–1582. doi: 10.1634/stemcells.2005-0399. [DOI] [PubMed] [Google Scholar]
- Jiang X, Kalajzic Z, Maye P, Braut A, Bellizzi J, Mina M, et al. Histological analysis of GFP expression in murine bone. J Histochem Cytochem. 2005;53:593–602. doi: 10.1369/jhc.4A6401.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
µ-CT images and H&E staining of calvarial defect repair by implantation of different doses of TRCs (a magnification of ×200).