Abstract
Gene transfer vectors may cause clonal imbalance and even malignant cell transformation by insertional upregulation of proto-oncogenes. Lentiviral vectors (LV) with their preferred integration in transcribed genes are considered less genotoxic than gammaretroviral vectors (GV) with their preference for integration next to transcriptional start sites and regulatory gene regions. Using a sensitive cell culture assay and a series of self-inactivating (SIN) vectors, we found that the lentiviral insertion pattern was approximately threefold less likely than the gammaretroviral to trigger transformation of primary hematopoietic cells. However, lentivirally induced mutants also showed robust replating, in line with the selection for common insertion sites (CIS) in the first intron of the Evi1 proto-oncogene. This potent proto-oncogene thus represents a CIS for both GV and LV, despite major differences in their integration mechanisms. Altering the vectors' enhancer–promoter elements had a greater effect on safety than the retroviral insertion pattern. Clinical grade LV expressing the Wiskott–Aldrich syndrome (WAS) protein under control of its own promoter had no transforming potential. Mechanistic studies support the conclusion that enhancer-mediated gene activation is the major cause for insertional transformation of hematopoietic cells, opening rational strategies for risk prevention.
Introduction
Gene vectors based on retroviruses are widely used for stable genetic modification of somatic cells.1 Improvements of retroviral vector technology have led to the design of self-inactivating (SIN) vectors in which the enhancer–promoter sequences of the long terminal repeats (LTRs) are deleted and instead the gene of interest is expressed from an internal promoter. Another major progress was the conversion of complex lentiviruses, such as the human immunodeficiency virus type 1, into efficient gene vectors that combine advantages of the SIN design with the potential to stably transduce nondividing cells.2 However, of all retroviruses investigated to date, lentiviruses and derived vectors show the greatest preference to integrate within active transcription units. In contrast to gammaretroviral vectors (GV), lentiviral vectors (LV) have no preference to integrate in a 10 kilobase window surrounding the transcriptional start sites and also no preference to integrate near regulatory gene regions that coincide with DNAse 1 hypersensitive sites.3,4 In freshly transduced primary human hematopoietic cells, insertions near proto-oncogenes are at least twice less likely for SIN-LV compared to LTR-driven GV.5
Considering the complex organization of individual genes within the genome, the relatively global perspective of bioinformatic “integrome” studies can hardly predict the functional consequences of vector insertions. Illustrating this issue, in the few cases of cell transformation observed to date when using LTR-driven GV, the activation of cellular proto-oncogenes (LMO2, MDS-EVI1, CYCLIND2, or BMI1) occurred from many types of genomic locations: distant upstream or downstream, upstream promoter-proximal, introns, and across gene borders.6,7,8 Randomly isolated cell clones transduced by SIN-LV showed that the likelihood to activate a neighboring cellular gene largely depends upon the type of the vector's internal promoter, and similar observations were made with SIN-GV.9 Recently, two model systems that used pretransformed target cells showed evidence for insertional transformation by LV containing strong enhancer–promoter sequences located in the LTRs,10,11 whereas SIN-LV containing the same enhancer–promoter sequences as an internal promoter did not accelerate tumorigenesis. Based upon the comparison of LTR-proficient LV and GV, evidence was obtained that the GV insertion pattern induces a 10-fold increased risk of cell transformation.10 The actual risk induced by SIN-LV containing strong internal enhancer–promoter sequences remained unknown.
We have previously used a simple cell culture model to quantify the risk of hematopoietic cell transformation by GV dependent on their design. This assay uses dose-escalated gene transfer into primary murine hematopoietic cells isolated from untreated adult C57BL6 mice, and thus operates in a genetic background lacking a pre-existing transforming lesion. Cell transformation is detected by culturing cells post-transduction under myeloid differentiation conditions, such that a replating step in limiting dilution suppresses residual self-renewal capacity unless insertional upregulation of cellular proto-oncogenes occurs. Animal experiments with a tumor end point are not required, and a quantification of transforming events is greatly facilitated when compared with presently available murine in vivo models. This approach, here referred to as in vitro immortalization (IVIM) assay, is relatively specific for upregulation of Evi1 or its close relative Prdm16 (refs. 12,13,14), both of which have been shown to be clinically relevant as inducers of clonal imbalance in the context of a gene therapy trial to treat patients suffering from an X-linked form of chronic granulomatous disease.6 Activating rearrangements in the human EVI1 gene are a major driving force of acute myeloid leukemia,15 underscoring the potential clinical relevance of the IVIM assay. As described earlier, the IVIM assay is able to quantify the incidence of mutants on the basis of the initial number of transduced cells and the clonal characterization of the mutants that show robust replating after limiting dilution. We show that the risk of insertional transformation by SIN-LV, just as previously reported for SIN-GV, depends upon the type of the internal promoter. Owing to the high sensitivity of the assay, we describe Evi1 as the first transforming common insertion sites (CIS) shared by SIN-GV and SIN-LV, and formally define the enhancer as the driving force of transformation in this model. SIN-LV designed to correct a genetic disease using a cellular promoter were unable to trigger cell transformation in this model.
Results
Lower incidence of transformation induced by SIN-LV compared to SIN-GV
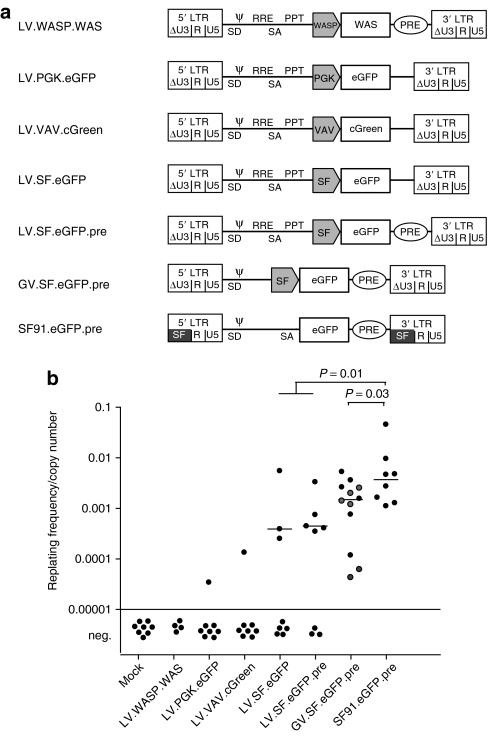
To address whether the clinically relevant SIN-LV may transform primary cells by insertional mutagenesis even in the absence of pre-established transforming lesions, we here performed a direct head-to-head comparison with GV, using the IVIM assay (experimental outline in Supplementary Figure S1).13,14 As a positive control, we chose LTR-driven GV (SF91.eGFP.pre) with the enhancer–promoter of spleen focus-forming virus (SFFV), which exhibits high activity in hematopoietic cells and is known to trigger leukemia by insertional mutagenesis in mice.16,17 To study the impact of vector backbone, we tested SIN-GV (GV.SF.eGFP.pre) and SIN-LV (LV.SF.eGFP and LV.SF.eGFP.pre) containing the same internal enhancer–promoter of SFFV (Figure 1a).
Figure 1.
Transforming activity of integrating vectors depends on vector background and content. (a) Vectors tested in the in vitro immortalization (IVIM) assay in the first set of experiments. We compared the IVIM frequency of three SIN-LV with different internal promoters (SFFV, PGK, and VAV) with an LTR-driven GV (SF91.eGFP.pre) and a SIN-GV (GV.SF.eGFP.pre). The latter represents the gammaretroviral equivalent of LV.SF.eGFP.pre. We also tested a SIN-LV that expresses WASP from the WAS promoter (LV.WASP.WAS). (b) Results of the IVIM assay: plotted are the replating frequencies corrected for the mean copy number as measured in the DNA of mass cultures taken 4 days after transduction. In none of the assays performed (n = 8), we obtained replating clones from untransduced cultures (mock), whereas SF91.eGFP.pre transduced cells always led to immortalized clones (n = 8). When transducing cells with the LV.SF.eGFP (n = 8) or LV.SF.eGFP.pre (n = 8), on average, every second assay developed replating clones (reduced incidence of immortalization, P = 0.0058 Fisher's exact test). In comparison, we have plotted results obtained with the GV.SF.eGFP.pre including previously published data indicated in gray,13 and the positive control, SF91.eGFP.pre. Horizontal bars indicate the median of all positive assays for a given vector. eGFP, enhanced green fluorescent protein; LTR, long terminal repeat; neg., negative; PGK, phosphoglycerate kinase; WAS, Wiskott–Aldrich syndrome.
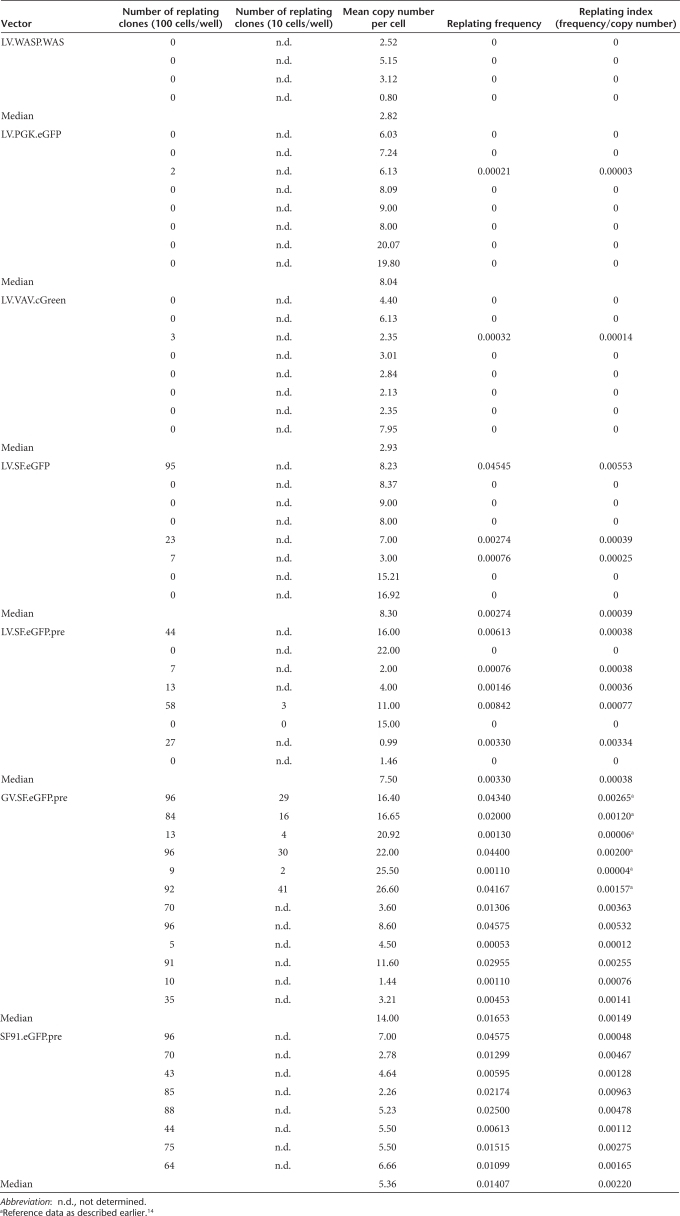
When exposing 100,000 cells per transduction in the IVIM assay, GV vectors with the SFFV enhancer–promoter induced replating in all experiments, and Southern blots revealed a maximum of two molecularly distinct clones per 100,000 transduced cells for SIN-GV.13,14 The average incidence of independent mutants obtained with this vector was thus 1–2 × 10−5. LV with a similar design and the same internal enhancer–promoter (SFFV) reduced the incidence of mutants to 5 × 10−6: replating clones occurred in every second assay, which resulted in a significant (approximately threefold) reduction of immortalizing events as compared to similarly designed SIN-GV (Fisher's exact test P = 0.0058) (Figure 1b). Importantly, a single insertion was sufficient for transformation by SIN-LV according to quantitative PCR detecting vector integrations (Table 1, clone S1).
Table 1.
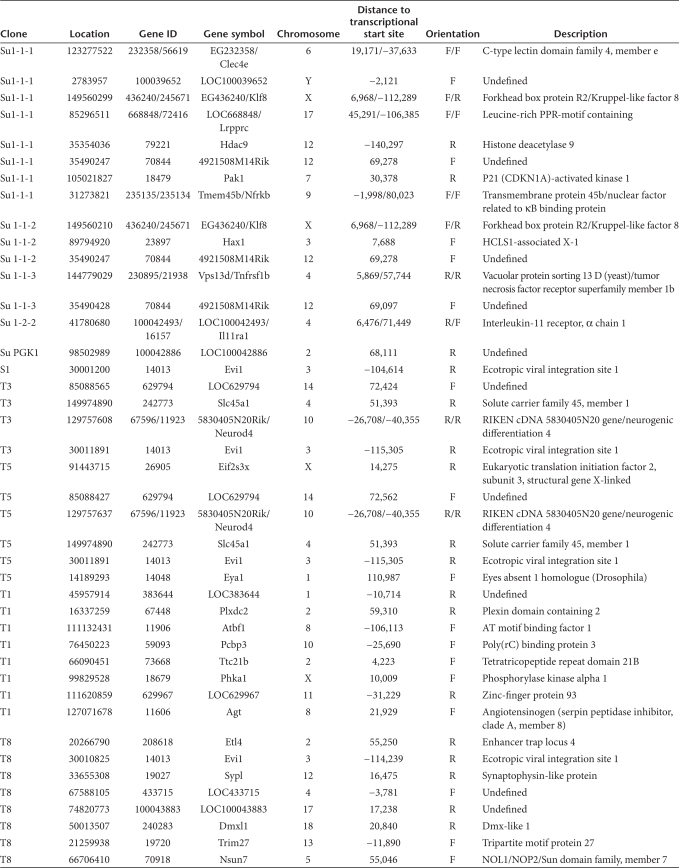
Insertion sites recovered from lentiviral immortalized clones transduced with the vector LV.SF.eGFP.pre (T1, T3, T5, T8, and S1) or LV.SF.eGFP (Su1-1-1, Su1-1-2, Su1-1-3, and Su1-2-2)
Lower fitness of mutants induced by SIN-LV
Next to the incidence of replating mutants, the assay also reports their “fitness” or self-renewal capacity. As mutants expand before the replating step, the frequency of positive wells in limiting dilution analysis reflects the self-renewing capacity of the clone.13,14 Assuming a positive correlation of the number of insertions with the risk of activating mutations, we correct the replating efficiency by the post-transduction copy number to calculate a replating index.13,14 To determine the average replating index of the mutants, we ignored assays with a negative result in the replating step (because no mutants were detectable in these cases). Interestingly, LV-induced mutants tended to have a lower replating index than GV harboring identical internal SFFV enhancer–promoters (approximately fourfold reduction, Figure 1b and Table 2). The post-transcriptional regulatory element derived from the woodchuck hepatitis virus, frequently used to boost titers and transgene expression in LV,18 had no significant effect (compare LV.SF.eGFP and LV.SF.eGFP.pre in Figure 1b and Table 2). Irrespective of the backbone (LV and GV), SIN vectors induced a significantly lower replating index than the LTR-driven GV (GV SIN versus GV LTR P = 0.03, LV SIN versus GV LTR P = 0.011 Wilcoxon two-sample test, Figure 1b).
Table 2.
Replating activity as a function of vector design
CIS in the Evi1 proto-oncogene shared by SIN-LV and SIN-GV
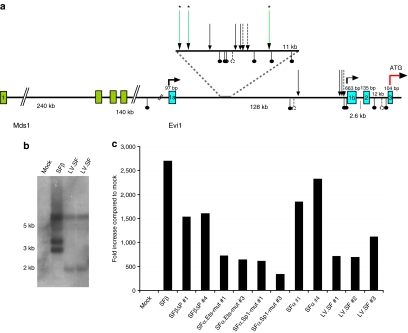
To investigate whether the clones induced by LV shared CIS with the mutants induced by GV, we analyzed integration sites by ligation-mediated PCR. CIS in independent clones indicate selection for transformation.19 In three of the six lentiviral clones examined, insertions occurred in a relatively small region of 11 kilobases (9%) of the first intron of the Evi1 proto-oncogene (Figure 2a). All insertions within this region retrieved from immortal clones were in reverse orientation, irrespective of the vector type used (Figure 2a). To our knowledge, this is the first demonstration of a shared transforming CIS for these only remotely related retroviruses (murine leukemia virus–based SIN-GV and human immunodeficiency virus type 1–based SIN-LV).
Figure 2.
Evi1 is a common insertion site for both gammaretroviral and lentiviral vectors. (a) Insertion sites identified in Evi1: in three of six independent lentiviral clones analyzed, we detected one insertion site in the first intron of Evi1 (long arrows with asterisks). The insertions were found within a region comprising about 11 kilobases in which most of the gammaretroviral vector (GV) insertions were detected so far. Previously reported insertion sites detected in in vitro immortalization clones are indicated as short hatched arrows (GV.SF.eGFP.pre) and short arrows (SF91.eGFP.pre).14 GV insertions recovered in leukemias and dominant clones in vivo in the mouse are indicated below the horizontal line.37 (b) Northern blot analysis showing strong upregulation of Evi1 transcript in all clones analyzed compared to the expression levels in mock-treated cells (left lane). (c) Evi1 expression levels in immortalized clones that were obtained and expanded in this study. All clones show strong upregulation of Evi1 mRNA when compared to the level in mock cells (grown for 2 weeks as mass culture).
Quantitative reverse transcription–PCR and northern blots revealed strongly increased Evi1 expression starting from the second alternative exon 1b, but not from exon 1a. Interestingly, Mds-Evi1 fusion transcripts were downregulated compared to controls in all clones tested. Northern blots showed a common larger band irrespective of the type of vector used, matching the full-length Evi1 transcript starting from exon 1b (BCO76620). Shorter transcripts were also induced whose sizes differed depending on the vector used (Figure 2b). An alternative transcript terminating with exon 8 (AK051098) could be amplified from immortalized clones. Expression of this shorter transcript in murine lineage-negative cells was not transforming (n = 3), in contrast to the established transforming activity of the full-length Evi1 transcript.12 It remains to be determined whether the different splice pattern observed dependent on the type of integrated vector sequences contributed to the observed differences in cell fitness.
Real-time reverse transcription–PCR indicated a >500-fold induction of Evi1 mRNA in clones induced by LV, similar to data obtained after insertional transformation with GV (Figure 2c).13,14 This supports the conclusion that the lower incidence of mutants obtained with LV compared to GV is primarily related to the integration pattern.
SIN-LV equipped with cellular promoters are less genotoxic
In a further series of experiments, we addressed whether the replacement of the SFFV enhancer–promoter by cellular promoters would reduce the transforming risk of LV insertions. We tested SIN-LV containing cellular constitutive promoters derived from the VAV and PGK genes. Replating cells could only be observed with a very low frequency, indicating a low incidence and also a low fitness of potential mutants (Figure 1b). Sustained growth was not observed. Insertion site analysis revealed untransduced cells or, in one clone, an insertion in a region of low gene density on chromosome 2 (Table 1), suggesting background replating activity. Thus, the choice of the promoter was more important to prevent the induction of mutants than the difference between the GV and LV insertion pattern.
Furthermore, we assessed whether experimental conditions could be adapted to test clinical vectors expressing potentially therapeutic genes. The SIN-LV named WASP.WAS is a potentially therapeutic construct expressing the Wiskott–Aldrich syndrome (WAS) protein under control of a fragment of its own promoter. WAS patients suffer from a severe inherited immunodeficiency syndrome. WAS promoter native regulatory sequences are weak compared to other promoters such as phosphoglycerate kinase (PGK) or SFFV,20 but may preserve natural gene regulation. Preclinical murine studies did not reveal potential induction of leukemia in the long-term follow-up.21 In the IVIM assay, up to five copies of this vector per cell did not induce replating, in contrast to the control SIN-LV containing the SFFV enhancer–promoter (Figure 1b).
Vector enhancer but not promoter sequences explains the induction of mutants in the IVIM assay
In cell clones transduced by GV and LV, dysregulation of neighboring cellular genes depended on the nature of the vector's enhancer–promoter.22 The mechanisms underlying the induction of potential transformation remained unknown. Using the IVIM assay, we addressed whether the insertional transformation depended on the enhancer or rather active promoters. To this end, we chose LTR-driven GV that give the highest incidence of transformation in our model,14 and are also more likely than SIN vectors to reveal a potential role of the promoter in proto-oncogene activation. The SFFV enhancer–promoter consists of a highly condensed cluster of transcription factor binding sites (TFBS) surrounding the so-called core element, and an extended basal promoter region with CAAT-TATA boxes (Figure 3a).16 We first generated LTR-GV in which we deleted the entire enhancer, the promoter, or both (constructs ΔE, ΔP, and ΔEP, respectively; Figure 3b). In comparison to the parental construct, ΔP did not reduce the incidence of immortalization but lowered the mutants' fitness (Figure 3c). This suggests that the LTR-contained promoter activity is not required for immortalization. In contrast, ΔE, and accordingly also ΔEP, did not induce replating, revealing the enhancer as the primary determinant of transformation in the IVIM assay. This is consistent with studies performed using replication-competent retrovirus as insertional mutagens to trigger leukemia or lymphoma in vivo,19 and also fits to the molecular analysis of transforming events induced by vectors in preclinical models and clinical trials.6,7,8
Figure 3.
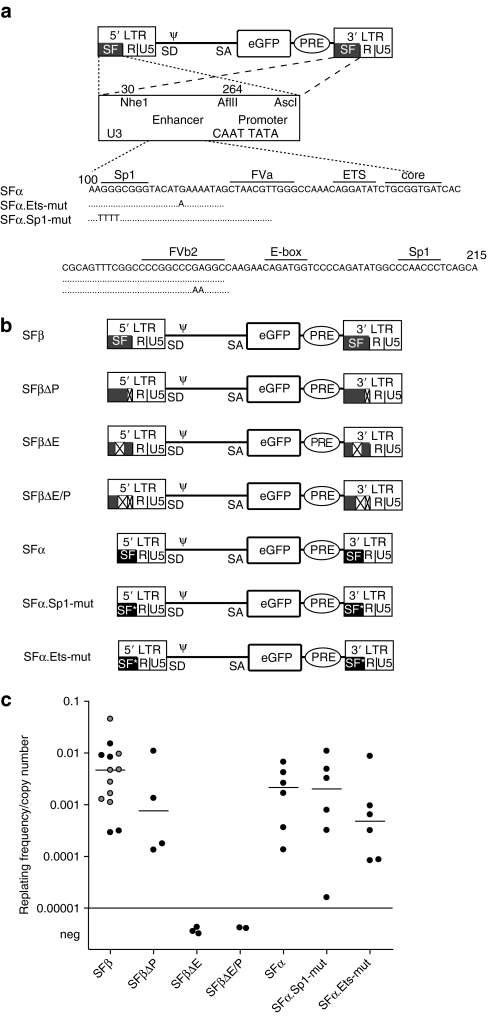
Enhancer sequences are the driving force of replating activity. (a) LTR-driven GV were generated that carried deletions of the complete enhancer (NheI-AflII, 30–264), or promoter (AflII-AscI, 264–413), or the enhancer and promoter. The deletions were introduced in the vector SFβ91.eGFP.pre. After retroviral transduction, the deletions were present in both LTRs. Further vectors had specific mutations in transcription factor binding sites, either in the Ets binding site at position 139, or both Sp1 sites at positions 102 and 220. Those mutations were introduced in the SFα, an LTR with a deletion of the imperfect direct repeat of the enhancer array.23 (b) Vector constructs used in the IVIM assays in the second set of experiments. (c) Replating index derived in the IVIM assays performed with enhancer deletion and enhancer mutation vectors: the deletion of the SFFV enhancer (SFβ91.ΔE) from the viral LTR leaving the promoter intact abrogated the immortalization ability of the SFβ91.eGFP.pre as did the complete deletion of the viral enhancer–promoter (SFβ91.ΔE/P). The mutation of an Ets site reduced the replating frequency/copy number ~10 times (P = 0.03). The data points shown for the SFβ91.eGFP.pre contain those obtained in this experimental set (black) and data points that were obtained in the previous experiments as shown in Figure 1 (gray). eGFP, enhanced green fluorescent protein; LTR, long terminal repeat; neg, negative.
Single-point mutations in the enhancer alter the risk of insertional transformation
To address whether the fitness of the clones depended on individual TFBS within the complex enhancer array, we chose three well-characterized mutants.16 In the above studies, the SFFV enhancer–promoter contains an imperfect direct repeat of the enhancer array (SFβ in Figure 3). In construct SFα, this imperfect direct repeat is deleted, which attenuates the enhancer. In construct mut-Sp1 based on SFα, two Sp1 TFBS at the edges of the ~115 base pair enhancer array were destroyed, further weakening the enhancer by ~40%.23 In construct mut-Ets based on SFα, the TFBS upstream of the core element is mutated to bind only GATA but not ETS transcription factors, which attenuates enhancer activity in myeloid but not erythroid cells.23 Although exchanging SFβ for SFα had no significant effect on the rate and degree of immortalization, the additional destruction of the ETS TFBS (SFα.mut-Ets) led to a significant reduction of the replating index in comparison to SFβ (Wilcoxon two-sample test P = 0.04) (Figure 3c). In contrast to the mutation of the ETS TFBS, destroying both TFBS for SP1 (SFα.mut-Sp1) had no clear effect in the IVIM assay. This may suggest that SP1 binding to the retroviral enhancer is not necessary for long-distance enhancer interactions with neighboring cellular promoter. None of these three enhancer mutations (SFα, SFα.mut-Ets, or SFα.mut-Sp1) was sufficient to suppress the induction of replating clones.
A closer look at Figures 1b and 3c reveals that the replating index of cells transduced by identical vectors may vary in the IVIM assay. This variance is potentially caused by well-documented intrinsic differences in the fitness of primary hematopoietic cells24 and the variable nature of the integration sites, thus precluding a better resolution of the impact of individual enhancer mutations. Nonetheless, the results obtained with the enhancer mutants indicate that gradual differences of enhancer activity determine the fitness of the transformed clones (Figure 3c).
Discussion
The present study, summarizing results of 90 IVIM experiments conducted with a panel of 13 different vectors, supports several important conclusions. First, the enhancer sequences contained in semi-randomly integrating LV and GV are the major cause of transformation (Figure 3), which is consistent with observations from animal models and clinical studies.6,7,8,25,26,27 Second, despite their lack of preference for promoter-proximal regions or DNAse1 hypersensitive sites, SIN-LV may still hit genomic regions from which enhancer-mediated proto-oncogene upregulation may occur (Figure 2), and accordingly, SIN-LV may trigger cell transformation of hematopoietic cells depending on the type of internal enhancer–promoter used (Figure 1). Third, if vectors contain strong enhancers that cannot be effectively shielded by genetic insulator elements, the lentiviral integration pattern is safer than the gammaretroviral (Figure 1). Finally, our results also suggest that the woodchuck hepatitis virus, a post-transcriptional enhancer of vector titer and transgene expression,18 has no major role to play in the potential transformation of hematopoietic cells.
In a tumor-prone mouse model, the insertion pattern of LV was calculated to be approximately tenfold safer than that of GV.10 As our experimental conditions reflect two independent oncogenic parameters (incidence and fitness of the mutants), our study indicates a similar magnitude (approximately threefold reduced incidence, approximately fourfold reduced fitness). At first glance, the threefold reduced incidence detected in the IVIM assay is compatible with global integrome studies of LV and GV, which indicated an approximately twofold higher risk of GV to hit promoter-proximal regions. However, the genomic organization of the Evi1 gene is quite complex, as indicated in Figure 2, with transcription potentially initiated from three different positions: exon 1 of the upstream Mds gene, and exons 1a or 1b of Evi1. In the mutants obtained in the IVIM assay, GV and LV insertions cluster in a region of only 11 kilobases that is far upstream of exon 1b. This region would be intronic if transcription is initiated in exon 1a or starts from Mds1. The actual risk of transformation in such loci can hardly be predicted based on global analyses of integration pattern, and requires functional models such as provided in the IVIM assay.
In the tumor-prone mouse model, only LV with active LTRs lead to insertional transformation of hematopoietic cells, whereas SIN-LV, even when containing the strong internal SFFV enhancer–promoter, did not trigger transformation above the background levels of this model.10 This indicates a relatively high sensitivity of the IVIM assay to detect insertional transformation events. The tumor-prone model selected for proto-oncogene activation by enhancer interactions or a splice mechanism, which implies a leaky termination at the 3′ LTR.10 It is thus possible that differences in the regulation of splicing and transcriptional termination contribute to the differential insertional risk profile of the tested vectors. Of note, human immunodeficiency virus type 1 and gammaretroviruses show substantial differences in these post-transcriptional events.28 Another recent study showed that LV equipped with active LTRs can transform an established interleukin-3 dependent B cell line to growth-factor independence, and again, gene activation involved a splice event between the lentiviral major splice donor and cellular exons.11 SIN vectors cannot create such fusion transcripts involving the major splice donor due to its location upstream of the internal promoter, and approaches to improve transcriptional termination have been described.29
Others have recently used a modification of the IVIM assay to suggest that the addition of insulator elements in SIN-LVs can reduce their genotoxic risk.30 Although they reported that SIN-LV equipped with an internal retroviral enhancer–promoter can transform murine hematopoietic cells, the exact integration events and the incidence of mutants were not determined. In line with our data obtained with SIN-GV,13 this report supports the rationale to equip LV with insulator sequences to reduce their genotoxic potential.30
To our knowledge, the present study is the first direct comparison of the genotoxic risk induced by SIN-GV and SIN-LV in an experimental model that was able to detect insertional transformation induced by those vectors. Modifications of the IVIM assay may allow to broaden the spectrum of oncogenic events that can be monitored with similar high sensitivity, and to test the impact of different vectors in a human cell background and/or other cell lineages. The synopsis of the currently available data already provides compelling evidence that for new clinical trials, appropriate vector design will substantially lower the frequency of insertional adverse events.
Materials and Methods
Vector design. Vectors LV.SF.eGFP.pre (RRL.PPT.eGFP.pre), GV.SF.eGFP.pre (SRS.SF.eGFP.pre), RSF91.eGFP.pre, LV.VAV.cGreen (RRL.PPT.VAV.cGreen), and LV.WASP.WAS (RRL.PPT.WASP.WAS.pre) have been described.20,31,32,33 The vector pCL1.PPT.SFFVeGFP (LV.SF.eGFP) was kindly provided by Helmut Hanenberg (Department of Pediatric Oncology, Hematology and Immunology, Children's Hospital, Heinrich Heine University, Düsseldorf, Germany).34 pRRLsin18.ppt.hPGKeGFP (LV.PGK.eGFP), originally conceded by Luigi Naldini (TIGET, Milano, Italy), encodes the eGFP under the control of the human PGK promoter and a PRE. The latter was deleted for the present study. LTR-driven GV with deletions of the enhancer, promoter, or enhancer–promoter were constructed as indicated in Figure 3.
Production of vector supernatants. Gammaretroviral and lentiviral supernatant production was performed using 293T cells as previously described, using ecotropic or VSVg envelopes.31,32,33,34,35 Cells were maintained in Dulbecco's modified Eagle's Medium supplemented with 10% fetal calf serum, 100 U/ml penicillin/streptomycin, and 2 mmol/l glutamine. Some of the VSVg-pseudotyped supernatants were concentrated by ultracentrifugation. Viral titers determined on HT1080 and SC-1 cells by flow cytometry were in the range of 106 to 107 IU/ml in unconcentrated supernatants and in the range of 108 to 109 in the concentrated ones.
Northern blot. Standard procedures were used for northern blots.31 Specific probes were directed against Evi1 exons 4–8.
Isolation of lineage-negative bone marrow cells and retroviral transduction. Lineage-negative (Lin−) bone marrow cells of untreated C57BL6/J mice (Charles River Laboratories, Wilmington, MA) were transduced as previously described.36 Briefly, Lin− cells were isolated from complete bone marrow by magnetic sorting using lineage-specific antibodies (Lineage Cell depletion kit; Miltenyi, Bergisch Gladbach, Germany) and cryopreserved in aliquots. Before retroviral transduction, Lin− cells were prestimulated for 2 days at a density of 1–5 × 105 cells/ml in StemSpan medium (Stem Cell Technologies, Vancouver, British Columbia, Canada) containing 50 ng/ml mSCF, 100 ng/ml hFlt-3 ligand, 100 ng/ml hIL-11, 10 ng/ml mIL-3 (PeproTech, Heidelberg, Germany), 1% penicillin/streptomycin, and 2 mmol/l glutamine. Cells were transduced on two following days (days 3 and 4, Figure 3) or on four following days (days 3–6, Figure 1) using 105 cells per initial transduction. Virus preloading was carried out on RetroNectin-coated (10 µg/cm2; Takara, Otsu, Japan) suspension culture dishes by spinoculation for 30 minutes at 4 °C. The culture volume was increased from 500 µl medium for 1 × 105 cells by 250 µl on following days, reaching a total volume of 1.25 ml on day 6. DNA samples for real-time PCR analysis (copy number) and flow cytometry (FACSCalibur; Becton Dickinson, Heidelberg, Germany) were taken 4 days after the last transduction.
IVIM assay. After retroviral transduction, cells were expanded as mass cultures for 2 weeks in Iscove's modified Dulbecco's medium containing 50 ng/ml mSCF, 100 ng/ml hFlt-3 ligand, 100 ng/ml hIL-11, 10 ng/ml mIL-3, 10% fetal calf serum, 1% penicillin/streptomycin, and 2 mmol/l glutamine. During this time, density was adjusted to 5 × 105 cells/ml every 3 days. After mass culture expansion for 14 days, cells were plated into 96-well plates at a density of 100 cells/well or 10 cells/well.14 Two weeks later, the positive wells were counted, and the frequency of replating cells was calculated based on Poisson statistics using L-Calc software (Stem Cell Technologies). Selected clones were expanded for further characterization.
TaqMan real-time PCR analysis. Quantitative PCR was performed on an Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA) using the QuantiTect SYBR Green Kit (Qiagen, Hilden, Germany) as previously described.14 Quantitative PCR detected eGFP, cGreen, or woodchuck hepatitis virus.33 To measure vector copy numbers, the vector-specific signal was normalized to the signal of the housekeeping gene Flk1. Results were quantified using the efficiency-corrected comparative CT method. To evaluate mRNA expression, RNA was extracted from expanded clones using RNAzol (Wako Chemicals, Steinbach, Germany) and the RNeasy micro kit (Qiagen). Reverse transcription was performed with 0.5–2 µg RNA using PowerScript MLV reverse transcriptase (Becton Dickinson), and real-time PCR for Evi1 expression as described.17
Statistical analysis. To determine whether the replating frequencies/vector copy number was significantly different, the values were log-transformed, and a Wilcoxon two-sample test was performed. Plotted are the untransformed data. Data from experiments are expressed as mean ± SEM. P < 0.05 was considered significant. Incidence of positive assays for each vector was calculated by Fisher's exact test or Wilcoxon two-sample test.
SUPPLEMENTARY MATERIALFigure S1. Primary lineage negative bone marrow (BM) cells harvested from untreated C57BL6 mice are expanded in cytokine-supplemented medium and transduced at defined numbers (100,000 cells per transduction) with γ retroviral or lentiviral vector supernatants.
Supplementary Material
Primary lineage negative bone marrow (BM) cells harvested from untreated C57BL6 mice are expanded in cytokine-supplemented medium and transduced at defined numbers (100,000 cells per transduction) with γ retroviral or lentiviral vector supernatants.
Acknowledgments
This work was supported by grants of the German Ministry for Research and Education (projects TreatID and iGene), the Deutsche Forschungsgemeinschaft (SPP1230 Ba1837 and excellence cluster REBIRTH), the European Union (FP6 CONSERT LSHB-CT-2004-5242 and FP7), the National Cancer Institute (5R01CA107492-02), the Food and Drug Administration of the United States, Ministerio de Ciencia e Innovación (SAF2005-00058), Instituto de Salud Carlos III (CIBER-ER), Genoma España (FANCOGENE), Fundación Marcelino Botín, and the French Muscular Dystrophy Association.
REFERENCES
- Kay MA, Glorioso JC., and , Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- Naldini L, Blömer U, Gage FH, Trono D., and , Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and , Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove PW, Kepes S, Hanawa H, Obenauer JC, Pei D, Cheng C, et al. Globin lentiviral vector insertions can perturb the expression of endogenous genes in beta-thalassemic hematopoietic cells. Mol Ther. 2008;16:525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhoven M, Stephen SL, Knight S, Gevers EF, Robinson IC, Takeuchi Y, et al. Insertional gene activation by lentiviral and gammaretroviral vectors. J Virol. 2009;83:283–294. doi: 10.1128/JVI.01865-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Jenkins NA., and , Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knöss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora G, Laricchia-Robbio L., and , Senyuk V. EVI1 and hematopoietic disorders: history and perspectives. Gene. 2006;368:1–11. doi: 10.1016/j.gene.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Baum C, Itoh K, Meyer J, Laker C, Ito Y., and , Ostertag W. The potent enhancer activity of the polycythemic strain of spleen focus-forming virus in hematopoietic cells is governed by a binding site for Sp1 in the upstream control region and by a unique enhancer core motif, creating an exclusive target for PEBP/CBF. J Virol. 1997;71:6323–6331. doi: 10.1128/jvi.71.9.6323-6331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Schambach A, Brugman MH, Wicke DC, Knoess S, Li Z, et al. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia. 2008;22:1519–1528. doi: 10.1038/leu.2008.118. [DOI] [PubMed] [Google Scholar]
- Zufferey R, Donello JE, Trono D., and , Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Kool J, Berns A., and , van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- Charrier S, Dupré L, Scaramuzza S, Jeanson-Leh L, Blundell MP, Danos O, et al. Lentiviral vectors targeting WASp expression to hematopoietic cells, efficiently transduce and correct cells from WAS patients. Gene Ther. 2007;14:415–428. doi: 10.1038/sj.gt.3302863. [DOI] [PubMed] [Google Scholar]
- Marangoni F, Bosticardo M, Charrier S, Draghici E, Locci M, Scaramuzza S, et al. Evidence for long-term efficacy and safety of gene therapy for Wiskott-Aldrich syndrome in preclinical models. Mol Ther. 2009;17:1073–1082. doi: 10.1038/mt.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruggi G, Porcellini S, Facchini G, Perna SK, Cattoglio C, Sartori D, et al. Transcriptional enhancers induce insertional gene deregulation independently from the vector type and design. Mol Ther. 2009;17:851–856. doi: 10.1038/mt.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlers A, Zipfel PF, Schwieger M, Ostertag W., and , Baum C. In vivo analysis of retroviral enhancer mutations in hematopoietic cells: SP1/EGR1 and ETS/GATA motifs contribute to long terminal repeat specificity. J Virol. 2002;76:303–312. doi: 10.1128/JVI.76.1.303-312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurier F, Gan OI, McKenzie JL, Doedens M., and , Dick JE. Lentivector-mediated clonal tracking reveals intrinsic heterogeneity in the human hematopoietic stem cell compartment and culture-induced stem cell impairment. Blood. 2004;103:545–552. doi: 10.1182/blood-2003-05-1558. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Düllmann J, Kamino K, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger A, Monks J., and , Proudfoot NJ. The retroviruses human immunodeficiency virus type 1 and Moloney murine leukemia virus adopt radically different strategies to regulate promoter-proximal polyadenylation. J Virol. 2001;75:11735–11746. doi: 10.1128/JVI.75.23.11735-11746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach A, Galla M, Maetzig T, Loew R., and , Baum C. Improving transcriptional termination of self-inactivating gamma-retroviral and lentiviral vectors. Mol Ther. 2007;15:1167–1173. doi: 10.1038/sj.mt.6300152. [DOI] [PubMed] [Google Scholar]
- Ramezani A, Hawley TS., and , Hawley RG. Combinatorial incorporation of enhancer-blocking components of the chicken beta-globin 5'HS4 and human T-cell receptor alpha/delta BEAD-1 insulators in self-inactivating retroviral vectors reduces their genotoxic potential. Stem Cells. 2008;26:3257–3266. doi: 10.1634/stemcells.2008-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach A, Bohne J, Chandra S, Will E, Margison GP, Williams DA, et al. Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells. Mol Ther. 2006;13:391–400. doi: 10.1016/j.ymthe.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Schambach A, Mueller D, Galla M, Verstegen MM, Wagemaker G, Loew R, et al. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 2006;13:1524–1533. doi: 10.1038/sj.gt.3302807. [DOI] [PubMed] [Google Scholar]
- Almarza E, Río P, Meza NW, Aldea M, Agirre X, Guenechea G, et al. Characteristics of lentiviral vectors harboring the proximal promoter of the vav proto-oncogene: a weak and efficient promoter for gene therapy. Mol Ther. 2007;15:1487–1494. doi: 10.1038/sj.mt.6300213. [DOI] [PubMed] [Google Scholar]
- Leurs C, Jansen M, Pollok KE, Heinkelein M, Schmidt M, Wissler M, et al. Comparison of three retroviral vector systems for transduction of nonobese diabetic/severe combined immunodeficiency mice repopulating human CD34+ cord blood cells. Hum Gene Ther. 2003;14:509–519. doi: 10.1089/104303403764539305. [DOI] [PubMed] [Google Scholar]
- Schambach A, Galla M, Modlich U, Will E, Chandra S, Reeves L, et al. Lentiviral vectors pseudotyped with murine ecotropic envelope: increased biosafety and convenience in preclinical research. Exp Hematol. 2006;34:588–592. doi: 10.1016/j.exphem.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Li Z, Schwieger M, Lange C, Kraunus J, Sun H, van den Akker E, et al. Predictable and efficient retroviral gene transfer into murine bone marrow repopulating cells using a defined vector dose. Exp Hematol. 2003;31:1206–1214. doi: 10.1016/j.exphem.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Geiger H, Li Z, Brugman MH, Chambers SM, Shaw CA, et al. Retroviral vector insertion sites associated with dominant hematopoietic clones mark “stemness” pathways. Blood. 2007;109:1897–1907. doi: 10.1182/blood-2006-08-044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primary lineage negative bone marrow (BM) cells harvested from untreated C57BL6 mice are expanded in cytokine-supplemented medium and transduced at defined numbers (100,000 cells per transduction) with γ retroviral or lentiviral vector supernatants.