Abstract
Two proteins are encoded by the mammalian retrotransposon long interspersed nuclear element 1 (LINE-1 or L1); both are essential for retrotransposition. The function of the protein encoded by the 5′-most ORF, ORF1p, is incompletely understood, although the ORF1p from mouse L1 is known to bind single-stranded nucleic acids and function as a nucleic acid chaperone. ORF1p self-associates by means of a long coiled-coil domain in the N-terminal region of the protein, and the basic, C-terminal region (C-1/3 domain) contains the nucleic acid binding activity. The full-length and C-1/3 domains of ORF1p were purified to near homogeneity then analyzed by gel filtration chromatography and analytical ultracentrifugation. Both proteins were structurally homogeneous and asymmetric in solution, with the full-length version forming a stable trimer and the C-1/3 domain remaining a monomer. Examination of the full-length protein by atomic force microscopy revealed an asymmetric dumbbell shape, congruent with the chromatography and ultracentrifugation results. These structural features are compatible with the nucleic acid binding and chaperone activities of L1 ORF1p and offer further insight into the functions of this unique protein during LINE-1 retrotransposition.
Long interspersed nuclear element 1, known as LINE-1 or L1, is a highly successful retrotransposon of mammalian genomes, comprising 17% of the human genome and 19% of the mouse genome, and likely generating an additional 8–13% in the form of short interspersed nuclear elements (SINEs) and processed pseudogenes (1). L1 belongs to a larger group of mobile elements known as the non-LTR retrotransposons, which are widely distributed throughout eukaryotes (2).
Non-LTR retrotransposons replicate by reverse transcription of an RNA intermediate, employing a unique mechanism known as target-site primed reverse transcription, or TPRT. In mammalian L1s, two element-encoded proteins are required for retrotransposition: ORF2p, which provides the crucial enzymatic activities for TPRT, endonuclease, and reverse transcriptase (reviewed in ref. 3); and ORF1p, which is a single-stranded nucleic acid binding protein (4, 5) that also acts as a nucleic acid chaperone (6).
The calculated mass of the protein encoded by ORF1 in the retrotransposition-competent element, L1spa (7), is 42,921 Da, and a 43-kDa ORF1p is detected in extracts prepared from a subset of mouse cells by Western blotting. Immunocytochemistry with the same anti-ORF1p Ab reveals a punctate cytoplasmic staining pattern in the two embryonal carcinoma cell types, F9 and JC44, where ORF1p is detected by Western blotting (8), as well as in prepachytene germ cells of the testes (9) and ovary (10). p43 is coenriched with full-length L1 RNA in ribonucleoprotein particles that can be fractionated from F9 cells (11, 12). Taken together, these observations suggest that L1 RNA is bound with many molecules of ORF1p during the cytoplasmic phase of the L1 replication cycle, perhaps playing a structural role in protecting the RNA and organizing the replication intermediates. A second role for ORF1p likely occurs during the TPRT reaction, where the nucleic acid chaperone activity of this protein may facilitate the strand transfers that are required to prime reverse transcription as well as the reverse transcriptase reaction itself (6).
To fully understand the role(s) of ORF1p in L1 retrotransposition, detailed information about the structure of this unusual protein is required. Although mouse ORF1p is homologous to other mammalian ORF1ps, as well as the ORF1ps from LINE-like elements in teleost fish (3), there are no structures available for any of these proteins. Furthermore, the L1 ORF1ps lack conserved sequence motifs, except for a long coiled-coil domain in the N-terminal half of the protein. In the case of mouse ORF1p, the coiled-coil region is both necessary and sufficient for protein–protein interaction by either two-hybrid or GST pull-down assay (13). The only other structural feature known for the mouse ORF1p is that the C-terminal, basic domain of the protein is responsible for nucleic acid binding and annealing activity (13).
An increased understanding of ORF1p structure will provide significant insights into the role of this unique protein in L1 retrotransposition. Here, we use biochemical and biophysical approaches to demonstrate that ORF1p exists as a stable trimer in solution, thus forming a distinctive structure for a known nucleic acid chaperone and single-strand nucleic acid binding protein. Parallel studies of the isolated C-1/3 domain demonstrate that this region is quantitatively monomeric in solution, supporting previous results that the coiled-coil domain mediates ORF1p self-association. Hydrodynamic analysis of the ORF1p trimer suggests that it forms a highly anisotropic, elongated conformation in solution. Indeed, direct visualization of ORF1p by atomic force microscopy clearly reveals an elongated dumbbell-like structure. Taken together, these results suggest a structural basis for the nucleic acid chaperone activity of ORF1p during L1 retrotransposition.
Materials and Methods
Protein Purification. The His-tagged ORF1p from L1spa was purified from baculovirus-infected SF9 cells by using nickel-agarose chromatography as described (6) and briefly summarized below. After cell lysis and centrifugation, the supernatant containing ORF1p was incubated with Ni-NTA Agarose (Qiagen, Valencia, CA). The resin was washed extensively, and then ORF1p was eluted with 250 mM imidazole. ORF1p was further purified by ammonium sulfate precipitation and fractionation on a Sephacryl 300 column (Amersham Biosciences). ORF1p concentration was determined by using an extinction coefficient of 16,050 M–1·cm–1.
The C-1/3 nucleic acid binding domain of mouse L1 ORF1p was expressed in Escherichia coli as an N-terminal His-tagged version to facilitate purification. Briefly, a 1-liter culture was induced with 1 mM IPTG at OD600 = 0.75. After3hof induction at 30°C, the cells were pelleted, resuspended, and sonicated, and the lysate was centrifuged. The supernatant containing C-1/3 ORF1p was recovered and incubated with TALON CellThru Resin (Clontech). After extensive washing, C-1/3 was eluted from the resin, precipitated with 85% saturating ammonium sulfate, and chromatographed on Sephacryl 200 (Amersham Biosciences). Finally, peak fractions containing C-1/3 ORF1p were pooled, chromatographed on a High-Trap-SP-FF column (Amersham Biosciences), and eluted with a NaCl gradient. The concentration of C-1/3 ORF1p was determined by using an extinction coefficient of 10,810 M–1·cm–1.
Purity of both proteins was assessed to be ≥95% by SDS/PAGE with Coomassie blue staining. Previous work has demonstrated that the purified full-length and C-1/3 proteins maintain nucleic acid binding and chaperone activities (4, 6, 13).
Chemical Cross-Linking. L1 ORF1p at concentrations of 129.5, 12.95, and 1.295 nM was incubated with 80, 40, and 20 μM Sulfo-EGS {ethylene glycol bis[sulfosuccinimidyl-succinate] (Pierce)}, respectively. After 16 min at room temperature, the reactions were quenched by the addition of glycine to 50 mM. Samples were denatured and analyzed by standard SDS/PAGE with 10% polyacrylamide gels. Proteins were transferred to nitrocellulose and the various forms of ORF1p were detected by staining or by Western blotting with an anti-ORF1 Ab (11).
Analytical Ultracentrifugation. Sedimentation velocity and sedimentation equilibrium analyses were carried out by using a Beckman XL-A centrifuge equipped with an AN-60 Ti rotor. Centrifugation conditions were 25 mM Hepes (pH 7.5), 250 mM NaCl, and 0.1 mM EDTA at 4°C. Protein concentrations ranged from 16.2 to 1.6 μM for ORF1p and from 41.2 to 4.1 μM for C-1/3. Velocity data were analyzed by using the method of Van Holde and Weischet as implemented in ultrascan (14, 15). Equilibrium data were analyzed by nonlinear least-squares parameter estimation with the program nonlin (16, 17). The degree of protein hydration was assumed to be 0.28 g of water/g of protein (18).
Atomic Force Microscopy. ORF1p was diluted to 10–500 nM in 25 mM Tris/350 mM KCl (pH 7.6), deposited (5 μl) onto freshly cleaved mica, and adsorbed to the surface for 5 min in a closed chamber at 100% humidity. For imaging in air, the samples were then rinsed with 1 ml of 5% ethanol in distilled water, dried in a stream of N2, and imaged in tapping mode by using oxidatively sharpened silicon cantilevers (nominal force constant 35 N/m, resonance frequency ≈300 kHz) with a Nanoscope IIIa (Digital Instruments, Santa Barbara, CA). For imaging in buffer, the samples were placed immediately in the fluid cell of the atomic force microscope and imaged by using silicon nitride cantilevers (nominal force constant 0.6 N/m, resonance frequency ≈40 kHz) in tapping mode. Images were processed by “flatten” filter, and scan streaks were removed. Reported sizes are averages of at least 60 molecules.
Results
As a step in the final purification of ORF1p, the protein was chromatographed through Sephacryl 300. Because the predicted molecular mass of this protein including the His-tag is ≈47 kDa, it was surprising to find that the majority of highly purified ORF1p eluted near ferritin, with an apparent molecular mass of 430 kDa. A small percentage of ORF1p was in the void volume, but none was detected in the region expected for a 47-kDa protein (data not shown).
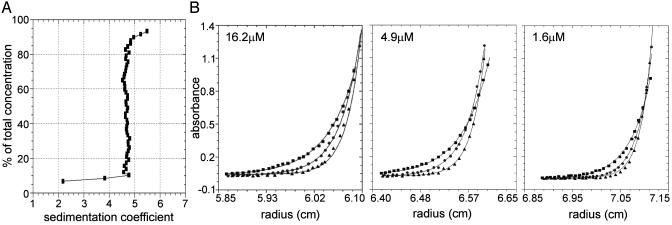
To address whether ORF1p recovered from the size-exclusion column represented a polydisperse aggregate or a structurally homogeneous ensemble, the highly purified protein was analyzed by analytical ultracentrifugation. Sedimentation velocity analysis indicated that ORF1p is essentially homogeneous with respect to molecular size and shape, with no evidence for dynamic self-association (Fig. 1A). Specifically, the vast majority of the population has a sedimentation coefficient of ≈4.7 S. The material sedimenting between 2.2 and 4.7 S likely reflects trace contaminants and/or breakdown products, whereas the material sedimenting between 4.7 and 5.5 S may reflect a small amount of higher-order complex.
Fig. 1.
Analysis of L1 ORF1p by analytical ultracentrifugation. (A) Van Holde–Weischet integral distribution plot (boundary fraction versus S20,w) derived from sedimentation velocity analysis. The S value at 50% of the boundary corresponds to the sedimentation coefficient, S20,w. The initial loading concentration of ORF1p was 16.2 μM. Data were collected at 280 nm and at a rotor speed of 50,000 rpm. (B) Sedimentation equilibrium analysis of ORF-1, plotted as absorbance versus radius. Shown are data from the three initial loading concentrations: 16.2 μM (scanned at 280 nm), 4.9 μM (scanned at 237 nm), and 1.6 μM (scanned at 230 nm). Symbols represent ORF-1 absorbance at each rotor speed (squares, 14,000 rpm; circles, 17,000 rpm; triangles, 20,000 rpm). Solid lines represent the best-fit model (trimer) from simultaneous analysis of all nine data sets. Square root of the variance for the analysis was 0.011 OD units. For clarity, only every fifth data point is shown.
A more rigorous examination of the ORF1p assembly state was carried out by using sedimentation equilibrium. Global analysis of three ORF1p concentrations at three rotor speeds resolved an apparent molecular mass of 131,668 ± 4,788 Da (Fig. 1B). Assuming a calculated monomer molecular mass of 46,968 Da, the ratio of apparent M/Mmonomer is 2.80 ± 0.10, suggesting that ORF-1 is purely trimeric under these conditions. The possibility that a ratio of 2.80 reflects a self-associating system (e.g., monomer–trimer or monomer–dimer–trimer) was not supported by the data; analyses by these models either failed to converge or gave physically meaningless results. In addition, analyses of the individual datasets failed to show a concentration-dependent increase in molecular mass. Finally, a dynamic equilibrium would generate curvature in the sedimentation velocity integral distribution plot. The actual plot was largely linear, with the exception of low molecular mass material (Fig. 1 A). Hence, the slight decrease in ratio from 3.0 is likely due to these small contaminants or breakdown products.
The experimentally determined molecular mass and sedimentation coefficient were used to calculate the frictional coefficient of ORF1p (Table 1). Comparison of this value to that of a hydrated sphere of the same molecular mass resolved a frictional coefficient of 1.69, indicative of extreme asymmetry. When the trimeric ORF1p was modeled as a prolate ellipsoid, the major to minor axis ratio was calculated to be ≈13:1, with the major axis length ≈36 nm and a Stokes radius of 6.4 nm. If ORF1p is modeled as a rod, the major to minor axis ratio was calculated to be 14:1 with a major axis length of 34 nm. This structural asymmetry likely explains the anomalous behavior of ORF1p on the size-exclusion column.
Table 1. Results of sedimentation velocity analyses.
| ORF1p | C-1/3 domain | |
|---|---|---|
| S20,w, sec | 4.73 × 10-13 | 1.43 × 10-13 |
| f, g/sec | 7.20 × 10-8 | 5.40 × 10-8 |
| Rs, nm | 6.4 | 2.9 |
| F (f/fo) | 1.69 | 1.46 |
S20,w is the experimentally determined sedimentation coefficient corrected for 20°C in water; f is the frictional coefficient calculated from the experimentally determined molecular weight and sedimentation coefficient; Rs is the Stokes radius; and F (f/fo) is the ratio between the experimentally derived f and the calculated value of an ORF1p hydrated sphere.
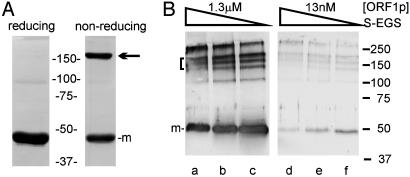
Consistent with the single species of ORF1p detected by analytical ultracentrifugation, only a single band was seen when the protein was examined by acrylamide gel electrophoresis under native conditions (data not shown). A single band was also detected when the protein was subjected to standard SDS/PAGE with reducing agent, yet a significant proportion of ORF1p remained in a slowly migrating form when the same protein preparation was separated under nonreducing conditions (Fig. 2A). The mobility of this form relative to the protein standards indicates that the highly stable ORF1p trimer remains intact in the presence of SDS and the absence of reducing agent. This finding further suggests that an intermolecular disulfide bond stabilizes the trimer.
Fig. 2.
SDS/PAGE analysis of ORF1p structure. (A) Coomassie-stained SDS-polyacrylamide (10%) gel to compare the behavior of the protein in standard reducing versus nonreducing conditions: 5.3 μg of ORF1p from the Sephacryl 300 column was boiled for 3 min in DTT-containing SDS sample buffer (reducing), and 3.4 μg was loaded in the same buffer without DTT and without heating (nonreducing). The position of the monomer is marked (m). The mobility of the more slowly migrating form (arrow) is consistent with a trimer. (B) SDS/PAGE analysis of ORF1p after cross-linking with Sulfo-EGS. Reactions contained ORF1p at 1.3 μM (lanes a–c) or 13 nM (lanes d–f) and 160 μM (lanes a and d), 80 μM (lanes b and e), or 40 μM (lanes c and f) bifunctional cross-linking reagent. Protein was transferred to nitrocellulose and detected by Western blot with anti-His (lanes a–c) or anti-ORF1p (lanes d–f) Ab and alkaline-phosphatase-conjugated secondary Ab as described (8). The position of the monomer is marked (m). Several additional bands are seen in B compared with A, ranging from dimers to higher-order aggregates. The forms in the bracket likely include trimers cross-linked via different amino acids; other studies have attributed different mobilities in SDS/PAGE to cross-linking at different residues (31). The relative mobility observed for each of the forms, except monomer, is extremely sensitive to the degree of cross-linking in the acrylamide gel (e.g., bisacrylamide:acrylamide ratio), indicative of shape effects.
The results of analytical ultracentrifugation are consistent with the behavior of ORF1p in size-exclusion chromatography, yet both of these methods required high protein concentrations (μM) relative to the concentrations of ORF1p present in RNA binding or nucleic acid chaperone assays, where activity occurs in the low nanomolar range (4, 6). Therefore, chemical crosslinking was used to probe the structure of ORF1p in increasingly dilute conditions. The forms of ORF1p detected after treatment of the protein with Sulfo-EGS and resolution of the products by SDS/PAGE were indistinguishable over a 10,000-fold dilution range, from 13 μM to 1.3 nM. The results obtained when ORF1p was incubated with crosslinking reagent at 1.3 μM and 13 nM are shown in Fig. 2B.
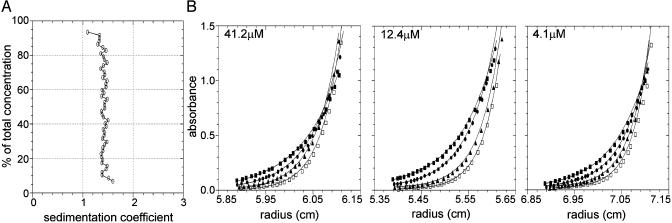
To further explore the basis of ORF1p assembly, we also examined the properties of its isolated C-1/3 nucleic acid binding domain by analytical ultracentrifugation. Sedimentation velocity analysis indicated that the C-1/3 domain of ORF1p was a homogeneous population with respect to molecular size and shape, with no evidence for self-association or heterogeneity. The resolved sedimentation coefficient was 1.43 S and the resolved frictional coefficient was 1.46 (Fig. 3A and Table 1), suggesting that this domain deviates from the model of a hydrated, rigid, compact sphere.
Fig. 3.
Analysis of the C-1/3 domain of L1 ORF1p by analytical ultracentrifugation. (A) Van Holde–Weischet integral distribution plot (boundary fraction versus S20,w) derived from sedimentation-velocity analysis. The initial loading concentration was 41.2 μM. (B) Sedimentation equilibrium analysis of C-1/3, plotted as absorbance versus radius. Shown are initial loading concentrations of 41.2 μM (scanned at 285 nm), 12.4 μM (scanned at 236 nm), and 4.1 μM (scanned at 230 nm) as indicated. Symbols represent C-1/3 absorbance at each rotor speed (squares, 30,500 rpm; circles, 35,000 rpm; triangles, 40,000 rpm; open squares, 45,000 rpm). Solid lines represent the best-fit model (monomer) from simultaneous analysis of all 12 data sets. Square root of the variance for the analysis was 0.011 OD units. For clarity, only every fifth data point is shown.
Global analysis of sedimentation equilibrium data collected at four rotor speeds and three protein concentrations of the C-1/3 domain resolved an apparent molecular mass of 16,413 ± 425 Da (Fig. 3B). Because the predicted molecular mass of this protein is 17,327 Da, the C-1/3 domain is apparently purely monomeric in solution, consistent with the velocity results demonstrating a homogeneous population. Analysis of any subset of the data did not change this result, nor was there any evidence for concentration dependence in the apparent molecular mass.
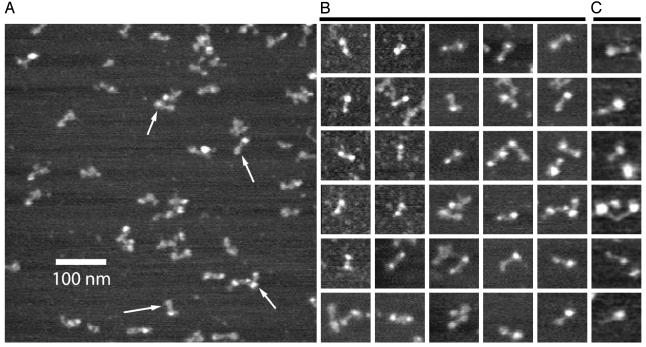
Hydrodynamic asymmetry can arise for a variety of reasons other than literal deviations from a spherical shape; thus, full-length ORF1p was directly visualized by atomic force microscopy, both in air and under buffer solution. The images revealed an asymmetric “dumbbell” shape (Fig. 4) with an average length of 32 nm (Fig. 4D), in good agreement with the sedimentation velocity estimates of 34 nm for ORF1p when modeled as a rod. Essentially identical structures were found when the sample was imaged either dry or in buffer (compare B and C of Fig. 4). Approximately half of the length of the molecule appears in the rigid bar-like region of the dumbbell, with a relatively large globular domain (diameter 10.7 nm) at one end and a smaller globular domain (diameter 7.8 nm) at the other. In many images, the dumbbell appears to be multipartite at one end or the other, consistent with our identification of the dumbbells as the ORF-1 trimer revealed by the analytical ultracentrifugation experiments.
Fig. 4.
Atomic force microscopy of L1 ORF1p. (A) Image in air of a representative field; arrows indicate ORF1p trimer dumbbells. (B) Collage of 30 dumbbells imaged in air. Each image is 100 nm2. (C) Collage of six dumbbells imaged under buffer.
Discussion
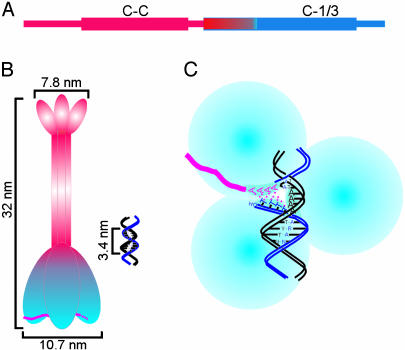
The ORF1 protein from mouse L1 forms a stable trimer in solution that has a dumbbell appearance by atomic force microscopy. There is a predicted coiled-coil domain (stablecoil, www.pence.ca/software/stablecoil/latest) in the sequence of L1spa ORF1p that is ≈120 aa in length, extending between residues 70 and 191, as depicted in Fig. 5A. Additional analysis of this sequence with multicoil (20) reveals a high probability of trimer formation in the predicted coiled-coil domain (data not shown). This coiled-coil region was previously shown to be both necessary and sufficient for self-association of L1 ORF1p by using two-hybrid and GST pull-down assays (13). Hence, we conclude that the trimerization documented in this study is mediated strictly via the coiled-coil domain. The absence of any detectable association of the C-1/3 domain in the analytical ultracentrifuge provides further support for this conclusion. We further hypothesize that the “bar” of the dumbbell as imaged corresponds to the triple-helical, coiled-coil region of a homotrimer of ORF1p. Given a typical length of 1.5 Å per residue in a coiled-coil, the length of the bar in the dumbbell is expected to be ≈180 Å or 18 nm. This length corresponds well to the structures visualized by atomic force microscopy (average lengths of 32 nm; Fig. 4). Because there are substantially more residues C-terminal to the coiled-coil region than N-terminal to it, the larger of the two dumbbell ends likely is the C-terminal, nucleic acid binding domain, with the smaller end the N-terminal region that is expanded in this fusion protein. Taken together, these data support the model for the solution structure of ORF1p illustrated in Fig. 5B, where the hydrodynamic asymmetry of the full-length protein that was detected by the analytical ultracentrifugation experiments is due to true shape asymmetry in the form of a rigid coiled-coil trimer. The hydrodynamic asymmetry of the C-1/3 domain, on the other hand, is less readily modeled from this analysis because it could represent either an anisotropic conformation of this domain (as drawn in Fig. 5B) or a global decrease in packing density. The resolution of our data are not sufficient to exclude either of these possibilities.
Fig. 5.
Model of the structure of ORF1p from mouse L1. (A) Domain structure of ORF1p. The length of the protein is represented as a thin rectangle, with thicker rectangles representing the N-terminal coiled-coil domain (C-C) and the C-terminal conserved domains. The black rectangle on the N terminus represents the His-tag. The sequence of ORF1p changes abruptly from acidic (red) to basic (blue) within the conserved domain; the C-1/3 domain (C-1/3) includes the basic region of the conserved domain through the C terminus of the protein. (B) Schematic trimer structure of ORF1p: the coiled-coil domain holds the three monomers together, forming the bar region seen in the atomic force micrographs (this region is likely helical, although that is not apparent in this drawing). The larger of the two ends contains the C-terminal, nucleic acid binding domain (13). This trimer is shown bound to RNA (red); for simplicity, only one trimer is depicted, but ORF1p likely coats the entire 7-kb L1 RNA to form a ribonucleoprotein particle (11). A DNA target site is shown, drawn to the same scale. (C) Cartoon modeling the strand-exchange reaction required for first strand cDNA synthesis by TPRT. The nucleic acid chaperone activity of ORF1p melts the DNA and then facilitates formation of the RNA:DNA hybrid (6). This view is rotated 90° from the view in B, looking at the bottom surface of the nucleic acid binding domain of ORF1p. Each subunit of the trimer contains one single-stranded nucleic acid binding interface (shaded crescent), which is shown bound with one of the DNA target strands (the nicked strand with HO-TTTT and the intact strand with AAAA; ref. 19) or the polyA tail of the L1 RNA (red). The double-stranded regions of the target are not bound (5). The protein is drawn to scale, as is the B-form target DNA.
The ORF1p coiled-coil trimer is remarkably stable. Because there was no evidence for dissociation of the ORF1p trimer in either the analytical ultracentrifugation or the cross-linking experiments, the dissociation constant of ORF1p to monomers must be at least in the low nM to pM range. One component of the high stability of the ORF1p trimer is likely due to the long length of its coiled-coil domain, 120 aa. Another component appears to be a disulfide cross-link between two cysteine residues, based on the data presented in Fig. 2, which demonstrate that the ORF1p multimer is resistant to dissociation by SDS when reducing agent is omitted from SDS/PAGE sample buffer. Intriguingly, there is just one cysteine residue in this mouse ORF1p sequence, so only two of the three monomers in the trimer can be disulfide cross-linked. This cysteine lies three heptads upstream of the C-terminal end of the coiled-coil domain and is common among the known active mouse L1s, but it is not conserved in L1s from other mammals.
The finding that ORF1p from mouse L1 is a stable trimer in solution was surprising because the only functions known for this protein are single-strand nucleic acid binding and nucleic acid chaperone activities (4–6). These activities have not been linked previously to any protein known to form a homotrimer via a coiled-coil domain. Proteins that multimerize via coiled-coil domains are a well known and functionally diverse group (21, 22); the subset of these proteins that function as homotrimers include those involved in membrane fusion (23) and carbohydrate recognition (24), as well as one transcription factor (25). Only the latter, heat shock transcription factor (HSF), binds nucleic acid. In contrast to the known activities of L1 ORF1p, however, it recognizes a specific sequence in double-stranded DNA to activate transcription after cellular stress (26). To our knowledge, this is the only other nucleic acid binding protein that has been demonstrated to multimerize via a coiled-coil domain and function as a homotrimer, although recent data suggest that hantavirus nucleocapsid (HVNC) is an RNA binding protein that functions as a homotrimer (27, 28). Like ORF1p, the coiled-coil domain of HVNC resides in the N terminus; however, unlike L1 ORF1p, the nucleic acid recognition domain appears to be in the middle of HVNC, with binding occurring preferentially to a double-stranded RNA hairpin (29). Thus, the structure and function of L1 ORF1p is unique among known proteins.
Why is ORF1p from mouse L1 a homotrimer? The low-resolution structure of ORF1p determined here evokes the known high-resolution structures for HSF and the carbohydrate recognition proteins such as tetranectin (24). All of these proteins have three globular ligand-recognition domains held in place by a single, extended coiled-coil domain, fixing the spatial orientation of the region of the protein involved in specific recognition and binding. This may also be the case for L1 ORF1p, allowing the protein to form unique, spatially constrained contacts with its ligand, single-strand nucleic acid. We propose that ORF1p performs at least two distinct functions during retrotransposition of L1, one to coat the RNA (Fig. 5B), as suggested by cofractionation of ORF1p with RNA (11, 30), and a second during the TPRT reaction (Fig. 5C; ref. 6), which is responsible for simultaneously converting the L1 RNA to cDNA and linking it covalently to genomic DNA at the insertion site. Both of these functions could be facilitated by the coiled-coil trimer structure demonstrated here. The well known propensity for coiled-coils to form higher-order structures may enhance coating when ORF1p encounters RNA. Later in the retrotransposition cycle, during TPRT, the ORF1p chaperone activity may facilitate melting of the duplex DNA target site and strand transfer to form the RNA:DNA duplex that acts as the substrate for reverse transcription. Intriguingly, this strand-exchange reaction involves three single strands of nucleic acid; contacts with all three could be achieved by a single ORF1p trimer, as depicted in Fig. 5C.
Acknowledgments
We thank M. Churchill, D. Jones, R. Hodges, and members of the Martin lab for helpful discussion. This work was supported by National Institutes of Health Grants GM40367 (to S.L.M.), GM63808 (to D.K.), and DK61933 (to D.L.B.).
Abbreviations: C-1/3, C-1/3 domain of ORF1p; LINE-1 or L1, long interspersed nuclear element 1; TPRT, target-site primed reverse transcription.
References
- 1.Consortium, M. G. S. (2002) Nature 420, 520–562.12466850 [Google Scholar]
- 2.Malik, H. S., Burke, W. D. & Eickbush, T. H. (1999) Mol. Biol. Evol. 16, 793–805. [DOI] [PubMed] [Google Scholar]
- 3.Moran, J. V. & Gilbert, N. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol., Washington, DC), pp. 836–869.
- 4.Kolosha, V. O. & Martin, S. L. (2003) J. Biol. Chem. 278, 8112–8117. [DOI] [PubMed] [Google Scholar]
- 5.Kolosha, V. O. & Martin, S. L. (1997) Proc. Natl. Acad. Sci. USA 94, 10155–10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin, S. L. & Bushman, F. D. (2001) Mol. Cell. Biol. 21, 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naas, T. P., DeBerardinis, R. J., Moran, J. V., Ostertag, E. M., Kingsmore, S. F., Seldin, M. F., Hayashizaki, Y., Martin, S. L. & Kazazian, H. H., Jr. (1998) EMBO J. 17, 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin, S. L. & Branciforte, D. (1993) Mol. Cell. Biol. 13, 5383–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Branciforte, D. & Martin, S. L. (1994) Mol. Cell. Biol. 14, 2584–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trelogan, S. A. & Martin, S. L. (1995) Proc. Natl. Acad. Sci. USA 92, 1520–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin, S. L. (1991) Mol. Cell. Biol. 11, 4804–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolosha, V. O. & Martin, S. L. (1995) J. Mol. Biol. 270, 2868–2873. [DOI] [PubMed] [Google Scholar]
- 13.Martin, S. L., Li, J. & Weisz, J. A. (2000) J. Mol. Biol. 304, 11–20. [DOI] [PubMed] [Google Scholar]
- 14.van Holde, K. E. & Weischet, W. O. (1978) Biopolymers 17, 1387–1403. [Google Scholar]
- 15.Demeler, B., Saber, H. & Hansen, J. C. (1997) Biophys. J. 72, 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, M. L., Correia, J. J., Yphantis, D. A. & Halvorson, H. R. (1981) Biophys. J. 36, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, M. L. & Frasier, S. G. (1985) Methods Enzymol. 117, 301–342. [Google Scholar]
- 18.Pessen, H. & Kumosinski, T. F. (1985) Methods Enzymol. 117, 219–255. [DOI] [PubMed] [Google Scholar]
- 19.Feng, Q., Moran, J. V., Kazazian, H. H. J. & Boeke, J. D. (1996) Cell 87, 905–916. [DOI] [PubMed] [Google Scholar]
- 20.Wolf, E., Kim, P. S. & Berger, B. (1997) Protein Sci. 6, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burkhard, P., Stetefeld, J. & Strelkov, S. V. (2001) Trends Cell Biol. 11, 82–88. [DOI] [PubMed] [Google Scholar]
- 22.Lupas, A. (1996) Trends Biochem. Sci. 21, 375–382. [PubMed] [Google Scholar]
- 23.Malashkevich, V. N., Singh, M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 8502–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen, B. B., Kastrup, J. S., Rasmussen, H., Holtet, T. L., Graversen, J. H., Etzerodt, M., Thogersen, H. C. & Larsen, I. K. (1997) FEBS Lett. 412, 388–396. [DOI] [PubMed] [Google Scholar]
- 25.Peteranderl, R., Rabenstein, M., Shin, Y. K., Liu, C. W., Wemmer, D. E., King, D. S. & Nelson, H. C. (1999) Biochemistry 38, 3559–3569. [DOI] [PubMed] [Google Scholar]
- 26.Peteranderl, R. & Nelson, H. C. (1992) Biochemistry 31, 12272–12276. [DOI] [PubMed] [Google Scholar]
- 27.Alfadhli, A., Steel, E., Finlay, L., Bachinger, H. P. & Barklis, E. (2002) J. Biol. Chem. 277, 27103–27108. [DOI] [PubMed] [Google Scholar]
- 28.Alfadhli, A., Love, Z., Arvidson, B., Seeds, J., Willey, J. & Barklis, E. (2001) J. Virol. 75, 2019–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu, X., Severson, W., Villegas, N., Schmaljohn, C. S. & Jonsson, C. B. (2002) J. Virol. 76, 3301–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hohjoh, H. & Singer, M. F. (1996) EMBO J. 15, 630–639. [PMC free article] [PubMed] [Google Scholar]
- 31.Sorger, P. K. & Nelson, H. C. (1989) Cell 59, 807–813. [DOI] [PubMed] [Google Scholar]