Abstract
Insertional mutagenesis by long terminal repeat (LTR) enhancers in γ-retrovirus-based vectors (GVs) in clinical trials has prompted deeper investigations into vector genotoxicity. Experimentally, self-inactivating (SIN) lentivirus vectors (LVs) and GV containing internal promoters/enhancers show reduced genotoxicity, although strong ubiquitously-active enhancers dysregulate genes independent of vector type/design. Herein, we explored the genotoxicity of β-globin (BG) locus control region (LCR), a strong long-range lineage-specific-enhancer, with/without insulator (Ins) elements in LV using primary hematopoietic progenitors to generate in vitro immortalization (IVIM) assay mutants. LCR-containing LV had ~200-fold lower transforming potential, compared to the conventional GV. The LCR perturbed expression of few genes in a 300 kilobase (kb) proviral vicinity but no upregulation of genes associated with cancer, including an erythroid-specific transcription factor occurred. A further twofold reduction in transforming activity was observed with insulated LCR-containing LV. Our data indicate that toxicology studies of LCR-containing LV in mice will likely not yield any insertional oncogenesis with the numbers of animals that can be practically studied.
Introduction
The lymphoproliferative and myelodysplastic disorders reported in patients undergoing successful gene therapy for X-linked severe combined immune deficiency1,2 and chronic granulomatous disease,3 respectively, from transactivation of flanking cellular genes by viral enhancer elements in γ-retrovirus-based vectors (GVs) and related observation in animal models have led to intense investigations into insertional oncogenesis and vector genotoxicity. The genotoxic potential of vector types,4,5,6 with their insertional preferences,7 and the role of vector design5,6,8 viral enhancers, transgenes9,10 cell-specific factors,11,12 and genes commonly shown to be dysregulated2,3,13 has been explored using innovative experimental systems that assess the risk of insertional activation or clonal dominance.4,5,8,14,15 Insertional site analyses of humans with adverse events following gene therapy have also shed important insight into the problem of vector genotoxicity.1,2,16,17 A self-inactivating (SIN) vector design that allows deletion of viral long-terminal repeat (LTR) transcriptional elements in the provirus and the use of internal cellular promoters6 reduces the risk of insertional mutagenesis in experimental systems.18
SIN lentivirus vectors (LVs) expressing β-globin or γ-globin genes for the correction of inherited hemoglobinopathies carry strong locus control region (LCR) enhancer elements, that transcriptionally activate β/γ-globin genes in erythroid cells.19 The LCR, located 50–60 kilobase (kb) upstream of the β-globin gene is imperative for high and therapeutic levels of β-globin gene expression.20 The LCR has been shown to be in an active chromatin configuration prior to erythroid lineage commitment in multipotent hematopoietic stem cells.21 LCR-containing LV has been reported to integrate near putative oncogenes in primary human cells,22 although expression of the proto-oncogenes was not analyzed in this study. Recently, some global perturbation of gene expression in colony forming units-spleen with the LCR driven LV was reported in microarray analysis.23 It was also reported that the chicken BG hypersensitive site-4 (cHS4) insulator (Ins) in GV results in reduced perturbation of gene expression in microarray analysis.24 These studies suggest that a careful analysis of the relative genotoxicity of LCR-containing LV compared to conventional LV and GV carrying strong viral enhancers, its activation potential of genes flanking the provirus and the effect of the Ins elements in blocking activation is necessary, until site-specific integration or gene targeting approaches become efficient and clinically available for gene therapy for hemoglobinopathies.
Therefore, in this study, using a recently established cell-culture assay system, we analyzed the ability of LV containing the LCR, or the LCR flanked by cHS4 Ins, to generate in vitro immortalization (IVIM) clones from primary murine hematopoietic cells; and performed a detailed integration site and expression analysis of genes in a 300 kb region (150 kb upstream and 150 kb downstream) of the proviruses in the IVIM assay. Our data suggests significant reduction in the genotoxic frequency and fitness of IVIM clones with uninsulated erythroid-specific SIN LVs compared to SIN LVs carrying strong internal enhancer/promoter elements. The risk of clonal dominance by enhancer-mediated gene activation was abrogated in vectors flanked by the cHS4 Ins element. This study would help evaluate and design safer gene therapy vectors for treatment of inherited disorders including severe hemoglobinopathies.
Results
Evaluation of the genotoxicity from SIN lentiviral vectors with erythroid-specific promoter and transcriptional regulatory elements
To assess whether SIN lentiviral vectors with lineage specific promoters and strong globin regulatory elements pose a reduced risk of insertional mutagenesis, we generated a series of LV carrying the minimal human BG promoter, and the hypersensitive site 2, 3, and 4 of the LCR,25 LCR-β-enhanced green fluorescent protein (GFP) cDNA. We placed the cHS4 5′ 250 base pair (bp) core, or a 650 bp cHS4 fragment (comprising of the 250 bp core and the 3′ 400 bp of cHS4 Ins), or the full-length cHS4 Ins in the 3′ LTR to generate three additional insulated vectors, LCR-β-GFPC, LCR-β-GFP650, and LCR-β-GFP-I, respectively. These vectors were compared to a GV SF91-eGFP-wPre,5 which carried the spleen focus forming virus LTRs (termed SF GV in these studies) and a LV carrying an internal spleen focus forming virus enhancer/promoter (SF LV) as positive controls, known to generate IVIM mutants from primary hematopoietic progenitor cells with a high probability in this assay.5,6 A vector carrying only the BG promoter without the LCR (β-GFP LV) and mock transductions were negative controls (Supplementary Figure S1).
Murine bone marrow lineage-negative (Lin−) cells were transduced with each of these vectors using previously optimized protocols for gene transfer of GV5 and LV,25 by exposing 100,000 cells per IVIM assay to GV or LV supernatant. At day 5 after transduction, cells were expanded for 2 weeks in bulk culture, and then plated in 96-well plates in limiting dilutions for generation of IVIM clones at 2 weeks (experimental design shown in Supplementary Figure S2). With the LCR-containing LV, the minimum cells/well that gave rise to IVIM cell growth was 100 cells/well, and this cell dose was subsequently used for all experiments. After 2 weeks, wells with abundant cells, occupying the majority of the well were scored and expanded for three additional weeks for analysis of surface phenotype and morphology, integration site and expression.
IVIM and replating frequency of erythroid-specific vectors
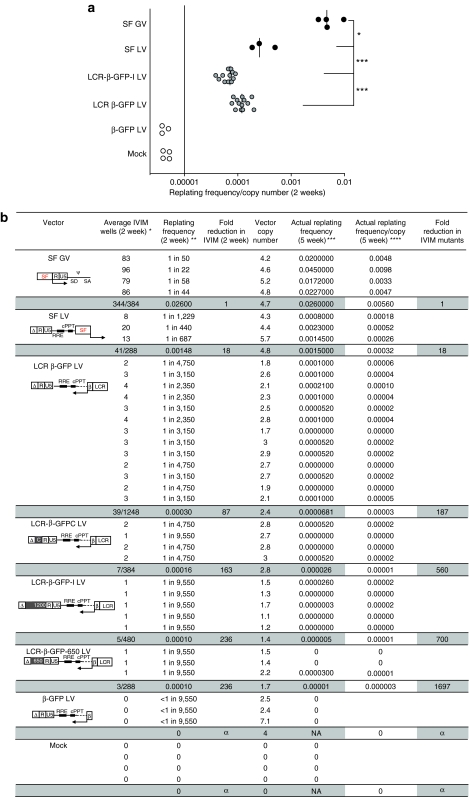
Transgene copy number was assessed in mass cultures 7 days after viral exposure by real-time PCR using primers for the ψ region (packaging signal) of the lentivirus. SF GV had an average copy number of 4.6, with an average of 86 positive clones detected in a 96 well plate, indicating higher transformation events. Typically >100,000-fold expansion was seen with IVIM clones derived from SF GV and SF LV, with individual clones typically exceeding 1 × 107 cells/well at 5 weeks. We observed the previously reported incidence of IVIM assay mutants with the SF GV of 2–3 × 10−5 cells at 2 weeks after replating (Figure 1). The replating frequency of SF LV was 18-fold <SF GV, when normalized for vector copies/cell. The replating frequency of LCR-containing vectors was ~50-fold <the SF GV at 2 weeks, and only 1–2 IVIM positive wells were scored per experiment. The number of IVIM clones derived from insulated vectors was nearly half of those derived from the uninsulated vectors (Figure 1a). The IVIM clonal expansion evident at 2 weeks with LCR vectors was not robust: only one third of the clones derived from LCR-containing LV (insulated and uninsulated LV) were able to expand by 5 weeks, and their expansion was nearly half that seen with the SF GV/LV derived IVIM clones (Figure 1b). Two thirds of the clones that showed initial expansion at 2 weeks after plating differentiated and did not expand further (small numbers of macrophages were evident in the wells at 5 weeks). Clones derived from the SF LV had the same expansion and replating potential/“fitness” as those derived from SF GV at 2 and 5 weeks. Due to this difference between the transformation characters of the clones, we assessed the replating frequency both at 2 weeks and 5 weeks (Figure 1b). The average replating frequency of the LCR-containing LV was ~200-fold lower at 5 weeks. The insulated LCR-containing LV had a significantly lower IVIM potential than the uninsulated LCR-β-GFP LV (P < 0.05), although at this low frequency, and low copy number in the insulated LV clones, the effect may be minimal. Notably, no IVIM mutants were generated in mock cultures or cultures transduced with the β-GFP LV that lacked the LCR, showing that this assay is extremely sensitive at detecting low levels of transformation, as well as predicting the fitness of the transformed mutants. The Ins elements appeared to significantly lower replating efficiency, and no differences were seen between LV with different Ins fragments. All insulated LV carried the cHS4 core, known to have enhancer blocking activity from the CTCF binding sites.26 It is to be noted that the overall number of IVIM clones with the insulated and uninsulated LV were too few, and that at this low frequency and fitness, the effect of the Ins sequences may be minimal.
Figure 1.
Frequency of generation of IVIM clones from lineage-negative primary hematopoietic cells scored at 2 weeks and at 5 weeks (actual replating frequency). (a) Two-week replating frequency (x axis) per vector copy. The y axis shows a schematic representation of the vectors. The regions of the 5′LTR are represented as U3, R, and U5. δ represents self-inactivating long terminal repeat (LTR) U3 enhancer deletion. The detailed vector maps are shown in Supplementary Figure 1. The in vitro immortalization (IVIM) clonal frequency of SF GV, a vector driven by the spleen focus forming virus (SFFV) LTR, and SF LV, carrying an internal SFFV promoter/enhancer was compared to LV carrying the lineage specific β-globin promoter and LCR enhancer (LCR-β-GFP). The replating frequency of three LCR-β-GFP insulated LV carrying different cHS4 insulator fragments in the U3 deletion (shown as a filled box) are combined and plotted. Rev-response element and central polypurine tract are indicated. Arrows indicate the orientation of gene transcription from the provirus. An enhancer-less vector, carrying only the β-promoter and mock transductions were negative controls. All vectors were compared with the SF GV (SF91.eGFP.pre) for their genotoxic potential using Student's “t”-test (unpaired and two tailed). Open circles represent the replating frequency/copy number from independent transductions. Open circles below the horizontal line indicate independent transductions, which did not give rise to any replating clones. Replating frequency was normalized for the mean copy number in the pooled cell population prior to replating. Median is indicated by the black line. P-values are *P < 0.05, ***P < 0.001 indicated in the graph. (b) Frequency of IVIM assay mutants following replating at 2 weeks and 5 weeks. Vector diagrams are as in a. Each row represents an individual experiment. Gray bars summarize results of a vector type, providing the number of wells with IVIM clones versus the total wells plated. Replating frequency was assessed at 2 weeks, as per the standard IVIM assay,5 and cells in each positive well expanded for 3 weeks for analysis. Only 12/36, 2/7, 2/5, and 1/3 clones identified at 2 weeks with the LCR-β-GFP, LCR-β-GFPC, LCR-β-GFP-I, and LCR-β-GFP650, respectively, expanded further and sufficient cells were available for analysis. The actual replating frequency was therefore calculated at 5 weeks. Only select clones from SF GV or SF LVs were chosen for expansion and all expanded robustly. *Number of wells showing clonal growth when 100,000 Lin− cells, transduced, expanded, and replated as 100 cells/well in 96-well plates. **L-Calc Software was used to calculate the replating frequency using # wells showing IVIM mutants, total number of wells plated. ***Assuming a direct correlation between in vitro immortalization potential and copy number. ****With LCR-containing vectors, only one third of the wells that showed expansion at 2 weeks could be expanded at 5 weeks to get sufficient cells for RNA, FACS, and DNA analysis. All wells picked for expansion with the SFFV driven GV or LV vectors were expanded robustly at 5 weeks, and expansion of SF driven vectors was 4.5 ± 1.2 higher. Note: Only select clones were expanded with the SF GV or LV vectors. NA, not applicable; RRE, Rev-response element; cPPT, central polypurine tract; α, infinity.
Phenotype of dominant clones
We next assessed the phenotype of the clones which showed growth fitness under limiting dilution conditions with the SIN-β-LCR vectors. Clones which were dominant in growth over others, and which could be expanded over a million cells were subjected to further analysis. These clones were checked for transgene expression and the copy number (data not shown). The robust growing clones were tested for expression of Sca1+, cKit, myeloid, lymphoid, and erythroid markers. Representative fluorescence activated cell sorter plots are shown in Supplementary Figure S3. IVIM mutants generated from LCR-containing LV did not express the erythroid marker TER-119, but expressed progenitor cell markers, Sca1 and cKit, and a megakaryocytic lineage marker CD41 and a myeloid marker CD11b (Supplementary Figure S3). Levels of Gr-1 and B220 expression was observed in clones tested. Clonal phenotype did not differ significantly between LCR-β-GFP uninsulated vector and the vectors with Ins elements in the SIN deletion. The GV IVIM mutants have been well characterized; and random sampling of 6–8 clones per IVIM assay reconfirmed previously reported results.5
Insertion site analysis and gene expression pattern on dominant clones
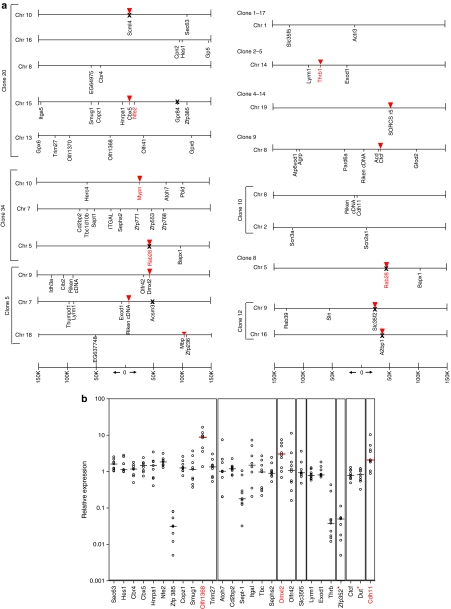
Analysis on LCR-β-GFP IVIM clones was performed on wells that could be expanded up to 5 weeks by ligation-mediated PCR for characterizing insertion sites. We analyzed insertion sites from 10 LCR-β-GFP clones. All identified insertions were blasted against the National Center for Biotechnology Information mouse build 37 genome database (http://www.ncbi.nlm.nih.gov); genes present 150 kb upstream and downstream of the integration site are shown in Figure 2a. The number of vector insertions was between 1 and 5 using a single enzyme for ligation-mediated PCR. As the SF RV transduced clones have been extensively characterized at a molecular level,6 those were not analyzed.
Figure 2.
Insertional site analysis and expression of surrounding genes in clones derived from LCR β-GFP LV. (a) Schematic representation of the genomic annotation scale ±150 kb around the viral integration site is shown at the bottom. Inverted triangles (red) illustrate the integration site in the clones near genes. In gene bare regions, the insertion site is at 0, according to the scale beneath the clones. The recovered genes surrounding the integration site is marked in black. Genes highlighted in red indicate genes implicated in cancer. ‘X' represents genes analyzed for expression by quantitative reverse-transcriptase PCR but were not expressed. (b) Quantitative RT-PCR analysis on the surrounding gene expression in an immortalized clone with a specific insertion site. Expression of genes flanking the provirus was determined using real-time RT-PCR using the TaqMan low-density custom arrays. Expression was first normalized to the level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA expression. Alteration in the surrounding gene expression in an immortalized clone with a specific insertion site was calculated by comparing it against other immortalized clones containing vector insertions at other locations. The SDS RQ Manager 1.2 Software (Applied Biosystems, Foster City, CA) was used to calculate relative expression levels. This type of analysis was done to insure that the effect of gene expression was specific to the unique insertion site and not due to chance. This real-time RT-PCR data from individual genes thus analyzed was plotted as relative gene expression (log10 variations) and compared against other immortalized, unrelated clones containing vector insertions at other locations (shown as empty circles). The horizontal black line represents the median level of expression of the immortalized clone compared against expression from other clones with unrelated insertion sites. The horizontal red line and genes in red indicate median level of expression from significantly upregulated genes. y axis represents expression levels as relative mRNA quantity after normalization for the level of glyceraldehyde-3-phosphate dehydrogenase, and plotted as log10 variations from the median level in all analyzed clones. The genes marked with * are located at 435 kb (Zfp352) and 206 kb (Dut) from insertion site.
Interestingly, two clones (clone 34 and clone 8) shared a common integrant in Rab28, suggesting that the cell divided after the first transduction, and subsequent insertions probably occurred with the second transduction in clone 34. Rab28 expression was undetectable in both clones. Gene hits for 47 genes were analyzed by quantitative reverse-transcriptase PCR. The gene hits and the neighboring genes include genes involved in protein coding, chromatin binding, DNA, and metal ion binding; regulation of Rab GTPase activity; and functions in the olfactory receptor area. In clone 2–5, the insertion was in the ThrB1 gene, which is a negative regulator of transcriptional activity. Clone 9 was inserted in the CCCTC-binding factor (CTCF) region, which is a conserved transcriptional repressor. The relative expression of the genes surrounding the provirus in each clone was compared to the expression of these genes in all the nine other clones and is depicted in Figure 2b. Insertion near Nfe2 (+28.3 kb), an erythroid-specific transcription factor did not affect Nfe2 expression. Genes that have been associated to be increased in cancers (http://atlasgeneticsoncology.org/Genes/Geneliste.html) are marked in red (Figure 2a) and expression of all these genes, but one, was analyzed and was unaffected. Expression of 3 of 47 genes located within 25 kb of the insertion site was dysregulated: Olfr1368 (functions in olfactory receptor activity; −25 kb), Dmxl2 (functions in Rab GTPase binding; +43 kb) and cadherin-11 (involved in mesenchymal cell differentiation; +8.4 kb) were dysregulated, relative to their expression in other clones without the provirus at that site. Of note, genes known to be involved in cancer were not upregulated, although most of these were either located >25 kb from the insertion site, or perhaps disrupted due to the vector insertion.
Analysis of surrounding gene expression from single copy uninsulated and insulated clones from the same integration site
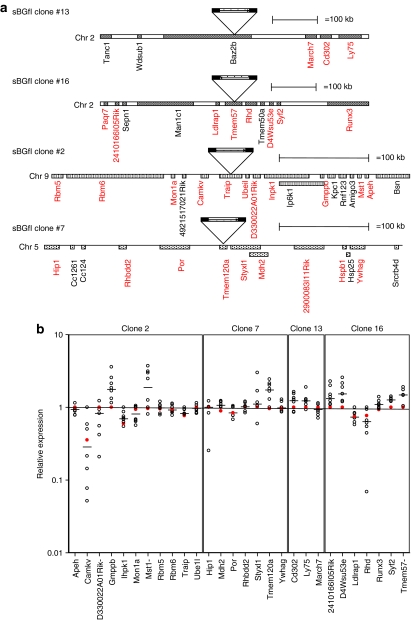
We next analyzed the effect of the Ins in an erythroid milieu and at the same insertion site. We generated a LCR-β-promoter β-globin LV with the 1.2 kb cHS4 in the 3′LTR that was flanked by Lox-P sites (sBGfI). Twenty-five mouse erythroleukemia (MEL) cell clones carrying a single copy of sBGfI were transduced with a nonintegrating LV-encoding Cre recombinase. Cells were cloned. Four clones that had the Ins removed from both SIN-LTR but had the rest of the provirus retained. Integrity of the provirus and Ins was confirmed in these clones via genomic Southern blot analysis (data not shown) and integration sites around the provirus were mapped (Figure 3a). Real-time reverse-transcriptase–PCR low-density array was used to analyze 47 genes flanking 500 kb of the four proviruses. The relative expression of genes in the uninsulated provirus in one clone was compared to the insulated counterpart (shown in red in Figure 3b), and to all other clones. There was no difference in expression of any of the genes flanking the provirus, including two genes known to be involved with cancer, Apeh and Runx3 (located >100 kb from insertion sites). Genes with minimal or no expression are not shown. From these data, the cHS4 did not seem to affect expression of flanking genes, although in the IVIM assay the generation of IVIM clones by insulated vectors was significantly lower than by uninsulated LV. Therefore, it is likely that the IVIM is a more sensitive assay to detect genotoxicity of vectors that have a very low activation potential, and the number of integrants analyzed by the removing the Ins from the same integration sites may have been too few. Alternatively, the Ins may have a minimal effect in a vector system that has inherently low genotoxicity.
Figure 3.
Gene expression pattern in murine erythroleukemia cell clones with the cHS4 insulated (sBGfI) or its floxed uninsulated provirus (sBG) at the same integration site. (a) Schematic representation of the proviral integration sites in single copy murine erythroleukemia clone carrying insulated vectors. The insertion site and the orientation of vector insertion is marked as a triangle. The dark shading indicates the insulator in the provirus. The insulator was flanked with lox-P sites and single integrant clones derived were subjected to cre-mediated recombination. Clones with the insulator removed at the same integration site as the parent clone were analyzed further by linear amplification-mediated PCR to map the surrounding genes in ±250 kb distance around the provirus. The genes marked in red indicate those analyzed for expression by TaqMan low-density array. The position of surrounding genes is drawn to scale (as indicated). (b) Quantitative reverse-transcriptase PCR analysis on the expression of the surrounding genes in individual clones are shown as empty circles. Red open circles indicate the expression level of the insulated clone compared to the expression from the uninsulated clone from the same integration site and also to other clones with unrelated integration sites. The horizontal black line indicates median level of expression from analyzed genes. y axis represents expression levels as relative mRNA quantity after normalization for the level of glyceraldehyde-3-phosphate dehydrogenase, and plotted as log10 variations from the median level in all analyzed clones.
Discussion
Insertional mutagenesis still remains a concern for SIN GV and LV, especially in clinical settings where high levels of transgene expression necessitate strong internal enhancer-promoter sequences. The LCR is one such enhancer that is necessary for high-level globin gene expression. The safety of the LCR is presumed to be in its erythroid-specific enhancement of transgene expression, and its expression in a lineage where enucleation imparts a natural safety feature.27 However, the LCR has been reported to be in an active configuration in multilineage progenitors21 and could exert genotoxicity prior to lineage commitment. Herein, the IVIM assay was used to assess genotoxicity in Lin− primary hematopoietic progenitors with the rationale that if LCR was activated prior to lineage commitment, it would transform and confer a proliferative advantage and immortalize the hematopoietic progenitors and thus result in their clonal expansion. Erythroid cells are derived from the common myelo-erythroid progenitors; and transformation of lineage committed progenitors with proto-oncogenes has been shown to result in leukemia.28
We demonstrate that the LCR does dysregulate gene expression, although the frequency of IVIM of hematopoietic progenitor cells is significantly lower compared with GV. Indeed, the LV without the LCR was unable to immortalize any Lin− cells in this assay, a result similar to that seen with mock transduced negative controls. Ins sequences further lowered the frequency of immortalized clones, with the greatest effect seen with either the full-length cHS4 Ins or a 650 bp fragment, which combines the canonical 5′ 250 bp core with 400 bp of 3′cHS4 sequence. We have recently shown that the 3′400 bp has Ins functions similar to the 5′ 250 bp core (P. Arumugam, F. Urbinati, C.S. Velu, T. Higashimoto, H.L. Grimes, and P. Malik, unpublished results). The cHS4 Ins has been well characterized for its enhancer blocking activity,29 and recently, found to block adjacent gene expression in lymphoid cell lines or an experimental system designed to test enhancer activation.15,30 The results in the four MEL clones with the “floxed” Ins were probably not informative due to analysis of a limited number of clones with a vector with relatively low transactivation potential. Here, the IVIM assay was more sensitive at detecting differences.
Of the significantly dysregulated genes, we observed increased expression of cadherin-11, Dmxl2, and Olfr 1368. Dmxl2 (+43 kb from the insertion site) functions as a scaffold protein and is involved in Rab GTPase binding.31 Olfr1368 (−25 kb downstream of the insertion site) is involved in G-protein coupled receptor activities.32 Cdh11 (+8.4 kb) belongs to a classical type-2 cadherin family and is involved in homophilic cell-cell adhesion and is shown to be elevated in aggressive human breast cancers.33 These genes are involved in cell signaling and could confer a proliferative advantage to hematopoietic progenitor cells. The other gene insertions that could be relevant to potential immortalization were Rab 28, chromobox homolog 5 and myopalladin. Rab28 (+45 kb), a member of the ras-oncogene family, involved in nucleotide binding and GTPase mediated signal transduction,34 Rab28 expression was not increased and the gene was likely disrupted. Myopalladin (+34 kb) distance from insertion site is involved in cellular morphogenesis and act as a scaffold that regulate actin organization.35 Other interacting molecular partners also play a role: chromobox homolog 5 (at +12.6 kb distance) is one of the critical factors in transcription/chromatin program and is sufficient to promote Hox and Meis independent immortalization of myeloid progenitors in stem cell maintenance.36 Notably, a single insertion (confirmed by quantitative PCR) into Thrb1 (−69.9 kb) in clone 2–5 downregulated expression of Thrb1, a negative regulator of transcription and implicated in cancer.37 This suggests that lack of regulation of transcription was important in immortalization of this clone. On the other hand, we did not observe increased expression of an erythroid-specific transcription factor Nfe2 (+28 kb). Lack of expression of Nfe2 in an immortalized clone in our study could be due to the higher order chromatin structure, where the gene was physically distinct to be activated or the increased distance from the insertion. Similar results were observed by insertional activations in T-cell clones where no direct correlation was observed between insertional activation and gene distances.38
In summary, our studies show the sensitivity of the IVIM assay of primary progenitors in illustrating the genotoxicity of vectors with lineage-specific enhancers as compared to GV. Although the LCR causes some perturbation of gene expression, confirmed by the total lack of IVIM clones in LV lacking the LCR, the frequency and fitness of IVIM clones was low, with no obvious increase in expression of genes known to be involved in cancer. Dysregulation of genes involved in cell signaling was observed. The use of primary Lin− hematopoietic progenitors, a short turn around time, and ease of comparison of several vectors simultaneously, the high number of progenitors that can be subjected to vector insertions and the robustness of the IVIM assay make it a useful and sensitive assay to study vector safety. Animal toxicology studies using GVs show one in 8–10 mice develop insertional oncogenesis after secondary transplants (1.5 years).39 Our data on LCR-containing LV indicate that toxicology studies in mice will likely not yield insertional oncogenesis with the lineage specific vectors with the numbers of animals that can be practically studied.
Materials and Methods
Vectors. The cloning of the SF GV (SF91-eGFP-wPRE) has been described previously.5 The LV SF carries the spleen focus forming virus U3 promoter/enhancer that drives eGFP expression.40 All LCR-containing LV were obtained by cloning different Ins fragments into a unique NheI/EcoRV site in the U3 3′LTR region of the sBG, as described.41 This plasmid carried the human β-globin gene and the hypersensitive site 2, 3, and 4 fragments of the human BG LCR, as previously described.25 pBGlo plasmid was digested with SnaBI and SmaI and ligated to remove one EcoRI site. pG-Bsd (kind gift of Dr Boris Fehse, Frankfurt, Germany) was digested with NcoI and EcoRI to isolate eGFP-Blasticidin fusion gene. eGFP-Blasticidin fragment was ligated to pBGlo plasmid digested with NcoI and EcoRI to replace β-globin gene to create pBGFP/Blast. pBGFP/Blast was digested with NcoI and XhoI to remove eGFP-Blasticidin fragment, and it was ligated to pLCR-BGlo plasmid digested with NcoI and XhoI to create pLCR-BGFP/Blast. For generating lentiviral expression vectors, sBG, sBGC, sBG650, and sBG-I plasmids were digested with BlpI and XhoI. pLCR-BGFP/Blast was digested with BlpI and XhoI to remove LCR, BG promoter and eGFP-Blast and ligated to the lentiviral backbones to generate LCR-β-GFP, LCR-β-GFPC, LCR-β-GFP650, and LCR-β-GFP-I. For lentiviral vector without LCR, pBGFP/Blast plasmid was digested with NotI and blunt end was generated by Klenow (New England Biolabs, MA) as recommended by the manufacturer. This plasmid was subsequently digested with XhoI to isolate BG promoter and eGFP-Blasticidin fragment. DNA fragment containing BG promoter and eGFP-Blasticidin was ligated to sSIN plasmid digested with SmaI and XhoI. To obtain the sBGfI plasmid, we cloned the Ins fragment into a pLox plasmid (kind gift from Dr Vesa Kaartinen, Childrens Hospital, LA), to obtain the pflox-Ins. This flox-Ins plasmid was then digested with XbaI and EcoRV restriction enzymes and cloned into NheI and EcoRV sites of the sBG vector.
GV was produced by transient transfection of 293T cells with packaging constructs coding for the gag-pol proteins and the ecotropic envelope. Viral titers were determined on HeLa cells and were in the range of 106–107 infectious units/ml. Lentiviral vectors were produced by transient cotransfection of 293T cells, as previously described.25 Virus was titrated in limiting dilutions on MEL.25 All experiments were performed using thawed vector stocks of known titers.
Isolation and transduction of Lin− cells. Lin− bone marrow cells of untreated C57Bl6/J mice were obtained from a single cell suspension of bone marrow resuspended in Iscove's modified Dulbecco's medium containing 2% heat-inactivated fetal bovine serum. Low-density cells were enriched by equilibrium centrifugation over a cushion of Ficoll-Histopaque (Sigma, St Louis, MO) at a density of 1.077 g/ml, washed and resuspended in Iscove's modified Dulbecco's medium containing 2% heat-inactivated fetal bovine serum. Lin− cells were isolated from bone marrow cells by magnetic sorting using biotin-labeled lineage specific antibodies Gr-1, CD11b, CD45R/B220, CD3e, TER-119 (BD Pharmingen, San Diego, CA) according to manufacturer's instructions. Freshly isolated Lin− cells were prestimulated for 2 days for retroviral transduction in Stemspan medium (Stem cell technologies, Vancouver, BC) containing 50 ng/ml mSCF, 100 ng/ml hIL-11 and 10 ng/ml mIL-3 (R&D Biosystems, Minneapolis, MN), 1% penicillin/streptomycin, and 2 mmol/l glutamine at a density of 1–5 × 105/ml. Cells were transduced on day 4 and day 5 postisolation using an MOI of 20 × 2. Virus preloading was carried out on retronectin coated (10 µg/cm2, Takara, Otsu, Japan) suspension culture dishes by spinoculation for 30 minutes at 4 °C. 1 × 105 cells were cultured in a single well of a 24-well plate with 500 µl on day 4 and 1 ml on day 5 to account for increasing cell numbers. On day 5, the cells were transferred to fresh retroviral preloaded plates for second round of transduction.
For LV transduction, Lin− cells were prestimulated overnight with Stemspan medium containing 50 ng/ml mSCF, 100 ng/ml hIL-11, and 10 ng/ml mIL-3, 1% penicillin/streptomycin and 2 mmol/l glutamine. On day 2, these cells were transduced with concentrated lentiviral supernatants SF (internal) LV, LCR-β-GFP LV, LCR-β-GFPC LV, LCR-β-GFP650 LV, LCR-β-GFP–I LV, and β-GFP LV at an MOI of 20, twice at 12-hour intervals, in Stemspan medium containing cytokines and expanded as mass cultures from day 3.
IVIM mutants assay. After a 5 day transduction, the Lin− cells were expanded as mass culture for 2 weeks in Stemspan medium containing 10% FBS, 50 ng/ml mSCF, 100 ng/ml hIL-11, and 10 ng/ml mIL-3, 1% penicillin/streptomycin, and 2 mmol/l glutamine, as described.5 During expansion as mass cultures, the cell density was adjusted to 5 × 105 cells/ml every 3 days. After 2 weeks of mass culture, the cells were plated into 96-well plates at a density of 100 cells/well. After 2 weeks, the positive wells were scored and expanded further into 24-well plates. The frequency of replating cells at the 2 week and 5 week time-point was calculated based on Poisson statistics using L-Calc software (Stem Cell Technologies, Vancouver, British Columbia, Canada).
Phenotypic characterization of dominant clones. Dominant clones were phenotypically characterized by staining for antibodies against cell surface markers CD11b, Gr-1, B220, TER-119, Sca1, CD41, F4/80, and cKit. Clonal morphology was analyzed on cytospins stained with Wright-Giemsa solution.
Quantitative real-time PCR. Transduction efficiency of retroviral and lentiviral vectors were assessed on bulk cultures at day 7 after vector exposure by quantitative real-time PCR analysis. Genomic DNA (50 ng) from a single copy MEL clone was diluted with untransduced DNA to generate copy number standards ranging from 1 copy/cell to 0.016 copies/cell. The primers and the probes for quantitation were designed using the Primer Express Sofware from Applied Biosystems (Foster City, CA). The primers were designed to recognize the ψ region of the provirus as previously described.42 For GV, the primers were designed to recognize the eGFP region. The PCR mixture was thermo cycled according to the thermal cycler protocol for 96 well plates in Applied Biosystems 7900HT Fast Real-Time PCR System Base Unit.
Integration site analysis. Ligation-mediated PCR was performed as described,5 to determine integration sites used in this assay. Approximately 100 ng of genomic DNA was used as starting material and digested with TSP 509I restriction enzyme followed by a cycle primer extension using lvLTR1; 5′-[bio]GAACCCACTGCTTAAGCCTCA-3′. After DNA enrichment of the biotinylated DNA a ligation was performed using the following linker primers to create the polylinker cassette; Linker 1; 5′-GACCCGGGAGATCTGAATTCAGTGGCACAGCAGTTAGG-3′; and Linker 2; 5′-CCTAACTGCTGTGCCACTGAATTCAGATCTCCCG-3′. Exponential PCRs followed using the following vector designed primers, lvLTRII; 5′-AGCTTGCCTTGA-GTGCTTCA-3′; and lvLTRIII; 5′-AGTAGTGTGTGCCCGTCTGT-3′. All PCR conditions have been previously described.5 The inserts obtained were isolated from a 2% agarose gel and purified using a gel extraction kit (Qiagen, Valencia, CA). DNA was precipitated overnight and a sequencing reaction was performed using a linker specific primer and the Big Dye Terminator v1.1 sequencing kit (Applied Biosystems). Samples were submitted to be read by the Cincinnati Children's Hospital Medical Center Sequencing Core.
Generation of clones with and without the Ins at the same integration site. sBGfI clones were derived from sBGfI transduced MEL cells, screened for the presence of Ins and β-globin by semiquantitative PCR and confirmed with a Southern blot analysis. The transgene expression was confirmed by fluorescence activated cell sorter analysis. These cells were transduced with varying dilutions of a nonintegrating CMV-mER-Cre LV. Two days post-transduction, the mutant estrogen receptor-Cre fusion gene was induced with tamoxifen, to MEL cells. On the day following tamoxifen treatment, sBGfI cells were washed and expanded in culture for 12–14 days. After expansion, cells were cloned. Genomic DNA was extracted from clones and screened by PCR for the presence of the Ins and BG. The clones which were double positive, BG+ Ins+, and those that were BG+ Ins−, screened by semiquantitative PCR were confirmed by Southern blot for size and presence and absence of BG and Ins, and expanded for further analysis. Integration sites of individual clones were mapped by linear amplification-mediated PCR and genes located 250 kb upstream and 250 kb downstream of the insertion site were mapped. RNA was extracted from the clones, reverse transcribed and 47 genes around the proviral integration sites, that had validated primer and probe sets for real-time quantitative reverse-transcriptase PCR were quantified using a custom TaqMan low-density arrays.
Real-time reverse-transcriptase–PCR analysis of gene expression by TaqMan low-density array. cDNAs were reverse transcribed from total RNA samples (1–2 µg) from sBGfI clones and dominant clones from IVIM assay, using the “high capacity cDNA reverse transcription kit” (Applied Biosystems). Custom low-density arrays were designed based upon genes located around a 500 kb or a 300 kb distance of the provirus integration sites for the sBGfI/sBG clone pairs or IVIM assay mutant clones, using specific primers designed and validated by the manufacturer on an ABI PRISM 7900 HT (Applied Biosystems). TaqMan PCRs were carried out using custom 7900 TaqMan low-density arrays. Expression of each gene (150 kb–250 kb window on either side of the provirus) was determined in triplicate reactions on the same array in the test clone and in other control clones. Gene expression profiling was measured as relative expression after normalization for the level of glyceraldehyde-3-phosphate by using the comparative cycle of threshold method of relative quantification using the RQ Manager software (Applied Biosystems). The expression values are plotted as log10 variation from the median levels.
Statistical analysis. Data from experiments are expressed as mean ± SEM. P < 0.05 was considered significant. All vectors were compared with the SF GV (SF91.eGFP.pre) for their genotoxic potential using Student's “t”-test (unpaired and two tailed).
SUPPLEMENTARY MATERIALFigure S1. Schematic representation of the proviral forms of the vectors. A SFFV LTR driven gammaretroviral vector SF91-eGFP-wPre (SF GV) and a SIN lentiviral vector (SF LV), with internal enhancer-promoter elements derived from spleen focus-forming virus (SFFV) were used as a positive control for insertional mutagenesis assay in primary murine hematopoietic stem cells. A series of SIN LV vectors with hβ-globin promoter and the locus control region, (LCR-β-GFP LV) and LCR-β-GFP vectors which additionally incorporate either the 5apostrope;250bp core insulator fragment of cHS4 (LCR β-GFPC LV), the 5apostrope;250bp core element combined with the 3apostrope;400 sequence of the cHS4 insulator(LCR-β GFP650 LV) or the full-length 1.2Kb cHS4 insulator (LCR-β-GFP-I) in the U3 deletion of the 3apostrope;LTR are shown. A SIN-β vector, devoid of LCR element (β-GFP LV) was used as a control vector. A SFFV LTR driven gammaretroviral vector SF91-eGFP-wPre (SF GV) and a SIN lentiviral vector (SF LV), with internal enhancer-promoter elements derived from spleen focus-forming virus (SFFV) were used as a positive control for insertional mutagenesis assay in primary murine hematopoietic stem cells. A series of SIN LV vectors with hβ-globin promoter and the locus control region, (LCR-β-GFP LV) and LCR-β-GFP vectors which additionally incorporate either the 5apostrope;250bp core insulator fragment of cHS4 (LCR β-GFPC LV), the 5apostrope;250bp core element combined with the 3apostrope;400 sequence of the cHS4 insulator(LCR-β GFP650 LV) or the full-length 1.2Kb cHS4 insulator (LCR-β-GFP-I) in the U3 deletion of the 3apostrope;LTR are shown. A SIN-β vector, devoid of LCR element (β-GFP LV) was used as a control vector.Figure S2. Experimental design of the IVIM assay. Lineage-negative cells were isolated from the bone marrow of C57BL6 mice, prestimulated and transduced with GV and LV twice at an MOI of 20. For GV transduction, lineage-negative cells were prestimulated for two days and transduced for two days, and at day 5, cells were expanded in bulk for 2 weeks. For LV transductions, lineage-negative cells were prestimulated overnight before transduction, and transduced twice 12 hours apart. Limiting dilution analysis was done on day 19 at 100 cells/well in a 96-well plate for all vectors. Two weeks later, wells which were nearly full of cells were scored as positive and expanded further for three weeks in 24-well plates for molecular analysis. Limiting dilution analysis showed a minimum of 10 cells/well with GV gave rise to IVIM clones, and 100 cells/well for LCR-LV gave rise to IVIM clones.Figure S3. Surface phenotype of IVIM mutant clones. A. Representative FACS plots for one clone are shown for Sca1+, cKit+, B220 and Gr-1. Each clones was also analyzed for CD11b, TER-119 and CD41 surface expression. B. Quantification of lineage marker expression is shown for all the LCR-LV clones. The clones expressed high levels of Sca1 and cKit+. Varying levels of CD11b expression was observed. Erythroid marker TER-119 expression was negligible, very low levels of Gr-1 and B220 expression was observed. There was a significant decrease in the expansion potential of the clones with insulators. Clonal phenotype did not differ significantly between LCR-β-GFP uninsulated vector and the vectors with insulator elements in the SIN deletion.
Supplementary Material
Schematic representation of the proviral forms of the vectors. A SFFV LTR driven gammaretroviral vector SF91-eGFP-wPre (SF GV) and a SIN lentiviral vector (SF LV), with internal enhancer-promoter elements derived from spleen focus-forming virus (SFFV) were used as a positive control for insertional mutagenesis assay in primary murine hematopoietic stem cells. A series of SIN LV vectors with hβ-globin promoter and the locus control region, (LCR-β-GFP LV) and LCR-β-GFP vectors which additionally incorporate either the 5apostrope;250bp core insulator fragment of cHS4 (LCR β-GFPC LV), the 5apostrope;250bp core element combined with the 3apostrope;400 sequence of the cHS4 insulator(LCR-β GFP650 LV) or the full-length 1.2Kb cHS4 insulator (LCR-β-GFP-I) in the U3 deletion of the 3apostrope;LTR are shown. A SIN-β vector, devoid of LCR element (β-GFP LV) was used as a control vector. A SFFV LTR driven gammaretroviral vector SF91-eGFP-wPre (SF GV) and a SIN lentiviral vector (SF LV), with internal enhancer-promoter elements derived from spleen focus-forming virus (SFFV) were used as a positive control for insertional mutagenesis assay in primary murine hematopoietic stem cells. A series of SIN LV vectors with hβ-globin promoter and the locus control region, (LCR-β-GFP LV) and LCR-β-GFP vectors which additionally incorporate either the 5apostrope;250bp core insulator fragment of cHS4 (LCR β-GFPC LV), the 5apostrope;250bp core element combined with the 3apostrope;400 sequence of the cHS4 insulator(LCR-β GFP650 LV) or the full-length 1.2Kb cHS4 insulator (LCR-β-GFP-I) in the U3 deletion of the 3apostrope;LTR are shown. A SIN-β vector, devoid of LCR element (β-GFP LV) was used as a control vector.
Experimental design of the IVIM assay. Lineage-negative cells were isolated from the bone marrow of C57BL6 mice, prestimulated and transduced with GV and LV twice at an MOI of 20. For GV transduction, lineage-negative cells were prestimulated for two days and transduced for two days, and at day 5, cells were expanded in bulk for 2 weeks. For LV transductions, lineage-negative cells were prestimulated overnight before transduction, and transduced twice 12 hours apart. Limiting dilution analysis was done on day 19 at 100 cells/well in a 96-well plate for all vectors. Two weeks later, wells which were nearly full of cells were scored as positive and expanded further for three weeks in 24-well plates for molecular analysis. Limiting dilution analysis showed a minimum of 10 cells/well with GV gave rise to IVIM clones, and 100 cells/well for LCR-LV gave rise to IVIM clones.
Surface phenotype of IVIM mutant clones. A. Representative FACS plots for one clone are shown for Sca1+, cKit+, B220 and Gr-1. Each clones was also analyzed for CD11b, TER-119 and CD41 surface expression. B. Quantification of lineage marker expression is shown for all the LCR-LV clones. The clones expressed high levels of Sca1 and cKit+. Varying levels of CD11b expression was observed. Erythroid marker TER-119 expression was negligible, very low levels of Gr-1 and B220 expression was observed. There was a significant decrease in the expansion potential of the clones with insulators. Clonal phenotype did not differ significantly between LCR-β-GFP uninsulated vector and the vectors with insulator elements in the SIN deletion.
Acknowledgments
This work was supported by NIH grants RO1-HL70135-01 (PM), U54 HL06-008 (PM), P01HL073104 project 2(PM), and NCI 5R01CA107492-02 (CB). The authors declare that they have no competing financial interests.
REFERENCES
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knöss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, et al. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- Maruggi G, Porcellini S, Facchini G, Perna SK, Cattoglio C, Sartori D, et al. Transcriptional enhancers induce insertional gene deregulation independently from the vector type and design. Mol Ther. 2009;17:851–856. doi: 10.1038/mt.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods NB, Bottero V, Schmidt M, von Kalle C., and , Verma IM. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440:1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- Pike-Overzet K, de Ridder D, Weerkamp F, Baert MR, Verstegen MM, Brugman MH, et al. 2006Gene therapy: is IL2RG oncogenic in T-cell development Nature 443E5;discussion E6discussion E7 [DOI] [PubMed] [Google Scholar]
- Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- Felice B, Cattoglio C, Cittaro D, Testa A, Miccio A, Ferrari G, et al. Transcription factor binding sites are genetic determinants of retroviral integration in the human genome. PLoS ONE. 2009;4:e4571. doi: 10.1371/journal.pone.0004571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calmels B, Ferguson C, Laukkanen MO, Adler R, Faulhaber M, Kim HJ, et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrie PC, Huo Y, Stolitenko RB., and , Russell DW. A rapid and quantitative assay for measuring neighboring gene activation by vector proviruses. Mol Ther. 2008;16:534–540. doi: 10.1038/sj.mt.6300398. [DOI] [PubMed] [Google Scholar]
- Ryu BY, Evans-Galea MV, Gray JT, Bodine DM, Persons DA., and , Nienhuis AW. An experimental system for the evaluation of retroviral vector design to diminish the risk for proto-oncogene activation. Blood. 2008;111:1866–1875. doi: 10.1182/blood-2007-04-085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzwaelder K, Howe SJ, Schmidt M, Brugman MH, Deichmann A, Glimm H, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metais JY., and , Dunbar CE. The MDS1-EVI1 gene complex as a retrovirus integration site: impact on behavior of hematopoietic cells and implications for gene therapy. Mol Ther. 2008;16:439–449. doi: 10.1038/sj.mt.6300372. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR., and , Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Li Q, Peterson KR, Fang X., and , Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez G, Griffiths SD, Ford AM, Greaves MF., and , Enver T. Activation of the beta-globin locus control region precedes commitment to the erythroid lineage. Proc Natl Acad Sci USA. 1992;89:10618–10622. doi: 10.1073/pnas.89.22.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imren S, Fabry ME, Westerman KA, Pawliuk R, Tang P, Rosten PM, et al. High-level beta-globin expression and preferred intragenic integration after lentiviral transduction of human cord blood stem cells. J Clin Invest. 2004;114:953–962. doi: 10.1172/JCI21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove PW, Kepes S, Hanawa H, Obenauer JC, Pei D, Cheng C, et al. Globin lentiviral vector insertions can perturb the expression of endogenous genes in beta-thalassemic hematopoietic cells. Mol Ther. 2008;16:525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- Li CL, Xiong D, Stamatoyannopoulos G., and , Emery DW. Genomic and functional assays demonstrate reduced gammaretroviral vector genotoxicity associated with use of the cHS4 chromatin insulator. Mol Ther. 2009;17:716–724. doi: 10.1038/mt.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveetil G, Scholes J, Carbonell D, Qureshi N, Xia P, Zeng L, et al. Successful correction of the human beta-thalassemia major phenotype using a lentiviral vector. Blood. 2004;104:3445–3453. doi: 10.1182/blood-2004-04-1427. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG., and , Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- von Kalle C, Baum C., and , Williams DA. Lenti in red: progress in gene therapy for human hemoglobinopathies. J Clin Invest. 2004;114:889–891. doi: 10.1172/JCI23132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, et al. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu BY, Persons DA., and , Nienhuis AW. The influence of chromatin insulator on interactins between globin regulatory elements and cellular promoters in erythroid cells. Mol Ther. 2006;13:S406. [Google Scholar]
- Kawabe H, Sakisaka T, Yasumi M, Shingai T, Izumi G, Nagano F, et al. A novel rabconnectin-3-binding protein that directly binds a GDP/GTP exchange protein for Rab3A small G protein implicated in Ca2+-dependent exocytosis of neurotransmitter. Genes Cells. 2003;8:537–546. doi: 10.1046/j.1365-2443.2003.00655.x. [DOI] [PubMed] [Google Scholar]
- Hague C, Hall RA., and , Minneman KP. Olfactory receptor localization and function: an emerging role for GPCR heterodimerization. Mol Interv. 2004;4:321–322. doi: 10.1124/mi.4.6.4. [DOI] [PubMed] [Google Scholar]
- Tamura D, Hiraga T, Myoui A, Yoshikawa H., and , Yoneda T. Cadherin-11-mediated interactions with bone marrow stromal/osteoblastic cells support selective colonization of breast cancer cells in bone. Int J Oncol. 2008;33:17–24. [PubMed] [Google Scholar]
- Brauers A, Schürmann A, Massmann S, Mühl-Zürbes P, Becker W, Kainulainen H, et al. Alternative mRNA splicing of the novel GTPase Rab28 generates isoforms with different C-termini. Eur J Biochem. 1996;237:833–840. doi: 10.1111/j.1432-1033.1996.0833p.x. [DOI] [PubMed] [Google Scholar]
- Otey CA, Rachlin A, Moza M, Arneman D., and , Carpen O. The palladin/myotilin/myopalladin family of actin-associated scaffolds. Int Rev Cytol. 2005;246:31–58. doi: 10.1016/S0074-7696(05)46002-7. [DOI] [PubMed] [Google Scholar]
- Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SY. Thyroid hormone receptor mutations in cancer. Mol Cell Endocrinol. 2003;213:23–30. doi: 10.1016/j.mce.2003.10.051. [DOI] [PubMed] [Google Scholar]
- Recchia A, Bonini C, Magnani Z, Urbinati F, Sartori D, Muraro S, et al. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc Natl Acad Sci USA. 2006;103:1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Schambach A, Brugman MH, Wicke DC, Knoess S, Li Z, et al. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia. 2008;22:1519–1528. doi: 10.1038/leu.2008.118. [DOI] [PubMed] [Google Scholar]
- Schambach A, Bohne J, Chandra S, Will E, Margison GP, Williams DA, et al. Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells. Mol Ther. 2006;13:391–400. doi: 10.1016/j.ymthe.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Urbinati F, Arumugam P, Higashimoto T, Perumbeti A, Mitts K, Xia P, et al. 2009Mechanism of Reduction in Titers From Lentivirus Vectors Carrying Large Inserts in the 3'LTR Mol TherEpub ahead of print). [DOI] [PMC free article] [PubMed]
- Arumugam PI, Scholes J, Perelman N, Xia P, Yee JK., and , Malik P. Improved human beta-globin expression from self-inactivating lentiviral vectors carrying the chicken hypersensitive site-4 (cHS4) insulator element. Mol Ther. 2007;15:1863–1871. doi: 10.1038/sj.mt.6300259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the proviral forms of the vectors. A SFFV LTR driven gammaretroviral vector SF91-eGFP-wPre (SF GV) and a SIN lentiviral vector (SF LV), with internal enhancer-promoter elements derived from spleen focus-forming virus (SFFV) were used as a positive control for insertional mutagenesis assay in primary murine hematopoietic stem cells. A series of SIN LV vectors with hβ-globin promoter and the locus control region, (LCR-β-GFP LV) and LCR-β-GFP vectors which additionally incorporate either the 5apostrope;250bp core insulator fragment of cHS4 (LCR β-GFPC LV), the 5apostrope;250bp core element combined with the 3apostrope;400 sequence of the cHS4 insulator(LCR-β GFP650 LV) or the full-length 1.2Kb cHS4 insulator (LCR-β-GFP-I) in the U3 deletion of the 3apostrope;LTR are shown. A SIN-β vector, devoid of LCR element (β-GFP LV) was used as a control vector. A SFFV LTR driven gammaretroviral vector SF91-eGFP-wPre (SF GV) and a SIN lentiviral vector (SF LV), with internal enhancer-promoter elements derived from spleen focus-forming virus (SFFV) were used as a positive control for insertional mutagenesis assay in primary murine hematopoietic stem cells. A series of SIN LV vectors with hβ-globin promoter and the locus control region, (LCR-β-GFP LV) and LCR-β-GFP vectors which additionally incorporate either the 5apostrope;250bp core insulator fragment of cHS4 (LCR β-GFPC LV), the 5apostrope;250bp core element combined with the 3apostrope;400 sequence of the cHS4 insulator(LCR-β GFP650 LV) or the full-length 1.2Kb cHS4 insulator (LCR-β-GFP-I) in the U3 deletion of the 3apostrope;LTR are shown. A SIN-β vector, devoid of LCR element (β-GFP LV) was used as a control vector.
Experimental design of the IVIM assay. Lineage-negative cells were isolated from the bone marrow of C57BL6 mice, prestimulated and transduced with GV and LV twice at an MOI of 20. For GV transduction, lineage-negative cells were prestimulated for two days and transduced for two days, and at day 5, cells were expanded in bulk for 2 weeks. For LV transductions, lineage-negative cells were prestimulated overnight before transduction, and transduced twice 12 hours apart. Limiting dilution analysis was done on day 19 at 100 cells/well in a 96-well plate for all vectors. Two weeks later, wells which were nearly full of cells were scored as positive and expanded further for three weeks in 24-well plates for molecular analysis. Limiting dilution analysis showed a minimum of 10 cells/well with GV gave rise to IVIM clones, and 100 cells/well for LCR-LV gave rise to IVIM clones.
Surface phenotype of IVIM mutant clones. A. Representative FACS plots for one clone are shown for Sca1+, cKit+, B220 and Gr-1. Each clones was also analyzed for CD11b, TER-119 and CD41 surface expression. B. Quantification of lineage marker expression is shown for all the LCR-LV clones. The clones expressed high levels of Sca1 and cKit+. Varying levels of CD11b expression was observed. Erythroid marker TER-119 expression was negligible, very low levels of Gr-1 and B220 expression was observed. There was a significant decrease in the expansion potential of the clones with insulators. Clonal phenotype did not differ significantly between LCR-β-GFP uninsulated vector and the vectors with insulator elements in the SIN deletion.