Abstract
Current anti-HIV-1 strategies reduce replication through targeting of viral proteins and RNA; meanwhile, targeting at the level of the integrated provirus has been less explored. We show here that mobilization-competent vectors containing small noncoding RNAs targeted to transcriptionally active regions of the human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR) can take advantage of integrated virus and modulate HIV-1 replication. Transcriptional silencing of HIV-1 correlates with an increase in silent-state epigenetic marks including histone and DNA methylation, a loss of nuclear factor-κB (NF-κB) recruitment, and requires Argonaute 1 (Ago-1), histone deacetylase 1 (HDAC-1), and DNA methyltransferase 3a (DNMT3a) localization to the LTR. Long-term suppression of the virus was observed for 1 month with no evidence of viral resistance. These data show that RNA-directed transcriptional silencing of HIV-1 can be delivered by a mobilization-competent vector, suggesting that this system could be used to target long-term selective pressures on conserved promoter elements to evolve less pathogenic variants of HIV-1.
Introduction
The human immunodeficiency virus type 1 (HIV-1) is a lentivirus that causes a persistent viral infection through the ability to integrate into the genome of the infected cell. Clearance of HIV-1 by the immune system is inefficient and latently infected cells exist undetected. As a result, two areas of focus for treatment of HIV-1 have emerged: (i) prevention of HIV-1 infection through the development of a vaccine and/or (ii) therapeutic approaches which select for less pathogenic variants of HIV-1 and prolonged survival of afflicted individuals. Significant effort has been expended toward the development of a vaccine; however, progress seems to be several years, if not several decades, away from successful implementation.1,2
Genetic-based therapies represent an emerging therapeutic paradigm for the treatment of HIV-1, as they have the potential to treat patients who have developed multidrug resistance and may also represent a long-term treatment alternative to highly active antiretroviral therapy. One such emerging genetic-based therapeutic is described here and is based on utilizing small noncoding RNAs to transcriptionally regulate the activity of HIV-1.
Small noncoding RNAs have been shown, when targeted to gene promoter loci, to result in directed epigenetic modifications that correlate with transcriptional gene silencing (TGS) (reviewed in ref. 3). TGS is mechanistically operative through small noncoding RNA direction of silent-state epigenetic modifications including histone and DNA methylation to the homology containing targeted promoter. This form of gene silencing may provide for longer-lasting suppression of HIV-1 expression than RNA interference (RNAi)-based post-TGS methodologies, as epigenetic changes are involved in the silencing instead of mRNA slicing (reviewed in ref. 4). RNA-mediated TGS may represent a therapeutic that places an indirect (chromatin remodeling) pressure on the virus and as such may prove more efficacious at suppressing viral replication while avoiding the emergence of resistant mutants.5,6,7
RNA-based therapeutics are proving to be extremely potent with regards to suppressing HIV-1, but the issue of delivery to target cells remains enigmatic.4 Mobilization-competent vectors represent a genetic-based therapeutic approach to deliver RNA-based modalities to those cells infected and targeted by HIV-1 (refs 8,9) Importantly, mobilization-competent vectors depend on HIV-1 for their expression, packaging, and spread to new cells.4 To date, there have been no observations concerning the spread and antiviral activity of HIV-1-mobilized lentiviral vectors carrying RNA modalities which target HIV-1 in a TGS fashion.
To more thoroughly determine the efficacy of small RNA-directed TGS to suppress HIV-1 replication from the context of a mobilization-competent vector, we generated HIV-2-based vectors with various U6 expressed antisense RNAs targeted to the long terminal repeat (LTR)/promoter of HIV-1. Data presented here show that mobilization-competent vectors can deliver small RNAs which are capable of modulating transcriptional silencing of the HIV-1 LTR. We show that the vector-mediated TGS occurs via epigenetic modifications that ultimately remodel the local chromatin in a manner that impedes nuclear factor-κB (NF-κB) recruitment and hinders viral transcription. Importantly, HIV-2 vectors containing small antisense RNAs targeting HIV-1 in a TGS manner retain mobilization and antiviral efficacy over time.
Results
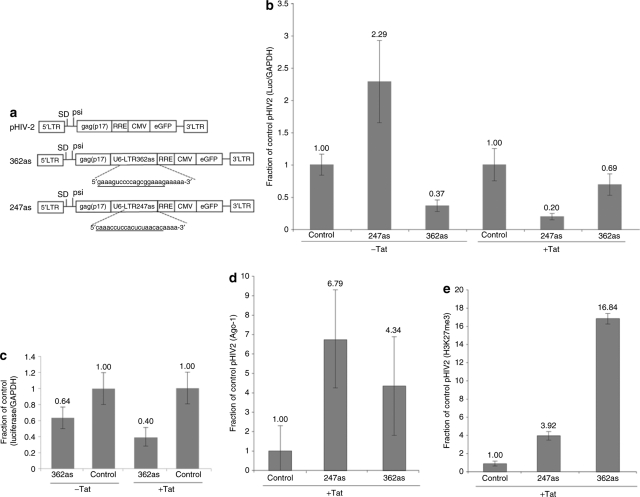
There has been little investigation into the ability to maintain vector mobilization and antiviral potential of small noncoding RNAs expressed by mobilization-competent vectors. To this effect, we introduced several small, noncoding, antisense RNAs previously shown to suppress HIV-1 in a transcriptional manner into the pHIV2 vector (Figure 1a).10 The mobilization-competent vector pHIV2 has previously been shown to mobilize in the presence of HIV-1 and the degree of mobilization is directly linked to the number of anti-HIV-1 ribozymes within the vector.9 HIV-1 has been shown to be susceptible to RNA-mediated TGS when siRNAs or antisense RNAs are generated to specifically target the LTR.10,11 To determine the ability of the various anti-HIV-1 RNAs to modulate LTR transcriptional activity from the pHIV2 vector, we co-transfected the various mobilization-competent vectors (Figure 1a) with or without an HIV-1 Tat expression plasmid in the HeLa-based TZM-bl cell line, in which luciferase expression is controlled by an integrated HIV-1 LTR (Supplementary Materials and Methods).12 In the presence of Tat-mediated activity, the anti-HIV-1 RNAs were capable of suppressing LTR transcriptional activity. Interestingly, only the LTR-362as expressing mobilization-competent vector (362as) was capable of suppressing basal-level LTR expression of luciferase in the absence of HIV-1 Tat (Figure 1b). This observed suppression by 362as was transcriptional in nature as determined by nuclear run-on analysis (Figure 1c, Supplementary Materials and Methods). It is unclear why in the absence of HIV-1 Tat that 247as modulated an increase in LTR activity. A possible explanation for this observation is that in the absence of suitable Tat-induced substrate, 247as mediates an off-target effect against the noncoding RNA C10orf76 resulting in cell-wide gene activation.13 Another alternative is the observation that 247as, but not 362as, may target a regulatory antisense transcript, which overlaps the 3′LTR specifically at the 247as but not the 362as targeted loci.14
Figure 1.
Mobilization-competent vector-mediated transcriptional silencing of HIV-1 LTR expression. (a) HIV-2 mobilization-competent vectors were generated to contain various small RNAs specifically targeted to the LTR/promoter regions of HIV-1b (Supplementary Materials and Methods).10 (b) Transcriptional silencing of LTR-expressed luciferase. TZM-bl cells were transfected with the respective pHIV2 vectors +/− Tat. The results from triplicate-treated cultures are shown with the standard deviations. (c) Nuclear run-on analysis of TZM-bl cells treated with either 362as or pHIV2 (control) +/− Tat. The results from triplicate RT-PCR reactions are shown with the standard errors of the mean. (d,e) Chromatin immunoprecipitation assays were performed on TZM-bl cells treated with the vectors in the presence of Tat for (d) Ago-1 and (e) H3K27me3. The results from triplicate-treated cultures are shown with the standard deviations.
In human cells, small noncoding RNAs designed to target gene promoter regions can modulate TGS.10,11,15,16,17,18,19,20,21,22,23,24 Small RNA-directed TGS requires Argonaute 1 and may also require Argonaute 2 (Ago-1 and Ago-2, respectively),17,19 and a low-copy promoter-associated RNA at the targeted loci.16,25 The resulting small noncoding RNA-targeted promoter exhibits a reduction in transcriptional activity20 that correlates with the markers of silenced chromatin, specifically histone H3 lysine-9 dimethylation (H3K9me2) and histone H3 lysine-27 tri-methylation (H3K27me3) (reviewed in refs 3,26). To determine whether the various anti-HIV-1 small noncoding RNAs, 247as or 362as, were modulating TGS in a manner similar to previous observations, chromatin immunoprecipitation (ChIP) assays were performed. Both the 247as and 362as expressing mobilization-competent vectors were capable of modulating a robust increase in Ago-1 (Figure 1d) and H3K27me3 (Figure 1e) enrichment at the targeted LTR, similar to previous observations with different small RNA-targeted gene promoters.10,16,17,19 These data suggest that the observed silencing modulated by 247as and 362as is operative via a TGS-based mechanism.
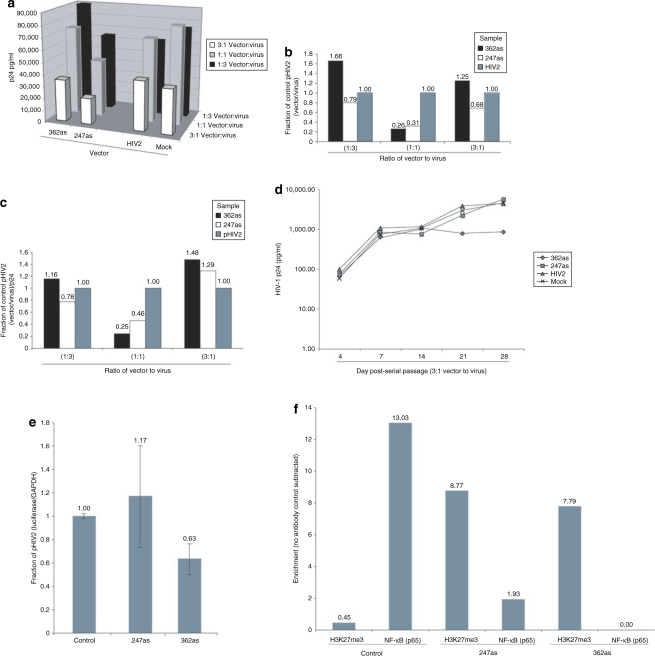
While TZM-bl cells appear susceptible to the effects of both 247as and 362as, the ability of the respective anti-HIV-1 mobilization-competent HIV-2 vectors to target HIV-1 in the context of a virally infected cell remains unknown. To determine the efficacy of the various mobilization-competent HIV-2 vectors (Figure 1a) to suppress HIV-1, co-transfection experiments that varied the amount of vector to virus DNA transfected were performed with the HIV-1 molecular clone HX10 (ref. 27). Similar to previous observations,10 both the 247as and 362as containing mobilization-competent HIV-2 vectors exhibited suppression of HIV-1 p24 production relative to mock and the pHIV2-vector treated cultures (Figure 2a). Of note, the greatest observable silencing correlated with those ratios of vector to virus which contained the greatest amount of vector, i.e., the 3:1 ratios (Figure 2a).
Figure 2.
The effects of vector treatment on HIV-1b expression. (a) Lentiviral vectors were co-transfected with the HIV-1 molecular clone HX10 in triplicate at varying ratios. Culture supernatants were collected, pooled together, and assayed by enzyme-linked immunosorbent assay for p24 expression. The effects of varied ratios of vector or virus on vector mobilization were determined. (b) Shed viral particles from the co-transfections were assessed by qRT-PCR for either virus or vector. Ratios of either (b) copies of vector/virus or (c) vector/virus ratios standardized to HIV-1 p24 pg/ml are shown. (d) Supernatants from 3:1 vector to virus ratio transfections were used to infect Jurkat cells, and a p24 analysis was carried out over the month-long serial passage. (e) Supernatants from day 28 of the serial passage were used to infect TZM-bl cells and luciferase expression determined 48 hours later. The results from triplicate-treated cultures are shown with the standard deviations. (f) Supernatants from day 28 post-serial passage were passaged to TZM-bl cells and a ChIP assay was performed for H3K27me3 or NF-κB (p65) enrichment, specifically at the targeted LTR. The results from a single experiment are shown.
Previous observations suggested that the mobilization frequency of the respective HIV-2 vector correlated directly with the number of anti-HIV-1-specific ribozymes in the vector, with those vectors containing the greatest number of ribozymes failing to effectively mobilize.9 Ribozymes have the potential to target not only the HIV-1 genome but the mRNA of the vector as well (reviewed in ref. 28), possibly explaining the loss of mobilization in vectors containing greater numbers of anti-HIV-1 ribozymes. However, it has remained unknown whether the same conditions apply to other forms of small RNA-mediated targeting. To determine to what extent the various mobilization-competent vectors can be spread by HIV-1, we measured vector RNA to virus RNA ratios in shed particles following co-transfections with the same vector to virus ratios performed previously. Interestingly, when the vector to virus ratios were assessed in the supernatants 4 days following the co-transfection, the 1:3 vector to virus samples appear to contain the greatest amount of vector (Figure 2b). However, when these numbers were further standardized to p24 to account for the differential output between samples, the 362as and 247as vectors showed increased packaging in both the 1:3 and 3:1 vector to virus samples (Figure 2c). These data indicate that packaging efficiency was not affected by the amount of antisense RNA present; rather, packaging of the vectors was increased when an excess of wild-type virus was present (1:3) or when an excess of the vector genome was present (3:1), indicating simple availability and concentration of either the packaging elements or vector genome was responsible for the differential levels (Figure 2b,c).
Previous experiments carried out with mobilization-competent HIV-2 vectors did not assess the ability of the mobilization-competent vectors to mobilize >1 passage and observed an overall loss of antiviral potential after 1 serial passage.9,29 The loss of antiviral efficacy in the previously tested mobilization-competent vectors may have been the result of the relatively inefficient activity of the anti-HIV-1 ribozymes and self-targeting of the vector.9 To determine the ability of the 247as or 362as vector to modulate virus infection, long-term serial passage of the vector/virus containing supernatants (Figure 2a) was carried out for 4 weeks in Jurkat cells. Relative to the control pHIV2-treated cultures, only the 362as vector was capable of stably reducing HIV-1 expression as determined by p24 expression (Figure 2d).
The observed long-term suppression of HIV replication by the 362as vector (Figure 2d) is hypothesized to be the result of 362as targeting to the LTR. To determine the ability of 362as to maintain LTR targeting following 28 days of serial passage, culture supernatants were collected from day 28 (Figure 2d), standardized to p24, exposed to TZM-bl indicator cells, and 48 hours later, total RNA was isolated and luciferase expression was determined relative to GAPDH. Interestingly, the 362as-treated cultures demonstrated a reduction in luciferase expression whereas the 247as and control-treated cultures did not (Figure 2e). Moreover, the observed suppression of LTR activity correlated with directed histone methylation and a loss of active forms of the transcription factor NF-κB at the target loci in cultures exposed to the same passaged vector/virus containing supernatants from day 28 (Figure 2f). Interestingly, the 247as vector containing supernatants mediated an enrichment of histone methylation, but failed to suppress NF-κB localization to the LTR (Figure 2f). It should be noted that the primers used in the ChIP assays are nonspecific and do not distinguish between the 5′ or 3′ LTRs, nor between the TZM-bl LTR-Luc construct or any integrated wild-type HIV-1 introduced on inoculation with the day 28 samples. These data suggest that the 362as mobilization-competent vector continues to functionally suppress LTR activity following several passages with HIV-1 and that NF-κB enrichment at the target loci is an important requirement for viral production, possibly explaining the reduction in p24 expression at day 28 post-serial passage in the 362as-treated cultures but not the 247as cultures (Figure 2d).
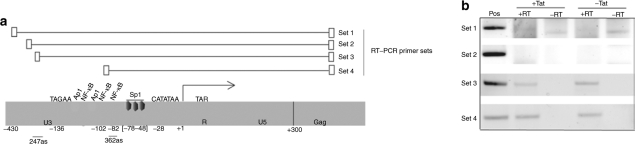
Both the 362as and 247as vectors appear to be retaining some level of LTR targeting during the month-long serial passage; however, only 362as was effective in also reducing viral output as measured by p24. Given these observations along with the previous observation that the 247as vector demonstrated suppression of LTR activity in the presence of HIV-1 Tat while the 362as vector demonstrated suppression +/− Tat (Figure 1b), we chose to further investigate the molecular mechanism that might explain the differential activity of these small antisense RNAs. These observations might be explained by differential transcription across the LTR in the TZM-bl cells, as transcription across the target site is required for TGS in human cells.16,25 To determine the extent of upstream transcription spanning the LTR, an RT-PCR screen was carried out using four different sets of primers spanning different regions of the LTR (Figure 3a, Supplementary Materials and Methods). In this screen, TZM-bl cells were treated with or without HIV-1 Tat and then cellular mRNA assessed by RT-PCR for transcription across various regions of the HIV-1 LTR. Interestingly, transcription appears to be present in the absence of Tat and to initiate upstream of the 247as target site, thus encompassing both the 247as and 362as targeted loci (Figure 3b). Similar observations of basal-level transcription at the HIV-1 LTR in the absence of Tat have been detected previously by others.30 Overall, these data suggest that in the context of TZM-bl cells the HIV-1 LTR is actively transcribed in the absence of Tat from the upstream TAGAA30 (Figure 3a), suggesting that the differential activity of 362as relative to 247as is not the result of LTR transcription but rather the target loci.
Figure 3.
Transcriptional analysis of HIV-1 LTR in TZM-bl cells. (a) The LTR from prototype HIV-1 virus LAI is shown schematically with the various primer sets used to assess transcriptional activity across the LTR. (b) TZM-bl cells were transfected with either pTatDsRed (+Tat) or with the control pcDNA3.1(+) (−Tat). The cultures were collected 48 hours later and cellular mRNA collected and assessed using the various primer sets (1–4) to determine relative transcription across the 5′ LTR. Sets 1–4 are shown from either RT converted (+RT) or cellular mRNA alone (−RT).
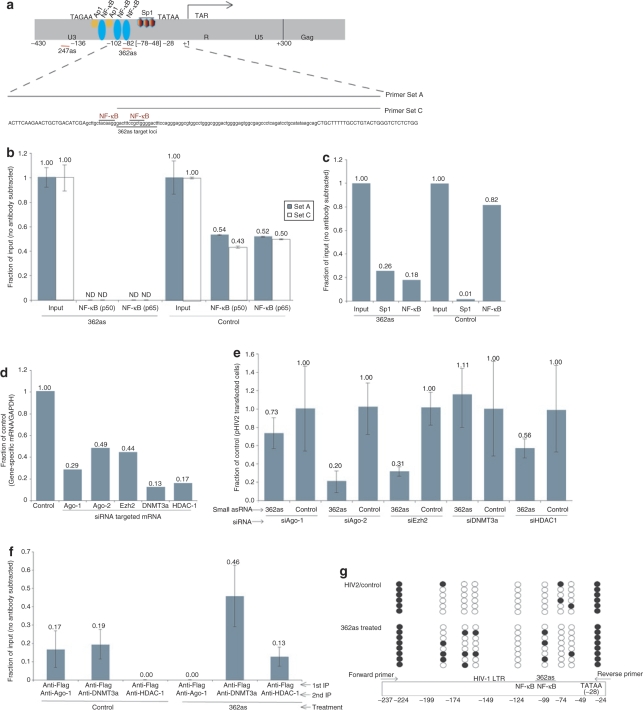
The various mobilization-competent vectors exhibit differing levels of LTR regulation in TZM-bl cells. While both 247as and 362as direct epigenetic remodeling components to the LTR, the 362as target loci overlaps a significant portion of one of the NF-κB sites closest to the transcriptional start site (Figure 4a). The observed suppressive activity of 362as might in part be the result of directing silent-state epigenetic marks to this loci that result in an inability of NF-κB to be recruited to the LTR, thus impeding LTR activity. To determine what effect 362as targeting may have on NF-κB localization, cultures were transfected with the 362as or control pHIV2 vectors and ChIP assays performed 48 hours later. Treatment with 362as resulted in a loss of detectable NF-κB at both NF-κB-binding sites in the LTR (Figure 4b). When the SP1 site was assessed by ChIP, an increase in SP1 was observed (Figure 4c). Interestingly, Sp1 have been found to be enriched at histone deacetylase 1 (HDAC-1) regions undergoing transcriptional suppression.31,32 These data suggest that 362as treatment results in an epigenetic remodeling of the local chromatin toward a silent state and that this event produces an unfavorable environment for the recruitment of NF-κB to the LTR.
Figure 4.
362as treatment results in directed epigenetic changes at the HIV-1 LTR. (a) The 247as and 362as target loci are shown schematically with the target LTR. The 2 transcriptional start sites,30 TAGAA or TATAA, and 3 NF-κB, Sp1 and Ap1 transcription factor-binding sites are also shown. (b) Treatment of TZM-bl cells with 362as results in a loss of NF-κB p50 and p65 at the targeted loci whereas the control-treated HIV-2 samples do not. (c) Treatment of TZM-bl cells with 362as results in a gain of SP1 and loss of NF-κB (p65) at the targeted loci relative to the control-treated HIV-2 samples. The results of a single experiment are shown. (d) Expression levels of various protein components known to be involved in RNAi and/or epigenetic gene regulation were suppressed by RNAi. Data represents the average knockdown from cells transfected in triplicate and then pooled to isolate total RNA. (e) DNMT3a, Ago-1 and HDAC-1 are required for 362as-mediated transcriptional silencing of LTR gene expression. The averages from triplicate-treated cultures are shown with the respective standard error of the means relative to the control-treated cultures for each treatment. (f) Treatment of cultures with 362as resulted in an enrichment of DNMT3a and HDAC-1 at the targeted LTR loci. Results from duplicate treated cultures are shown with the respective ranges. (g) Treatment of cultures with 362as results in an enrichment of DNA methylation at the targeted loci relative to control-treated cultures as determined by bisulfite sequence analysis from anti-Flag (first IP)-eluted samples shown in e.
Previous observations in human cells have shown that Ago-1 is required for the initiation of TGS.17,19 Moreover, DNA methyltransferase 3a (DNMT3a) has been shown to co-immunoprecipitate with the small RNAs at the targeted promoters10 as well as to co-immunoprecipitate with Enhancer of zeste 2 (Ezh2)33 and HDAC-1.34 Recently, HDAC-1 has also been shown to be required for small RNA-directed TGS in human cells.22 Ezh2 is the histone methyltransferase involved in generating the H3K27me3 epigenetic mark which has been observed in the silencing event. To determine the requirement of these factors in the observed TGS induced by 362as treatment, we first suppressed the various proteins with previously validated siRNAs (Figure 4d) followed by treatment with 362as or control plasmids. Interestingly, only Ago-1, DNMT3a, and HDAC-1 appeared to be required for TGS in this system (Figure 4e). These data suggest that 362as-mediated TGS of the LTR in TZM-bl cells requires Ago-1, HDAC-1, and DNMT3a, and further support the notion that small noncoding RNA-directed TGS in human cells is mechanistically distinct when compared to traditional Ago-2-mediated post-TGS.
To verify that indeed DNMT3a and HDAC-1 are involved and associated in 362as-mediated TGS of LTR activity, cultures were transfected with a flag-tagged DNMT3a construct and either 362as or control plasmids. After 48 hours a flag-specific immunoprecipitation assay was performed followed by a second immunoprecipitation for either DNMT3a or Ago-1 or HDAC-1 (ref. 35). This assay allows for direct analysis of two different protein components at the small noncoding RNA-targeted loci. A significant increase in both DNMT3a and HDAC-1 was observed at the LTR following 362as treatment relative to controls (Figure 4f). These data support previous observations of TGS10 and highlight the importance of DNMT3a and HDAC-1 in small noncoding RNA-mediated TGS.
To determine whether DNMT3a can direct DNA methylation at the 362 target site, DNA isolated from the flag-specific immunoprecipitation described above was bisulfate treated to examine methylation at the CpG motif flanked by the two NF-κB sites. Methylation-specific PCR primers were used to amplify a fragment of the LTR surrounding the 362as target site and the PCR product was cloned and sequenced. Of eight clones isolated and sequenced from 362as-treated cellular DNA, three showed methylation at the 362as target site (Figure 4g). In comparison, none of the six clones sequenced from the control-treated cellular DNA showed methylation at the 362as target site. There was also a noted enrichment of methylation at other CpG motifs in the 362as-treated cells as compared to the control cells (Figure 4g).
Discussion
RNAi has emerged as a potent antiviral agent which can exhibit specific targeted suppression of HIV-1 (ref. 36). However, traditional RNAi-based approaches have been designed to directly target the viral transcript in a post-transcriptional manner, invariably selecting for RNAi escape variants.5,6 Consequently, for an effective RNAi-based approach to treat HIV-1 infection, multiple sites and/or conserved regions of the viral genome will need to be targeted for successful suppression of replication.
Previous observations have defined the target potential of conserved regions in the LTR which are susceptible to small noncoding RNA-mediated TGS.10,11,37,38 We show here that (i) long-term suppression of HIV-1 p24 expression can be obtained by a small noncoding RNA when partially targeted to NF-κB loci in the LTR, and (ii) that HIV-2-based mobilization-competent vectors can be employed to deliver small noncoding RNAs capable of modulating TGS by commandeering HIV-1 to co-package both the vector and wild-type virus genomes. The data generated here also suggest that targeting the promoter/LTR of HIV-1 with small noncoding RNAs in a transcriptional manner employs a different mechanism from RNAi-based post-TGS, specifically through directed epigenetic modifications that involve the activity of HDAC-1, Ago-1, and DNMT3a. A direct link among DNMT3a enrichment at the LTR and increased DNA methylation at the LTR and 362as target site was also observed. The role of DNA methylation in HIV-1 LTR activity when assessed in more biologically relevant peripheral blood mononuclear cells remains unclear;39 however, methylation has previously been implicated in transcriptional regulation of the HIV-1 LTR in vitro,40,41,42,43 and more specifically, methylation of the CpG motif located at the 362as target site has also been shown to inhibit NF-κB binding in vitro.41
The data presented here support previous observations from our group and others that the small noncoding RNA-directed transcriptional silencing complex involves HDAC-1, Ago-1, and DNMT3a.10,19,22,44 Importantly, epigenetic control of gene expression has the potential to result in longer-term, if not permanent, silencing of the targeted gene expression3 as well as to function as an indirect viral targeted approach.
Mobilization-competent vectors alone (devoid of an antiviral therapy) have been shown to act in a parasitic manner and alter HIV replication, likely acting through the sequestering of viral proteins needed for replication.8,45,46,47 Here, we observe that addition of a small noncoding RNA-mediated TGS cassette reduces viral replication as compared to the empty vector, placing selective pressure upon HIV-1 through both co-packaging and the small noncoding RNA-directed epigenetic changes to the LTR (Figure 5). It remains to be seen whether viral resistance can or will emerge after long-term targeting of epigenetic changes to the LTR by 362as and how this might affect the NF-κB binding site and/or localization. Importantly, others have also found that the NF-κB site in the HIV-1 LTR is susceptible to small RNA-directed TGS and serves as a good target for control of viral transcriptional activity.22 The use of mobilization-competent vectors to deliver small noncoding RNAs that function to transcriptionally silence HIV-1 may allow for the selection of less pathogenic variants of HIV-1. As the small noncoding RNA can place a selective pressure on the conserved promoter elements, mutations within these elements may result in escape variants with reduced fitness. The evolution of HIV-1 under selective pressure through targeting of conserved promoter elements and the more indirect pressure of induced chromatin remodeling at the LTR remains to be examined. Furthermore, the idea that the parasitic nature of the vector, whereby a co-infection of two selfish organisms occurs, could potentially result in reduced fitness for both the vector and HIV-1 (the Prisoner's Dilemma48) remains to be determined. Therefore, it may be possible to steer the evolutionary direction of HIV-1 through control of HIV-1 gene expression or improvement of the selective advantage of the vector in combination with current antiretroviral drugs.
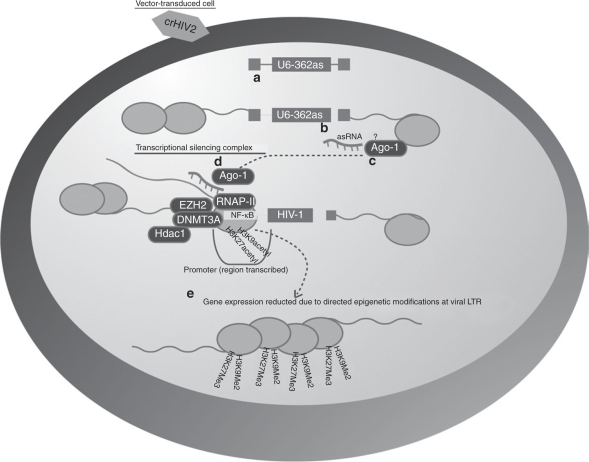
Figure 5.
Model for 362as-mediated transcriptional control of HIV-1 gene expression. (a) Once integrated into the genome of the target cell, the mobilization-competent vector can (b) express a small noncoding RNA which may then (c) interact directly with Ago-1 and localize to the elongating transcript (d) which contains the target loci for the small noncoding RNA. The small noncoding RNA and Ago-1 might also localize at the targeted site with DNMT3a, HDAC-1, and possibly Ezh2. The result of this complex localizing at the targeted promoter is histone, DNA methylation, and transcriptional gene silencing.
Materials and Methods
ChIP assays H3K27 and Ago-1. TZM-bl cells (~2 × 106) were co-transfected with HIV-2-based vectors pHIV2 (control), 247as, or 362as (1.8 µg) and the HIV-1 Tat expression plasmid pTatDSred Tat (0.2 µg) (a gift from J.J. Rossi, the Beckman Research Institute at the City of Hope, Durate, CA). The cultures were collected 72 hours later and ChIP assay performed for trimethyl-Histone H3 (Lys27) (Upstate, Lake Placid, NY) or Ago-1 (Upstate) as described.10,19 The NF-κB and Sp1-specific ChIPs consisted of TZM-bl cells (4 × 106) transfected with 4 µg of 362as or pHIV2 (control). Cells were collected 48 hours later and ChIP assays preformed for using antibodies against NF-κB p50 (Invitrogen, Carlsbad, CA), p65 (Invitrogen), or Sp1 (Abcam, Cambridge, MA).
Sequential ChIPs. TZM-bl cells (4 × 106) were transfected with 4 µg of 362as or pHIV2 (control). Cells were collected 48 hours later and a sequential ChIP was performed as previously described35 with only one aspect altered, as the immunoprecipated complexes isolated from the first antibody incubation were diluted in lysis buffer rather than elution buffer. The initial ChIP was performed using an anti-Flag antibody (Invitrogen), followed by secondary immunoprecipitations using anti-Ago-1 (Upstate), HDAC-1 (Abcam), and DNMT3a (ProSci, San Diego, CA) antibodies.
DNA methylation analysis. DNA isolated from the initial Flag-DNMT3A IP in the sequential ChIP was bisulfate treated using the EZ DNA methylation kit (Zymo Research, Orange, CA) to analyze DNA methylation at the LTR. Methylation-specific PCR (Platinum Taq Supermix; Invitrogen) was performed using the following primers LTR methylated forward 5′-GTT TGT ATG GGA TGG ATG ATT C-3′ and LTR methylated reverse 5′-TAT ATA CAA AAT CTA AAA ACT CGC C-3′ (IDT, Coralville, IA) designed using MethPrimer49 to yield a 214 bp product. The resulting PCR products were gel purified using the Minielute Gel Purification Kit (Qiagen, Valencia, CA) and cloned into pCR2.1-TOPO (Invitrogen). Inserts were sequenced and then analyzed for methylation at all of the CpG sites found within the amplicon.
Vector/virus co-transfections. The 362as, 274as, and pHIV2 (Control) vector plasmids were transfected into HEK293T cells with the HIV-1 proviral clone pHX10 at various vector to virus ratios. HEK293T cells (~5 × 105) were transfected in triplicate using Lipofectamine 2000 (Invitrogen) with the following vector to virus ratios: 1:3 (250 ng vector: 750 ng virus), 1:1 (500 ng vector: 500 ng virus), and 3:1 (750 ng vector: 250 ng virus). In addition, mock transfections were also performed in triplicate using 250 ng, 500 ng, and 750 ng of pHX10 plasmid for the 1:3, 1:1, and 3:1 ratios respectively, and 72 hours after transfection, the supernatants from the triplicate transfections were pooled and centrifuged at 3,000g to remove cellular debris. Cleared supernatants were sterile filtered through a 0.45 µm filter (Millipore, Billerica, MA) and stored at −80 °C. For isolation of viral/vector RNA, 1 ml of filtered supernatant was concentrated by centrifugation at 20,800g for 1 hour. Viral RNA was isolated as described below and viral replication was assayed by p24 enzyme-linked immunosorbent assay.
Jurkat serial passage. Jurkat cells (5 × 105) were incubated with filtered supernatants from the 3:1 ratio vector/virus co-transfection for 362as, 274as, pHIV2 (control), and Mock. These represented the optimal vector to virus RNA packaged for the respective vectors. Initial infection was standardized at 1,000 pg p24 for each sample. Cells were split 2:3 at day 4 and cleared supernatants and cell pellets were collected at day 7. On day 7, 200 µl of the supernatant from each culture was used to infect 5 × 105 newly plated Jurkat cells. Cells were again split after 4 days and samples collected after day 7. This procedure was repeated for 28 days to complete a month-long passage. Viral replication was assayed by p24 enzyme-linked immunosorbent assay RETROtek HIV-1 p24 antigen enzyme-linked immunosorbent assay (ZeptoMetrix, Buffalo, NY).
RNA isolation and analysis. Viral/vector RNA and total cellular RNA were isolated according to manufacturer's instructions using the QIAamp Viral RNA Mini kit and the RNeasy Mini kit, respectively, automated by the Qiacube (Qiagen). All RNA samples subject to quantitative RT-PCR (qRT-PCR) (viral, vector, and total cellular RNA) were prepared according to the following procedure. Isolated RNA in nuclease-free water was DNase treated using Turbo DNA-free (Ambion, Austin, TX) according to manufacturer's instructions for 30 minutes at 37 °C. Following treatment, samples were standardized and subject to reverse transcription (RT)-PCR using the iScript cDNA synthesis kit (BioRad, Hercules, CA) according to instructions. Controls not subject to RT did not receive the RT enzyme. All qRT-PCR was carried out using an Eppendorf Mastercycler ep Realplex. The qRT-PCR was carried out using SYBR GreenER qPCR SuperMix (Invitrogen) and the following primers to detect and amplify the respective DNA during PCR: luciferase forward: 5′-CCT GGA ACA ATT GCT TTT AC-3′; luciferase reverse: 5′-GTT TCA TAG CTT CTG CCA AC-3′; GAPDH forward: 5′-AGG GGT CAT TGA TGG CAA CAA TAT CCA-3′; GAPDH reverse: 5′-TTA CCA GAG TTA AAA GCA GCC CTG GTG-3′; GFP forward (vector): 5′-AGC AAA GAC CCC AAC GAG AA-3′; GFP reverse (vector): 5′-GGC GGC GGT CAC GAA-3′; HIV-1 forward: 5′-TGA GAC AAC ATC TGT TGA GGT GGG-3′; HIV-1 reverse: 5′-GGC TGT ACT GTC CAT TTA TCA GGA-3′; HIV-1 LTR (ChIP) forward: 5′-CAC ACA AGG CTA CTT CCC TGA-3′; and HIV-1 LTR (ChIP) reverse: 5′-GGC CAT GTG ATG AAA TGC TA-3′. Thermal-cycling parameters started with 2 minutes at 50 °C, 8 minutes at 95 °C, followed by 40 cycles of 95 °C for 15 seconds, 55 °C for 15 seconds and 68 °C for 15 seconds. Specificity of the PCR products was verified by melting-curve analysis.
Knockdown of RNAi and epigenetic pathways. siRNAs (IDT) against the various epigenetic or RNAi-related factors: Ago-1 5′CGUUGCCA AUGGGCAGUGCUU3′, Ago-2 is described in,50 Ezh2 5′-UAA GAU UUC CGU UCU UUC CUU-3′, DNMT3a 5′-GAA CUC AAA GAA GAG CCG GUU-3′, HDAC-1 5′-UGG UUC AAA GUU AAG AAC GUU-3′, and control R854 5′-AAU UCU UUG GCC UGA AUA AAA-3′ were initially validated by transfecting 293T cells in triplicate with 50 nmol/l of each siRNA (Lipofectamine RNAiMax; Invitrogen). Cells from the triplicate transfections were pooled together, and the total RNA was isolated as described above from these samples. RNA was subject to DNase treatment and cDNA conversion as previously described, followed by qRT-PCR using the following primers (IDT) Ago-1 forward 5′-GCA CTG CCC ATT GGC AAC GAA-3′, Ago-1 reverse 5′- CAT TCG CCA GCT CAC AAT GGC T-3′, Ago-2 forward 5′-GGC CCA GTA TTT CAA GGA CA-3′, Ago-2 reverse 5′-TTT CTG CTC CTG TCC GAC TT-3′, DNMT3A forward 5′-GCC TCA ATG TTA CCC TGG AA-3′, DNMT3A reverse 5′-CAG CAG ATG GTG CAG TAG GA-3′, HDAC-1 forward 5′-AGC CTA GTG CGG TGG TCT TA-3′, HDAC-1 reverse 5′-TGA AGC AAC CTA ACC GAT CC-3′, Ezh2 forward 5′-TTC ATG CAA CAC CCA ACA CT-3′, Ezh2 reverse 5′-CTC CCT CCA AAT GCT GGT AA-3′, CCR5 forward 5′-CTG CCT CCG CTC TAC TCA CT-3′, and CCR5 reverse 5′-GCT CTT CAG CCT TTT GCA GT-3′ for the siRNA target. Expression was standardized to GAPDH. To assess the role of the various RNAi and epigenetic factors in the observed TGS, TZM-bl cells (~2 × 105) were transfected with the siRNAs targeted to the various epigenetic-related or RNAi-related factors (50 nmol/l, Lipofectamine RNAiMax; Invitrogen). Six hours later, the siRNA-treated cells were again transfected with either 362as or control (HIV-2) vectors (0.2 µg). Cellular mRNA was isolated 48 hours later and assessed for luciferase mRNA expression relative to GAPDH. The averages from triplicate-treated cultures are shown with the respective standard error of the means relative to the control-treated cultures for each treatment.
Supplementary MaterialMaterials and Methods.
Supplementary Material
Acknowledgments
This project is funded by R01 HL083473-02 and UCSD Center for AIDS Research (NIAID 5 P30 AI36214) to K.V.M. as well as the Stein Endowment Fund at The Scripps Research Institute.
REFERENCES
- Klausner RD, Fauci AS, Corey L, Nabel GJ, Gayle H, Berkley S, et al. Medicine. The need for a global HIV vaccine enterprise. Science. 2003;300:2036–2039. doi: 10.1126/science.1086916. [DOI] [PubMed] [Google Scholar]
- Zinkernagel RM.The challenges of an HIV vaccine enterprise Science 20043031294–1297.author reply [DOI] [PubMed] [Google Scholar]
- Hawkins P., and , Morris KV. RNA and transcriptional modulation of gene expression. Cell Cycle. 2007;7:602–607. doi: 10.4161/cc.7.5.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV.Therapeutic potential of siRNA-mediated transcriptional gene silencing Biotechniques 20067–13.Suppl [DOI] [PubMed]
- Boden D, Pusch O, Lee F, Tucker L., and , Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, et al. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhout EM, Ooms M, Vink M, Das AT., and , Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukovsky AA, Song J., and , Naldini L. Interaction of human immunodeficiency virus-derived vectors with wild-type virus in transduced cells. J Virol. 1999;73:7987–7092. doi: 10.1128/jvi.73.8.7087-7092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV., and , Looney DJ. Characterization of human immunodeficiency virus (HIV)-2 vector mobilization by HIV-1. Hum Gene Ther. 2005;16:1463–1472. doi: 10.1089/hum.2005.16.1463. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen Z, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2005;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D., and , Kelleher A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J RNAi Gene Silencing. 2005;1:66–78. [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B., and , Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg MS, Barichievy S, Schaffer L, Han J., and , Morris KV. An RNA targeted to the HIV-1 LTR promoter modulates indiscriminate off-target gene activation. Nucleic Acids Res. 2007;35:7303–7312. doi: 10.1093/nar/gkm847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S, Halin M, Lefort S, Audet B, Vaquero C, Mesnard JM, et al. Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology. 2007;4:71–87. doi: 10.1186/1742-4690-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Tommasi S, Li M, Li H, Yanow S, Pfeifer GP, et al. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol Ther. 2005;12:179–183. doi: 10.1016/j.ymthe.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Han J, Kim D., and , Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D, et al. Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nat Chem Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV., and , Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE., and , Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Pulukuri SM., and , Rao JS. Small interfering RNA directed reversal of urokinase plasminogen activator demethylation inhibits prostate tumor growth and metastasis. Cancer Res. 2007;67:6637–6646. doi: 10.1158/0008-5472.CAN-07-0751. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA, et al. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem. 2008;283:23353–23363. doi: 10.1074/jbc.M709651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AH, Schuebel KE, Herman JG., and , Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Feng Y, Pan L, Wang Y, Xu X, Lu J, et al. The proximal GC-rich region of p16(INK4a) gene promoter plays a role in its transcriptional regulation. Mol Cell Biochem. 2007;301:259–266. doi: 10.1007/s11010-007-9427-4. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV. siRNA-mediated transcriptional gene silencing: the potential mechanism and a possible role in the histone code. Cell Mol Life Sci. 2005;62:3057–3066. doi: 10.1007/s00018-005-5182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner L, Fisher A, Jagodzinski L, Mitsuya H, Liou RS, Gallo RC, et al. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retroviruses. 1987;3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- Morris KV., and , Rossi JJ. Anti-HIV-1 gene expressing lentiviral vectors as an adjunctive therapy for HIV-1 infection. Curr HIV Res. 2004;2:185–191. doi: 10.2174/1570162043484906. [DOI] [PubMed] [Google Scholar]
- Morris KV, Grahn RA, Looney DJ., and , Pedersen NC. Characterization of a mobilization-competent simian immunodeficiency virus (SIV) vector containing a ribozyme against SIV polymerase. J Gen Virol. 2004;85:1489–1496. doi: 10.1099/vir.0.19106-0. [DOI] [PubMed] [Google Scholar]
- Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K., and , Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Lee JH, Park JG., and , Lee YI. Trichostatin A, a histone deacetylase inhibitor, activates the IGFBP-3 promoter by upregulating Sp1 activity in hepatoma cells: alteration of the Sp1/Sp3/HDAC1 multiprotein complex. Biochem Biophys Res Commun. 2002;296:1005–1012. doi: 10.1016/s0006-291x(02)02001-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y., and , Dufau ML. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J Biol Chem. 2002;277:33431–33438. doi: 10.1074/jbc.M204417200. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2005;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Godin N, Kasai M., and , Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S, Chen L-F, Kwon H, Ruiz-Jarabo C, Verdin E., and , Greene W. NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Chung C, Witke W., and , Looney DJ. Inhibition of HIV-1 Replication by siRNA Targeting Conserved Regions of gag/pol. RNA Biology. 2004;1:114–117. doi: 10.4161/rna.2.1.1198. [DOI] [PubMed] [Google Scholar]
- Omoto S., and , Fujii YR. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol. 2005;86:751–755. doi: 10.1099/vir.0.80449-0. [DOI] [PubMed] [Google Scholar]
- Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, et al. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pion M, Jordan A, Biancotto A, Dequiedt F, Gondois-Rey F, Rondeau S, et al. Transcriptional suppression of in vitro-integrated human immunodeficiency virus type 1 does not correlate with proviral DNA methylation. J Virol. 2003;77:4025–4032. doi: 10.1128/JVI.77.7.4025-4032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarik DP, Cook JA., and , Pitha PM. Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. EMBO J. 1990;9:1157–1164. doi: 10.1002/j.1460-2075.1990.tb08222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarik DP, Duckett C, Kim SU, Perez VL, Griffis K, Guenthner PC, et al. DNA CpG methylation inhibits binding of NF-κB proteins to the HIV-1 long terminal repeat cognate DNA motifs. New Biol. 1991;3:969–976. [PubMed] [Google Scholar]
- Bednarik DP, Mosca JD., and , Raj NB. Methylation as a modulator of expression of human immunodeficiency virus. J Virol. 1987;61:1253–1257. doi: 10.1128/jvi.61.4.1253-1257.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Hamano A, Koiwa T., and , Watanabe T. 5′ long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology. 2006;3:69. doi: 10.1186/1742-4690-3-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S, Pisano DG., and , Serrano M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7:2601–2608. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- An DS, Morizono K, Li Q-X, Mao SH, Lu S., and , Chen ISY. An Inducible human immunodeficiency virus type 1 (HIV-1) vector which effectively suppresses HIV-1 replication. J Virol. 1999;73:7671–7677. doi: 10.1128/jvi.73.9.7671-7677.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbeau P., and , Wong-Staal F. Anti-HIV effects of HIV vectors. Virology. 1998;243:268–274. doi: 10.1006/viro.1998.9089. [DOI] [PubMed] [Google Scholar]
- Dropulic BMH, Pitha PM. A conditionally replicating HIV-1 vector interferes with wild-type HIV-1 replication and spread. Proc Natl Acad Sci USA. 1996;93:11103–11108. doi: 10.1073/pnas.93.20.11103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PE, Chao L. Prisoner's dilemma in an RNA virus. Nature. 1999;398:441–443. doi: 10.1038/18913. [DOI] [PubMed] [Google Scholar]
- Li L-C., and , Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G., and , Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.