Abstract
The human Rad17–Rfc2-5 and Rad9–Rad1–Hus1 complexes play crucial roles in the activation of the ATR-mediated DNA damage and DNA replication stress response pathways. In response to DNA damage, Rad9 is recruited to chromatin in a Rad17-dependent manner in human cells. However, the DNA structures recognized by the Rad17–Rfc2-5 complex during the damage response have not been defined. Here, we show that replication protein A (RPA) stimulates the binding of the Rad17–Rfc2-5 complex to single-stranded DNA (ssDNA), primed ssDNA, and a gapped DNA structure. Furthermore, RPA facilitates the recruitment of the Rad9–Rad1–Hus1 complex by the Rad17–Rfc2-5 complex to primed and gapped DNA structures in vitro. These findings suggest that RPA-coated ssDNA is an important part of the structures recognized by the Rad17–Rfc2-5 complex. Unlike replication factor C (RFC), which uses the 3′ primer/template junction to recruit proliferating cell nuclear antigen (PCNA), the Rad17–Rfc2-5 complex can use both the 5′ and the 3′ primer/template junctions to recruit the Rad9–Rad1–Hus1 complex, and it shows a preference for gapped DNA structures. These results explain how the Rad17–Rfc2-5 complex senses DNA damage and DNA replication stress to initiate checkpoint signaling.
The maintenance of genomic stability requires cells to duplicate and segregate their genomes precisely and coordinately. In response to DNA damage or replication stress, cells activate a complex signaling pathway to halt genome duplication and segregation, to stabilize the replication forks that encounter interference, and to promote DNA repair or apoptosis (1). Two large protein kinases, ATM and ATR, are the central components of the damage response pathway. Whereas ATM primarily responds to double-stranded DNA breaks (DSB), ATR is important for the responses to DSB as well as a variety of types of DNA damage that interfere with DNA replication (2). We recently showed that ATRIP, the regulatory partner of ATR, specifically recognizes RPA-coated single-stranded DNA (ssDNA) and thereby localizes the ATR–ATRIP complex to the sites of DNA damage (3). Independently of the ATR–ATRIP complex, the Rad17–Rfc2-5 and the Rad9–Rad1–Hus1 complexes (referred below as the Rad17 and the Rad9 complexes, respectively), two protein complexes required for the ATR-mediated response, may also function in the sensing of DNA damage (4–6). However, how these complexes recognize DNA damage and how they interact with ATR–ATRIP on DNA have not been biochemically elucidated.
The Rad17 complex is a replication factor C (RFC)-like protein complex in which Rfc1 is substituted by Rad17, a protein homologous to all five subunits of RFC (7, 8). The Rad9 complex is a ring-shape protein complex that resembles PCNA (9). During DNA replication, RFC specifically recognizes the 3′-primer/template junction and enables PCNA, the sliding clamp of DNA polymerases, to encircle DNA (10). Analogously, Rad17 is required for the recruitment of Rad9 complexes to DNA damage in vivo (4–6). A recent biochemical study (11) reported that the recombinant human Rad17 complex could recruit the Rad9 complex to nicked or gapped plasmids, the same DNA structures used by RFC to load PCNA. The Rad24 complex, the budding yeast counterpart of the human Rad17 complex, was also shown to recruit the PCNA-like Ddc1–Mec3–Rad17 complex onto gapped plasmids (12). Although these studies provided evidence that the Rad17 complex could bring the Rad9 complex to DNA in vitro, they did not reveal any difference between the DNA structure specificities of the Rad17 complex and RFC. Why the function of the Rad17 complex is regulated by DNA damage remains unanswered.
RPA-coated ssDNA is a common structure generated at the sites of DNA damage, and it plays an important role in the recruitment of the ATR–ATRIP complex to DNA lesions (3). RPA is known to facilitate the specific binding of RFC to the primer/template junction, and it stimulates the loading of PCNA by RFC in vitro (13). Whether RPA has a role in regulating the activity and/or DNA-structure specificity of the Rad17 complex is not known. In budding yeast, rfc4-2, a mutant allele of the RFC4 gene, is defective for the checkpoint responses and sensitive to hydroxyurea (HU)-induced replication blocks (14). Interestingly, the HU sensitivity of the rfc4-2 mutant can be suppressed by specific mutant alleles of the RFA1 gene (14), suggesting that RPA and the RFC-like checkpoint complex might function in concert during the checkpoint response.
In this study, we found that the yeast rfa1-t11 mutant is defective for the recruitment of Ddc1 to DNA damage in vivo. In vitro, human RPA stimulates the loading of the Rad17 complex onto linear ssDNA, primed templates and gapped templates. Furthermore, RPA stimulates the ability of Rad17 to recruit the Rad9 complex to primed or gapped templates. Unlike RFC, the Rad17 complex can use both the 5′ and the 3′ primer/template junctions to recruit the Rad9 complex, and it exhibits a clear preference to the DNA templates with single-stranded gaps. The unique DNA-structure specificity of the Rad17 complex might explain how it recognizes various types of DNA damage in vivo. These findings suggest that RPA is not only important for the recruitment of the ATR–ATRIP complex to DNA damage, but is also critical for damage recognition by the Rad17 complex.
Materials and Methods
Yeast Strains. The yeast strain carrying wild-type RFA1, DDC1-2HA, Gal-HO, and a single HO cleavage site (Y2326) was derived from yJK8-1 provided by D. Toczyski (University of California at San Francisco). To generate the rfa1-t11 mutant in this strain background, the promoter and a 5′ portion of the RFA1 gene was cloned into pRS406, and the rfa1-t11 mutation (G104A) was introduced by site-directed mutagenesis. The plasmid carrying the rfa1-t11 mutation was then linearized with SpeI and transformed into Y2326 cells. The rfa1-t11 mutation in the resultant yeast strain (Y2327) was verified by sequencing the PCR products derived from the RFA1 locus.
Expression and Purification of the Protein Complexes. The expression and purification of the Rad17 and Rad9 complexes were performed essentially as described (8). The human RPA was expressed in Escherichia coli and purified as described in ref. 15.
DNA Templates. To generate the DNA templates used in the DNA-binding assays, ≈5 pmols of single-stranded pCR-Script DNA was incubated with 25 pmols of various biotinylated DNA oligomers in 100 μl of annealing buffer (10 mM Tris·HCl, pH 7.5/100 mM NaCl). The mixture of circular ssDNA and oligomers was heated at 95°C for 3 min and cooled down slowly. The unannealed oligomers were then removed with Qiagen (Valencia, CA) PCR purification columns. In the experiments using preattached DNA templates, the DNA templates were incubated with Dynal (Great Neck, NY) beads coated with streptavidin for 30 min. After the incubation, the beads were washed twice with annealing buffer to remove unattached DNA.
DNA-Binding Assays for the Rad17 Complex. Approximately 0.5 pmol of ssDNA or primed ssDNA, 0.5 pmol of the purified Rad17 complex, and various amounts of RPA were incubated in 500 μl of binding buffer (40 mM Tris·HCl, pH 7.5/150 mM NaCl/10 mM MgCl2/100 μg/ml BSA/1 mM DTT/1 mM ATP/10% glycerol) at room temperature for 30 min. Subsequently, streptavidin-coated Dynal beads were added to the reactions, and incubations were continued for another 30 min. After the incubation, the beads were retrieved and washed three times with binding buffer containing 250 mM NaCl and 0.25 mM ATP. Finally, the proteins associated with the beads were released by boiling in denaturing sample buffer and analyzed by SDS/PAGE and immunoblotting.
Recruitment Assay of the Rad9 Complex. To remove the contaminating insect Rad17 complex, the Rad9 complex purified from insect cells needs to be precleared with RPA-coated gapped DNA template. Approximately 0.5 pmol of the Rad9 complex, 0.5 pmol of RPA, and 0.5 pmol of the gapped DNA template attached to beads were incubated in 500 μl of binding buffer at room temperature for 30 min. After the incubation, the beads were separated from the soluble fractions, and the Rad9 complex remaining in the soluble fractions was used in the recruitment assays.
To test the recruitment of Rad9 complex to various DNA templates, ≈0.5 pmol of the Rad17 complex, 0.5 pmol of RPA, and 0.5 pmol of various DNA templates attached to beads were added to the precleared Rad9 complex in 500 μl of binding buffer. After a 30-min incubation, the beads were retrieved and washed three times with binding buffer containing 250 mM NaCl and 0.25 mM ATP. The proteins associated with the beads were released in denaturing sample buffer and analyzed by immunoblotting.
Results and Discussion
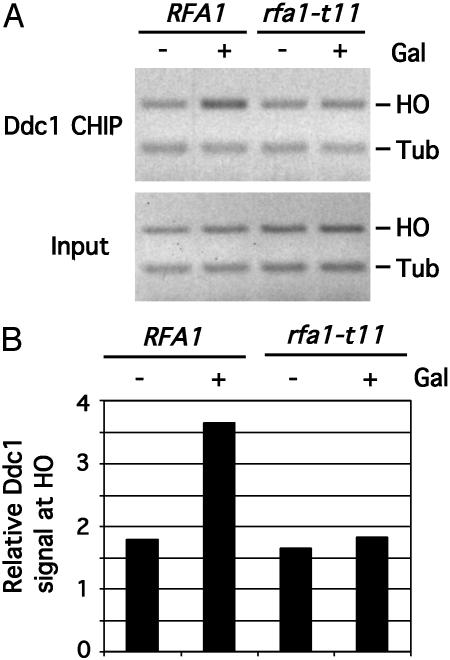
RPA Is Required for the Recruitment of the Ddc1 Complex to DNA Damage in Vivo. In budding yeast, a single DSB can be generated at a specific site of the genome by the HO endonuclease, and the recruitment of checkpoint proteins to this site can be monitored by chromatin immunoprecipitation (4, 5). It has been shown that Ddc1, Mec3, and Rad17, the three components of the yeast proliferating cell nuclear antigen (PCNA)-like checkpoint complex, are specifically recruited to the HO-induced breaks in vivo (4, 5). Furthermore, the recruitment of the Ddc1–Mec3–Rad17 complex to HO breaks requires Rad24, the yeast counterpart of human Rad17, but is independent of Mec1, the yeast homologue of ATR (4, 5). To examine whether RPA is involved in the recruitment of the Ddc1 complex to DNA damage, we sought to analyze the binding of Ddc1 to HO breaks in the rfa1-t11 mutant. The protein encoded by rfa1-t11 binds ssDNA at the site of HO-induced breaks as well as wild-type RPA complex (3), but is partially defective for the checkpoint responses (14, 16, 17).
The rfa1-t11 mutant cells expressing hemagglutinin-tagged Ddc1 and their isogenic cells with wild-type RFA1 were first synchronized with nocodazole in G2/M before galactose was added to the cultures to induce the expression of HO endonuclease. After a 4-hour incubation in galactose-containing medium, the cells were collected and analyzed by chromatin immunoprecipitation by using antibodies to hemagglutinin. In the control RFA1 cells, Ddc1 is specifically recruited to the HO breaks as described (Fig. 1A and ref. 4). The recruitment of Ddc1 to the HO breaks was reduced in the rfa1-t11 cells (Fig. 1). Furthermore, because the rfa1-t11 mutant displays no defects in DNA replication and the cells were arrested outside of S phase, the diminished Ddc1 recruitment is unlikely to be a consequence of altered DNA replication. Thus, RPA is required for the efficient recruitment of the PCNA-like checkpoint complex in vivo.
Fig. 1.
Defective recruitment of Ddc1 to DNA damage in the rfa1-t11 mutant. (A) rfa1-t11 mutant cells carrying DDC1-GFP, Gal-HO, and a single HO cleavage site, and the isogenic RFA1 cells were synchronized at G2/M with nocodazole for 3 h at 30°C. The cells were then cultured in the presence or absence of 2% galactose for another 4 h and were analyzed by chromatin immunoprecipitation with antibodies to GFP. HO, PCR products derived from a locus 0.5 kb from the HO-induced break. Tub, PCR products derived from a control locus at TUB1. (B) Quantification of the Ddc1 signal at the HO locus relative to the TUB1 locus from the samples shown in A.
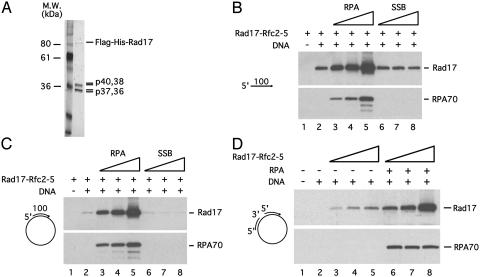
RPA Stimulates the Binding of the Rad17 Complex to ssDNA and Primed ssDNA. Because the recruitment of Ddc1 to DNA damage depends on Rad24, we asked whether RPA regulates the recruitment and/or function of the RFC-like checkpoint complex. The human Rad17–Rfc2-5 complex was expressed in insect cells and affinity purified (Fig. 2A and ref. 8). The purified Rad17 complex was incubated with a 100-nucleotide, ssDNA oligomer that was biotinylated at the 3′ end (Fig. 2B). After the incubation, the ssDNA–protein complexes were retrieved with streptavidin beads and the ssDNA-bound proteins were analyzed by immunoblotting. In the absence of RPA, the Rad17 complex can clearly associate with ssDNA (Fig. 2B, lane 2). In the presence of increasing amounts of RPA, the amounts of Rad17 on ssDNA increased proportionally (Fig. 2B,lanes3–5). In sharp contrast, the single-strand DNA-binding protein (SSB) from E. coli slightly inhibited the binding of Rad17 complex to ssDNA (Fig. 2B, lanes 6–8), showing that the stimulation of Rad17–ssDNA association by RPA is specific and not simply due to the elimination of secondary structures of ssDNA. Thus, the Rad17 complex has a higher affinity to RPA-coated ssDNA than to naked ssDNA.
Fig. 2.
RPA stimulates the binding of the Rad17 complex to ssDNA, primed ssDNA, and gapped DNA templates. (A) A silver-stained gel of the recombinant Rad17 complex purified from insect cells. (B) RPA stimulates the binding of the Rad17 complex to ssDNA. Purified Rad17 complex (0.5 pmol) was incubated with a 3′ biotinylated, 100-nucleotide ssDNA oligomer (0.5 pmol) in the absence of RPA, or in the presence of various amounts of RPA (0.5, 1, and 2 pmol). The ssDNA-bound Rad17 and RPA70 were then retrieved by streptavidin beads and detected by immunoblotting with the respective antibodies. (C) RPA stimulates binding of the Rad17 complex to primed ssDNA. A 3′ biotinylated, 100-nucleotide ssDNA oligomer was annealed to circular, single-stranded pCR-Script DNA. The resultant DNA structure was incubated with the purified Rad17 complex in the absence or presence of RPA, and bound proteins were recovered as in B. (D) RPA stimulates binding of the Rad17 complex to gapped DNA template. Two 100-nucleotide ssDNA oligomers, one of which is biotinylated at the 3′ end, were annealed to single-stranded pCR-Script DNA. The resultant DNA structure contains a 200-nucleotide single-stranded gap between the two annealed oligomers. The gapped DNA structure was incubated with various amounts of the Rad17 complex (0.25, 0.5, and 1 pmol) in the absence or presence of RPA (0.5 pmol). The ssDNA-bound Rad17 and RPA70 were detected as above.
Because RFC uses the primer/template junction to recruit PCNA, we asked whether RPA could stimulate the binding of the Rad17 complex to DNA structures that contain both single-stranded and double-stranded regions. A 3′ biotinylated, 100-nucleotide oligomer was annealed to circular single-stranded pCR-Script DNA to form a primed ssDNA template. Surprisingly, although the resultant DNA structure contains a large single-stranded region, the Rad17 complex bound very weakly to this template in the absence of RPA (Fig. 2C, lane 2). The weak association of Rad17 with this template might be due to the lack of free ssDNA ends on this template. Furthermore, the Rad17 complex does not display a high affinity to the 5′ primer/template junction on this template. However, in the presence of RPA, the binding of Rad17 to this primed template was significantly stimulated (Fig. 2C, lanes 3–5).
Inhibition of lagging strand synthesis could result in the presence of a gapped template. To mimic this DNA structure, a second circular DNA template with two annealed primers 200-nucleotide apart was tested for association with the Rad17 complex (Fig. 2D). Although this DNA template has both 5′ and 3′ primer/template junctions, it also only weakly associated with Rad17 in the absence of RPA (Fig. 2D, lanes 3–5). Therefore, neither the 5′ nor the 3′ primer/template junction can recruit the Rad17 complex efficiently. In the presence of RPA, the binding of Rad17 to this DNA template was significantly stimulated at multiple Rad17 complex concentrations (Fig. 2D, lanes 6–8). Hence, RPA stimulates the binding of Rad17 complex to primed ssDNA in the absence of free ssDNA ends.
ssDNA is present at recessed DNA breaks (18). Although the Rad17 complex can bind to ssDNA with free ends, RPA can stimulate this binding and thereby facilitates the recruitment of Rad17 complex to DNA breaks. Furthermore, increased amounts of ssDNA are generated at stalled replication forks (19). Our data suggest that RPA might play an important role in recruiting the Rad17 complex to stalled replication forks even in the absence of ssDNA ends. The binding of the Rad17 complex to RPA-coated ssDNA may represent the initial step of its recognition of DNA damage. It is possible that, once recruited to the sites of DNA damage, the Rad17 complex can then associate with a DNA structure that can support efficient loading of the Rad9 complex
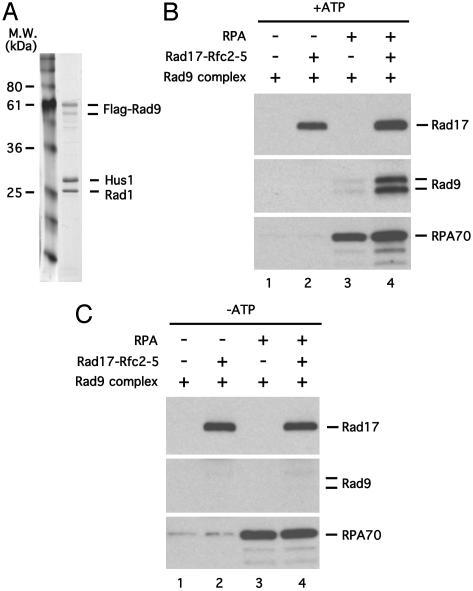
RPA Stimulates the Recruitment of Rad9 Complex by the Rad17 Complex in Vitro. To investigate how the Rad17 complex recruits the Rad9 complex to DNA, we coexpressed Rad9, Rad1, and Hus1 in insect cells and affinity purified the trimeric complex (Fig. 3A and ref. 8). The binding of the Rad9 complex to the double-primed circular DNA template was examined in the absence or presence of the Rad17 complex and RPA. In these reactions, the DNA templates were first attached to streptavidin beads and then incubated with RPA, the Rad17 complex, and the Rad9 complex. In contrast to the previous experiments, some Rad17 complexes associated with primed ssDNA templates even in the absence of RPA when the DNA is bound to the beads before initiation of the binding reaction. It is possible that the attachment of the DNA template to the beads interferes with the annealing between primers and circular ssDNA, thereby generating ssDNA ends. Alternatively, it is possible that the Rad17 complex has a weak affinity for both the DNA and the beads themselves, allowing it to remain associated throughout the experiment. It should be noted that, although some Rad17 complex bound alone, it was further stimulated for binding by the presence of RPA (Fig. 3B, lanes 2 and 4).
Fig. 3.
A Rad17- and RPA-dependent recruitment of Rad9 complex in vitro. (A) A silver-stained gel of the recombinant Rad9 complex purified from insect cells. (B) RPA is required for the efficient recruitment of the Rad9 complex to gapped DNA template. The Rad9 complex purified from insect cells was precleared with beads carrying gapped DNA template coated with RPA (see Materials and Methods). The precleared Rad9 complex was then incubated with the Rad17 complex, RPA, and the gapped DNA template attached to streptavidin beads as indicated. The Rad17, Rad9, and RPA70 bound to the gapped DNA were detected by immunoblotting with the respective antibodies. (C) The recruitment of Rad9 complex to gapped DNA is ATP-dependent. The recruitment of Rad9 complex to gapped DNA was tested as in B, except that ATP was omitted from the binding buffer.
In the absence of the Rad17 complex and RPA, no Rad9 was detected on the DNA template (Fig. 3B, lane 1). When the Rad17 complex alone was added to the reaction, the binding of Rad9 was not significantly stimulated (Fig. 3B, lane 2). This result indicates that the bound Rad17 is not active. However, when RPA was included in the reactions, the amounts of Rad9 on DNA was significantly increased even in the absence of the Rad17 complex (data not shown). This Rad17-independent recruitment of the Rad9 complex could be due to trace amounts of contaminating insect Rad17 complex in the Rad9 preparation used, or it could be due to a direct recruitment of the Rad9 complex by the RPA-coated ssDNA. To distinguish these possibilities, we sought to deplete the potential contaminating insect Rad17 complex from the purified Rad9 complex. Because human Rad17 complex can associate with gapped DNA template efficiently in the presence of RPA, we reasoned that the contaminating insect Rad17 complex might also bind to RPA-coated gapped DNA template and therefore used this protein–DNA structure to preclear the Rad9 complex. After the preclearance, the remaining Rad9 complex can no longer associate with the DNA template efficiently in the absence of the Rad17 complex even in the presence of RPA (Fig. 3B, lane 3). This result shows that RPA-coated ssDNA alone is not sufficient to recruit the Rad9 complex. In the presence of both the Rad17 complex and RPA, however, the binding of Rad9 complex to the DNA template was clearly observed (Fig. 3B, lane 4). Therefore, as we observed in vivo, the Rad9 complex is recruited to the DNA template in a Rad17- and RPA-dependent manner in vitro.
The recruitment of PCNA by RFC is ATP-dependent (13, 20). Furthermore, it has been shown that the Rad17 complex associates with the Rad9 complex in an ATP-dependent manner (11). To address whether ATP is required for the recruitment of the Rad9 complex by the Rad17 complex in our reactions, we carried out the Rad9 recruitment reactions in the absence of ATP. In the absence of ATP, Rad9 was not detected on the DNA template even in the presence of both RPA and the Rad17 complex (Fig. 3C). Therefore, the in vitro recruitment of Rad9 by the Rad17 complex is an ATP-dependent reaction. This result is consistent with the finding that the ATP-binding motif in yeast Rad24 is required for its checkpoint function in vivo (21).
The Structure Requirements for the Recruitment of the Rad9 Complex. RFC recognizes the 3′ primer/template junction to recruit PCNA. However, the yeast Ddc1 complex can be recruited to 5′-to-3′ recessed DSB in vivo (4, 5) where only recessed 5′ ends are present. Therefore, the structure specificity must be different between RFC and the RFC-like checkpoint complex.
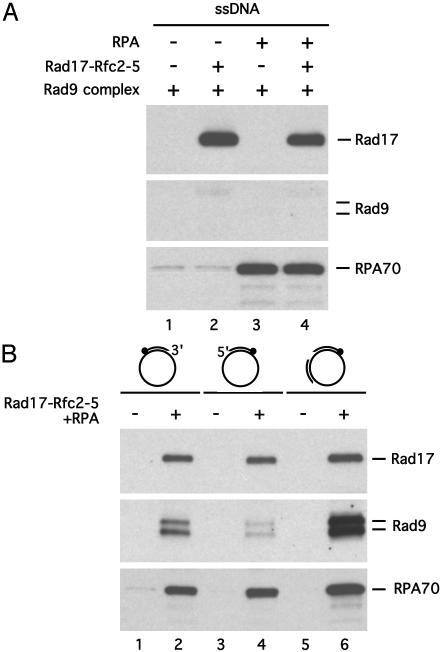
To determine the structure specificity of the Rad17 complex, we first tested whether the Rad17 complex can recruit the Rad9 complex onto ssDNA. A 100-nucleotide, biotinylated ssDNA oligomer was incubated with the precleared Rad9 complex, the Rad17 complex, and RPA in the presence of ATP (Fig. 4A). No Rad9 was detected on ssDNA (Fig. 4A), suggesting that primer/template junctions are needed for the recruitment of Rad9 complex by the Rad17 complex. To further reveal the type of primer/template junction that is required for the Rad9 recruitment, we generated three different primed ssDNA structures and tested them for the recruitment of Rad9 complex (Fig. 4B). The first DNA structure contains an annealed primer whose biotinylated 5′ end was attached to the beads. This DNA template offers only free 3′ primer/template junctions, a typical structure used by RFC to recruit PCNA. The Rad9 complex was clearly recruited to this DNA template in the presence of the Rad17 complex and RPA (Fig. 4B, lane 2). Hence, the Rad17 complex can recognize the 3′ primer/template junction to recruit the Rad9 complex. Instead of a 3′ primer/template junction, the second DNA structure we tested contains a free 5′ primer/template junction. This structure is not typically used by RFC to recruit PCNA. However, some Rad9 was detected on the template in the presence of the Rad17 complex and RPA (Fig. 4B, lane 4). Thus, unlike RFC, the Rad17 complex can use the 5′ primer/template junction to recruit the Rad9 complex. Our third DNA structure contains two annealed primers and both 5′ and 3′ primer/template junctions. Furthermore, this structure contains a 200-nucleotide gap with primer/template junctions on both sides. The recruitment of Rad9 to this DNA structure was significantly more efficient than those to the other two structures (Fig. 4B, lane 6). Therefore, the Rad17 complex displays a preference for a gapped structure for the recruitment of the Rad9 complex.
Fig. 4.
Structural requirements for the recruitment of the Rad9 complex in vitro. (A) ssDNA does not support the recruitment of the Rad9 complex. A 3′ biotinylated, 100-nucleotide ssDNA oligomer bound to strepavidin beads was incubated with the precleared Rad9 complex, the Rad17 complex, and RPA, and bound protein was assayed as in Fig. 3 B and C. The presence of Rad17, Rad9, and RPA70 on ssDNA was examined by immunoblotting with the respective antibodies. (B) The Rad9 complex is recruited by the Rad17 complex and RPA to the DNA structures containing free 5′ or 3′ primer/template junctions, or single-stranded gaps. The DNA structure with a free 3′ primer/template junction was generated by annealing a 5′ biotinylated oligomer to single-stranded pCR-Script DNA. The DNA structures with a free 5′ primer/template junction or single-stranded gaps were generated as in Fig. 2. All of the resultant DNA structures were first attached to streptavidin beads and then incubated with the precleared Rad9 complex, the Rad17 complex, and RPA as indicated. The Rad17, Rad9, and RPA70 associated with the various DNA structures were analyzed by immunoblotting with the respective antibodies.
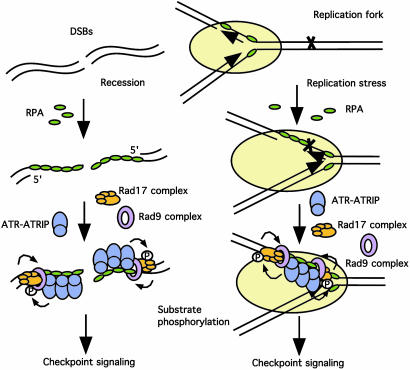
The unique structure specificity of the Rad17 complex revealed by our experiments sheds light on its function in damage recognition. First, recessed 5′ ends are present at enzymatically processed DSB and unprotected telomeres (Fig. 5 and refs. 18 and 22). These structures might be recognized only by the Rad17 complex but not by RFC. Second, when the DNA synthesis on the leading strand is blocked, a 3′ primer/template junction will be generated. The Rad17 complex might compete with RFC for the binding to this junction and recruit the Rad9 complex to the stalled replication forks. Third, when the DNA synthesis on the lagging strand is inhibited, gaps between Okazaki fragments will be generated. These gaps are likely good substrates for the Rad17 complex to recruit the Rad9 complex (Fig. 5). Furthermore, when DNA synthesis is blocked by aphidicolin in Xenopus extracts, the amounts of DNA polymerase α (Polα) on chromatin is substantially increased (23). This result indicates that Polα/primase complexes are recruited to the ssDNA at stalled forks and might continue to synthesize short RNA or RNA-DNA fragments. This action might create more substrates for the Rad17 complex to recruit the Rad9 complex. Moreover, gaps are also generated by DNA repair processes such as nucleotide excision repair (24). The recognition of gapped DNA by the Rad17 complex might explain its role in the activation of ATR-mediated checkpoint in response to UV-induced DNA damage.
Fig. 5.
A model for RPA's functions in checkpoint signaling. In response to double strand breaks or unprotected telomeres (not shown), ssDNA is generated by the action of nucleases leaving 3′ ssDNA extensions. ssDNA is also generated at the site of stalled replication forks. RPA binds to this DNA, and the RPA–ssDNA complex is critical for the recruitment of the ATR–ATRIP complex to these sites. RPA–ssDNA in the context of a 5′ end of duplex DNA, a3′ end of duplex DNA, or both a 5′ and 3′ end (gapped) substrate is capable of recruiting the Rad17 complex and subsequently recruiting the Rad9 complex to these molecules in an ATP-dependent manner. The interactions among the ATR–ATRIP, Rad17, and Rad9 complexes on DNA are important for the phosphorylation of ATR substrates and the initiation of checkpoint signaling.
Previous studies (4–6) have suggested that both the ATR– ATRIP complex and the Rad17 complex are involved in the sensing of DNA damage. Furthermore, the two complexes carry out their sensor functions independently of each other. We have shown that the RPA–ssDNA complexes play an important role in recruiting the ATR–ATRIP complex to the site of DNA damage (Fig. 5). Here, our results reveal that RPA is also required for the efficient recruitment of Rad9 complex by the Rad17 complex (Fig. 5). Therefore, RPA-coated ssDNA is a key structure recognized by both the ATR–ATRIP and the Rad17 complexes. It should be noted that the binding of both the Rad17 and Rad9 complexes and the ATR–ATRIP complexes to damage sites may be further stabilized by additional factors present at replication forks or recruited to DSB.
Our experiments also bring us a step closer to the reconstitution of an ATR-, Rad17-, and Rad9-dependent checkpoint reaction in vitro. It is still unclear how the Rad17 and Rad9 complexes interact with the ATR–ATRIP complex on damaged DNA and regulate the function of ATR–ATRIP. The establishment of a Rad17-dependent recruitment of Rad9 complex to DNA will allow us to further characterize the functional interactions among the checkpoint complexes on DNA.
Acknowledgments
We thank D. Toczyski, J. Hurwitz, and Aziz Sancar for reagents. L.Z. is a Fayez Sarofim fellow of the Damon Runyon Cancer Research Fund. S.J.E. is funded by a grant from the National Institutes of Health (GM44664) and is an investigator with the Howard Hughes Medical Institute and a Welch Professor of Biochemistry.
Abbreviations: DSB, double-stranded DNA breaks; ssDNA, single-stranded DNA; RFC, replication factor C; PCNA, proliferating cell nuclear antigen.
References
- 1.Zhou, B. B. & Elledge, S. J. (2000) Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 2.Abraham, R. T. (2001) Genes Dev. 15, 2177–2196. [DOI] [PubMed] [Google Scholar]
- 3.Zou, L. & Elledge, S. J. (2003) Science 300, 1542–1548. [DOI] [PubMed] [Google Scholar]
- 4.Kondo, T., Wakayama, T., Naiki, T., Matsumoto, K. & Sugimoto, K. (2001) Science 294, 867–870. [DOI] [PubMed] [Google Scholar]
- 5.Melo, J. A., Cohen, J. & Toczyski, D. P. (2001) Genes Dev. 15, 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou, L., Cortez, D. & Elledge, S. J. (2002) Genes Dev. 16, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green, C. M., Erdjument-Bromage, H., Tempst, P. & Lowndes, N. F. (2000) Curr. Biol. 10, 39–42. [DOI] [PubMed] [Google Scholar]
- 8.Lindsey-Boltz, L. A., Bermudez, V. P., Hurwitz, J. & Sancar, A. (2001) Proc. Natl. Acad. Sci. USA 98, 11236–11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffith, J. D., Lindsey-Boltz, L. A. & Sancar, A. (2002) J. Biol. Chem. 277, 15233–15236. [DOI] [PubMed] [Google Scholar]
- 10.Tsurimoto, T. & Stillman, B. (1990) Proc. Natl. Acad. Sci. USA 87, 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bermudez, V. P., Lindsey-Boltz, L. A., Cesare, A. J., Maniwa, Y., Griffith, J. D., Hurwitz, J. & Sancar, A. (2003) Proc. Natl. Acad. Sci. USA 100, 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majka, J. & Burgers, P. M. (2003) Proc. Natl. Acad. Sci. USA 100, 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waga, S. & Stillman, B. (1998) Mol. Cell. Biol. 18, 4177–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, H. S. & Brill, S. J. (2001) Mol. Cell. Biol. 21, 3725–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henricksen, L. A., Umbricht, C. B. & Wold, M. S. (1994) J. Biol. Chem. 269, 11121–11132. [PubMed] [Google Scholar]
- 16.Umezu, K., Sugawara, N., Chen, C., Haber, J. E. & Kolodner, R. D. (1998) Genetics 148, 989–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pellicioli, A., Lee, S. E., Lucca, C., Foiani, M. & Haber, J. E. (2001) Mol. Cell 7, 293–300. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. E., Moore, J. K., Holmes, A., Umezu, K., Kolodner, R. D. & Haber, J. E. (1998) Cell 94, 399–409. [DOI] [PubMed] [Google Scholar]
- 19.Sogo, J. M., Lopes, M. & Foiani, M. (2002) Science 297, 599–602. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, G., Gibbs, E., Kelman, Z., O'Donnell, M. & Hurwitz, J. (1999) Proc. Natl. Acad. Sci. USA 96, 1869–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naiki, T., Shimomura, T., Kondo, T., Matsumoto, K. & Sugimoto, K. (2000) Mol. Cell. Biol. 20, 5888–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lydall, D. & Weinert, T. (1995) Science 270, 1488–1491. [DOI] [PubMed] [Google Scholar]
- 23.Michael, W. M., Ott, R., Fanning, E. & Newport, J. (2000) Science 289, 2133–2137. [DOI] [PubMed] [Google Scholar]
- 24.Wood, R. D. (1996) Annu. Rev. Biochem. 65, 135–167. [DOI] [PubMed] [Google Scholar]