Abstract
Episomal vector with the capacity to deliver a large gene containing all the critical regulatory elements is ideal for gene therapy. Human artificial chromosomes (HACs) have the capacity to deliver an extremely large genetic region to host cells without integration into the host genome, thus preventing possible insertional mutagenesis and genomic instability. Duchenne muscular dystrophy (DMD) is caused by mutation in the extremely large dystrophin gene (2.4 Mb). We herein report the development of a HAC vector containing the entire human dystrophin gene (DYS-HAC) that is stably maintained in mice and human immortalized mesenchymal stem cells (hiMSCs). The DYS-HAC was transferred to mouse embryonic stem (ES) cells, and isoforms of the DYS-HAC-derived human dystrophin in the chimeric mice generated from the ES cells were correctly expressed in tissue-specific manner. Thus, this HAC vector containing the entire dystrophin gene with its native regulatory elements is expected to be extremely useful for future gene and cell therapies of DMD.
Introduction
Duchenne muscular dystrophy (DMD) is a recessive, fatal, X-linked disorder mainly caused by the absence of the dystrophin protein in skeletal muscle as well as in other tissues such as brain, retina, and smooth muscle, in which isoforms of dystrophin are normally expressed.1,2,3,4,5 Major current therapeutic approaches of DMD consist of (i) introducing (e.g., via viral or nonviral vectors) or repairing (e.g., via exon skipping) the genetic message, (ii) transplanting dystrophin-positive cells (e.g., via myoblast transplantation), or (iii) regulating synthesis of an endogenous gene product (e.g., via upregulation of utrophin gene expression).6,7 Several groups have reported various approaches to introduce genes for gene therapy.8,9 However, these vector systems have several clinical drawbacks including host genome integration, the limited ability to insert large genes, and an inappropriate expression. Random integration of the gene into the host genome may cause genomic instability and a misregulation of proximal gene loci adjacent to integration sites.10,11 Importantly, another possible problem is that an overexpression of a transgene can produce muscular dystrophy.12 Therefore, an exact dosage of the transgene is needed to achieve a therapeutic result in muscle gene therapy. Another approach of gene therapy is exon skipping. Exon skipping, aiming to reading frame restoration, is a highly promising approach to convert the DMD phenotype into the milder Becker phenotype. However, exon skipping is a sequence-specific therapy which is not applicable to patients who have mutations in promoter regions or essential functional domains, and in the case of antisense oligonucleotides, the readministration will also be necessary as these compounds have a limited half-life.7 For all these reasons, it would be desirable to target the dystrophin gene of DMD patients directly and permanently, independently from the nature of the mutation.
For more effective treatment of DMD, the development of a dystrophin expression vector that produces all isoforms of dystrophin may be required. Dystrophin gene encodes a number of tissue-specific isoforms of dystrophin including Dp427, Dp260, Dp140, Dp116, and Dp71 generated by transcription from at least seven promoters and also by alternative splicing.13 Until now, no episomal vector containing the entire dystrophin genomic region has been reported, because of its extremely large size (2.4 Mb).13,14
Human artificial chromosome (HAC) is a highly promising gene delivery tool that possesses several advantages over conventional gene delivery systems. Like native chromosomes, HAC vectors have the capacity to replicate and segregate autonomously without integration into the host genome.15,16,17,18,19,20,21,22,23,24 In addition, HACs have the capacity to carry large genomic loci with their regulatory elements.25 The ideal gene delivery vector for safe gene therapy should be structurally defined and should not contain extra genes. We therefore recently produced a HAC vector by deleting all of the endogenous genes on human chromosome 21. This HAC contains an enhanced green fluorescent protein (EGFP) gene for visualizing, the herpes simplex virus thymidine kinase (HSV-tk) gene for the elimination of itself and a loxP site for cloning of a transgene.
In this study, we developed a HAC vector containing the entire human dystrophin gene including its all transcriptional regulatory elements (DYS-HAC). We confirmed that the multiple tissue-specific isoforms of human dystrophin were produced in mice containing the DYS-HAC, and the DYS-HAC was stably maintained in mice and cultured human cells.
Results
Targeted integration of a loxP site into the proximal loci to the dystrophin gene on a human X chromosome fragment
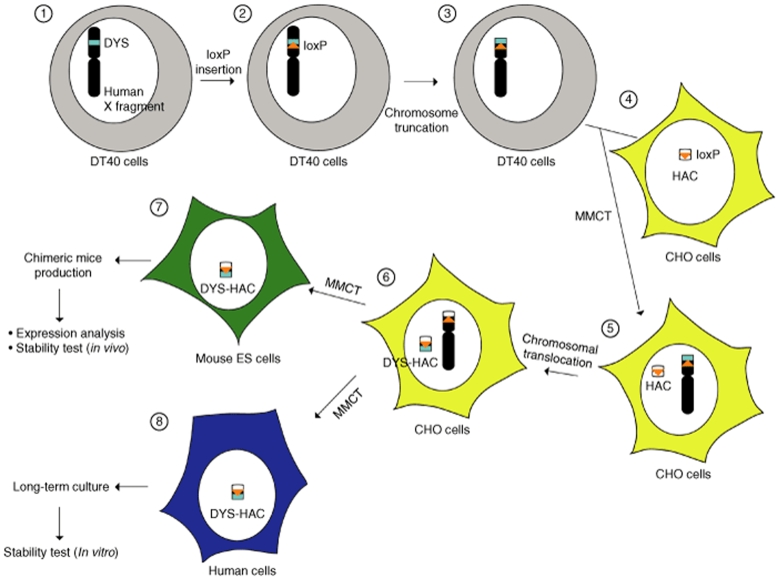
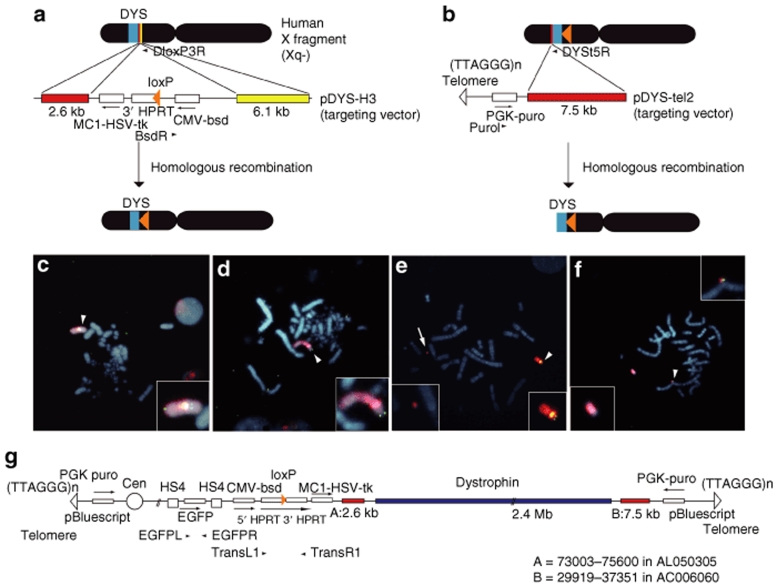
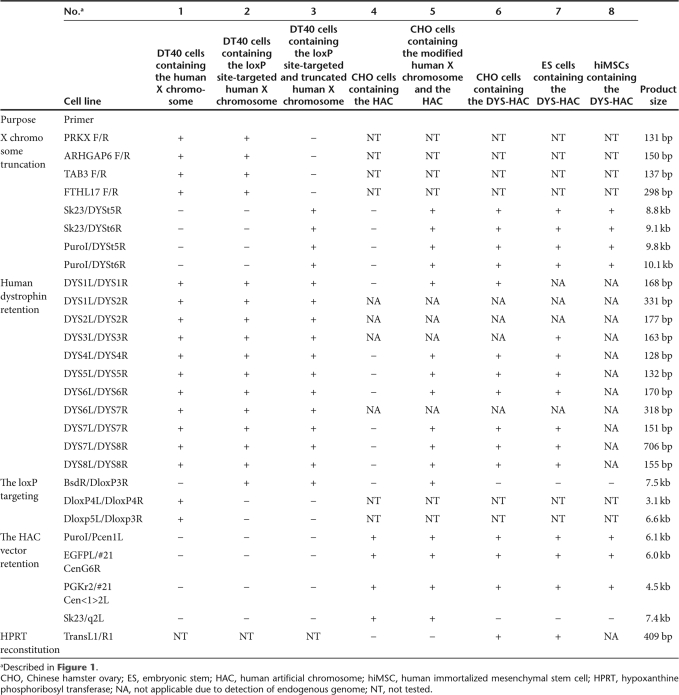
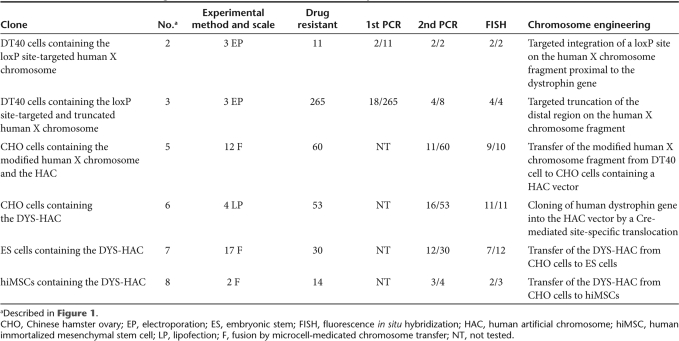
We performed the following experiments to develop a HAC containing the entire dystrophin gene using the chromosome engineering. The overall strategy for vector construction is shown schematically in Figure 1. We have previously established monochromosomal hybrids containing individual human chromosomes.26 In this study, a human X chromosome fragment was transferred from A9 cells into DT40 cells by microcell-medicated chromosome transfer (MMCT), because DT40 cells exhibit a high frequency of homologous recombination between an exogenous DNA template and its chromosomal counterparts.27 This human X chromosome fragment containing the dystrophin gene was originally derived from the human primary fibroblast cells by MMCT. Initially, to integrate a loxP site containing the drug-resistant marker blasticidin S deaminase (bsd) and the 3′ hypoxanthine phosphoribosyl transferase (HPRT) into the proximal loci to the human dystrophin gene, the loxP targeting vector, pDYS-H3 was introduced into the DT40 hybrid cells retaining a single copy of human X chromosome fragment tagged with the neomycin-resistant gene (Figure 2a). PCR analyses using the primers (BsdR/DloxP3R, DloxP4L/4R and DloxP5L/DloxP3R) showed that 2 of 11 blasticidin S Hydrochloride (BS)-resistant transfectants were correctly targeted (Tables 1 and 2). By fluorescence in situ hybridization (FISH), the digoxigenin-labeled human Cot1 DNA probe localized to an independently segregating X chromosome fragment and the biotin-labeled bsd probe localized at the proximal region on the human X chromosome fragment. Neither insertion nor translocation were observed in both clones (Figure 2c and Table 2). These results indicate that the loxP site was successfully targeted in the correct location. These two clones were used for a targeted truncation of the distal region of the dystrophin gene on this human X chromosome fragment.
Figure 1.
Schematic diagram of the construction of various cells containing the DYS-HAC vector. The human dystrophin is located on the short arm of the human X chromosome. Chromosome manipulation was carried out in homologous recombination-proficient DT40 cells. To clone human dystrophin gene into the human artificial chromosome (HAC) vector using the Cre-loxP mediated chromosomal translocation, a loxP was targeted to the proximal locus of the dystrophin gene on the human X chromosome. Extra genes on the distal of the dystrophin gene were deleted by telomere-associated chromosome truncation in the DT40 cells. The modified human X chromosome fragment was transferred into Chinese hamster ovary (CHO) hybrids containing the HAC including the loxP vector by microcell-medicated chromosome transfer (MMCT). The dystrophin gene (2.4 Mb) was cloned into the HAC vector in CHO cells using Cre-loxP mediated chromosomal translocation. From the CHO hybrids, the DYS-HAC vector was further transferred to human immortalized mesenchymal stem cells (hiMSCs) and mouse embryonic stem (ES) cells. The stability of the DYS-HAC was investigated in hiMSC cells. To study the expression of the human dystrophin gene on the DYS-HAC in vivo, the chimeric mice were produced from the ES cells containing the DYS-HAC.
Figure 2.
Construction of the DYS-HAC vector. (a) Strategy for the targeted integration of the loxP site into the proximal loci to the dystrophin gene on the human X chromosome fragment by the loxP targeting vector pDYS-H3. (b) Strategy for the targeted truncation of the distal region of the dystrophin gene on the X chromosome by the targeting vector pDYS-tel2. (c–f) Fluorescence in situ hybridization analyses of the modified chromosomes in the DT40 or the Chinese hamster ovary (CHO) hybrid cells. Chromosomal DNA was counterstained with DAPI. The digoxigenin-labeled human Cot1 DNA (red) detected the human chromosome fragment in the each hybrid. (c) The targeted loxP site on the human X chromosome fragment in DT40 backgrounds, hybridized to the biotin-labeled CMV-bsd probe (green), is indicated with an arrowhead. (d) The truncated site on the human X chromosome in DT40 backgrounds, hybridized to the biotin-labeled PGK-puro (green), is indicated with an arrowhead. (e) The dystrophin gene on the modified human X fragment in CHO backgrounds, hybridized to the bacterial artificial chromosome (BAC) (RP11-954B16) containing the part of the dystrophin genome (green), is indicated with an arrowhead. An arrow indicates the human artificial chromosome (HAC) and the insets show enlarged images of the HAC vector and the modified human X fragment. (f) The region of dystrophin gene on the DYS-HAC in CHO backgrounds, hybridized to the biotin-labeled BAC (RP11-954B16) containing a part of the human dystrophin genome (green), is indicated with an arrowhead. The insets show enlarged images of the DYS-HAC and the X/HAC fragment. (g) The map of the DYS-HAC vector. The DYS-HAC vector contains the enhanced green fluorescent protein (EGFP) gene, the herpes simplex virus thymidine kinase (HSV-tk) gene, and several selection markers (bsd, puromycin, and HPRT gene). Both telomeres of the DYS-HAC are artificial. The centromere of the DYS-HAC is derived from human chromosome 21.
Table 1.
Summary of the expected pattern in hybrid cell lines by PCR analyses
Table 2.
The results in the screening of the various clones isolated for this study
Targeted truncation of the distal region of the dystrophin gene on the human X chromosome
The terminal regions from the dystrophin gene on the X chromosome fragment were deleted in DT40 cells using the telomere-associated chromosome truncation, which is the combined use of gene targeting and telomere-mediated chromosome breakage to generate a defined truncation of a human chromosome.16,28 A targeting construct containing the region homologous to the distal region of the dystrophin gene, the selectable marker gene, puromycin and 1 kb of telomeric sequence repeats, pDYS-tel2, was transfected to the DT40 cells containing the loxP site-targeted human X chromosome fragment (Figure 2b). Two hundred and sixty-five puromycin-resistant transfectants were isolated and first screened by PCR analyses with primers for the detection of the distal region (PRKX F/R and FTHL17 F/R). Eighteen of 265 clones showed the expected patterns of marker retention. Next, four of eight clones randomly selected among them showed the expected pattern of marker retention by PCR analyses with all other primers (Figure 2b, Tables 1 and 2). FISH analysis with the biotin-labeled PGK-puro probe and the digoxigenin-labeled human Cot1 DNA probe revealed that the human X chromosome fragment was truncated at the distal region of the dystrophin gene and the fragment segregated independently without either host genome insertion or translocation in all four clones (Figure 2d). These results indicate that the genes on the distal from the dystrophin gene were deleted at the defined locus by the telomere-associated chromosome truncation strategy.
Transfer of the modified X chromosome fragment from the DT40 cells to the Chinese hamster ovary (CHO) hybrids containing the HAC vector
The truncated X chromosome fragment containing the human dystrophin gene and the 3′ HPRT-loxP was transferred from the DT40 cells into the CHO cells containing the HAC vector including the 5′ HPRT-loxP using MMCT. Sixty G418-resistant CHO clones were obtained from three independent DT40 clones containing the modified X chromosome fragment as donor cells. PCR analyses with primer sets for the detection of the HAC and the modified X chromosome showed that 11 of 60 CHO hybrids retained both the HAC vector and the modified X chromosome (Table 1). FISH analyses showed the HAC vector and the modified X chromosome to be independently present in 9 of 10 clones randomly selected among them (Figure 2e and Table 2). The efficiency of MMCT from DT40 cells to CHO cells was 5 × 10−6, which is comparable to our previous results.29
Cloning of the human dystrophin gene into the HAC vector by a Cre-mediated site-specific translocation
In order to clone only the human dystrophin gene into the HAC vector containing a 5′ HPRT-loxP using the Cre-loxP system, CHO hybrids containing the HAC and the modified human X chromosome including a 3′ HPRT-loxP were transfected with the Cre-recombinase expression vector. Intra- and intermolecular recombination between two loxP sites is efficiently promoted using Cre-recombinase.30 A loxP-mediated site-specific translocation reconstitutes a functional HPRT gene, which confers HAT-resistance. Fifty-three HAT-resistant CHO clones were obtained using two independent CHO hybrids. The translocation confirmed in 16 of 53 CHO clones by PCR analysis (Tables 1 and 2). In addition, FISH analysis confirmed that the bacterial artificial chromosome (BAC) probe signal containing the part of human dystrophin gene was hybridized with the DYS-HAC and the DYS-HAC was present as an independent minichromosme without integration into the host genome in all 11 clones randomly selected among them (Figure 2f and Table 2). These results indicate that the DYS-HAC was successfully developed by cloning the human dystrophin gene into the HAC vector.
Generation of mouse embryonic stem (ES) cells containing the DYS-HAC vector and production of chimeric mice
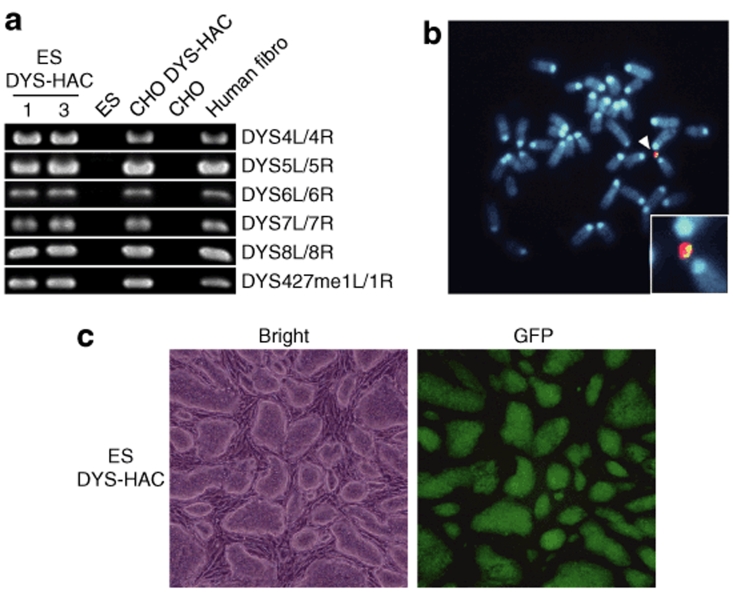
To investigate whether the DYS-HAC vector shows physiological expression of the human dystrophin gene, we produced the chimeric mice containing the DYS-HAC vector. First, the DYS-HAC was transferred from the CHO hybrids into mouse ES cells using MMCT technique. Thirty BS-resistant ES clones were obtained using three independent CHO cells containing the DYS-HAC vector as donor cells. In 12 of 30 drug-resistant ES hybrids, the DYS-HAC loci were detected and the expected patterns were showed by PCR (Figure 3a and Table 1). A FISH analysis showed that independently segregating the DYS-HAC was observed without insertion into host genome and the karyotype of mouse backgrounds was normal in 7 of 12 ES hybrids which showed the expected patterns by PCR (Figure 3b and Table 2). The efficiency of MMCT from CHO hybrids to ES cells was 1.8 × 10−7. The protein produced from the cytomegalovirus enhancer/chicken β-actin promoter driven EGFP gene on the DYS-HAC was detected in mouse ES cells containing the DYS-HAC by fluorescence microscopy (Figure 3c). Three independent ES hybrids containing the DYS-HAC clones were used to produce viable chimeric mice. Chimeric mice with up to 60% coat color chimerism were produced. The germline transmission of the DYS-HAC to the progency mice was not observed using the chimeric mice (data not shown).
Figure 3.
Transfer of the DYS-HAC vector into the mouse embryonic stem (ES) cells. (a) The detection of the human dystrophin gene of the DYS-HAC-transferred ES hybrids by PCR analyses using the human dystrophin-specific primers. The ES cells and the Chinese hamster ovary (CHO) cells were used as negative controls. The CHO cells containing the DYS-HAC and the human fibroblasts were used as positive controls. (b) A fluorescence in situ hybridization analysis in the ES cells is shown. The DYS-HAC vector, hybridized to the digoxigenin-labeled human Cot1 DNA probe (red), is indicated with an arrowhead. The dystrophin gene hybridized to the biotin-labeled bacterial artificial chromosome (RP11-954B16) containing the part of the human dystrophin genome (green). The inset shows enlarged images of the DYS-HAC vector. (c) The expression of the enhanced green fluorescent protein (EGFP) gene on the DYS-HAC was tracked in living cells by fluorescence microscopy.
Expression of the EGFP gene and the human dystrophin gene in various tissues of the chimeras
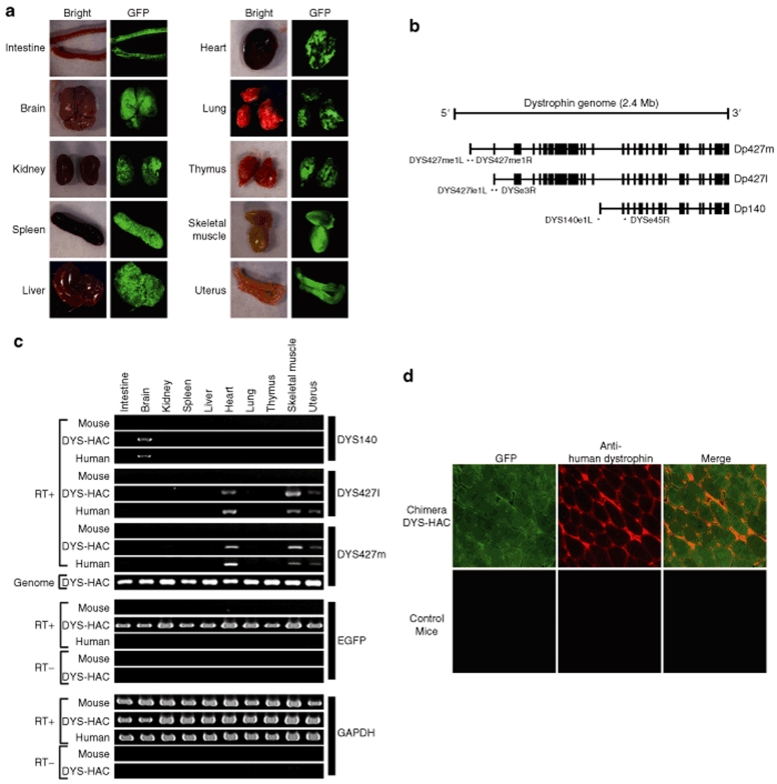
We examined the stability of the DYS-HAC and the human dystrophin gene expression from the DYS-HAC in four chimeric mice containing the DYS-HAC. The EGFP gene driven by chicken β-actin promoter on the DYS-HAC was expressed in all examined tissues of the chimeric mice including intestine, brain, kidney, spleen, liver, heart, lung, thymus, skeletal muscle, and uterus (Figure 4a), suggesting that the DYS-HAC was stably maintained in vivo. To investigate whether the human dystrophin gene on the DYS-HAC vector was appropriately expressed, total RNAs from various tissues of the chimeras were analyzed by reverse transcription–PCR using three pairs of specific primers to detect the human dystrophin tissue-specific transcripts. Using the DYS140e1L/DYSe45R primer sets, transcripts of human dystrophin from the DYS-HAC chimera were found mainly in brain as well as those from human used as control. Using the primer pairs of DYS427me1L/1R and DYS427le1L/DYSe3R, transcripts of human dystrophin from the DYS-HAC chimera were detected mainly in the heart, skeletal muscle, and uterus, as well as in those from normal human, whereas the genomic dystrophin were detected in all examined tissues using the DYS427me1L/1R primer sets (Figure 4b,c). An immunostaining analysis using human dystrophin-specific antibody showed the human dystrophin protein to be localized at the sarcolemmal membrane in muscle (Figure 4d).14,31,32 These results suggested that the isoforms of human dystrophin gene on the DYS-HAC vector were tissue-specifically expressed in the mouse genetic background, and the protein of the human dystrophin was localized at the correct locus.
Figure 4.
Expression of enhanced green fluorescent protein (EGFP) and human dystrophin in various tissues of the chimeric mice containing the DYS-HAC. (a) The EGFP gene on the DYS-HAC was expressed in all examined tissues of the chimeric mice (intestine, brain, kidney, spleen, liver, heart, lung, thymus, skeletal muscle, and uterus). (b) The positions of primers for detecting dystrophin isoforms are shown. The black boxes show exons. The sequence of the DYS427le1L primer is located at the Dp427l isoforms but not at the Dp427m isoforms. (c) The expression of the human dystrophin gene derived from the DYS-HAC in each tissue of the chimeric mice containing the DYS-HAC vector was detected by reverse transcription–PCR using three different sets of the human dystrophin-specific primers. EGFP and GAPDH were used as internal controls. cDNA from C57BL/6 mouse tissues and human cDNA were used as a negative and a positive control, respectively. DNA from the tissues of the chimera containing the DYS-HAC was used as a control. (d) An immunofluorescence analysis of muscle biopsies from the chimeras containing the DYS-HAC vector, stained with the anti-human dystrophin-specific antibody (red). C57BL/6 mouse muscle tissue was used as a negative control. The expression of the EGFP gene on the DYS-HAC was tracked by fluorescence microscopy. Representative photographs are shown.
Stability of the DYS-HAC vector in human cells
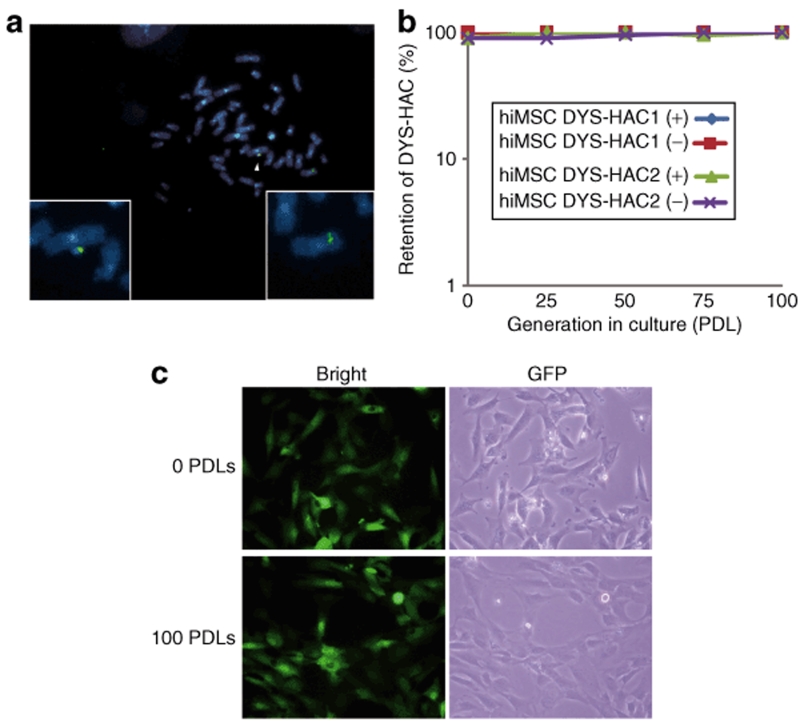
To investigate the stability of the DYS-HAC in human cell, the DYS-HAC was transferred into the human immortalized mesenchymal stem cells (hiMSCs).33 Fourteen hiMSC hybrids were selected with BS, and four clones randomly selected among them were screened by PCR and FISH analyses. In two of four clones, successful transfer was observed (Tables 1 and 2, Figure 5a). The stability of the DYS-HAC vector was tested in the human cell line using the two hiMSCs containing the DYS-HAC. The cell lines were split and maintained independently either in medium with BS or without any selection. The retention of the DYS-HAC vector in the two sublines (both with and without selection) was assessed in 100 interphases and 20 metaphases at 25, 50, 75, and 100 population-doubling levels by the FISH analysis. The retention rate of the DYS-HAC vectors in the hiMSCs background remained in almost 100% of interphases by 100 population-doubling levels even without the BS-selection (Figure 5b). No rearrangement was observed in the metaphases at 100 population-doubling levels (data not shown). Silencing of EGFP gene expression on the DYS-HAC was not observed in the hiMSCs containing the DYS-HAC at 100 population-doubling levels (Figure 5c). These results indicate that the DYS-HAC vector in the human cells is stable without integration into the host genomes even after long-term culture in vitro.
Figure 5.
Retention of the DYS-HAC in the human immortalized mesenchymal stem cells (hiMSCs). (a) A fluorescence in situ hybridization (FISH) analysis in the hiMSCs is shown. The dystrophin gene on the DYS-HAC, hybridized to the biotin-labeled bacterial artificial chromosome (BAC) (RP11-954B16) containing the part of human dystrophin genome (green), is indicated with an arrowhead. The insets show the DYS-HAC and the endogenous human X chromosome. (b) Retention of the DYS-HAC was determined by FISH using the biotin-labeled dystrophin BAC (RP11-954B16) after a long period of culture with and without selection. (c) The expression of the EGFP gene on the DYS-HAC was tracked in living cells by fluorescence microscopy. Representative photographs are shown. PDL, population-doubling level.
Discussion
We herein report the development of a HAC containing the entire dystrophin gene as a potential strategy for the treatment of DMD. The entire human dystrophin genome is unusually large and beyond the capability of viral vector systems.6 One important advantage of the HAC system report here is no apparent size limitation of inserted transgenes.34 We thus inserted the entire dystrophin gene within this HAC vector.
Dystrophin has a complicated regulation of expression system including proper levels of expression and important functional isoforms that require selective promoter usage and highly regulated RNA processing events. Previously reported vector systems drove expression of dystrophin by the heterologous promoters that produced only one dystrophin isoform, mainly the 427 kDa dystrophin isoform.8 Since the DYS-HAC vector used here contains the entire dystrophin gene with all known regulatory elements, we found evidence that at least three dystrophin isoforms were produced. Recently, Hoen et al. reported the development of a yeast artificial chromosome vector containing the entire human dystrophin gene that was successfully introduced into mice.35 However, yeast artificial chromosome vector systems require the random integration of the transgene into the host genome creating the possibility of alteration of gene regulation and genomic instability.36 In contrast, this study demonstrates that the DYS-HAC can be maintained in host cells as an independent minichromosome without integration into the host genome. The DYS-HAC contains the entire dystrophin gene with all its known native regulatory elements and its copy number is one similarly to native dystrophin subjected to X-inactivation. Therefore, unlike other vector systems in which dystrophin expression was driven by heterologous promoters resulting in an aberrant protein steady state levels,12 the dystrophin produced from the DYS-HAC vector is expressed at levels similar to native dystrophin expression levels and isoform production. More importantly, the DYS-HAC can be repeatedly used for transferring to different host cells, whereas the other systems are expected to cause different results in each host cell.
Regarding future DMD treatment strategies, the DYS-HAC may perhaps be introduced into several types of patient stem or progenitor cells for ex vivo therapy, e.g., induced pluripotent stem cells, mesoangioblasts, and mesenchymal stem cells.37,38,39,40,41 The DYS-HAC also contains a “suicide” gene encoding the HSV-tk gene as a backup system to specifically eliminate cells containing this vector. The cells containing the DYS-HAC can be specifically killed by ganciclovir, for example, if they induce a cellular abnormality such as carcinogenesis after transplantation. In addition, the mice obtained by crossing dystrophin-deficient mice with mice containing the DYS-HAC can also be used for functional and genetic studies. Therefore, the DYS-HAC vector will be useful not only for the treatment of DMD genetic disorders, but also for applications in animal transgenesis.
In conclusion, the DYS-HAC vector system reported herein is a unique nonintegrating vector maintaining the ability to incorporate exceptionally large regions of DNA and is highly stably maintained as an episomal element in host cells. Although the transferring efficiency of chromosome via MMCT need to be improved for cells with a limited life span, the current technology is sufficient for isolation of the immortalized stem cells with HACs transferred by ex vivo. When the HAC vector meets more sophisticated technologies of cellular differentiation to come in a near future, this nonintegrated episomal vector will facilitate various gene and cell therapies. Thus, the construction of the DYS-HAC is an initial step for the DMD-gene therapy. Future studies using DMD animal models with autologous genetically corrected stem cells containing the DYS-HAC vector will elucidate the efficacy of ex vivo gene therapy.
Materials and Methods
Cell culture. The DT40 hybrid cells were maintained in Roswell Park Memorial Institute medium 1640 (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Biowest, Nuaillé, France), 1% chicken serum (Invitrogen), 50 µmol/l 2-mercaptoethanol (Sigma, St Louis, MO) and the appropriate antibiotics. The DT40 hybrids containing a single copy of human X chromosome fragment (a long arm deletion of X chromosome) were produced by MMCT from mouse A9 cells containing its fragment (established by Dr Hanaoka; unpublished) and maintained with 1.5 mg/ml G418 (Invitrogen). The HPRT-deficient CHO (JCRB0218) hybrids containing the HAC vector were constructed previously and maintained in Ham's F-12 nutrient mixture (Invitrogen) plus 10% fetal bovine serum with 8 µg/ml BS (Funakoshi, Tokyo, Japan). The HAC contains no endogenous genes, with modifications from the original one,19 i.e., containing an EGFP gene and the HSV-tk gene. hiMSCs were maintained in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum.33 Mouse ES cell culture (TT2-F, isolated from TT2) was performed as described previously.26
Construction of targeting vectors. The targeting vector, pDYS-H3 for introducing 3′ HPRT-loxP/bsd/tk, was constructed as follows: two 2.6 kb and 6.1 kb fragments for homologous arms corresponding to human dystrophin locus in AL050305 were amplified by PCR using primers DloxP4L/4R (3.1 kb), 5′-TCCACACGCCCTTTGATAGGAATAGGAT-3′ and 5′- CATCTGAGGGTTTTGGTTCTCGGTTTTC-3′; DloxP5L/5R (6.6kb), 5′-TGGAGAGAGTGAAGAGGGAAGGAGGAAA-3′ and 5′-CA CCTGCAGAACTCTCCACCCAAAAGTA-3′, digested with either EcoRI or BglII and sub-cloned into the equivalent sites of pKO Scrambler V901 backbone vector (Lexicon Genetics, Woodlands, TX), respectively, which contained the following three selection/insertion cassettes between the EcoRI and BglII sites: a 2 kb fragment from pMC1-tk (Stratagene, La Jolla, CA) containing the HSV-tk gene, a 2.3 kb XbaI/AscI fragment from pKO SelectHPRT V820 (Lexicon Genetics) containing the part of human HPRT gene with loxP site inserted at XbaI site of intron B and a 1.3 kb XhoI fragment from pCMV/bsd (Invitrogen) encoding the bsd gene. For construction of the chromosome truncation vectors pDYS-tel2, a PCR product from the human dystrophin locus in AC006060 was amplified by PCR with the following primers DYSt5L/5R (8.8 kb), (5′-TAAGG ATCCCCCTCATTGTAGCGAAGAGCAGG-3′ and 5′- CTGAGCCCTCA CCAGAATCACCTTGATA-3′), digested with both EcoRI and BamHI (7.5 kb) and sub-cloned into the EcoRI/BamHI sites of pBS-TEL/Puro vector.16
MMCT. The modified X fragment containing the human dystrophin gene and the 3′ HPRT-loxP was transferred from the DT40 hybrids into the CHO hybrids containing the HAC vector by MMCT technology.34 Briefly, microcells were prepared by centrifuge of 1 × 109 DT40 cells attached on flasks (Nalge Nunc, Rochester, NY) coated with poly-L-lysine (Sigma) and were fused to 1 × 106 CHO cells by 47% polyethylene glycol 1000 (WAKO, Osaka, Japan). CHO hybrids were selected in 0.8 mg/ml G418 and picked for expansion. After construction of the HAC vector containing the entire human dystrophin gene (DYS-HAC) by the site-specific translocation in CHO hybrids, transfer of the DYS-HAC vector from CHO cells to hiMSCs or mouse ES cells was performed using standard procedures.26,42 The hiMSC hybrids were selected in 4 µg/ml BS and the ES hybrids were selected in 4 µg/ml BS.
Genomic PCR analyses. Genomic DNA from cell lines and chimeric tissue specimens was extracted using a genomic extraction kit (Gentra System, Minneapolis, MN). PCR analyses were carried out using standard techniques. The primer pairs for the detection of the chromosome truncation at the AC006060 locus were: Sk23/DYSt5R (8.8 kb), 5′-GGCCGCTCTAGAACTAGTGGATC-3′ and 5′-CTGAGCCCTCACCAGAATCACCTTGATA-3′; Sk23/DYSt6R (9.1 kb), 5′-CACACCACTTTTTGAGTCCTTTGCTGCT-3′; PuroI/DYSt5R (9.8 kb), 5′-GAGCTGCAAGAACTCTTCCTCACG-3′; PuroI/DYSt6R (10.1 kb). The primer pairs for the confirmation of deleted loci of distal region on the modified human X chromosome fragment by the telomere-associated chromosome truncation were: PRKX F/R (131 bp), 5′-CTTCACGGCATAAGGACATCTC-3′ and 5′-TTCAAAACCTTCCTACAGTGTGG-3′; ARHGAP6 F/R (150 bp), 5′-CCACTGGGAATTTCATCACC-3′ and 5′-ATTTTTGACCAGCAGAT GGC-3′; TAB3 F/R (137 bp), 5′-TGGCACAAAGTTTTATCTGTGC-3′ and 5′-CCATTCCAAAATCCATTTGG-3′; FTHL17 F/R (298 bp), 5′-TGGCCC TGGAGAACTTCTT-3′ and 5′-ATGGTCTTGACTTGCTCGTG-3′. The primer pairs for the detection of the region of human dystrophin gene were: DYS1L/1R (168 bp), 5′- TGCTCTGGCTCATGTGTTTGC-3′ and 5′-AGCTCCCCTTTCGCATGATTC-3′; DYS1L/2R (331 bp), 5′- CCA TGCCAGCTGTTTTTCCTG-3′; DYS2L/2R (177 bp), 5′- CGAAAGGGG AGCTGTTGGAAT-3′; DYS3L/3R (163 bp), 5′-AACAACTGAAC AGCCGGTGGA-3′ and 5′-GGGGTGGTGGGTTGGATTTT-3′; DYS4L/ 4R (128 bp), 5′-GCAAGAGCAACAAAGTGGCCTA-3′ and 5′-AGCTTCTT CCAGCGTCCCTCA-3′; DYS5L/5R (132 bp), 5′-ACCTTCAGAA CCGGAGGCAAC-3′ and 5′-AGGGACCCTCCTTCCATGACTC-3′; DYS6L/6R (170 bp), 5′-TGGAACGCATTTTGGGTTGTT-3′ and 5′-AAAA CAATGCGCTGCCTCAAA-3′; DYS6L/7R (318 bp), 5′-AAACTCAA GCCTGCCCCACTC-3′; DYS7L/7R (151 bp), 5′-TTTGCATCCTTTT GGCGTGAT-3′; DYS7L/8R (706 bp); DYS8L/8R (155 bp), 5′-GCTGCTAGC AATGCCACGATT-3′ and 5′-GGATGGGCTGGGAATCCATAG-3′. The primer pairs for the detection of the loxP site targeting were: BsdR/DloxP3R (7.5 kb), 5′-GCTCAAGATGCCCCTGTTCT-3′ and 5′-CACCTGCAGAACTCTCCACCCAAAAGTA-3′; DloxP4L/4R (3.1 kb), 5′-TCCACACGCCCTTTGATAGGAATAGGAT-3′ and 5′-CATCTGAGGGTTTTGGTTCTCGGTTTTC-3′; DloxP5L/DloxP3R (6.6 kb), 5′-TGGAGAGAGTGAAGAGGGAAGGAGGAAA-3′. The primer pairs for the detection of the markers on the HAC vector were: PuroI/ Pcen1L (6.1 kb), 5′-ACTGCTGCCATGCAGACAGTTGTGCTTT-3′; EGFPL/#21CenG6R (6.0 kb), 5′-CCTGAAGTTCATCTGCACCA-3′ and 5′-CCCGGCCAGATTCAGATTTTTATTAGGG-3′; PGKr2/#21Cen <1>2L (4.5 kb), 5′-ATCTGCACGAGACTAGTGAGACGTGCTA-3′ and 5′-AAATGCATCACCATTCTCCCAGTTACCC-3′; Sk23/ q2L (7.4 kb), 5′-TCATGCCACAATCAATCTCCCAAGTAGC-3′. The primer pairs for the detection of the HPRT gene reconstruction were: TransL1/R1 (409 bp), 5′-TGGAGGCCATAAACAAGAAGAC-3′ and 5′-CCCCTTGACCCAGAAATTCCA-3′.
Reverse transcription–PCR analyses. Total RNA was prepared from chimeric tissue specimens using ISOGEN (Nippon Gene, Tokyo, Japan). Prepared RNA was treated with RNase-free DNase I (WAKO) and cleaned up using RNeasy columns according to the manufacturer's instructions (Qiagen, Hilden, Germany). First-strand cDNA synthesis was carried out using an oligo-(dT)20 primer and the SuperScript III reverse transcriptase (Invitrogen). Human tissue cDNAs from the multiple tissue cDNA panel I and II (Clontech, Palo Alto, CA) were used as a control. PCR was performed with the cDNA using AmpliTaq Gold (Perkin Elmer, Waltham, MA). Amplifications were performed with an annealing temperature of 58 °C for 30–35 cycles and then the amplified fragments were resolved by electrophoresis on a 2% agarose gel, followed by staining with ethidium bromide. The primer sequences were as follows: for dystrophin, DYS 427me1L/DYS427me1R (211 bp), 5′-TTCCCCCTACAGGACTCAGA-3′ and 5′-TCTTCCCACCAAAGCATTTT-3′; DYS427le1L/DYSe3R (150 bp), 5′-CTCATGATGAAAGAGAAGATGTTCAA-3′ and 5′-CTGTCAGGCC TTCGAGGA-3′; DYS 140e1L/DYS e45R (189 bp), 5′-TGCTGGCTGC TCTGAACTAA-3′ and 5′-GGCTTCCCAATTTTTCCTGT-3′; for EGFP, EGFPL/R (479 bp), 5′-CCTGAAGTTCATCTGCACCA-3′ and 5′-TGCTC AGGTAGTGGTTGTCG; for GAPDH (mouse and human), RPC1/2, 5′-CCATCTTCCAGGAGCGAGA-3′ and 5′-TGTCATACCAGGAAA TGAGC-3′. For DNA transfection, FISH, generation of chimeric mice containing the DYS-HAC and immunofluorescence staining, see the Supplementary Materials and Methods.
Supplementary MaterialSupplementary Materials and Methods.
Supplementary Materials
Acknowledgments
We thank Junya Toguchida (Kyoto University, Kyoto, Japan) for providing the hiMSCs; Glenn E. Morris (Keele University, Shropshire, UK) for providing the MANDYS106 antibody; Kazunori Hanaoka (Kitasato University, Kanagawa, Japan) for producing and providing the DT40 hybrids retaining a single copy of the human X chromosome fragment; our members, Yoshiteru Kai, Mitsuhiko Osaki, Junya Nakamura, Yuichi Iida, Saori Tsuji, Hitedoshi Yamada, Kei Hiramatsu, Haruaki Ninomiya for technical assistance; and Akihiro Kurimasa, Hiroyuki Kugoh, Toshiaki Inoue, Masaharu Hiratsuka, Tetsuya Ohbayashi and Motonobu Katoh for valuable discussions. We also thank David Gilley (Indiana University, Indianapolis, IN) for helpful suggestions. This study was supported by the 21st Century COE program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (M.O.), the Grant-in-Aid for Scientific Research (B) from Ministry of Education, Culture, Sports, Science and Technology of Japan (M.O.).
REFERENCES
- Koenig M, Monaco AP., and , Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Chelly J, Hamard G, Koulakoff A, Kaplan JC, Kahn A., and , Berwald-Netter Y. Dystrophin gene transcribed from different promoters in neuronal and glial cells. Nature. 1990;344:64–65. doi: 10.1038/344064a0. [DOI] [PubMed] [Google Scholar]
- Zhao J, Uchino M, Yoshioka K, Miyatake M., and , Miike T. Dystrophin in control and mdx retina. Brain Dev. 1991;13:135–137. doi: 10.1016/s0387-7604(12)80123-9. [DOI] [PubMed] [Google Scholar]
- Byers TJ, Kunkel LM., and , Watkins SC. The subcellular distribution of dystrophin in mouse skeletal, cardiac, and smooth muscle. J Cell Biol. 1991;115:411–421. doi: 10.1083/jcb.115.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perloff JK, de Leon AC., Jr, and , O'Doherty D. The cardiomyopathy of progressive muscular dystrophy. Circulation. 1966;33:625–648. doi: 10.1161/01.cir.33.4.625. [DOI] [PubMed] [Google Scholar]
- Quenneville SP., and , Tremblay JP. Ex vivo modification of cells to induce a muscle-based expression. Curr Gene Ther. 2006;6:625–632. doi: 10.2174/156652306779010651. [DOI] [PubMed] [Google Scholar]
- Foster K, Foster H., and , Dickson JG. Gene therapy progress and prospects: Duchenne muscular dystrophy. Gene Ther. 2006;13:1677–1685. doi: 10.1038/sj.gt.3302877. [DOI] [PubMed] [Google Scholar]
- Rando TA. Non-viral gene therapy for Duchenne muscular dystrophy: progress and challenges. Biochim Biophys Acta. 2007;1772:263–271. doi: 10.1016/j.bbadis.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Wells DJ. Therapeutic restoration of dystrophin expression in Duchenne muscular dystrophy. J Muscle Res Cell Motil. 2006;27:387–398. doi: 10.1007/s10974-006-9081-6. [DOI] [PubMed] [Google Scholar]
- O'Connor TP., and , Crystal RG. Genetic medicines: treatment strategies for hereditary disorders. Nat Rev Genet. 2006;7:261–276. doi: 10.1038/nrg1829. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Zhu X, Hadhazy M, Groh ME, Wheeler MT, Wollmann R., and , McNally EM. Overexpression of gamma-sarcoglycan induces severe muscular dystrophy. Implications for the regulation of Sarcoglycan assembly. J Biol Chem. 2001;276:21785–21790. doi: 10.1074/jbc.M101877200. [DOI] [PubMed] [Google Scholar]
- Muntoni F, Torelli S., and , Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C., and , Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K., and , Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Shinohara T, Notsu T, Tomizuka K, Yoshida H, Takeda S, et al. Efficient modification of a human chromosome by telomere-directed truncation in high homologous recombination-proficient chicken DT40 cells. Nucleic Acids Res. 1998;26:3447–3448. doi: 10.1093/nar/26.14.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci C, Ascenzioni F, Auriche C, Piccolella E, Guerrini AM., and , Donini P. Use of a human minichromosome as a cloning and expression vector for mammalian cells. Hum Mol Genet. 1999;8:1417–1424. doi: 10.1093/hmg/8.8.1417. [DOI] [PubMed] [Google Scholar]
- Grimes BR, Schindelhauer D, McGill NI, Ross A, Ebersole TA., and , Cooke HJ. Stable gene expression from a mammalian artificial chromosome. EMBO Rep. 2001;2:910–914. doi: 10.1093/embo-reports/kve187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Ayabe F, Norikane S, Okada T, Masumoto H, Horike S, et al. Construction of a novel human artificial chromosome vector for gene delivery. Biochem Biophys Res Commun. 2004;321:280–290. doi: 10.1016/j.bbrc.2004.06.145. [DOI] [PubMed] [Google Scholar]
- Larin Z., and , Mejia JE. Advances in human artificial chromosome technology. Trends Genet. 2002;18:313–319. doi: 10.1016/S0168-9525(02)02679-3. [DOI] [PubMed] [Google Scholar]
- Oshimura M., and , Katoh M. Transfer of human artificial chromosome vectors into stem cells. Reprod Biomed Online. 2008;16:57–69. doi: 10.1016/s1472-6483(10)60557-3. [DOI] [PubMed] [Google Scholar]
- Basu J., and , Willard HF. Human artificial chromosomes: potential applications and clinical considerations. Pediatr Clin North Am. 2006;53:843, viii–853. doi: 10.1016/j.pcl.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Nishii K, Okazaki T., and , Ikeno M. Human artificial chromosomes constructed using the bottom-up strategy are stably maintained in mitosis and efficiently transmissible to progeny mice. J Biol Chem. 2006;281:26615–26623. doi: 10.1074/jbc.M603053200. [DOI] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuki Y, Hoshiya H, Kai Y, Abe S, Takiguchi M, Osaki M, et al. Correction of a genetic defect in multipotent germline stem cells using a human artificial chromosome. Gene Ther. 2008;15:617–624. doi: 10.1038/sj.gt.3303091. [DOI] [PubMed] [Google Scholar]
- Tomizuka K, Yoshida H, Uejima H, Kugoh H, Sato K, Ohguma A, et al. Functional expression and germline transmission of a human chromosome fragment in chimaeric mice. Nat Genet. 1997;16:133–143. doi: 10.1038/ng0697-133. [DOI] [PubMed] [Google Scholar]
- Buerstedde JM., and , Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- Itzhaki JE, Barnett MA, MacCarthy AB, Buckle VJ, Brown WR., and , Porter AC. Targeted breakage of a human chromosome mediated by cloned human telomeric DNA. Nat Genet. 1992;2:283–287. doi: 10.1038/ng1292-283. [DOI] [PubMed] [Google Scholar]
- Kazuki Y, Kimura M, Nishigaki R, Kai Y, Abe S, Okita C, et al. Human chromosome 21q22.2-qter carries a gene(s) responsible for downregulation of mlc2a and PEBP in Down syndrome model mice. Biochem Biophys Res Commun. 2004;317:491–499. doi: 10.1016/j.bbrc.2004.03.069. [DOI] [PubMed] [Google Scholar]
- Baubonis W., and , Sauer B. Genomic targeting with purified Cre recombinase. Nucleic Acids Res. 1993;21:2025–2029. doi: 10.1093/nar/21.9.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH., Jr, and , Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Zubrzycka-Gaarn EE, Bulman DE, Karpati G, Burghes AH, Belfall B, Klamut HJ, et al. The Duchenne muscular dystrophy gene product is localized in sarcolemma of human skeletal muscle. Nature. 1988;333:466–469. doi: 10.1038/333466a0. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Aoyama T, Nakayama T, Nakamata T, Hosaka T, Nishijo K, et al. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem Biophys Res Commun. 2002;295:354–361. doi: 10.1016/s0006-291x(02)00661-7. [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Tomizuka K, Shinohara T, Kazuki Y, Yoshida H, Ohguma A, et al. Manipulation of human minichromosomes to carry greater than megabase-sized chromosome inserts. Nat Biotechnol. 2000;18:1086–1090. doi: 10.1038/80287. [DOI] [PubMed] [Google Scholar]
- t Hoen PA, de Meijer EJ, Boer JM, Vossen RH, Turk R, Maatman RG, et al. Generation and characterization of transgenic mice with the full-length human DMD gene. J Biol Chem. 2008;283:5899–5907. doi: 10.1074/jbc.M709410200. [DOI] [PubMed] [Google Scholar]
- Pravtcheva DD., and , Wise TL. A postimplantation lethal mutation induced by transgene insertion on mouse chromosome 8. Genomics. 1995;30:529–544. doi: 10.1006/geno.1995.1274. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K., and , Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Sampaolesi M, Blot S, D'Antona G, Granger N, Tonlorenzi R, Innocenzi A, et al. Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 2006;444:574–579. doi: 10.1038/nature05282. [DOI] [PubMed] [Google Scholar]
- Porada CD, Zanjani ED., and , Almeida-Porad G. Adult mesenchymal stem cells: a pluripotent population with multiple applications. Curr Stem Cell Res Ther. 2006;1:365–369. doi: 10.2174/157488806778226821. [DOI] [PubMed] [Google Scholar]
- Dezawa M, Ishikawa H, Itokazu Y, Yoshihara T, Hoshino M, Takeda S, et al. Bone marrow stromal cells generate muscle cells and repair muscle degeneration. Science. 2005;309:314–317. doi: 10.1126/science.1110364. [DOI] [PubMed] [Google Scholar]
- Kugoh HM, Hashiba H, Shimizu M., and , Oshimura M. Suggestive evidence for functionally distinct, tumor-suppressor genes on chromosomes 1 and 11 for a human fibrosarcoma cell line, HT1080. Oncogene. 1990;5:1637–1644. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.