Abstract
Spliceosome-mediated RNA trans-splicing has emerged as an exciting mode of RNA therapy. Here we describe a novel trans-splicing strategy, which targets highly abundant pre-mRNAs, to produce therapeutic proteins in vivo. First, we used a pre-trans-splicing molecule (PTM) that mediated trans-splicing of human apolipoprotein A-I (hapoA-I) into the highly abundant mouse albumin exon 1. Hydrodynamic tail vein injection of the hapoA-I PTM plasmid in mice followed by analysis of the chimeric transcripts and protein, confirmed accurate and efficient trans-splicing into albumin pre-mRNA and production of hapoA-I protein. The versatility of this approach was demonstrated by producing functional human papillomavirus type-16 E7 (HPV16-E7) single-chain antibody in C57BL/6 mice and functional factor VIII (FVIII) and phenotypic correction in hemophilia A mice. Altogether, these studies demonstrate that trans-splicing to highly abundant albumin transcripts can be used as a general platform to produce therapeutic proteins in vivo.
Introduction
Although frequently encountered in flatworms, trypanosomes and nematodes, trans-splicing between two separate pre-mRNAs to produce a chimeric mRNA is a relatively rare event in mammalian cells.1,2,3 Nonetheless, the ability to harness the natural RNA splicing machinery to reprogram a message, combined with the advantages that such a mechanism confers, has brought spliceosome-mediated RNA trans-splicing (SMaRT) technology to the forefront of RNA therapeutic and diagnostic applications.4,5,6
Unlike conventional gene therapies that utilize cDNA or minigene delivery to provide a therapeutic effect, delivery of an engineered pre-trans-splicing molecule (PTM, Figure 1) depends on the expression of the targeted pre-mRNA. While trans-splicing to an endogenous pre-mRNA has been successful for a number of disease models including cystic fibrosis, hemophilia A, tauopathies, spinal muscular atrophy, among others,7,8,9,10,11,12,13 achieving therapeutic levels of a trans-spliced product in vivo has been hampered by the relatively low abundance of certain targeted pre-mRNA(s). In other work, we have shown that efficiency of trans-splicing is directly proportional to the available target level (unpublished data), therefore finding a means to increase the endogenous target level can help to maximize the success of trans-splicing in vivo.
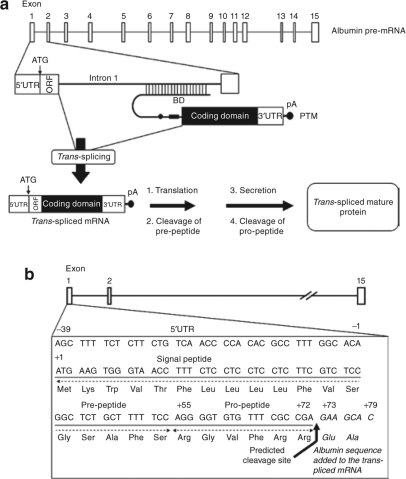
Figure 1.
Overview of the albumin trans-splicing strategy. (a) Trans-splicing between the PTM and the albumin pre-mRNA results in a chimeric mRNA containing the signal peptide and initiator ATG from albumin fused to the coding domain from the PTM. Translation of this mRNA leads to the expression and secretion of the trans-spliced mature protein. BD, binding domain and pA, polyadenylation signal. (b) Nucleotide sequence of the mouse albumin exon 1 showing the 5′UTR, signal peptide, predicted signal peptide cleavage site (indicated by arrow) and the albumin sequence that is added into the trans-spliced mRNA (in italic).
To significantly improve the efficiency of trans-splicing, we have developed a novel strategy whereby highly abundant albumin pre-mRNA is targeted for trans-splicing. In humans, albumin represents ~54% of the protein mass in plasma (35–50 mg/ml), and is the most abundantly produced protein in the human liver.14,15 Indeed, the albumin pre-mRNA is present at concentrations that dwarf most liver primary transcripts. Even modest trans-splicing efficiency (1–5% of albumin pre-mRNA) would potentially result in the expression of protein in the microgram to milligram range, a level that would provide therapeutic benefit for a wide range of diseases. This strategy would minimally reduce albumin levels, which would have few if any negative consequences.14,16,17 Therefore, we conjectured that highly abundant transcripts, such as albumin, represent an ideal target for conversion and for future therapeutic applications of SMaRT.
We tested this conjecture and here we describe the development of this novel strategy for targeting highly abundant albumin pre-mRNA to produce therapeutic proteins in vivo. We have obtained proof-of-concept using three different models: production of human apoA-I protein and synthesis of single-chain monoclonal antibody specific for the human papillomavirus type-16 E7 (HPV-E7) oncoprotein in C57BL/6 mice, and production of functional factor VIII (FVIII) and phenotypic correction in hemophilia A mice. The results presented here demonstrate that trans-splicing to highly abundant transcripts such as albumin can be used as a general platform to produce therapeutic proteins in vivo.
Results
Trans-splicing into albumin: a general strategy
The primary goal of this study was to demonstrate that trans-splicing to a highly abundant albumin pre-mRNA target could be used as a general strategy to produce therapeutic levels of proteins in plasma in vivo. This trans-splicing strategy targeting mouse albumin (mAlb) pre-mRNA is illustrated in Figure 1a. The albumin exon 1 is 118 nucleotides from the transcription initiation site to the first exon/intron junction (Figure 1b). The 5′UTR sequence extends from nucleotides −39 to −1 (numbering relative to the albumin open reading frame (ORF)), and nucleotides +1 through +72 encode the N-terminal signal peptide.18,19 Any PTM trans-splicing to albumin exon 1 is expected to produce a chimeric mRNA that contains 118 nucleotides from albumin mRNA, which encode the first 26 amino acids of unprocessed albumin, followed by the PTM coding sequence. The trans-spliced chimeric mRNA contains the initiating ATG from albumin in frame with the gene of interest. This mRNA is predicted to translate in the liver as a precursor protein and the N-terminal signal peptide should be cleaved, to produce the mature protein that contains two additional amino acids in-frame and immediately upstream of the gene of interest (Figure 1b).
Trans-spliced albumin-human apoA-I chimeric protein is functionally active
Before developing PTMs for specific therapeutic applications, we sought to determine whether the presence of the additional amino acids contributed by albumin would inhibit or impair the function of the trans-spliced product. We chose human apoA-I (hapoA-I), the major component of high-density lipoprotein (HDL), as our initial model because of its athero-protective properties and its ability to reduce plaque size when administered intravenously as a therapeutic agent.20 The albumin-targeting strategy described here was predicted to result in the production of hapoA-I protein in hepatocytes, which are the primary site of synthesis of endogenous apoA-I.21 To test this strategy, we created a chimeric cDNA to precisely recapitulate the sequences produced by trans-splicing to endogenous albumin and tested for function. Properly cleaved and secreted, chimeric mouse albumin-hapoA-I (mAlb-hapoA-I) protein predicted to contain two amino acid changes (Asp, Glu → Glu, Ala) at the N-terminus relative to wild-type (wt) hapoA-I. Transfection of HEK293 cells with pc3.1-mAlb-hapoA-I chimeric cDNA plasmid or wt hapoA-I cDNA (pc3.1-hapoA-I) followed by western blot analysis confirmed >90% of the mature hapoA-I protein in the supernatant, thereby demonstrating that the mAlb-hapoA-I chimeric product could be processed and secreted (Supplementary Figure S1).
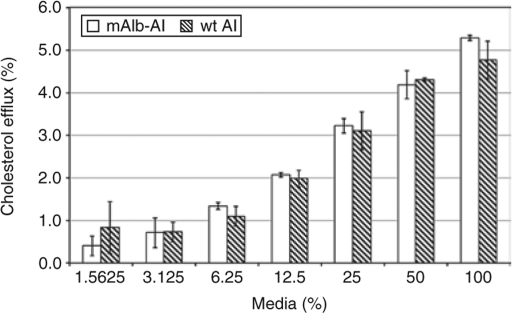
The effect of the altered amino acids on hapoA-I function was evaluated using a cholesterol efflux functional assay.22 HeLa cells stably expressing an ATP-binding cassette transporter (ABC1) were loaded with 3H cholesterol. After equilibrating for 24 hours, cells were incubated with a serial dilution of supernatant from cells transfected with the pc3.1-mAlb-hapoA-I chimeric cDNA plasmid or with the wt hapoA-I plasmid. The amount of ABC1-mediated efflux observed with mAlb-hapoA-I chimeric protein was similar to that of wt hapoA-I (Figure 2). These data indicate that the two additional amino acids at the N-terminus did not adversely affect the function of the chimeric hapoA-I protein.
Figure 2.
Trans-spliced albumin-hapoA-I protein is functionally active. Cholesterol efflux assay was performed as described previously22 by analyzing the supernatant collected from cells transfected with pc3.1-mAlb-hapoA-I chimeric cDNA or pc3.1-hapoA-I wt cDNA expression plasmid. Values are plotted as a mean value and the standard error of the mean. mAlb-AI, mouse albumin-hapoA-I chimeric cDNA plasmid, and wt AI, wt human apoA-I cDNA plasmid.
Trans-splicing to endogenous mouse albumin pre-mRNA produced human apoA-I protein in vivo
The first step in PTM development is the design of a binding domain (BD), a sequence complementary to a region of the targeted pre-mRNA that modulates both trans-splicing specificity and efficiency. Using a fluorescence-based high-throughput screen, several lead candidate BDs to mouse albumin intron 1 were isolated from an initial pool of >106 candidates.23 The lead BD, 97C2, obtained from this screen was cloned into a PTM containing a trans-splicing domain (TSD: spacer, branch point, polypyrimidine tract and 3′ splice site) followed by the coding sequence for human apoA-I and its 3′UTR (Figure 3a). The entire cassette was cloned into a modified pcDNA3.1(−) vector, which contains a MAZ4 transcriptional pause site upstream of a CMV-IE or ApoE/hAAT promoter to reduce read-through transcription.24,25 A negative control splicing-defective PTM1 (SM-PTM1) containing a point mutation at the 3′ splice site (cag → cat) was also constructed using pc3B-97C2-(CMV) as the starting plasmid.
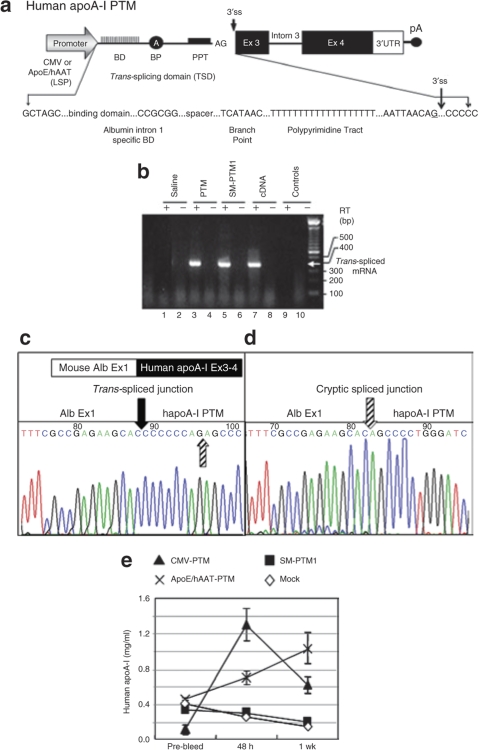
Figure 3.
Trans-splicing into endogenous albumin pre-mRNA and production of hapoA-I protein in vivo. (a) Schematic diagram of a prototype hapoA-I PTM. The PTM consists of mouse albumin intron 1-specific binding domain (BD), spacer, branch point (BP), polypyrimidine tract (PPT), 3′ splice site followed by the human apoA-I coding sequence including intron 3 and its 3′UTR and the polyA site. Underlined nt “G” at the splice junction is the nt modified to create the splice mutant PTM1 (SM-PTM1). Human apoA-I PTM (pc3B-97C2) either with the CMV or ApoE/hAAT liver-specific promoter or SM-PTM1 or saline (Mock) was HD-injected into C57BL/6 mice. Serum and liver tissue was collected at the indicated time points and analyzed by enzyme-linked immunosorbent assay (ELISA) and RT-PCR. (b) Endpoint RT-PCR followed by sequence analysis confirmed the correct trans-splicing between the mouse albumin target and the functional hapoA-I PTM (c) and (d) cryptic trans-splicing with the SM-PTM1. (e) In vivo production of hapoA-I protein. Plasma was analyzed for the production of hapoA-I protein by ELISA as described in Materials and Methods. Values are plotted as a mean value and the standard error of the mean. SM-PTM1, splicing-defective PTM1; CMV-PTM, pc3B-97C2-(CMV) and ApoE/hAAT-PTM, pc3B-97C2-(LSP). Solid arrow indicates correct splice junction (c) and striped arrows indicate, cryptic splice junction (d).
To demonstrate PTM-mediated trans-splicing to the endogenous mouse albumin pre-mRNA and production of functional mAlb-hapoA-I chimeric protein in vivo, 50 µg of the plasmid encoding hapoA-I PTM (pc3B-97C2) containing either the cytomegalovirus (CMV) or ApoE/hAAT liver-specific promoter/enhancer or SM-PTM1 or saline were hydrodynamically (HD) injected into the tail vein of 8-week-old C57BL/6 mice.26,27 Serum and liver tissue was collected at 48 hours and 1 week after injection and analyzed for trans-splicing. Endpoint RT-PCR analysis using specific primers spanning the trans-spliced junction produced the predicted 390 bp product in mice that received the functional PTM (Figure 3b, lane 3) which co-migrated with the positive control (lane 7). No product was detected in mice treated with saline (lane 1) or in samples where reverse transcription (RT) was omitted (lanes 2, 4, 6, 8 and 10). Sequencing of the RT-PCR product confirmed accurate trans-splicing between the hapoA-I PTM and the endogenous albumin exon 1 (Figure 3c). The SM-PTM1 did not trans-splice at the mutated 3′ splice site, but rather to a cryptic 3′ splice site located eight nucleotides downstream (Figure 3d) in the PTM coding domain resulting in a truncated trans-spliced product (Figure 3b, lane 5).
Next we analyzed sera by enzyme-linked immunosorbent assay for the production of hapoA-I protein and confirmed the production of mAlb-hapoA-I chimeric protein in mice that received the functional PTM (Figure 3e). In contrast, mice that received either saline or the SM-PTM1 showed no significant activity over prebleed (Figure 3e). An increase in protein expression was observed at 48 hours for the CMV-driven hapoA-I PTM, followed by an apparent silencing of the promoter. Expression from the ApoE/hAAT liver-specific promoter continued to increase throughout the duration of the experiment. Real-time quantitative RT-PCR (qRT-PCR) analysis further confirmed expression of the PTM and trans-splicing in these mice (Supplementary Figure S2).
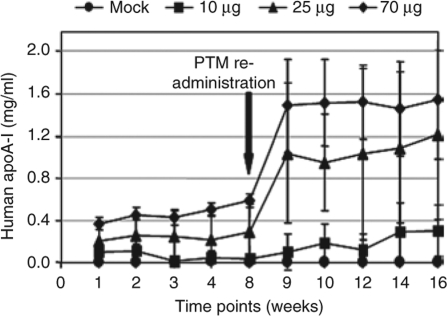
To increase and sustain expression levels of the PTM for subsequent long-term studies, we cloned the hapoA-I PTM containing the ApoE/hAAT promoter into a minicircle vector engineered to be devoid of bacterial sequences that have been proposed to be responsible for transgene silencing.28,29,30 Three different doses of the hapoA-I minicircle PTM (10, 25, and 70 µg) were HD-injected into C57BL/6 mice and mAlb-hapoA-I chimeric protein expression was measured weekly by enzyme-linked immunosorbent assay using a hapoA-I-specific antibody (Figure 4). A persistent, dose response was observed for all groups receiving the PTM. After 8 weeks, minicircle PTM DNA was readministered at the same dose as the first injection. This produced a three- to fivefold increase in mAlb-hapoA-I chimeric protein which continued for an additional 8 weeks (Figure 4), suggesting the absence of an immune response to either the hapoA-I protein or the minicircle PTM. This was further confirmed by analyzing the sera for liver toxicity which showed a transient increase in liver enzymes, alanine aminotransferase and aspartate aminotransferase as reported previously (refs. 27,31 and data not shown). In addition, we also performed an enzyme-linked immunosorbent assay on serum from mice expressing human apoA-I protein and were unable to detect any mouse anti-HDL antibodies (data not shown).
Figure 4.
Re-administration of minicircle PTM results in increased human apoA-I protein in vivo. C57BL/6 mice HD-injected with 10 µg to 70 µg of the minicircle encoding hapoA-I PTM. A same dose of the minicircle PTM was readministered after 8 weeks of the initial injection. Serum was collected at the indicated time points and analyzed by enzyme-linked immunosorbent assay. Values are plotted as a mean value and the standard error of the mean.
The results described above demonstrate the albumin-targeting strategy can be used to produce functional human apoA-I protein in vivo. Additionally, the studies above show that the albumin-targeting strategy can be mediated by different delivery vehicles, such as minicircle DNA vectors, which can be used for long-term expression of the PTM.
Trans-splicing to albumin pre-mRNA produced HPV-E7-specific single-chain antibodies in vivo
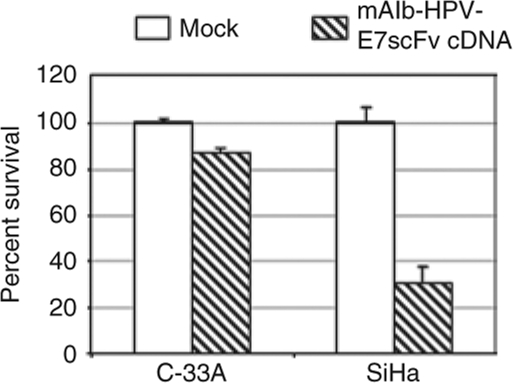
To evaluate the new strategy to produce proteins not normally produced in the liver, we tested the ability of albumin trans-splicing to produce a single-chain monoclonal antibody (mAb). We tested the production of a mAb specific for the HPV-E7 oncoprotein. Similar to hapoA-I, we first tested the effect of the albumin sequence on mouse albumin-HPV-E7 single-chain antibody (mAlb-HPV-E7scFv) function based on the ability of the chimeric protein to downregulate HPV-E7 expression in cervical cancer cells.32 SiHa cells, an HPV-E7 positive cervical cancer cell line, and a matched negative control cell line, C-33A, were transfected with the pc3.1-mAlb-HPV-E7scFv chimeric cDNA plasmid or pcDNA3.1(−) control vector and cell viability was determined by an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. SiHa cell proliferation was inhibited by ~75% compared to ~15% inhibition observed with C-33A cells which confirmed the production of functional mAlb-HPV-E7scFv (Figure 5).
Figure 5.
Expression of the functional mAlb-HPV-E7scFv in vitro. SiHa, an HPV-E7 positive cervical cancer cell line, and a control cell line, C-33A, were transfected with the pc3.1-mAlb-HPV-E7scFv cDNA plasmid or pcDNA3.1(−) control vector (Mock). Five days after transfection, number of viable cells was determined by a colorimetric MTT assay in quadruplicate and the background absorbance subtracted. Results are an average of two independent transfections.
To demonstrate the ability of the HPV-E7scFv PTM to trans-splice to the mouse albumin pre-mRNA and produce a functional mAlb-HPV-E7scFv protein in vivo, 100 µg of pc3B-97C2-HPV-E7scFv PTM or pc3.1-mAlb-HPV-E7scFv chimeric cDNA or saline was HD-injected into the tail vein of C57BL/6 mice. Serum was collected at different time points and analyzed by western blotting. The mAlb-HPV-E7scFv chimeric protein was detected in plasma as early as 8 hours after injection in mice receiving either the cDNA or PTM (data not shown). Although expression levels began to drop, expression and secretion of the mAlb-HPV-E7scFv remained detectable in mice receiving the active PTM at 24 hours (Supplementary Figure S3a). Real-time qRT-PCR analysis confirmed the presence of the PTM and trans-spliced mRNA in these mice (Supplementary Figure S3b). Although preliminary, these data extended the albumin trans-splicing strategy to proteins not normally produced in the liver, suggesting that the approach could be easily generalized.
Trans-splicing of FVIII to albumin pre-mRNA
To demonstrate that the albumin strategy can be used as a general platform for correction of genetic disorders, we decided to test whether trans-splicing of FVIII sequences to albumin pre-mRNA could produce circulating FVIII in vivo. Contrary to the therapeutic effects of hapoA-1 and single-chain mAbs, which require long-term expression, the therapeutic effect of FVIII, a decrease in clotting time, can be ascertained soon after expression of the protein in hemophilia A mice. Moreover, previous studies have demonstrated successful correction of the coagulation deficiency in hemophilia A mice by conventional SMaRT, trans-splicing the normal FVIII sequences (exons 16 to 26) to endogenous FVIII pre-mRNA using plasmids or viral vectors.8
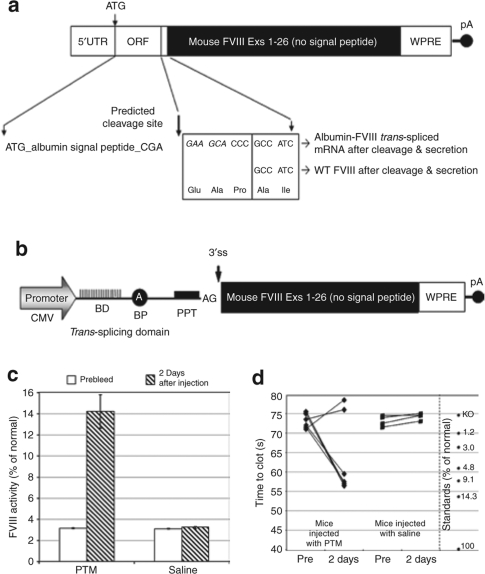
Trans-splicing of FVIII to albumin pre-mRNA would result in a chimeric product with the signal peptide derived from albumin. Therefore as a first step we assessed the effect of the amino acid alterations using a chimeric mouse albumin-FVIII cDNA containing sequences identical to those expected from trans-splicing between mouse albumin pre-mRNA and a mouse FVIII PTM (Figure 6a). Testing of this chimeric cDNA (pc3.1-mAlb-FVIII) in vitro and in hemophilia A mice along with the wt FVIII cDNA showed no major difference in FVIII activity levels between the two constructs (Supplementary Figure S4). This demonstrated that a chimera between mouse albumin and FVIII is capable of producing functional FVIII.
Figure 6.
Trans-splicing into albumin restores coagulant activity in hemophilia A mice. Schematic diagrams of (a) mouse albumin-FVIII trans-spliced chimeric mRNA and (b) an albumin-targeted mouse FVIII PTM. BD, binding domain; BP, branch point; PPT, polypyrimidine tract; pA, polyadenylation signal; 3′ ss, 3′ splice site. Hemophilia A mice were HD-injected with 100 µg of the pc3B-97C2-FVIII PTM or saline. Plasma samples were collected at 2 days after injection and analyzed by the Coatest assay. Values are plotted as a mean value and the standard error of the mean. (c) Mouse FVIII activity was assayed using the Coatest SP4 FVIII kit and (d) clotting times were assessed using the activated partial thromboplastin time assay as described in Materials and Methods. Five out of seven mice treated with PTM showed a significant reduction in clotting times compared to prebleed values (P = 0.02) and to saline-treated mice (P = 0.01).
To test whether trans-splicing between a FVIII PTM and albumin pre-mRNA could generate circulating FVIII in hemophilia A mice, the hapoA-I PTM was modified to encode exons 1 to 26 of mouse FVIII (B-domain deleted), excluding the N-terminal signal peptide necessary for secretion of processed FVIII. The PTM plasmid used for this study contains a CMV promoter, a woodchuck post-transcriptional regulatory element sequence in the 3′UTR and the SV40 polyadenylation signal (Figure 6b). Without the signal peptide sequence it was presumed that no FVIII activity could be generated by this PTM. Similar to hapoA-I, the signal peptide acquired from albumin during trans-splicing would provide the necessary signals for processing and secretion of FVIII. The hemophilia A mice were HD-injected with 100 µg of pc3B-97C2-FVIII PTM or saline as a negative control. A dose of 100 µg was chosen as a starting point because this amount was similar on a molar basis to that used for the human apoA1 studies described above. Plasma was collected at 2 days after injection and assayed using the Coatest FVIII SP4 kit (Chromogenix, Milan, Italy). Background activity in the plasma of naive hemophilia A mice ranged from ~2.5 to 3.0% of normal. Mice that received the functional PTM showed significant levels of FVIII activity with a mean value of 14.2 ± 1.6% (n = 7; P = 0.0004) whereas mice that received saline showed no significant activity over prebleed levels (Figure 6c). Endpoint RT-PCR analysis of the liver total RNA and sequencing confirmed accurate trans-splicing between the FVIII PTM and albumin exon 1 (data not shown).
To rule out any possibility that the PTM was being translated in the absence of trans-splicing and creating residual FVIII activity, or that protein trans-complementation was occurring between a light chain protein from the PTM and a heavy chain protein expressed in the hemophilia A mouse, a splicing-defective PTM was generated by mutating the splice acceptor site of pc3B-97-C2-FVIII. Pretesting of this mutant PTM in vitro showed that the PTM was indeed nonfunctional (data not shown). The same splicing-defective PTM was HD-injected into hemophilia A mice and plasma was collected at 2 days after injection. Coatest analysis of the plasma showed no FVIII activity above prebleed levels (prebleed = 2.8 ± 0.07% and 2 days = 2.8 ± 0.11%; n = 4). These results confirm that the FVIII activity generated by the functional PTM is through trans-splicing of the PTM into albumin pre-mRNA and not due to protein trans-complementation.
To confirm coagulant activity in PTM-treated mice, plasma was analyzed using the activated partial thromboplastin time assay. Five out of seven samples showed significantly reduced clotting times with an average of 74 seconds for the prebleed samples and 57 seconds for the samples taken at 2 days after injection (Figure 6d). Statistical comparisons showed there to be a significant difference between the prebleed and PTM-treated samples (P = 0.02) and between the PTM and saline-treated mice (P = 0.01). The average values for the normal and knockout mouse pooled plasma were 41 and 75 seconds, respectively. The two plasma samples that did not have a reduced clotting time could have been compromised during collection and analysis as Coatest and qRT-PCR assays clearly showed the presence of FVIII activity (Figure 6c) and high levels of PTM and trans-spliced FVIII mRNA (data not shown). Additionally we recently performed a similar study with a PTM in minicircle form (see below) and here all PTM-treated mice showed a significant difference in clotting times compared to saline-treated mice (Supplementary Figure S5).
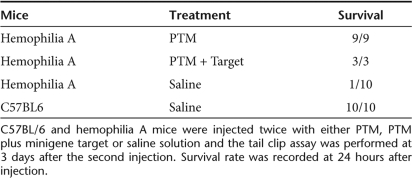
To demonstrate that the FVIII produced through trans-splicing to albumin can correct the bleeding disorder in hemophilia A mice we performed a tail clip survival study. For this assay it is necessary to have sustained FVIII production and so we used the same FVIII PTM described above but in a minicircle DNA vector (p2ф97C2-FVIII-LSP). Hemophilia A mice were injected twice with either 50 µg of PTM alone (p2ф97C2-FVIII-LSP) or with 50 µg of PTM plus 50 µg of minigene target at day 0 and day 3. Wild-type C57BL/6 mice and hemophilia A mice injected twice with saline served as the positive and negative controls, respectively. Following tail clip mice were checked for survival after 24 hours. As expected all of the normal C57BL/6 mice (n = 10) survived the challenge, while 9 out of 10 hemophila A mice treated with saline died within 24 hours of the tail clip (Table 1). Remarkably all the hemophilia A mice treated with PTM alone (n = 9) or PTM plus minigene target (n = 3) survived the tail clip. These studies are the first demonstration that a FVIII PTM targeted to an endogenous, heterologous gene can be used to generate functional circulating FVIII and correct the bleeding phenotype in a murine model of hemophilia A.
Table 1.
Assessment of phenotypic correction in hemophilia A mice by the tail clip assay
Discussion
In this report we describe the development of a novel SMaRT strategy for targeting highly abundant albumin pre-mRNA to produce high levels of therapeutic proteins and antibodies. We demonstrate that this approach has broad applications and can be easily adapted to produce proteins of interest in vivo. Proof-of-concept was confirmed by the production of functional (i) human apoA-I protein, (ii) HPV-E7 single-chain monoclonal antibody in normal mice, and (iii) circulating FVIII levels up to 11% (above background) of normal and phenotypic correction in hemophilia A mice. Furthermore, using minicircle, a viable and attractive nonviral delivery system, we demonstrated that the PTMs can be readministered to further increase and maintain transgene expression; a feature that may be vital for any gene therapy application.
Since each trans-splicing reaction inhibits one cis-splicing reaction, any predicted decrease in plasma albumin levels are expected to be minimal. Based on our previous results, in vivo trans- splicing has been on the order of 3–6%.8,33 Given the normal range of albumin is relatively wide (35–50 mg/ml),14,15 a modest 1–5% reduction through trans-splicing should not have any negative consequences on albumin levels. Indeed, patients diagnosed with known deficiencies in albumin, such as analbuminemia, a rare inherited disorder characterized by extremely low (1/1,000th of normal) or undetectable albumin levels, exhibit mild symptoms primarily due to compensatory increases in other serum proteins including apolipoproteins.16,17
The SMaRT strategy described here offers a number of advantages over the cDNA or the “gene-specific” SMaRT approaches previously used for therapeutic applications.7,8,9,10,11,12,13 A major challenge of gene-specific trans-splicing in some indications is to overcome the relatively low level of certain target pre-mRNAs available for trans-splicing. Through our own studies it has become apparent that some of the disease-related pre-mRNAs targeted for trans-splicing are expressed at very low levels, which could hinder the ability to elicit a robust and consistent therapeutic response. In conditions where only a minimal level of repair is required to alleviate the prominent phenotype (i.e., achieving ~5% FVIII expression in hemophilia A patients34 or ~8% cystic fibrosis transmembrane conductance regulator gene expression in cystic fibrosis patients35), gene-specific trans-splicing in the respective disease models has shown considerable promise. However, potential patient to patient variability suggests that the success of any gene-specific trans-splicing strategy may be limited in scope within a given patient population. The alternative has been to develop a general trans-splicing strategy whereby a single, highly abundant pre-mRNA such as albumin can be used to produce many liver-specific secreted proteins. Since only the signal peptide is acquired from albumin we have the ability to deliver nearly a complete gene containing all of its inherent active sites, binding motifs to modulate its own activity, and still maintain endogenous regulation in hepatocytes. Doing so has the potential to benefit a much larger patient population as many disorders, such as hemophilia A, can be attributed to any one of several polymorphisms scattered throughout the FVIII gene. Application of this albumin-targeting strategy would be particularly beneficial for the treatment of hemophilia A as the FVIII pre-mRNA target available for trans-splicing appears to be in particularly low abundance (unpublished data). Targeting the albumin transcript would circumvent this deficiency. Although, it is difficult to make a precise comparison between the current strategy and repair of defective FVIII transcripts, i.e., comparison of different PTMs targeted to different genes and using very disparate delivery systems, we can say that the current strategy produced the highest individual values of FVIII in hemophilia A mice we have ever observed, and the survival rate of mice following tail clip challenge was greater. In our earlier work where we targeted the FVIII transcript for repair8 using adenoviral delivery we achieved a survival rate of 8 out of 10 mice following a tail clip challenge. With the current strategy i.e., trans-splicing FVIII into albumin using minicircle delivery we were able to achieve a survival rate of 100% with a larger group of mice (n = 12).
Another significant advantage of this novel trans-splicing approach is that it provides a greater level of cell-type and organ-specific expression than that afforded by only using promoter elements driving expression of full length cDNAs. This has implications for disease in which ectopic expression is detrimental to the cell or where off target expression can exacerbate cellular immune responses to the transgene. For example, the new trans-splicing approach may be particularly beneficial for hemophilia A because FVIII expression will be restricted to hepatocytes, and prevented in cells with antigen presenting capability where the albumin target is either absent or extremely low. This would likely have the effect of reducing the cellular immune responses to FVIII, which has slowed progress in achieving a gene therapy for hemophilia A.
For noninherited conditions such as infectious disease, the albumin-targeting strategy offers a new dimension for treatment. As we have demonstrated here, this strategy has the capacity to provide rapid, cost-effective, in vivo production of antibodies against a broad spectrum of infectious agents. If necessary, antibody expression can be engineered to extinguish and thus provide only acute passive immunity without the issues inherent in long-term expression of biologics. With the appropriate delivery systems and further optimization in trans-splicing efficiency, we predict this strategy will achieve sufficiently high plasma concentrations of the antibody to improve efficacy and minimize an autoimmune response. For the proof-of-principle studies discussed herein we were able to produce a functional scFv, the smallest fragment that retains full binding potential for an antigen.36 In addition, this targeting strategy can be applied to produce both heavy and light chains to produce antibodies as a means to broaden its applications.
Clearly the ability to produce functional therapeutic proteins by albumin-targeted trans-splicing is not limited to inherited conditions or antibody production, but can readily be applied to the treatment of acquired diseases including atherosclerosis and cardiovascular disease. ApoA-I is the major component of HDL and plays an important role in the reverse cholesterol transport pathway by promoting the efflux of excess cholesterol from peripheral cells and tissues for transfer to the liver for excretion.37 Numerous in vitro and in vivo studies38,39,40,41 including a clinical trial42 have demonstrated the protective effects of apoA-I and HDL against plaque development. Epidemiological data have shown that even an increase of 1 mg/dl in HDL correlates with a risk reduction of cardiovascular disease of 2–3%.43,44 In the present study, using an albumin-targeted PTM we have shown the production of functional human apoA-I protein in normal mice, and that re-administration of minicircle PTM resulted in an increase in hapoA-I protein, arguing against an immune reaction to either transgene or minicircle DNA. These data suggest that with optimization of minicircle DNA vectors, albumin-targeted PTMs could form the basis of a clinical product.
One potential limitation of the trans-splicing approach into albumin is that the chimeric trans-spliced protein contains 1–2 additional amino acids (depending on the target species) on the N-terminus of the expressed proteins. This could potentially create a novel epitope and may result in an immune response to any expressed proteins. In the current study, no immune response to human apoA-I in mice was observed (data not shown), but this issue has to be carefully examined whenever this kind of treatment is tried in human subjects where a single additional amino acid would be present. In addition, the current approach may not be suitable to all diseases. For example, since the trans-spliced product is expressed only when the target pre-mRNA is expressed selection of target pre-mRNA, level of therapeutic product and disease has to be carefully examined.
Finally, while we have selected albumin pre-mRNA as the primary target for trans-splicing, the use of other highly abundant transcripts including casein, myosin and fibroin has the potential to dramatically expand the use of this trans-splicing strategy for a wide range of therapeutic and nontherapeutic applications. We believe this albumin-targeted trans-splicing strategy represents an innovative and exciting new means of producing physiologically and/or clinically effective levels of therapeutic proteins in vivo with broad applications in gene and RNA therapy.
Methods
Albumin-human apoA-I plasmid. The mouse albumin-hapoA-I chimeric cDNA construct (mAlb-hapoA-I) was created using a combination of annealed oligos and PCR amplification of albumin exon 1 sequence using the following primers: 5′-ATGAAGTGGGTAACCTT TCTCCTCCTCCTCTTCGTCTCCGGCTCTGCTTTTTCC AGGGGTGTGTTTCGCC GAGAAGCACCC-3′ and 5′- GGGTGCTTC TC GGCGAAACACACCCCTGGAAAAAGCAGAGC CGGAGACGAA GAGGAGGAGG AGAAAGGTTACCCACTTCATG-3′. The hapoA-I coding sequence (exons 3, 4, and 3′ UTR) excluding the signal peptide necessary for secretion of processed hapoA-I was PCR amplified using a cDNA clone (ATCC no. MGC-1249) and primers Apo23 (5′-CCCCAGAGCCCCTGGGATCGAGTG-3′) and Apo5 (5′-CTAGAAGCTTCCCACTTTGGAAACGTTTATTCTGAGCACCGG-3′). The PCR product was blunt-ended, digested with HindIII, pre-ligated to the mouse albumin exon 1 PCR product then cloned between NheI and HindIII sites in pcDNA3.1(−) resulting in pc3.1-mAlb-hapoA-I. Plasmid encoding the wild-type (wt) hapoA-I including the signal peptide and 3′UTR was also constructed and used as a positive control (pc3.1-hapoA-I).
Human apoA-I PTMs. Human apoA-I PTM [pc3B-97C2-(CMV)] was modeled after CF-PTM24 (ref. 33). The CF-PTM contains the CMV promoter, the CFTR intron 9-specific BD, the TSD and the 3′ half of lacZ coding domain followed by the poly A signal. pc3B-97C2-(CMV) was generated by substituting the lacZ coding sequence with wt hapoA-I sequence and CFTR BD with mouse albumin intron 1-specific BD. The hapoA-I sequence (exons 3 and 4 plus the 3′UTR minus the signal peptide) was PCR amplified using a cDNA clone (ATCC no. MGC-1249) and primers Apo30 (5′-CTAGTTAATTAACAGCCCCCCAGAGCCCCTGGGATCGAG-3′) and Apo31 (5′-CTAGGGCGCGCCTTACTGGGTGTTGAGCTTCTTAGTGTACTC-3′), digested with PacI and AscI and cloned into the corresponding sites of pc3B-CF-PTM24. pc3B-97C2-(LSP) PTM with a liver-specific promoter was generated by substituting the CMV promoter with the human ApoE/hAAT promoter/enhancer. The liver-specific promoter was PCR amplified using a plasmid (pBS.sApoE.HCR.hAAT.hFIX.IntA.bpA) obtained from Mark Kay (Stanford University) and primers Apo57 (5′-CATGGCTAGCGACTAGAATTCGATATCAAGCTTATC-3′) and Apo65 (5′-CTAGGCTAGCGGTAT CGATCCACTCGAGTGGCCACTAGTTAGGCTCAGAGGC ACACAG-3′), digested with SpeI and NheI and cloned into the corresponding sites of pc3B-97C2-(CMV). A splicing-defective PTM1 (SM-PTM1) with a point mutation (cag→cat) at the splice junction of pc3B-97C2-(CMV) was created by PCR mutagenesis and used as a negative control.
HPV-E7scFv plasmids. The HPV-16 anti-E7 single-chain antibody (pHPV-E7scFv) DNA a generous gift from Feng Wang-Johanning (MD Anderson Cancer Center, The University of Texas) was used to PCR amplify the full length scFv ORF. The mouse albumin-HPV-E7scFv chimeric cDNA (pc3.1-mAlb-HPV-E7scFv) construct was derived from pc3.1-mAlb-hApoA-I. The HPV-E7scFv-specific PTM (pc3B-97C2-HPV-E7scFv) was generated by substituting the hapoA-I sequence with the HPV-E7scFv sequence in the pc3B-97C2-(CMV).
FVIII plasmids. FVIII PTMs were derived from the hapoA-I PTM [pc3B-97C2-(CMV)] that contains a CMV promoter, mouse albumin intron 1-specific BD, TSD, mouse FVIII coding sequence, and woodchuck post-transcriptional regulatory element followed by the poly A signal. pc3B-97C2-FVIII was generated by substituting the hapoA-I coding sequence with wt mouse FVIII sequence. The wt coding sequence for mouse FVIII exons 1–26 (minus the signal peptide and B-domain) was PCR amplified from an existing PTM using PfuUltra DNA polymerase (Stratagene, La Jolla, CA), digested with PacI and AscI and cloned into the corresponding sites of pc3B-97C2-(CMV). A splicing-defective PTM was created by mutating the splice acceptor site (cag→cct) in pc3B-97C2-FVIII PTM.
Minicircle PTMs. Mouse albumin-targeted minicircle producing plasmids were created by cloning in the entire PTM cassette from the promoter through polyA site of pc3B-97C2-(LSP) into a minicircle vector (p2фC31) obtained from Mark Kay (Stanford University). The PTM cassette was digested with ClaI and DraIII, blunted and cloned into XhoI digested and blunted minicircle vector to generate p2ф97C2-(LSP). The FVIII PTM in a minicircle DNA vector that was used in the tail clip assay was derived from p2ф97C2-(LSP). Briefly, the FVIII plasmid pc3B-97C2-FVIII was digested with NotI and blunted, digested with PacI to release the entire coding sequence, and then cloned into p2ф97C2-(LSP) to create p2ф97C2-FVIII-(LSP). Minicircles were produced as described29,30 with minor modifications. Quality of induction and minicircle purity was assayed using the QIAprep Spin Miniprep Kit (Qiagen). For animal experiments, large scale minicrcle DNA was isolated using the EndoFree Plasmid Mega Kit (Qiagen, Valencia, CA).
Cholesterol efflux assay. For the cholesterol efflux assay22 negative control and ABC1-stably transfected HeLa cells were grown to near confluency. Cells were loaded with 1 µCi/ml 3H cholesterol (New England Nuclear, Boston, MA) and equilibrated for 24 hours. Cells were washed 3× with serum-free media and incubated with a serial dilution of the supernatant from HEK293 cells transfected with the chimeric mAlb-hapoA-I cDNA construct or wt hapoA-I plasmid. Cells were allowed to efflux for 18 hours and media was collected and counted by liquid scintillation. The remaining counts in the cell fraction were determined after an overnight extraction with isopropanol. The percent efflux was calculated by dividing the counts in the efflux media by the sum of the counts in the media plus the cell fraction. Dulbecco's modified Eagle's medium containing bovine serum albumin was used as a blank and subtracted from the radioactive counts obtained in the presence of an acceptor in the efflux media.
Animals and animal procedures. C57BL/6 (stock no. 000664) mice were obtained from Jackson Laboratory (Jackson Labs, Bar Harbor, ME). Homozygous female and hemizygous hemophilia A mice (FVIII exon 16 knockout)45 were obtained from H. Kazazian at the University of Pennsylvania. The mice were intrabred and maintained at Biocon (Rockville, Maryland), and all the animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee. PTMs were delivered to mice using the well established hydrodynamic injection procedure.26,27 Blood from mice was collected before and after injection by retro-orbital plexus bleeding into 10 mmol/l EDTA for hapoA-I and HPV-E7 scFv samples or 3.2% sodium citrate at a ratio of 1 part sodium citrate to 9 parts blood for FVIII samples. The plasma samples were aliquoted, snap frozen in liquid nitrogen and stored at −80 °C. Mice were not anesthetized during the collection procedure. At the end of the study liver tissue was collected for RNA analysis.
Phenotypic correction of hemophilia A mice was assessed using the tail clip assay. Mice were injected twice HD with either 50 µg of minicircle PTM, or with 50 µg of PTM plus 50 µg of albumin minigene target at day 0 and day 3. Wild-type C57BL/6 mice and hemophilia A mice injected twice with saline served as the positive and negative controls, respectively. FVIII activity was assessed in all mice before and after injection. For the PTM-treated group mice with FVIII levels above background >1% was selected for the tail clip. Mice were placed on a temperature controlled warming plate and sedated with 2% isoflurane gas using an EZ Anesthesia collar device (Euthanex, Palmer, PA). The tail was prewarmed by immersing in saline at 37 °C for 2 minutes, and then inserted through a 1.5 mm diameter opening in a metal template until a snug fit was achieved, and then transected with a number 22 scalpel. Mice were returned to their cages and survival assessed at 24 hours after the tail clip.
Real-time qRT-PCR. Real-time qRT-PCR was performed essentially as described previously.33 Total RNA from liver tissue was isolated using RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. Residual plasmid and genomic DNA was removed by treating with DNAse I. First strand cDNA was synthesized using the SuperScript III FirstStrand synthesis Kit (Invitrogen, Carlsbad, CA). Approximately 2 µg of total cellular RNA was incubated with random hexamer and dNTPs at 65 °C for 5 minutes in a 10 µl reaction. After 2 minutes on ice, 10 µl of master mix consisting of buffer, MgCl2, DTT, RNase OUT, SuperScript III enzyme was added and incubated at 25 °C for 10 minutes then 55 °C for 50 minutes followed by heat inactivation at 85 °C for 5 minutes. Real-time quantitative PCR was carried out on an iCycler IQ system using intercalation of SYBR Green as fluorescence reporter (Bio-Rad, Hercules, CA). Briefly, 5 µl first strand cDNA (10 ng/µl) or control plasmid was incubated with 300 nmol/l each forward and reverse primer, 20 mmol/l Tris-HCl (pH 8.4), 50 mmol/l KCl, 3 mmol/l MgCl2, 0.2 mmol/l each dNTP, 1.25 U AccuStart Taq DNA polymerase, SYBR Green I dye and 10 nmol/l fluorescein well factor dye in a 25 µl reaction. Standard curves were created using tenfold serial dilutions for each run. The PCR cycling conditions were as follows: an initial 10 minutes denaturation step at 95 °C was followed by 50 cycles of denaturation and annealing/extension (95 °C for 15 seconds, 60 °C for 1 minute). Trans-splicing, total target mRNA and PTM levels were measured individually using specific primers as described below. Trans-splicing efficiency was calculated using the formula [trans-spliced mRNA/total target mRNA].
Human apoA-I PTM and trans-splicing levels were quantified using the primers huApoA1E3F (5′-CTCAAAGACAGCGGCAGAGAC-3′) and huApoA13R (5′-CCTCCTCCAGATCCTTGCTCA-3′), and AlbA1TSF2 (5′-ACCTTTCTCCTCCTCCTCTTCGT-3′) and AlbA1TSR1 (5′-ACATA GTCTCTGCCGCTGTCTTT-3′), respectively. FVIII PTM and trans-spliced mRNA were quantified using primers: mF725 (5′-GCCATACCA CATTTGTAGAGG-3′) and mF726 (5′-TGAAATTTGTGATGCTA TTGCT-3′); mF484 (5′-TCCTCCTCCTCTTCGTCTCC-3′) and mF485 (5′-TGTATGCAGCACACTGAGCA-3′), respectively.
Human apoA-I protein analysis. Serum levels of hapoA-I protein was quantified by enzyme-linked immunosorbent assay (ALerCHEK, Portland, ME; cat. no. A70101). The assay was performed according to the manufacturer's instructions in triplicate. Briefly, 100 µl diluted serum was incubated for 2 hours at room temperature. The plates were washed 4× with wash buffer and incubated with 100 µl HRP conjugated goat apoA-I antibody for 2 hours. After washing, 100 µl tetra-methyl-benzidine/peroxide substrate was added and incubated for 30 minutes at room temperature. The reaction was terminated with 100 µl 0.5 N sulfuric acid and the OD was read at 450 nm. Standard curves were generated for each assay using the hapoA-I protein supplied in the kit.
HPV-E7scFv in vitro analysis. For the growth assay, SiHa and C-33A cells (ATCC no. HTB-35 and HTB-31) were seeded at 20% confluency and transfected 24 hours later with pc3.1-mAlb-HPV-E7scFv chimeric cDNA plasmid or pcDNA3.1(−) vector plasmid using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Five days after transfection, cell viability was measured using an MTT assay (Sigma, St. Louis, MO; cat. no. TOX1) according into manufacturer's instructions. Samples were read at 570 nm in quadruplicate and the background absorbance subtracted.
HPV-E7scFv in vivo analysis. C57BL/6 mice were HD-injected with 100 µg PTM (pc3B-97C2-HPV-E7scFv) or the control pc3.1-mAlb-HPV-E7scFv cDNA plasmid. Serum and liver tissue was collected and snap frozen for analysis. Serum was collected at different time points, immunoprecipitated using the FLAGIPT-1 kit (Sigma; cat. no. F3165) and the production of chimeric mAlb-HPV-E7scFv was detected using the WesternBreeze Chromogenic Immunodetection System.
FVIII activity assay. Mouse FVIII activity was assayed using the Coatest SP4 FVIII kit from Chromogenix (Milan, Italy; cat. no. 82409463). Diluted plasma samples were analyzed in duplicate. The assay was performed according to the manufacturer's instructions with the exception that (i) volumes were scaled down four fold for analysis using 96-well plates, and (ii) standard curves were generated in the range of 1.2–20% by diluting pooled normal plasma from C57BL/6 mice in pooled plasma from naïve hemophilia mice (male and female).
Coagulation assay. Clotting times were assessed using the activated partial thromoboplastin time assay with soluble ellagic acid as an activator (Helena Laboratories, Beaumont, TX; cat. no. 5389), and the reaction was performed using the manual tilt method. Plasma samples were diluted 1 in 5 in Owren's Veronal buffer, mixed 1:1 with FVIII deficient substrate (Helena Laboratories, Beaumont, TX) and incubated at 37 °C for 2 minutes. Activated partial thromboplastin time–SA assay reagent (100 µl) was added to 100 µl of the test plasma and the mix incubated at 37 °C for 3 minutes. Prewarmed calcium chloride (100 µl) solution was added and the tube tilted back and forth in a 37 °C water bath and the time taken to form a clot was recorded. Samples were analyzed in duplicate.
Supplementary MaterialFigure S1. Mouse albumin-hapoA-I chimeric protein expressed, processed and secreted normally in cells.Figure S2. Assessment of PTM and trans-splicing level by real-time qRT-PCR.Figure S3. Expression of the trans-spliced mAlb-HPV-E7scFv in vivo.Figure S4. Mouse albumin-FVIII chimeric protein is functionally active.Figure S5. Trans-splicing into albumin restores coagulant activity in hemophilia A mice.
Supplementary Material
Mouse albumin-hapoA-I chimeric protein expressed, processed and secreted normally in cells.
Assessment of PTM and trans-splicing level by real-time qRT-PCR.
Expression of the trans-spliced mAlb-HPV-E7scFv in vivo.
Mouse albumin-FVIII chimeric protein is functionally active.
Trans-splicing into albumin restores coagulant activity in hemophilia A mice.
Acknowledgments
The hemophilia work was supported in part by a Phase II SBIR grant from the National Heart, Lung, and Blood Institute (grant no. R44-HL072687). We thank Laurent Humeau and Peter Donnelly for critical reading of the manuscript; John Stonik, Catherine Knapper, Boris Vaisman for assistance in experimental design and technical help; Lili Portilla for assistance with the CRADA. The apoA-I research was performed under a CRADA between NHLBI's Vascular Medicine Branch/Lipoprotein Metabolism Section and Intronn, Inc. We thank Paul McCray (University of Iowa) for his advice on performing the tail clip assay. J.W., S.G.M., P.D.J., G.J.M., and M.P. are employees of VIRxSYS Corporation (formerly with Intronn, Inc) and C.A.C. and K.W. were former Intronn, Inc employees. Spliceosome-mediated RNA trans-splicing technology was developed by Intronn, Inc., and majority of the work in this article was conducted at Intronn, Inc. In September 2007, VIRxSYS Corporation acquired all of Intronn, Inc., assets including intellectual property. M.A.G.-B is a co-corresponding author in this article and was a co-founder and consultant to Intronn, Inc., and is currently a consultant to VIRxSYS Corporation.
REFERENCES
- Caudevilla C, Serra D, Miliar A, Codony C, Asins G, Bach M, et al. Natural trans-splicing in carnithine octenyltransferase pre-mRNAs in rat liver. Proc Natl Acad Sci. 1998;95:12185–12190. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finta C., and , Zahhiropoulos PG. Intergenic mRNA molecules resulting from trans-splicing. J Biol Chem. 2002;277:5882–5890. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- Flouriot G, Brand H, Seraphin B., and , Gannon F. Natural trans-spliced mRNAs are generated from the human estrogen receptor-α (hER α) gene. J Biol Chem. 2002;277:26244–26251. doi: 10.1074/jbc.M203513200. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco MA. Messenger RNA reprogramming by spliceosome-mediated RNA trans-splicing. J Clin Invest. 2003;112:474–480. doi: 10.1172/JCI19462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenger BA., and , Gilboa E. Emerging clinical applications of RNA. Nature. 2002;418:252–258. doi: 10.1038/418252a. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco MA, Baraniak AP., and , Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang Q, Mansfield SG, Puttaraju M, Zhang Y, Zhou W, et al. Partial correction of endogenous DeltaF508 CFTR in human cystic fibrosis airway epithelia by spliceosome-mediated RNA trans-splicing. Nat Biotechnol. 2002;20:47–52. doi: 10.1038/nbt0102-47. [DOI] [PubMed] [Google Scholar]
- Chao H, Mansfield SG, Bartel RC, Hiriyanna S, Mitchell LG, Garcia-Blanco MA, et al. Phenotype correction of hemophilia A mice by spliceosome-mediated RNA trans-splicing. Nat Med. 2003;9:1015–1019. doi: 10.1038/nm900. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martin T, Garcia-Blanco MA, Mansfield SG, Grover AC, Hutton M, Yu Q, et al. Reprogramming of tau alternative splicing by spliceosome-mediated RNA trans-splicing: implications for tauopathies. Proc Natl Acad Sci. 2005;102:15659–15664. doi: 10.1073/pnas.0503150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coady TH, Shababi M, Tullis GE., and , Lorson CL. Restoration of SMN function: Delivery of a trans-splicing RNA re-directs SMN2 pre-mRNA splicing. Mol Ther. 2007;15:1471–1478. doi: 10.1038/sj.mt.6300222. [DOI] [PubMed] [Google Scholar]
- Puttaraju M, Jamison SF, Mansfield SG, Garcia-Blanco MA., and , Mitchell LG. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat Biotechnol. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- Tahara M, Pergolizzi RG, Kobayashi H, Krause A, Luettich K, Lesser ML, et al. Trans-splicing repair of CD40 ligand deficiency results in naturally regulated correction of a mouse model of hyper-IgM X-linked immunodeficiency. Nat Med. 2004;10:835–841. doi: 10.1038/nm1086. [DOI] [PubMed] [Google Scholar]
- Zayed H, Xia L, Yerich A, Yant SR, Kay MA, Puttaraju M, et al. Correction of DNA protein kinase deficiency by spliceosome-mediated RNA trans-splicing and sleeping beauty transposon delivery. Mol Ther. 2007;15:1273–1279. doi: 10.1038/sj.mt.6300178. [DOI] [PubMed] [Google Scholar]
- Peters T., Jr Serum albumin. Adv Protein Chem. 1985;37:161–245. doi: 10.1016/s0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- Anderson NL., and , Anderson NG. The human plasma proteome: History, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- Baldo-Enzi G, Baiocchi MR, Vigna G, Andrian C, Mosconi C., and , Fellin R. Analbuminemia: a natural model of metabolic compensatory systems. J Inherit Metab Dis. 1987;10:317–329. doi: 10.1007/BF01799973. [DOI] [PubMed] [Google Scholar]
- Watkins S, Madison J, Galliano M, Minchiotti L., and , Putnam FW. A nucleotide insertion and frame shift cause analbuminemia in an Italian family. Proc Natl Acad Sci. 1994;91:2275–2279. doi: 10.1073/pnas.91.6.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugaiczyk A, Law SW., and , Dennison OE. Nucleotide sequence and the encoded amino acids of human serum albumin mRNA. Proc Natl Acad Sci. 1982;79:71–75. doi: 10.1073/pnas.79.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minghetti PP, Ruffner DE, Kuang WJ, Dennison OE, Hawkins JW, Beattie WG, et al. Molecular structure of the human albumin gene is revealed by nucleotide sequence within q11–22 of chromosome 4. J Biol Chem. 1986;261:6747–6757. [PubMed] [Google Scholar]
- Sethi AA, Amar M, Shamburek RD., and , Remaley AT. Apolipoprotein AI mimetic peptides: possible new agents for the treatment of atherosclerosis. Curr Opin Investig Drugs. 2007;8:201–212. [PubMed] [Google Scholar]
- Wu AL., and , Windmueller HG. Relative contributions by liver and intestine to individual plasma apolipoproteins in the rat. J Biol Chem. 1979;254:7316–7322. [PubMed] [Google Scholar]
- Remaley AT, Stonik JA, Demosky SJ, Neufeld EB, Bocharov AV, Vishnyakova TG, et al. Apolipoprotein specificity for lipid efflux by the human ABCAI transporter. Biochem Biophys Res Commun. 2001;280:818–823. doi: 10.1006/bbrc.2000.4219. [DOI] [PubMed] [Google Scholar]
- Puttaraju M, Yang Y, Spindler JE, Wang J, Cote CA, Ke W, et al. High capacity screen to select optimal pre-trans-splicing molecules for trans-splicing applications. Mol Ther. 2004;9:S271–S272. [Google Scholar]
- Enriquez-Harris P, Levitt N, Briggs D., and , Proudfoot NJ. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991;10:1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, West S., and , Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Song YK., and , Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Hodges BL., and , Scheule RK. Hydrodynamic delivery of DNA. Expert Opin Biol Ther. 2003;3:911–918. doi: 10.1517/14712598.3.6.911. [DOI] [PubMed] [Google Scholar]
- Darquet AM, Cameron B, Wils P, Scherman D., and , Crouzet J. A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Ther. 1997;4:1341–1349. doi: 10.1038/sj.gt.3300540. [DOI] [PubMed] [Google Scholar]
- Chen ZY, He CY, Ehrhardt A., and , Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high level transgene expression in vivo. Mol Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- Chen ZY, He CY, Meuse L., and , Kay MA. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. GeneTher. 2004;11:856–864. doi: 10.1038/sj.gt.3302231. [DOI] [PubMed] [Google Scholar]
- Suda T., and , Liu D. Hydrodynamic gene delivery: its principles and applications. Mol Ther. 2007;15:2063–2069. doi: 10.1038/sj.mt.6300314. [DOI] [PubMed] [Google Scholar]
- Wang-Johanning F, Gillespie GY, Grim J, Rancourt C, Alvarez RD, Siegal GP, et al. Intracellular expression of a single-chain antibody directed against human papillomavirus type 16 E7 oncoprotein achieves targeted antineoplastic effects. Cancer Res. 1998;58:1893–1900. [PubMed] [Google Scholar]
- Puttaraju M, DiPasquale J, Baker CC, Mitchell LG., and , Garcia-Blanco MA. Messenger RNA repair and restoration of protein function by spliceosome mediated RNA trans-splicing. Mol Ther. 2001;4:105–114. doi: 10.1006/mthe.2001.0426. [DOI] [PubMed] [Google Scholar]
- Gan SU, Kon OL., and , Calne RY. Genetic engineering for haemophilia A. Expert Opin Biol Ther. 2006;6:1023–1030. doi: 10.1517/14712598.6.10.1023. [DOI] [PubMed] [Google Scholar]
- Chu CS, Trapnell BC, Curristin SM, Cutting GR., and , Crystal RG. Extensive posttranscriptional deletion of the coding sequences for part of nucleotide-binding fold 1 in respiratory epithelial mRNA transcripts of the cystic fibrosis transmembrane conductance regulator gene is not associated with the clinical manifestations of cystic fibrosis. J Clin Invest. 1992;90:785–790. doi: 10.1172/JCI115952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D., and , Celer V. The production and application of single-chain antibody fragments. Folia Microbiol (Praha) 2003;48:687–698. doi: 10.1007/BF02993480. [DOI] [PubMed] [Google Scholar]
- Lewis GF., and , Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ Res. 2005;96:1221–1232. doi: 10.1161/01.RES.0000170946.56981.5c. [DOI] [PubMed] [Google Scholar]
- Rubin EM, Krauss RM, Spangler EA, Verstuyft JG., and , Clift SM. Inhibition of early atherogenesis in transgenic mice by human apolipoprotein AI. Nature. 1991;353:265–267. doi: 10.1038/353265a0. [DOI] [PubMed] [Google Scholar]
- Plump AS, Scott CJ., and , Breslow JL. Human apolipoprotein A-I gene expression increases high density lipoprotein and suppresses atherosclerosis in the apolipoprotein E-deficient mouse. Proc Natl Acad Sci USA. 1994;91:9607–9611. doi: 10.1073/pnas.91.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duverger N, Kruth H, Emmanuel F, Caillaud JM, Viglietta C, Castro G, et al. Inhibition of atherosclerosis development in cholesterol-fed human apolipoprotein A-I-transgenic rabbits. Circulation. 1996;94:713–717. doi: 10.1161/01.cir.94.4.713. [DOI] [PubMed] [Google Scholar]
- Tangirala RK, Tsukamoto K, Chun SH, Usher D, Puré E., and , Rader DJ. Regression of atherosclerosis induced by liver-directed gene transfer of apolipoprotein A-I in mice. Circulation. 1999;100:1816–1822. doi: 10.1161/01.cir.100.17.1816. [DOI] [PubMed] [Google Scholar]
- Nissen SE, Tsunoda T, Tuzcu EM, Schoenhagen P, Cooper CJ, Yasin M, et al. Effect of recombinant ApoA-I Milano on coronary atherosclerosis in patients with acute coronary syndromes: a randomized controlled trial. JAMA. 2003;290:2292–2300. doi: 10.1001/jama.290.17.2292. [DOI] [PubMed] [Google Scholar]
- Castelli WP. Cholesterol and lipids in the risk of coronary artery disease--the Framingham Heart Study. Can J Cardiol. 1988;4 Suppl A:5A–10A. [PubMed] [Google Scholar]
- Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III), Final Report NIH Publication 02–5215; National Cholesterol Education Program, NHLBI, NIH; September 2002 [PubMed] [Google Scholar]
- Bi L, Lawler AM, Antonarakis SE, High KA, Gearhart JD., and , Kazazian HH., Jr Targeted disruption of the mouse factor VIII gene produces a model of haemophilia A. Nat Genet. 1995;10:119–121. doi: 10.1038/ng0595-119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mouse albumin-hapoA-I chimeric protein expressed, processed and secreted normally in cells.
Assessment of PTM and trans-splicing level by real-time qRT-PCR.
Expression of the trans-spliced mAlb-HPV-E7scFv in vivo.
Mouse albumin-FVIII chimeric protein is functionally active.
Trans-splicing into albumin restores coagulant activity in hemophilia A mice.