Abstract
Three combinatorial libraries of polymeric vectors were evaluated to investigate the functional roles of molecular weight (MW), cations, pH-sensitive moieties, and hydrophobic derivitization in polymer-mediated gene delivery. Four cationic and pH-sensitive moieties (imidazole, primary, secondary, and tertiary amino) and three hydrophobic residues (C4 butyl, C6 hexyl, and C8 octyl) were assessed in single and serially incremented, binary combinations. Three MWs were evaluated—10, 30, and 50 kDa. The highest levels of transfection, comparable to branched PEI (25 kDa), were achieved by 30 kDa and 50 kDa formulations containing primary amino and imidazole groups. Primary amino groups offered superior charge-neutralizing and size-condensing capacity, while imidazole groups appeared to bind with DNA via nonelectrostatically mediated interactions to produce stable polyplexes that were resistant to premature dissociation. Eight of the 10 highest-transfecting polymers possessed IC50 values greater than the maximum concentration of free polymers exposed to cells (200 µg/ml). The results herein have identified highly efficient polymeric formulations with superb toxicity profiles and have revealed the functional roles that the investigated pendant groups play in the transfection process. The reported polymeric system offers a versatile and robust platform upon which future structure–function studies may be based to create safer and more efficient polymeric vectors.
Introduction
Significant advances in molecular biology and genomic research have uncovered the genetic basis for a variety of diseases and have given rise to the use of gene therapy as a possible mode of treatment.1,2,3,4 Unfortunately, a primary obstacle preventing human gene therapy from becoming widely available is the lack of safe and efficient gene delivery systems.
A variety of vectors have been designed to overcome the known biological barriers that hinder safe and efficient delivery of exogenous genes. Most delivery systems developed to date fall under the general categories of viral, lipid-based, inorganic, and polymeric delivery systems.5,6,7,8 However, safety concerns and inefficient delivery profiles have restricted the advancement of any one of these systems toward a broader clinical application. In particular, polymeric systems have fallen short in their delivery efficiency, despite the diverse strategies for improving polymer-mediated gene delivery that have been reported in the past two decades.8 Many of the polymeric systems currently in development are based on design concepts that have been substantiated by a number of reports. Such design concepts include the use of cationic moieties to complex with DNA, pH-sensitive functional groups to promote intracellular trafficking, derivitization with carbohydrates to enhance biocompatibility and facilitate both cellular and nuclear targeting, and hydrophobic functionalization to promote cellular uptake.9,10,11,12,13,14 The wealth of information produced from these studies has reinforced the strategy of altering the chemical and structural characteristics of polymeric vectors to enhance and fine-tune their biophysical properties and transfection capacities. However, because many of these studies have been conducted under heterogeneous experimental conditions (e.g., the use of different polymeric backbones of different molecular weights (MWs) and polydispersities, diverse synthetic techniques, various cell lines transfected, different transfection protocols, etc.), it is difficult to directly correlate how the chemical and structural attributes of polymeric vectors influence transfection efficiency based solely on the analysis of existing literature. In the absence of clearly defined structure–function relationships, the rational modification of polymer vector designs remains a challenge.
This study aims to determine the structure–function relationships that govern one class of polymer-mediated gene delivery by systematically altering MW and side-chain composition in a combinatorial fashion. To explore (i) the effects of different charge groups and (ii) the hypothesized role of pH-sensitive moieties as buffering agents that facilitate intracellular trafficking and enhance transfection efficiency, we evaluated primary, secondary, tertiary amino, and imidazole groups.9,14,15,16 To understand whether the hydrophobic properties of linear hydrocarbon alkyl chains affect the transfection capacity of polymeric vectors, C4 butyl, C6 hexyl, and C8 octyl groups were also investigated. The effect of each functional group was systematically evaluated by forming single and binary combinations of all moieties at 75/25, 50/50, and 25/75 molar ratios to constitute a combinatorial library of polymeric vector candidates. Three such combinatorial libraries were synthesized, each representing a different MW (10, 30, and 50 kDa) and comprising the same polymer formulations. Characterization of the polymeric vectors included in vitro transfection capacity, polyplex size, zeta potential, relative binding strength, polyplex stability, buffering capacity, and cytotoxicity.
The results from this study have (i) revealed the structure–function relationships of different charge groups, pH-sensitive moieties, and hydrophobic residues; (ii) identified several polymeric vectors with transfection capacities as high as branched PEI (25 kDa) with more favorable cytotoxicity profiles; and (iii) established a robust system of polymers for future structure–function studies in a move toward safer and more efficient polymeric vector designs.
Results
Polymer synthesis and characterization
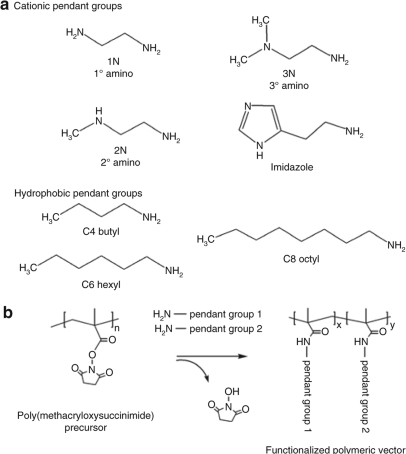
To isolate the effects of the various pendant groups (Figure 1a) on transfection and polyplex properties, pendant groups were conjugated in single and binary combinations to a polymer precursor of a single MW and polydispersity index, to form the functionalized polymeric vector (Figure 1b). Within the binary combinations, the following input molar ratios were evaluated: 25/75, 50/50, and 75/25. Conjugation efficiencies at all input ratios were nearly quantitative as shown in Supplementary Figure S1. Detailed characterization of the mono-functionalized polymer conjugates are provided in our previous report.17
Figure 1.
Chemical composition of polymeric vectors. (a) Cationic, pH-sensitive, and hydrophobic pendant groups. (b) General synthetic scheme for functionalizing pendant groups to the poly(methacryloxysuccimide) precursor to form mono- and bi-functionalized polymeric vectors (adapted from ref. 17).
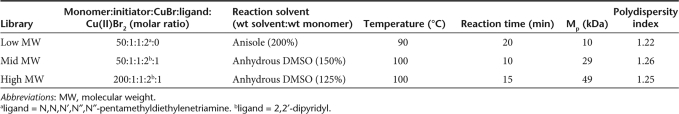
To evaluate the effects of polymer MW on transfection and polyplex properties, three identically formulated polymer libraries were synthesized, each representing a different MW. The MW and polydispersity index of the three libraries are summarized in Table 1.
Table 1.
Conditions and results for the synthesis of homopolymer precursor, poly(methacryloxysuccinimide)
Trends in transfection and biophysical properties
For organizational purposes, all transfection and biophysical characterization results are divided into two subpopulations of polymers and referred to hereafter as the cationic polymers and the alkylated polymers. The cationic polymers comprise all mono- and bi-functionalized polymer formulations containing only cationic pendant groups and the alkylated polymers comprise all formulations that contain an alkyl group. Due to water insolubility, not all alkyl combinations could be evaluated as vector candidates.
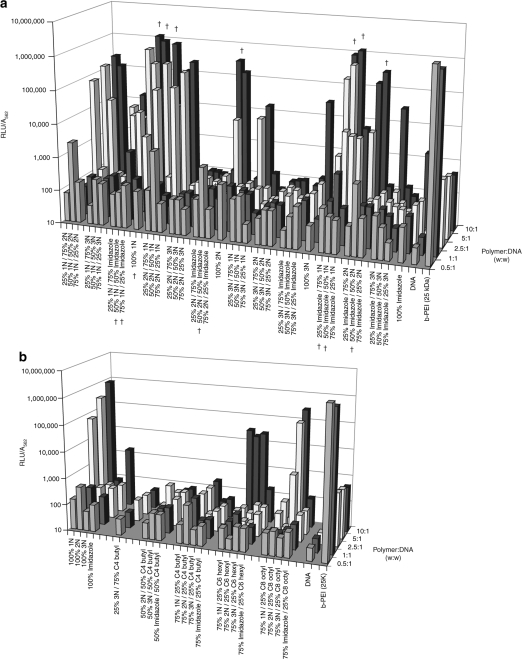
An exhaustive screen of the 30 kDa library of polymers revealed several interesting trends in transfection as a function of the pendant groups conjugated to the polymeric vector (Figure 2). Among the cationic polymers, those cofunctionalized with up to 50% imidazole or at least 50% 1N resulted in the highest transfection efficiencies at a polymer:DNA weight ratio of at least 5:1 (labeled with †), mediating levels as high as branched PEI (b-PEI). In general, polymers containing 2N, 3N, and alkyl groups mediated only moderate levels of transfection at their best.
Figure 2.
Transfection profile of 30 kDa polymeric vectors in NIH/3T3 cells. Transfection efficiency of (a) cationic polymers and (b) alkylated polymers is expressed as the relative light units (RLU) measured from luciferase reporter protein expression normalized to the absorbance measured at 562 nm (BCA assay). Each polymer was evaluated in triplicate at five polymer:DNA weight ratios: 0.5:1, 1:1, 2.5:1, 5:1, and 10:1. Samples labeled with a dagger (†) represent polymer formulations with the highest transfection capacities. For ease of viewing, error bars for each average transfection value have been omitted.
To understand how physicochemical changes to the polymeric vectors may give rise to the observed transfection trends, we evaluated the following biophysical properties as a function of the conjugated pendant groups: relative binding strength, polyplex stability, net surface charge, and effective diameter of polyplexes, and buffering capacity and cytotoxicity of free polymers.
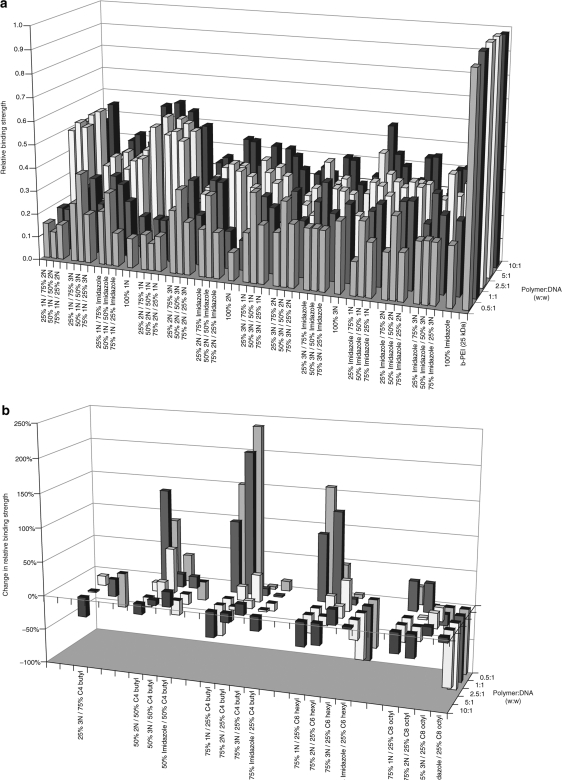
One of the principal design criteria for polymeric delivery systems is the ability to bind with DNA and protect it from enzymatic degradation.18,19,20 This binding interaction must be sufficiently stable to withstand competitive interactions from anionic species, such as endogenous proteins and nucleic acids, which can displace the bound DNA and result in its premature release.21,22 To assess the binding strength of polymer/DNA interactions and the stability of resultant polyplexes, we employed the EtBr fluorescence quenching and recovery assay.9,23,24 Among the cationic polymers, 1N-containing polymers generally possessed the strongest interactions with DNA with approximately 10% greater binding strengths over 2N- and imidazole-containing polymers and 25% greater strengths over 3N-containing polymers at the 5:1 and 10:1 polymer:DNA ratios (Figure 3a). To represent the effects of alkylation on the relative binding strength, the percent change in binding levels of alkylated polymers relative to their respective mono-functionalized cationic polymer are plotted in Figure 3b. For 1N-, 2N-, and imidazole-containing polymers, 25% alkylation generally reduced binding strengths at weight ratios of 2.5:1 and greater; whereas the opposite trend was observed among the 3N polymers.
Figure 3.
Relative binding strengths of 30 kDa polymeric vectors. The binding strength of (a) cationic polymers and (b) alkylated polymers was measured with the ethidium bromide fluorescence–quenching assay. Each polymer was evaluated in triplicate at five polymer:DNA weight ratios: 0.5:1, 1:1, 2.5:1, 5:1, and 10:1. The effects of alkylation on binding strength, shown in b, are represented as the percentage change in binding strength relative to the mono-functionalized cationic polymer. Results for the alkylated polymers are shown in reverse order of the polymer:DNA weight ratios for clarity. For ease of viewing, error bars for each average binding strength value have been omitted.
The binding stabilities of all polyplexes are shown in Figure 4a,b. In general, imidazole-based polyplexes were more resistant to dissociation compared to 1N-, 2N-, and 3N-based polyplexes. In particular, at polymer:DNA ratios of 5:1 and 10:1, imidazole-based polyplexes were on average 140%, 75%, and 50% more resistant to dissociation than 1N-, 2N-, and 3N-based polyplexes, respectively. The effects of alkylation on polyplex stability are represented as the percent change in polyplex dissociation relative to the mono-functionalized polymer (Figure 4b). In general, alkylation reduced the degree to which DNA dissociated from 1N-, 2N-, and 3N-containing polymers, indicating an enhancement in polyplex stability. This enhancement did not appear to follow any consistent trend as a function of hydrocarbon chain length or degree of conjugation. In contrast, alkylation decreased polyplex stability among polymers cofunctionalized with imidazole groups.
Figure 4.
Polyplex stability of 30 kDa polymeric vectors. The stability of polyplexes formed by (a) cationic polymers and (b) alkylated polymers was evaluated with the ethidium bromide fluorescence recovery assay performed immediately subsequent to the relative binding strength quenching assay. Each polymer was evaluated in triplicate at five polymer:DNA weight ratios: 0.5:1, 1:1, 2.5:1, 5:1, and 10:1. The effects of alkylation on polyplex stability, shown in b, are represented as the percentage change in polyplex dissociation relative to the mono-functionalized cationic polymer. Positive values represent an increase in dissociation and thus a decrease in polyplex stability due to alkylation and vice versa for negative values. Results for the alkylated polymers are shown in reverse order of the polymer:DNA weight ratios for clarity. For ease of viewing, error bars for each average polyplex stability value have been omitted.
Overall, the highest levels of transfection, comparable to those mediated by b-PEI, correlated with polymers with binding strengths sufficient to quench at least 40% of the EtBr/DNA fluorescence and polyplexes sufficiently stable to maintain at least 10% and at most 50% of the quenched fluorescence in the presence of competing polyanions.
DNA binding with polycationic vectors also functions to neutralize the anionic charge of DNA and condense its bulky structure to appropriate length scales for cellular internalization.25 Based on the trends observed from the zeta potential profiles of both cationic and alkylated libraries shown in Supplementary Figure S2, the charge-neutralizing capacities of the cations decrease in the order of 1N > 2N ~ 3N >> imidazole. Particularly worth noting is the markedly weaker charge-neutralizing ability of imidazole compared to 1N, 2N, and 3N. From Supplementary Figure S2a, the polymer:DNA ratio required to produce a positive zeta potential was at least 5:1 compared to the 2.5:1 ratio required by non–imidazole-containing polymers. Moreover, polymers containing 100% imidazole formed polyplexes that were barely neutralized (+1.3 mV) at the highest polymer:DNA ratio (10:1). In general, alkyl cofunctionalization resulted in a notably diminished charge-neutralizing power compared to their respective mono-functionalized polymers (Supplementary Figure S2b).
The relative size–condensing capacity of the cationic moieties decreased in the order of 1N > 3N > 2N > imidazole (data not shown). Polyplexes formed by cationic and alkylated polymers containing 1N, 3N, and 2N were generally <200, 250, 300 nm in size, respectively. Polyplexes larger than these size limits were typically polymers cofunctionalized with either alkyl or imidazole groups. In general, the highest-transfecting polymers produced polyplexes with at least a +10 mV zeta potential and less than 220 nm in size.
A strategy used for avoiding lysosomal degradation of endocytosed polyplexes is the incorporation of pH-sensitive moieties into polymeric vectors to act as buffering agents within endosomes.9,15,16,26 The buffering capacities for the cationic and alkylated polymers are shown in Supplementary Figure S3. All polymers possessed similar buffering capacities, with b-PEI possessing a nearly fivefold greater capacity. Closer examination of the buffering capacities revealed no obvious trends between formulations or salient correlations between transfection levels and buffering capacity.
The cytotoxic effect of both the polyplex and free polymer is one of the most important considerations in polymeric vector design.27,28 The in vitro cytotoxicity of each polymer was assessed in their free, uncomplexed form by the MTS cell viability assay. Because the cytotoxic effects of polymeric vectors are typically greater in their free form compared to their complexed state, the cell viability measurements reported herein represent a conservative cytotoxicity estimate of each polymeric vector.9,29 All cationic and alkylated polymers possessed IC50 values >200 µg/ml, with only a single exception (data not shown). The 100% 1N polymer formulation possessed an IC50 value of approximately 50 µg/ml. For comparison, the IC50 value of b-PEI was approximately 5 µg/ml.
Effects of MW on transfection and biophysical properties
Numerous reports have shown MW to be a critical structural attribute that can greatly influence the transfection capacity and biophysical properties of polymeric delivery systems.30,31,32,33 To investigate the effects of MW on this system of polymers, two additional libraries were evaluated—one at 10 kDa and the other at 50 kDa, each comprising the same polymer formulations as the previously described 30 kDa library.
In general, the trends in transfection within the 10 kDa and 50 kDa polymer libraries remained fairly consistent with those observed in the 30 kDa library (Supplementary Figure S4). However, the levels of transfection appeared to increase with increasing MW for the cationic polymers. Overall inspection of Supplementary Figure S4a shows that the 50 kDa polymer library is populated with a greater number of high-performing polymers that mediated higher levels of transfection—an order of magnitude greater in some cases—at a lower polymer:DNA ratio compared to the 10 kDa and 30 kDa polymer libraries.
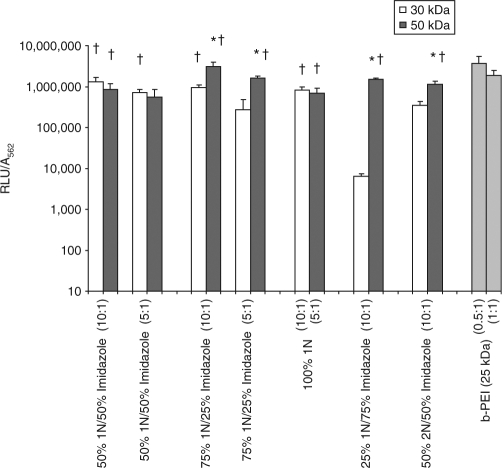
Figure 5 shows the highest-transfecting 30 kDa and 50 kDa polymers plotted alongside their 50 kDa and 30 kDa counterparts, respectively. The highest-transfecting polymers (labeled with †) mediated statistically comparable levels of transfection to b-PEI (25 kDa) at its optimal weight ratio with DNA (0.5:1). Labeled with an asterisk (*) are the formulations for which an increase in MW from 30 kDa to 50 kDa resulted in statistically higher levels of transfection.
Figure 5.
Transfection profiles of high-performing 30 kDa and 50 kDa polymeric vectors. High-performing polymers comprise those that mediated statistically comparable levels of transfection to b-PEI (25 kDa) at its optimal weight ratio with DNA (0.5:1) (labeled with a dagger (†)). Labeled with an asterisk (*) are the 50 kDa formulations that mediated statistically higher levels of transfection compared to their 30 kDa counterparts. The biophysical properties corresponding to each formulation are detailed in Table 2.
Biophysical characterization of the high-transfecting polymers, identified in Figure 5, uncovered some interesting trends amongst the formulations for which transfection efficiencies were enhanced due to an increase in MW (Table 2). In general, higher transfection efficiencies afforded by a higher MW corresponded with polyplexes with slightly greater zeta potentials and reduced effective diameters, although polyplexes as large as 400 nm were still able to mediate high levels of transfection. Interestingly, MW appeared to have little effect on the relative binding strength, yet significantly affected the stability of polyplexes, with over a threefold increase in stability afforded by some 50 kDa polymers compared to equivalent 30 kDa formulations.
Table 2.
Biophysical properties of high transfecting 30 kDa and 50 kDa polymers compared with branched PEI (25 kDa)
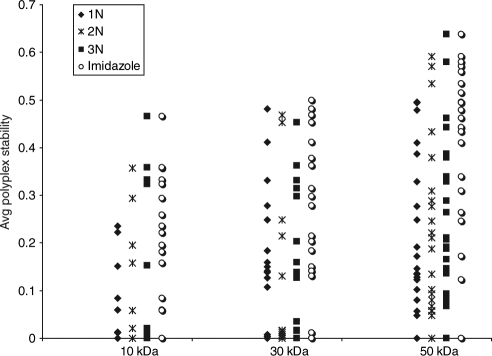
To further examine whether the trends in biophysical properties observed among the high-performing 50 kDa polymers were representative of the rest of the 50 kDa library, the relative binding strength, polyplex stability, and cytotoxicity profiles were characterized for the entire 50 kDa library. MW appeared to have little appreciable effect on the relative binding strength of all polymer formulations (data not shown). In contrast, MW appeared to affect the stability of polyplexes, specifically those formed by cationic polymers. To summarize the general effects of MW on polyplex stability, Figure 6 plots the polyplex stabilities of all 1N-, 2N-, 3N-, and imidzole-containing formulations, at the 5:1 and 10:1 weight ratios, among the cationic polymers. The upward trend as MW increases suggests that high MW polymers formed polyplexes that were more stable and resistant to dissociation. The cytotoxic effects of the free polymers increased only slightly with increasing MW. Supplementary Figure S5 shows the three polymer compositions for which an increase in MW resulted in an increase in cytotoxicity as indicated by a decrease in IC50 values.
Figure 6.
Effects of molecular weight on the polyplex stability of cationic polymers. Shown are the polyplex stabilities of 1N-, 2N-, 3N-, and imidazole-based polyplexes formed at the polymer:DNA weight ratios of 5:1 and 10:1.
Discussion
The results in this study have revealed (i) several important chemical and structural attributes of polymeric vectors that favor transfection and (ii) the structure–function relationships that govern their transfection-enhancing effect. In particular, polymers with an MW of 50 kDa containing ≥50% 1N or imidazole groups at ≤50% mediated the greatest levels of transfection.
The primary transfection-enhancing effect of 1N lies in its superior ability to efficiently complex with DNA. Figure 3a shows the relatively high binding strengths of 1N-containing polymers, which resulted in polyplexes that were less than 220 nm in size and carried a net positive zeta potential (Supplementary Figure S2a).
Interestingly, imidazole's main transfection-enhancing effect was not attributable to its oft-reported superior buffering capacity because high-transfecting, imidazole-containing polymers had similar buffering capacities to low-transfecting polymers (Supplementary Figure S3).9,15,16 Rather, imidazole functioned primarily to interact and bind with DNA. Resultant polyplexes were sufficiently stable to resist premature dissociation yet able to subsequently release the bound DNA, which is believed to be necessary in order for efficient gene expression to occur (Figure 4).22,34 Furthermore, imidazole's mode of interaction with DNA seemed to include nonelectrostatic interactions, a property that has also been observed by others and is supported here by several lines of evidence.9,35,36 First, Supplementary Figure S2a shows that imidazole-containing polymers have a weaker charge-neutralizing ability compared to 1N-, 2N-, or 3N-containing polymers. In principle, the zeta potential of polyplexes should be closely related to the binding strength of cationic polymers because the electrostatic interaction between cation and anionic DNA is the binding mode for which cations are employed in polymeric vector designs. Cationic polymers that induce polyplexes with a more positive zeta potential presumably bind to DNA more tightly.37 Thus, one might expect the binding strengths of the cationic pendant groups to trend in the same order as that for charge-neutralizing power (i.e., 1N > 2N ~3N >> imidazole). Figure 3a, however, suggests that imidazole-containing polymers bound DNA as strong as 2N-containing polymers and in fact, 15% stronger than 3N-containing polymers. The enhanced binding, yet inferior charge-neutralizing and size-condensing capacity of imidazole, compared to both 2N and 3N, suggest an additional, nonelectrostatic mode of binding mediated by imidazole groups.
A second line of evidence for the nonionic binding interactions of imidazole with DNA is provided by the results from the polyplex stability studies shown in Figure 4a. In this study, the stability of polyplex binding was evaluated by the degree to which polyplexes dissociated upon exposure to an excess of a competing polyanion. The use of an ionic species to effect polyplex dissociation assumes that any interaction that is of an electrostatic nature or weaker, between the polymer and DNA, will be disrupted by the competing electrostatics. The results from the stability studies revealed that imidazole-containing polymers dissociated less compared to 1N-, 2N-, and 3N-containing polymers, suggesting that (i) the binding interactions between DNA and imidazole-containing polymers were stronger than those between DNA and 1N-, 2N-, and 3N-containing polymers; and/or (ii) binding interactions between imidazole and DNA may be mediated by nonelectrostatic forces, which stabilized polyplexes and resulted in more efficient delivery to cells.
Higher MWs also appeared to further strengthen the transfection-enhancing effect of the imidazole interaction with DNA as evidenced by the statistically greater transfection efficiencies mediated by certain 50 kDa formulations compared to their 30 kDa equivalents. (Figure 5, Table 2) However, further evaluation of the intracellular trafficking mechanisms, cellular localization, and transport dynamics will be needed to fully resolve the spatial and temporal details of DNA dissociation from imidazole-based polymers. The information obtained from these studies could have important implications on the strategic use of imidazole groups in polymer vector design.
Also observed in this study were the structure–function relationships associated with the 2N, 3N, C4, C6, and C8 pendant groups. In general, polymers containing these pendant groups mediated only moderate to negligible levels of transfection, and 2N and 3N functioned primarily as cationic centers that allowed for fine tuning of a polyplex's charge-neutralizing and size-condensing power when cofunctionalized with other pendant groups (e.g., 1N, alkyls) (Figure 3a, Supplementary Figure S2a). Based on the collective results of all biophysical properties evaluated in the present work, there appeared to be little other transfection-enhancing ability offered by 2N and 3N, and their use merely as cationic centers did not appear to be advantageous, at least in this system of polymers.
Hydrophobic residues have been shown to enhance transfection presumably through favorable interactions with membrane components that result in cellular uptake.37,38 In this study, we evaluated both the effect of hydrocarbon chain length and degree of alkyl conjugation using the C4, C6, and C8 hydrophobic groups. Based on the binding strength and polyplex stability trends observed among the alkylated polymers (Figures 3b and 4b), hydrophobic moieties appeared to affect polyplex formation. Particularly noteworthy was the enhancement in polyplex stability due to alkylation of 1N-, 2N-, or 3N-based polymers. This result was surprising because alkylation tended to reduce the charge-neutralizing capacity of cationic polymers (Supplementary Figure S2b), which might imply a weaker interaction between alkylated polymers and DNA. However, the observed enhancement in polyplex stability suggests that alkylation may induce a hydrophobically driven conformational rearrangement of the polymer perhaps during polyplex formation. That is, hydrophobic residues may drive the polyplex to assume a more thermodynamically favorable conformation that collapses down on the DNA thereby preventing it from dissociating readily, even in the presence of a tenfold excess of a competing polyanion. Although enhanced polyplex stability appeared to be beneficial for imidazole-based polyplexes, this characteristic seemed to be detrimental for alkylated polymers. Because the dissociation properties for alkylated polymers did not appear to change with MW (data not shown), it was difficult to verify whether the inability for the polyplex to unravel explains the poor transfection profiles of the alkylated polymers. In light of other reports that have conjectured similar clustering effects of hydophobically derivitized polymeric vectors and their subsequent effect on transfection, it is clear that hydrophobic derivitization of polymers may be a strategy for enhancing transfection.10,37 However, further investigation will be needed to fully understand the underlying mechanisms by which hydrophobic moieties can augment the transfection process before they can be strategically incorporated into polymeric gene delivery systems.
An important general attribute worth noting is the favorable cytotoxicity profiles of the polymers examined in this study. Of the 114 total polymers evaluated, only 4 formulations possessed IC50 values less than the maximum concentration of free polymers exposed to cells (200 µg/ml) (Supplementary Figure S5). Eight of the ten highest-transfecting formulations identified possessed IC50 values >200 µg/ml. (Table 2) For comparison, the IC50 value of b-PEI was approximately 5 µg/ml. Considering that the majority of polymers within the three MW libraries maintained good cell viability, it is unlikely that cytotoxicity factors account for the transfection differential observed between the three libraries. More important, in light of the cytotoxic effects of b-PEI and many PEI-based vector designs, these favorable cytotoxicity profiles further reinforce the utility of the polymeric system reported herein as a viable, alternative platform for future studies of various polymeric chemical and structural features and their functional roles in gene delivery.
In conclusion, we have demonstrated the utility of combinatorial strategies for systematically evaluating the functional role of cationic, pH-sensitive, and hydrophobic pendant groups in the transfection process. In addition to reporting the polymeric chemical and structural features that most favor transfection and their purported mechanisms, this work introduces a simple and robust polymeric platform upon which future structure–function studies may be based. By standardizing the polymeric structures and synthetic protocols, the structure–function relationships that govern polymer-mediated gene delivery may be more clearly defined, potentially leading to safer and more efficient polymeric delivery systems.
Materials and Methods
General procedure for the synthesis and characterization of polymer precursor. All chemicals were purchased from Sigma-Aldrich (St Louis, MO) and used without further purification. Dowex Marathon MSC hydrogen ion exchange resin was dried in vacuo overnight before use.
The monomer, N-methacryloxysuccinimide was synthesized as previously reported.39 The polymer precursor, poly(methacryloxysuccinimide) (poly(MAOS)), was synthesized with a slight modification to two previously reported methods.40,41 To obtain a precursor MW of 10 kDa, the protocol reported by Shunmugam and Tew was adopted, whereas a modified version of the protocol by Godwin et al. was employed for synthesis of the precursor at 30 kDa and 50 kDa. Both protocols followed the same experimental setup with the only difference being the reaction temperature, transition metal ligand, and solvent used. Reaction conditions are summarized in Table 1. Briefly, a 5 ml pear-shaped flask with a flea magnetic stir bar was fitted with a rubber septum and flame dried while purging with N2(g), then cooled to ambient temperature. Cu(I)Br, Cu(II)Br2, and the ligand were added to the flask and resealed with the septum. Solvent that was purged with N2(g) for 10 minutes prior to use was injected into the flask and the contents were stirred under a nitrogen atmosphere for approximately 10 minutes until dissolved. N-methacryloxysuccinimide was added to the flask and allowed to dissolve for another 10 minutes. The initiator, ethyl-2-bromoisobutyrate, was directly injected through the septum into the flask with a Hamilton syringe and immersed into a thermostated oil bath. To stop the polymerization, the flask was quenched in an ice bath and the product was dissolved with anhydrous DMF at a 1:3.75 (wt/wt) ratio of monomer:DMF. Approximately 20 mg of Dowex hydrogen ion exchange resin was added to the redissolved solution and allowed to stir for approximately 10 minutes. The ion exchange resin was added to remove any active copper catalysts remaining in the solution.42 The final polymer was isolated as a white powder by precipitation into stirring acetone, filtered through a fine-fritted Buchner funnel, washed three times with fresh acetone, and dried overnight in vacuo. 1H NMR (DMSO-d6) δ: 1.44 (b,3H,CH3), 2.41 (b,2H,CH2), 2.78 (b,4H,CH2CH2).
The poly(MAOS) precursor was hydrolyzed to its sodium salt form for MW and polydispersity index characterization via gel permeation chromatography. Details of the hydrolysis and characterization methods are provided in our previous report.17 Table 1 summarizes the MW and polydispersity index values determined for each of the three precursors used in the present study.
General procedure for the synthesis and characterization of functionalized polymers. The seven pendant groups of interest, shown in Figure 1a, were conjugated to poly(MAOS) in single and binary—25/75, 50/50, and 75/25 input molar ratios—combinations following a previously reported protocol.17
To characterize the conjugation ratio of polymers that were bi-functionalized with two of the pendant groups in Figure 1a, a secondary set of bi-functionalized polymer conjugates were first synthesized, according to the same protocol, comprising each pendant group in Figure 1a cofunctionalized with benzylamine at 25/75, 50/50, and 75/25 molar input ratios.17 Owing to benzylamine's characteristic UV/Vis absorbance wavelength of 260 nm and 1H NMR chemical shift of ~7.2 ppm, the benzylamine content of the secondary set of polymer conjugates could be quantified using both analytical methods. The conjugation efficiency of the pendant groups in Figure 1a cofunctionalized together was then indirectly determined. Details of the UV/Vis analysis are provided in the Supplementary Materials and Methods.
Plasmid DNA (pCMV-luc) transfection. Prior to transfection assays, NIH/3T3 cells were cultured according to a protocol previously reported by our group.9 For transfection assays, NIH/3T3 cells were grown in clear and opaque, flat-bottomed tissue culture polystyrene 96-well plates (Costar, Corning, NY) at an initial density of 5,000 cells/well in 200 µl of phenol red-free, supplemented Dulbecco's modified Eagle's medium. Following 24 hours of incubation at 37 °C and 5% CO2, the growth medium was removed and replaced with 150 µl of polyplex transfection solution, which was prepared as described in the Supplementary Materials and Methods. Each 150 µl aliquot of transfection solution contained 3 µg of plasmid DNA. Cells were incubated with the polyplexes for 4 hours, after which the complex-containing medium was aspirated and replaced with 100 µl of phenol red-free, supplemented Dulbecco's modified Eagle's medium. Following 44 hours of additional incubation at 37 °C and 5% CO2, growth medium was aspirated and each well was rinsed with 200 µl of phosphate-buffered saline (pH 7.4).
To each well of the opaque 96-well plate, 100 µl of Bright-Glo working solution (Promega, Madison, WI), prepared according to the manufacturer's direction, was added. After 2 minutes of incubation at room temperature, luciferase expression was quantified as the relative light units measured in a Veritas Microplate Luminometer (Turner Biosystems, Sunnyvale, CA).
Total protein content was measured using the BCA Protein Quantitation Assay (Pierce Biotechnology, Rockford, IL). To each well of the clear 96-well plate, 20 µl of RIPA buffer (Pierce Biotechnology) was added and the plate incubated at room temperature for 10 minutes with gentle shaking. BCA working reagent (200 µl), prepared according to the manufacturer's directions, was then added to each well and gently shaken for 30 seconds at room temperature. Plates were incubated at 37 °C and 5% CO2 for 1 hour before being read at 562 nm on a Molecular Devices SpectraMax Plus384 UV/Vis spectrophotometer. Transfection results are expressed as the ratio of relative light units to absorbance at 562 nm.
Relative binding strength and polyplex stability assay. The relative degree of electrostatic binding between DNA and polymer in the absence and presence of a competing polyanion was measured using ethidium bromide (EtBr) (Fisher Scientific, Pittsburgh, PA) fluorescence quenching and recovery.23,24,36 Heparin sodium salt (4.38 mmol/l) dissolved in 10 mmol/l HEPES buffer (pH 7.2) was used as the competing polyanion.24 A tenfold molar excess of heparin, relative to DNA was employed in the polyplex stability studies to ensure that heparin competed with DNA and bound not only with uncomplexed free polymer that may be present in solution.23 A free polymer solution (180 µl) for each polymer:DNA ratio was prepared by combining the appropriate volume of 1.5 mg/ml polymer stock solution with 10 mmol/l HEPES buffer such that each 50 µl aliquot resulted in the intended wt:wt ratio of polymer and pCMV-luc. A stock solution of EtBr/DNA was made by combining equal volumes of pCMV-luc (150 µg/ml pCMV-luc in 10 mmol/l HEPES buffer) and EtBr (0.1095 mmol/l EtBr in 10 mmol/l HEPES buffer) stock solutions, resulting in a 4:1 ratio of phosphate groups to EtBr monomer units. All fluorescence readings were obtained in a Molecular Devices SpectraMax GeminiXS at λex = 535 nm and λem = 595 nm (automatic calibration, 10 seconds of mixing prior to reading, and automatic sensitivity of PMT detector).
First, 100 µl of EtBr/DNA solution was added to each well of a black polystyrene 96-well plate (Costar, Corning, NY) and the fluorescence was measured. An aliquot of 50 µl of free polymer solution was then added to each EtBr/DNA-containing well, gently shaken at room temperature on a microplate shaker for 15 minutes, and the fluorescence measured again. A 50 µl aliquot of 4.38 mmol/l heparin was added to each well, gently shaken at room temperature on a microplate shaker for 4 hours, and the fluorescence measured for a final time. The relative binding strength (BS) and polyplex stability (PS) were calculated from the following equations:
 |
 |
where FE/D is the corrected fluorescence of EtBr/DNA in the absence of polymeric vector, Fpolymer is the corrected fluorescence of EtBr/DNA in the presence of polymeric vector, Fhep is the corrected fluorescence of EtBr/DNA in the presence of polymeric vector and heparin.
All fluorescence measurements were corrected by subtracting background fluorescence due to free EtBr and due to volume effects. Background values from free EtBr were obtained from triplicate wells, on each 96-well plate, containing EtBr and 10 mmol/l HEPES buffer for the first two fluorescence readings and EtBr, HEPES buffer, and heparin for the final fluorescence measurement. Background values due to volume effects were determined from triplicate wells, on each 96-well plate, containing EtBr/DNA and HEPES buffer in volumes equal to those added to the polymer samples.
Preparation of polymer–DNA complex (polyplex) transfection solutions and free polymer cytotoxicity solutions, polyplex size and zeta potential measurement, buffering capacity assay, MTS cell viability assay, and statistical testing. Details of these procedures are provided in the Supplementary Materials and Methods.
Supplementary Material Figure S1. Conjugation profiles of bi-functionalized polymers. Figure S2. Surface-charge measurements of polyplexes formed by cationic and alkylated polymers. Figure S3. Buffering capacity of cationic and alkylated polymers in their free, uncomplexed form. Figure S4. Effects of molecular weight on the transfection profile of cationic and alkylated polymers in NIH/3T3 cells. Figure S5. Effects of molecular weight on the cytotoxicity of polymeric vectors. Supplementary Materials and Methods.
Supplementary Material
Conjugation profiles of bi-functionalized polymers.
Surface-charge measurements of polyplexes formed by cationic and alkylated polymers.
Buffering capacity of cationic and alkylated polymers in their free, uncomplexed form.
Effects of molecular weight on the transfection profile of cationic and alkylated polymers in NIH/3T3 cells.
Effects of molecular weight on the cytotoxicity of polymeric vectors.
Acknowledgments
This research was supported by a Biomedical Research Grant from the Whitaker Foundation. The authors have no disclosed conflicts of interest related to this work
References
- Burton E, Glorioso J., and , Fink D. Gene therapy progress and prospects: Parkinson's disease. Gene Ther. 2003;10:1721–1727. doi: 10.1038/sj.gt.3302116. [DOI] [PubMed] [Google Scholar]
- Factor P. Gene therapy for acute diseases. Mol Ther. 2001;4:515–524. doi: 10.1006/mthe.2001.0504. [DOI] [PubMed] [Google Scholar]
- Pawliuk R, Westerman K, Fabry M, Payen E, Tighe R, Bouhassira E, et al. Correction of sickle cell disease in transgenic mouse models by gene therapy. Science. 2001;294:2368–2371. doi: 10.1126/science.1065806. [DOI] [PubMed] [Google Scholar]
- von Laer D, Hasselmann S., and , Hasselmann K. Gene therapy for HIV infection: what does it need to make it work. J Gene Med. 2006;8:658–667. doi: 10.1002/jgm.908. [DOI] [PubMed] [Google Scholar]
- Coura R., and , Nardi N. A role for adeno-associated viral vectors in gene therapy. Genet Mol Biol. 2008;31:1–11. [Google Scholar]
- Flotte T. Gene therapy: The first two decades and the current state-of-the-art. J Cell Physiol. 2007;213:301–305. doi: 10.1002/jcp.21173. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zeng Q, Lu G., and , Yu A. Inorganic nanoparticles as carriers for efficient cellular delivery. Chem Eng Sci. 2006;61:1027–1040. [Google Scholar]
- Wong S, Pelet J., and , Putnam D. Polymer systems for gene delivery-past, present, and future. Prog Polym Sci. 2007;32:799–837. [Google Scholar]
- Chen D, Majors B, Zelikin A., and , Putnam D. Structure-function relationships of gene delivery vectors in a limited polycation library. J Controlled Release. 2005;103:273–283. doi: 10.1016/j.jconrel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Doody A, Korley J, Dang K, Zawaneh P., and , Putnam D. Characterizing the structure/function parameter space of hydrocarbon-conjugated branched polyethylenimine for DNA delivery in vitro. J Controlled Release. 2006;116:227–237. doi: 10.1016/j.jconrel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Hartmann L, Haefele S, Peschka-Suess R, Antonietti M., and , Borner H. Tailor-made poly(amidoamine)s for controlled complexation and condensation of DNA. Chem Eur J. 2008;14:2025–2033. doi: 10.1002/chem.200701223. [DOI] [PubMed] [Google Scholar]
- Masuda T, Akita H, Nishio T, Niikura K, Kogure K, Ijiro K, et al. Development of lipid particles targeted via sugar-lipid conjugates as novel nuclear gene delivery system. Biomaterials. 2008;29:709–723. doi: 10.1016/j.biomaterials.2007.09.039. [DOI] [PubMed] [Google Scholar]
- Neu M, Fischer D., and , Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J Gene Med. 2005;7:992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- Wang J, Gao S, Zhang P, Wang S, Mao M., and , Leong K. Polyphosphoramidate gene carriers: effect of charge group on gene transfer efficiency. Gene Ther. 2004;11:1001–1010. doi: 10.1038/sj.gt.3302248. [DOI] [PubMed] [Google Scholar]
- Midoux P., and , Monsigny M. Efficient gene transfer by histidylated polylysine pDNA complexes. Bioconjug Chem. 1999;10:406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- Putnam D, Gentry C, Pack D., and , Langer R. Polymer-based gene delivery with low cytotoxicity by a unique balance of side-chain termini. Proc Natl Acad Sci USA. 2001;98:1200–1205. doi: 10.1073/pnas.031577698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S., and , Putnam D. Overcoming limiting side reactions associated with an NHS-activated precursor of polymethacrylamide-based polymers. Bioconjug Chem. 2007;18:970–982. doi: 10.1021/bc0603790. [DOI] [PubMed] [Google Scholar]
- Abdelhady H, Allen S, Davies M, Roberts C, Tendler S., and , Williams P. Direct real-time molecular scale visualisation of the degradation of condensed DNA complexes exposed to DNase I. Nucleic Acids Res. 2003;31:4001–4005. doi: 10.1093/nar/gkg462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechardeur D, Sohn K, Haardt M, Joshi P, Monck M, Graham R, et al. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- Schaffer D., and , Lauffenburger D. Optimization of cell surface binding enhances efficiency and specificity of molecular conjugate gene delivery. J Biol Chem. 1998;273:28004–28009. doi: 10.1074/jbc.273.43.28004. [DOI] [PubMed] [Google Scholar]
- Huth S, Hoffman F, von Gersdorff K, Laner A, Reinhardt D, Rosenecker J, et al. Interaction of polyamine gene vectors with RNA leads to the dissociation of plasmid DNA-carrier complexes. J Gene Med. 2006;8:1416–1424. doi: 10.1002/jgm.975. [DOI] [PubMed] [Google Scholar]
- Schaffer D, Fidelman N, Dan N., and , Lauffenburger D. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Danielsen S, Maurstad G., and , Stokke B. DNA-polycation complexation and polyplex stability in the presence of competing polyanions. Biopolymers. 2005;77:86–97. doi: 10.1002/bip.20170. [DOI] [PubMed] [Google Scholar]
- Danielsen S, Strand S, Davies C., and , Stokke B. Glycosaminoglycan destabilization of DNA-chitosan polyplexes for gene delivery depends on chitosan chain length and GAG properties. Biochim Biophys Acta Gen Subj. 2005;1721:44–54. doi: 10.1016/j.bbagen.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Putnam D. Polymers for gene delivery across length scales. Nat Mater. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- Medina-Kauwe L, Xie J., and , Hamm-Alvarez S. Intracellular trafficking of nonviral vectors. Gene Ther. 2005;12:1734–1751. doi: 10.1038/sj.gt.3302592. [DOI] [PubMed] [Google Scholar]
- Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E., and , Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- Godbey W, Ku K, Hirasaki G., and , Mikos A. Improved packing of poly(ethylenimine)/DNA complexes increases transfection efficiency. Gene Ther. 1999;6:1380–1388. doi: 10.1038/sj.gt.3300976. [DOI] [PubMed] [Google Scholar]
- Kunath K, von Harpe A, Fischer D., and , Kissel T. Galactose-PEI-DNA complexes for targeted gene delivery: degree of substitution affects complex size and transfection efficiency. J Controlled Release. 2003;88:159–172. doi: 10.1016/s0168-3659(02)00458-3. [DOI] [PubMed] [Google Scholar]
- Godbey W, Wu K., and , Mikos A. Size matters: Molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45:268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Plank C, Mechtler K, Szoka F., and , Wagner E. Activation of the complement system by synthetic DNA complexes: a potential barrier for intravenous gene delivery. Hum Gene Ther. 1996;7:1437–1446. doi: 10.1089/hum.1996.7.12-1437. [DOI] [PubMed] [Google Scholar]
- Plank C, Zatloukal K, Cotten M, Mechtler K., and , Wagner E. Gene-transfer into hepatocytes using asialoglycoprotein receptor mediated endocytosis of DNA complexed with an artificial tetra-antennary galactose ligand. Bioconjug Chem. 1992;3:533–539. doi: 10.1021/bc00018a012. [DOI] [PubMed] [Google Scholar]
- Wolfert M, Dash P, Nazarova O, Oupicky D, Seymour L, Smart S, et al. Polyelectrolyte vectors for gene delivery: Influence of cationic polymer on biophysical properties of complexes formed with DNA. Bioconjug Chem. 1999;10:993–1004. doi: 10.1021/bc990025r. [DOI] [PubMed] [Google Scholar]
- Honore I, Grosse S, Frison N, Favatier F, Monsigny M., and , Fajac I. Transcription of plasmid DNA: Influence of plasmid DNA/polyethylenimine complex formation. J Controlled Release. 2005;107:537–546. doi: 10.1016/j.jconrel.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Dervan P., and , Edelson B. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr Opin Struct Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- Zelikin A, Trukhanova E, Putnam D, Izumrudov V., and , Litmanovich A. Competitive reactions in solutions of poly-L-histidine, calf thymus DNA, and synthetic polyanions: Determining the binding constants of polyelectrolytes. J Am Chem Soc. 2003;125:13693–13699. doi: 10.1021/ja036387y. [DOI] [PubMed] [Google Scholar]
- Thomas M., and , Klibanov A. Enhancing polyethylenimine's delivery of plasmid DNA into mammalian cells. Proc Natl Acad Sci USA. 2002;99:14640–14645. doi: 10.1073/pnas.192581499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takigawa D., and , Tirrell D. Disruption of phospholipid packing by branched poly(ethylenimine) derivatives. Macromolecules. 1985;18:338–342. [Google Scholar]
- Ferruti P, Fere A., and , Bettelli A. High polymers of acrylic and methacrylic esters of N-hydroxysuccinimide as polyacrylamide and polymethacrylamide precursors. Polymer. 1972;13:462–464. [Google Scholar]
- Godwin A, Hartenstein M, Muller A., and , Brocchini S. Narrow molecular weight distribution precursors for polymer-drug conjugates. Angew Chem Int Ed Engl. 2001;40:594–597. doi: 10.1002/1521-3773(20010202)40:3<594::AID-ANIE594>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Shunmugam R., and , Tew G. Efficient route to well-characterized homo, block, and statistical polymers containing terpyridine in the side chain. J Polym Sci A. 2005;43:5831–5843. [Google Scholar]
- Matyjaszewski K, Pintauer T., and , Gaynor S. Removal of copper-based catalyst in atom transfer radical polymerization using ion exchange resins. Macromolecules. 2000;33:1476–1478. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conjugation profiles of bi-functionalized polymers.
Surface-charge measurements of polyplexes formed by cationic and alkylated polymers.
Buffering capacity of cationic and alkylated polymers in their free, uncomplexed form.
Effects of molecular weight on the transfection profile of cationic and alkylated polymers in NIH/3T3 cells.
Effects of molecular weight on the cytotoxicity of polymeric vectors.