Abstract
The major Hsp70 of the mitochondrial matrix (Ssc1 in yeast) is critically important for the translocation of proteins from the cytosol, across the mitochondrial inner membrane, and into the matrix. Tim44, a peripheral inner membrane protein with limited sequence similarity to the J domain of J-type cochaperones, tethers Ssc1 to the import channel. Here we report that, unlike a J protein, Tim44 does not stimulate the ATPase activity of Ssc1, nor does it affect the stimulation by either a known mitochondrial J protein or a peptide substrate. Thus, we conclude that Tim44 does not function as a J protein cochaperone of Ssc1; rather, it tethers Ssc1 to the import channel through interactions independent of those critical for J protein function. However, a previously unstudied essential gene, PAM18, encodes an 18-kDa protein that contains a J domain and is localized to the mitochondrial inner membrane. Pam18 stimulates the ATPase activity of Ssc1; depletion of Pam18 in vivo disrupts import of proteins into the mitochondrial matrix. We propose that Pam18 is the J protein partner for Ssc1 at the import channel and is critical for Ssc1's function in protein import.
Most proteins of the mitochondrial matrix are synthesized on cytosolic ribosomes and must therefore be imported across the outer and inner mitochondrial membranes. Translocation across the inner membrane occurs through the inner membrane channel and is driven by the membrane potential and an import motor (1–3). Three critical components of this motor are the major Hsp70 molecular chaperone of mitochondria (mtHsp70; Ssc1 in yeast); the peripheral inner membrane protein Tim44, which tethers Ssc1 to the import channel; and the nucleotide release factor for Ssc1, Mge1. Multiple cycles of Ssc1 binding to and release from translocating polypeptide, driving the import process, are required for import of proteins into the mitochondrial matrix (1–3).
Ssc1, like other Hsp70s, contains a C-terminal domain that binds short segments of unfolded polypeptides rich in hydrophobic amino acids (4, 5). The N-terminal ATPase domain regulates this binding through interaction with adenine nucleotides. In turn, binding of the peptide segment to the C-terminal domain stimulates the ATPase activity of the N terminus. In the ATP-bound state, interaction with peptide substrate is unstable, with very fast on and off rates. In the ADP-bound state, the interaction is relatively stable, with slow on and off rates. Therefore, it is thought that the ATP-bound form of Hsp70 initiates interaction with polypeptide substrate, which is then stabilized by the hydrolysis of ATP. Exchange of ATP for bound ADP results in the release of peptide, thus completing the cycle.
Hsp70s rarely, if ever, function independently. Rather, they function with cochaperones. J proteins, named because of the presence of the signature J domain that interacts with the ATPase domain, stimulate Hsp70's ATPase activity, thus facilitating interaction with substrate polypeptides (6, 7). This activity is a critical feature of J protein function, because amino acid alterations in the J domain that decrease ATPase stimulation are functionally defective in vivo (8). For example, in the lumen of the endoplasmic reticulum, the ability of the membrane-associated J protein Sec63 to stimulate the ATPase activity of the luminal Hsp70 Kar2 is essential for translocation of proteins from the cytosol into the endoplasmic reticulum lumen (9–11).
No J protein of mitochondria has been reported to be involved in protein translocation. However, three mitochondrial J proteins have been studied in the yeast Saccharomyces cerevisiae. The soluble matrix J protein Mdj1 is known to function with Ssc1 in folding of imported and mitochondrially encoded proteins but does not play a significant role in import (12, 13). The J protein Jac1 functions with the minor Hsp70 of the matrix, Ssq1, in the biogenesis of Fe/S clusters (14, 15). Mdj2 is a J protein of the inner membrane. However, a deletion of MDJ2 has little or no phenotypic effect; thus, its function remains obscure (16).
Tim44, a mitochondrial inner membrane protein, has been considered as a possible J protein partner for Ssc1. Tim44, which serves as a tether of Ssc1 for the import channel, shows a weak similarity to the J domain of Sec63, and a deletion of this region disrupts the interaction of Tim44 with Ssc1 in mitochondria (17). Although controversial, some recent reports indicate that Tim44 interacts with the ATPase domain of Ssc1, as does the J domain of known J proteins (18, 19). However, regardless of whether it acts as a J protein partner of Ssc1, Tim44 regulates the interaction of Ssc1 with the import channel. Tim44 tethers ATP-bound Ssc1 to the import channel. When polypeptide substrates bind Ssc1[ATP], Ssc1 is released from Tim44 even in the absence of ATP hydrolysis, thus permitting the binding of another Ssc1[ATP] at the channel and a second cycle of interaction (20).
Because of the importance of the import of proteins into mitochondria for cell function, we set out to test the idea that Tim44, analogous to Sec63, functions as a J protein partner of Ssc1 in protein import. We found that Tim44 does not have activities associated with a prototypical J protein. Rather, a previously unstudied essential gene, PAM18, encodes a J protein of the inner membrane that is important for protein translocation in vivo and is a J protein partner of Ssc1.
Materials and Methods
Yeast Strains, Plasmids, and Genetic Techniques. PAM18 was obtained by PCR-amplifying genomic DNA from position –303 to +702 by using Pfu Turbo polymerase (Stratagene) and cloned into pRS316 (21). To obtain a null allele of PAM18, coding sequences from +71 to +542 were replaced with the TRP1 gene. A heterozygous PAM18/pam18 diploid of PJ53 (22) carrying PAM18 on a plasmid was sporulated. Δpam18 haploids carrying PAM18 on pRS316 were isolated by dissecting tetrads onto YP medium (1% yeast extract/2% peptone). Mutants of PAM18 were constructed by site-directed mutagenesis by using QuikChange protocol (Stratagene) and cloned into pRS315.
Pam18 was epitope-tagged at its C terminus with three tandem copies of the hemagglutinin (HA) peptide by first introducing a NotI site at its termination codon and then subcloning the HA tag as a NotI fragment from pGTEP1 (23). The tagged protein was functional; cells carrying the tagged protein as the only PAM18 protein grew as well as wild-type cells under a variety of conditions. For purposes of purification, the entire ORF of PAM18 was cloned into the glutathione S-transferase expressing vector pGEX-KG (24), thus forming a fusion protein with Pam18 at the C terminus. To modulate expression of Pam18, the entire ORF (+1 to +507) was cloned into pCM189 (25), placing PAM18 under control of a tetracycline-regulatable promoter, resulting in repression of transcription upon addition of the tetracycline derivative doxycycline.
Complete synthetic media lacking specific amino acids or containing 5-fluoroorotic acid were prepared as described (26). All chemicals were obtained from Sigma unless otherwise stated.
Fractionation of Mitochondria. Mitochondria were fractionated essentially as described in ref. 27. Briefly, 25 μg of mitochondria were swollen in 500 μl of EM buffer (10 mM Mops KOH, pH 7.2/1 mM EDTA) and incubated on ice for 15 min. Efficiency of disruption of the outer membrane was assessed by incubation with proteinase K at 50 μg/ml on ice for 15 min. As a control, sodium deoxycholate was added to 1% to disrupt the mitochondrial inner membrane. Mitochondria were separated into membrane and soluble fractions as described in ref. 28. Mitochondrial fractions were subjected to immunoblot analysis by using the ECL system (Amersham Pharmacia Biotech) according to the manufacturer's suggestion, by using polyclonal antibodies specific for Tim44, cytochrome b2, and Mge1 and a monoclonal antibody specific for HA.
Protein Purification. Pam18 was purified from bacterial strain C41 (29). Expression was induced for3hat37°C by addition of 1 mM isopropyl-β-d-thiogalactopyranoside. After affinity chromatography using glutathione agarose, biotinylated thrombin was added and the mixture was incubated for3hat20°C. The cleaved Pam18 product was recovered by removal of the glutathione agarose beads by centrifugation and removal of the thrombin by streptavidin/agarose (catalog no. 69022-3, Novagen).
Tim44His (20), Mdj1His (30), Ssc1His (31), and Ssq1His (31) were purified as described. All His-tagged proteins were previously shown to be able to rescue the absence of the normal protein in vivo.
ATPase Assay. Isolation of Hsp70:ATP complexes and ATPase assays were carried out essentially as described in refs. 20 and 32, with some changes. To obtain radiolabeled ATP:Ssc1 complexes, 50 μg of Hsp70 was incubated with 100 μCi of [α-32P]ATP (3,000 Ci/mmol, DuPont) in buffer A (25 mM Hepes-KOH, pH 7.4/100 mM KCl/11 mM Mg(OAC)2/10% glycerol) containing 25 μM ATP in 100 μl on ice for 30 sec. The complex was isolated immediately on a NICK column (Amersham Pharmacia Biotech) and frozen in aliquots at –70°C. For ATPase assays, aliquots were thawed on ice; time 0 was the time of shift to 23°C, when additions of peptide P5 (CALLLSAPRR), Tim44, Mdj1, and/or Pam18 were made to some samples. At the indicated times, 3-μl aliquots were removed and mixed with 1 μl of stop solution containing 4 M formic acid, 2 M LiCl, and 36 mM ATP. The mixture was spotted onto a polyethyleneimine-cellulose TLC plate and developed in 1 M formic acid and 0.5 M LiCl. After drying, the TLC plate was exposed to a PhosphorImager screen (Molecular Dynamics), and the amount of ATP and ADP was quantified on a PhosphorImager system (Molecular Dynamics). The percentage of hydrolysis at 0 time (≈20%) was subtracted from all time points.
Coimmunoprecipitation Assay. The Ssc1 coimmunoprecipitation assay was carried out essentially as described in ref. 20. Affinity-purified Ssc1 antibodies were crosslinked to protein A-Sepharose beads. Six micrograms of Ssc1 was incubated with Ssc1 antibody beads at 4°C for 1 h. Under this condition, ≈ 90% of Ssc1 was typically immunoprecipitated. After three washes, Tim44 (0.6 μg) was added. The mixture was incubated in a final volume of 150 μl at 23°C for 30 min at which point 1.5 μl of a 1mg/ml solution of peptide P5 was added to some reactions, and incubation continued for an additional 30 min. Tim44:Ssc1 complex formation was robust, forming at 0°C and under a variety of salt and buffer conditions (data not shown). The final concentrations of Ssc1, Tim44, and the peptide P5 were 0.6, 0.09, and 10 μM, respectively. After centrifugation, the beads were washed three times with mitochondrial lysis buffer (250 mM sucrose/80 mM KCl/20 mM Mops-KOH, pH 7.2/0.2% Triton X-100/1 mM PMSF) containing the appropriate nucleotide. Tim44 or Ssc1 bound to the beads was detected after SDS/PAGE by immunoblot analysis by using polyclonal antibodies specific for Tim44 and Ssc1.
Peptide Binding Assay. Briefly, fluorescence anisotropy assays for binding of peptide were carried out as described in refs. 20 and 32. Peptide P5 was labeled with fluorescein to generate F-P5 as described (33). Various concentrations of Ssc1 were incubated with F-P5 (10 nM) at 23°C in buffer A (25 mM Hepes-KOH, pH 7.4/100 mM KCl/11 mM Mg(OAC)2/10% glycerol). After binding reached equilibrium, anisotropy measurements were made with the Beacon 2000 Fluorescence Polarization system (Panvera, Madison, WI) at 23°C with excitation at 490 nm and emission at 535 nm.
Results
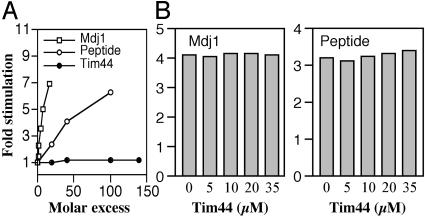
Tim44 Does Not Stimulate the ATPase Activity of Ssc1. To address the possibility that Tim44 acts as a J protein cochaperone at the import channel, we tested its ability to affect the ATPase activity of Ssc1 using single-turnover conditions. A complex between Ssc1 and radiolabeled ATP was isolated, and the rate of ADP formation was determined upon incubation in the presence and absence of Tim44. The basal rate of ATP hydrolysis by Ssc1 was 0.07 min–1, consistent with previous results (32). No effect on the ATPase activity of Ssc1 was observed upon addition of Tim44 over a range of concentrations, even at 35 μM, a 140 molar excess over Ssc1 (Fig. 1A). As a control, increasing concentrations of either the soluble mitochondrial J protein Mdj1 or a peptide substrate were added. Both stimulated the hydrolysis of ATP. For example, at a 10-fold molar excess (2 μM) of Mdj1, activity was stimulated 7-fold; at a 100-fold excess (20 μM), peptide activity was stimulated 6.5-fold.
Fig. 1.
Effect of Tim44 on the ATPase activity of Ssc1. Ssc1:ATP complex (≈0.2 μM) was incubated at 23°C in the presence of various concentrations of Mdj1 or peptide P5. The rate of conversion to ADP was measured and plotted as the fold stimulation over the basal rate determined in the absence of additional components. (A) Mdj1, peptide P5, or Tim44, as indicated. (B) Reactions with 1 μM Mdj1 (Left)or5 μM peptide P5 (Right) were carried out in the presence of indicated concentrations of Tim44.
Tim44 Interaction with Ssc1 Is Distinct from That of a J Protein. To assess whether Tim44 was interacting with Ssc1 at sites similar to or overlapping with those contacting a J protein, we tested the effect of increasing concentrations of Tim44 on the ability of Mdj1 to stimulate the ATPase activity of Ssc1. No effect on stimulation was seen when Tim44 was added at concentrations between 5 and 35 μM, while Mdj1 was held constant at 1 μM (Fig. 1B). Similar experiments were carried out in the presence of peptide substrates. Again, no effect was seen on peptide stimulation of ATPase activity upon addition of Tim44. These results indicate that Tim44 does not act as a J protein partner of Ssc1, nor does it interfere with binding of a substrate peptide.
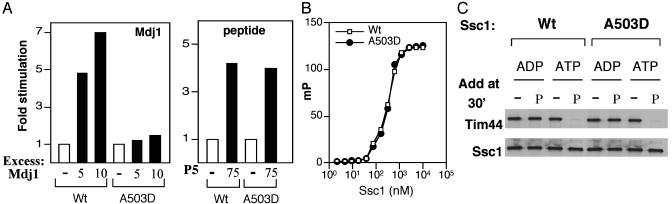
To further probe the differences between Tim44 and J protein interactions, we used a mutant that we isolated during a search for temperature-sensitive point mutations in SSC1. Ssc1A503D has a single amino acid alteration in the peptide-binding domain. The ATPase activity of A503D mutant protein was not stimulated by Mdj1 (Fig. 2A). However, interactions with Tim44 and a peptide substrate were normal. Ssc1A503D bound fluorescein-tagged peptide with the same affinity as wild-type protein, with a dissociation constant (Kd) of 0.3 μM (20), as measured by fluorescence anisotropy (32) (Fig. 2B). A peptide substrate stimulated the ATPase activity of Ssc1A503D nearly as well as wild-type protein (4.0- and 4.2-fold, respectively) at a peptide concentration of 15 μM (Fig. 2 A). Tim44 bound to both Ssc1[ATP] and Ssc1A503D[ADP], and, as with wild-type protein, peptide was able to stimulate release of Tim44 from a Ssc1A503D[ATP] complex (ref. 20 and Fig. 2C). Thus, interaction with Tim44 and peptide substrate, but not the J protein Mdj1, was normal for this mutant Ssc1 protein. Because Tim44 cannot compete with binding of Mdj1 or peptide and interacts normally with Ssc1 A503D, we conclude that Tim44 is not a J protein and does not bind to sites on Ssc1 that prevent its interaction with a known J protein.
Fig. 2.
Functional defect of Ssc1A503D. (A) ATPase activity. Ssc1:ATP and Ssc1A503D:ATP complexes (≈0.2 μM) were incubated at 23°C in the presence of Mdj1 or peptide P5. The basal rates of hydrolysis of the mutant and wild-type protein were indistinguishable. (B) Peptide binding measured by fluorescence anisotropy: Fluorescein-labeled F-P5 (10 nM) was incubated in the presence of the indicated concentrations of wild-type and A503D mutant protein and ADP. Anisotropy measurements were taken at 25°C; raw polarization values were fitted to a one-site binding (hyperbola) equation to determine the Kd. (C) Effect of peptide on Ssc1:Tim44 interaction. Tim44 (0.09 μM) was incubated with Ssc1 (0.6 μM) in the presence of either ATP or ADP, as indicated, for 30 min. In some cases, 10 μM peptide P5 was added, and incubation continued for an additional 30 min. The mixtures were immunoprecipitated by using Ssc1-specific antibodies as described in Materials and Methods. The presence of Tim44 and Ssc1 in the precipitate was assessed by immunoblot analysis by using antibodies specific for the respective proteins.
Essential J Protein of the Mitochondrial Inner Membrane, Pam18. Multiple binding and release cycles of Ssc1 with an incoming polypeptide chain are required for its import (1–3). It has been estimated that, on average, in vivo, three polypeptide chains pass through each import channel per minute (34). Thus, the turnover rate of ATP hydrolysis by Ssc1 must be sufficiently fast to allow numerous interaction cycles within this time frame. The turnover rate of Ssc1 observed, in vitro, in the presence of a high concentration of peptide substrate is one turnover per minute. This rate of hydrolysis is clearly not sufficient to drive import in vivo, because at this rate less than one cycle of interaction would occur for each imported polypeptide. Assuming that the rate of ATP hydrolysis of Ssc1 that we observe in our in vitro assays is reflective of the natural state of the protein, a cofactor for Ssc1 that stimulates ATP hydrolysis must exist.
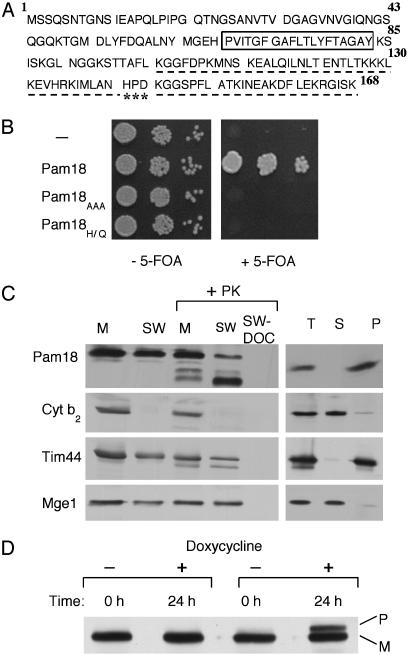
Because Tim44 does not stimulate the ATPase activity of Ssc1, we searched the yeast genome database (www.yeastgenome.org) for potential J protein partners for Ssc1. The ORF YLR008C (PAM18) was of interest because it is an essential gene that encodes a protein containing a predicted internal membrane spanning domain (Fig. 3A) and a J domain that is 57% identical to that of the J protein of the mitochondrial inner membrane Mdj2 (16). We confirmed that a deletion of PAM18 in the W303 strain background resulted in inviability, while deletion of MDJ2 had little phenotypic effect (data not shown).
Fig. 3.
In vivo function of the J protein Pam18. (A) Predicted amino acid sequence of Pam18. A box is drawn around the putative membrane-spanning region. The region having sequence identity to a J domain is underlined; the highly conserved HPD is indicated by ***. (B) Requirement in vivo of J domain of Pam18. Strains carrying plasmids expressing no protein (–), wild-type Pam18, and mutant Pam18, Pam18AAA or Pam18H141Q (Pam18HI/Q) were spotted as a 1- fold dilution series on media lacking (Left) or containing (Right) 5-fluoroorotic acid. Plates were incubated for 3 days at 30°C. (C) Fractionation of mitochondria. Mitochondria were purified from cells expressing Pam18 having an HA tag at the C terminus. Mitochondria were separated into membrane and soluble components (Right). Mitochondrial protein (T) and fractional equivalents of membrane pellet (P) and supernatant (S) were analyzed. Mitochondria (M) were subjected to swelling to disrupt the outer membrane (SW) (Left). Equivalent amounts were treated with proteinase K and in some cases treated with detergent (sodium deoxycholate), as indicated. The fractions were subjected to immunoblot analysis by using antibodies specific for the HA tag on Pam18, as well as an antibody specific for known proteins of the intermembrane space (cytochrome b2), inner membrane (Tim44), and matrix (Mge1). (D) Δpam18 cells carrying Pam18 under control of the tetracycline regulatable promoter were grown overnight in rich media, at which point they were subcultured in the presence or absence of the tetracycline analogue doxycycline. After 24 h whole-cell lysates were prepared and separated by SDS/PAGE, followed by immunoblot analysis by using antibodies specific for Hsp60.
To determine whether the J domain of Pam18 is important for its function, we constructed two mutants causing alterations in the J domain. Because the hallmark of a J domain is a highly conserved tripeptide, histidine, proline, and aspartic acid (HPD), we constructed a mutation that results in the replacement of the HPD with alanines (Pam18A3). In addition, we constructed a mutation that results in a substitution of a glutamine in place of the histidine (Pam18H141Q). To test whether these mutants could rescue the inviability of the Δpam18 strain, plasmids carrying the PAM18 mutants were transformed into a Δpam18 strain carrying a wild-type copy of PAM18 on a centromeric plasmid having the URA3 gene. When the resulting strains were plated on media containing 5-fluoroorotic acid, which selects for cells having lost the URA3-containing plasmid carrying the wild-type PAM18 gene, no viable cells were recovered (Fig. 3B). Thus, the J domain is essential for PAM18 function.
For Pam18 to be a J protein partner for Ssc1 in vivo, it must be localized in mitochondria such that its J domain can interact with Ssc1, which is in the matrix. To determine the cellular location of Pam18, mitochondria from cells expressing Pam18 with a HA tag at its C terminus (Pam18-HA) were isolated. These mitochondria contained a protein that reacted with the HA-specific antibody and was not present in mitochondria expressing Pam18 lacking the HA tag (data not shown). Mitochondria containing Pam18-HA were separated into membrane and soluble fractions, which were then subjected to immunoblot analysis by using HA-specific antibodies. Pam18-HA was present in the membrane fraction, as was Tim44 (Fig. 3C Right). To further localize Pam18, mitochondria having an intact inner membrane, but disrupted outer membrane, were formed by swelling mitochondria in a buffer of low osmotic strength. The swollen mitochondria were then treated with protease, as were intact mitochondria (Fig. 3C Left). Like Tim44, the intermembrane space protein cytochrome b2, and the matrix protein Mge1, Pam18 was not substantially affected by protease digestion of intact mitochondria, suggesting that Pam18 was not in the outer membrane. However, treatment of swollen mitochondria with protease resulted in the production of a predominant cleavage fragment containing the HA tag. These results suggest that Pam18 is located in the inner membrane. In addition, because the HA tag at the C terminus was resistant to protease attack, the J domain that is C-terminal to the putative membrane-spanning domain is predicted to be on the matrix side of the inner membrane, with the N terminus protruding into the intermembrane space. Such an orientation would make the J domain accessible to Ssc1 in the matrix.
To assess whether Pam18 has a role in mitochondrial protein import in vivo, PAM18 was placed under the control of the tetracycline-repressible promoter. Drug was added to turn off expression, and accumulation of the precursor form of a nuclear-encoded mitochondrial protein, Hsp60, was monitored. As is the case with most nuclear-encoded matrix proteins, the presequence of Hsp60 is cleaved by the matrix processing protease immediately upon traversing the inner membrane (35); the accumulation of the normally undetectable precursor form has been used previously as an in vivo indicator of function of the import process (28, 36). Cells were harvested 24 h after drug addition, when the growth of the tester strain had begun to slow compared with the control strain. The precursor form of Hsp60 accumulated in the strain in which Pam18 expression was repressed, but not in the control strain. This accumulation is indicative of a role for Pam18 in import of proteins into the mitochondrial matrix in vivo.
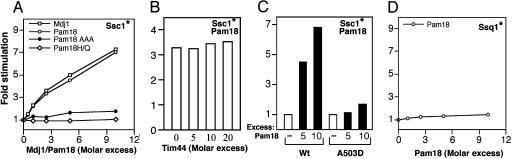
Pam18 Specifically Stimulates Ssc1 ATPase Activity. The results described in the previous section suggest that Pam18 acts as a J protein in mitochondrial import. We thus purified PAM18 wild-type and mutant proteins and tested their ability to stimulate the ATPase activity of Ssc1 using the single-turnover assay described above. Wild-type Pam18 stimulated the ATPase activity 7-fold when present at a 7-fold excess (Fig. 4A). Pam18 and Mdj1 had very similar concentration dependences for stimulation of Ssc1 ATPase activity, suggesting that the two proteins interact similarly with Ssc1. As was the case with Mdj1, addition of excess Tim44 did not inhibit the ability of Pam18 to stimulate Ssc1 (Fig. 4B), indicating that binding of Tim44 and Pam18 are not mutually exclusive.
Fig. 4.
Stimulation of Ssc1 ATPase activity by Pam18. Hsp70:ATP complexes were prepared, and the rate of hydrolysis was measured at 23°C in the presence of various concentrations of Mdj1, wild-type Pam18, or mutant Pam18, in the presence or absence of Tim44. The rate of conversion to ADP was measured and is plotted as the fold stimulation over the basal rate. In each panel, the Hsp70 for which ATP hydrolysis was measured is indicated by an asterisk. (A) Ssc1:ATP (≈0.2 μM) was incubated in the presence of various concentrations of wild-type Pam18, Mdj1, or the Pam 18 mutant proteins Pam18AAA or Pam18H141Q. (B) Reactions with 1 μM Pam18 were carried out in the presence of indicated concentrations of Tim44. (C) Ssc1:ATP and Ssc1A503D:ATP complexes (≈0.2 μM) were incubated in the presence of excess Pam18. (D) Ssq1:ATP complexes (0.1 μM) in the presence of various concentrations of Pam18.
To test whether Pam18 stimulates Ssc1 ATPase activity as a prototypical J protein, we used the J domain mutants described in the previous section, as well as the Ssc1 mutant that has a defective interaction with the known J protein Mdj1. Pam18 mutant proteins with altered J domains that were unable to function in vivo also failed to stimulate Ssc1's ATPase activity (Fig. 4A). In addition, we tested the ability of wild-type Pam18 to stimulate the ATPase activity of the A503D Ssc1 mutant (Fig. 4C). Like Mdj1 (Fig. 2D), Pam18 did not stimulate the mutant protein. Also, the stimulation by Pam18 was specific, because it failed to stimulate the ATPase activity of another Hsp70 of the mitochondrial matrix Ssq1 (Fig. 4D). Thus, we conclude that Pam18 is a J protein partner of Ssc1.
Discussion
The data presented here allow a better understanding of the mechanism of translocation of proteins across the mitochondrial inner membrane. First, while Tim44 tethers Ssc1 to the import channel, it does not function as a J protein cochaperone for mitochondrial Hsp70 Ssc1, nor does it bind to the same site(s) on Hsp70 as a J protein. Second, we have identified a J protein partner for Ssc1 in the inner membrane that stimulates Ssc1's ATPase activity. These results lead to a model in which Ssc1, tethered to the import channel, can be stimulated by a J protein in close proximity, thus ensuring rapid binding of the incoming polypeptide chain.
Determining the mechanism of action of Tim44 in protein translocation is important, because it is central to the initial interaction of a translocating protein with the import motor. The inability of Tim44 to stimulate the ATPase activity of Ssc1, even at concentrations 20-fold higher than that required for a 5-fold stimulation by the known J protein partner of Ssc1, Mdj1, argues strongly against Tim44 functioning as a J cochaperone of Ssc1. The original idea that Tim44 might function as a J-type cochaperone for Ssc1 was based on its limited sequence homology to the J domains of J proteins (17). It is also possible that the J-like region evolved from a true J domain to facilitate interaction between Ssc1 and Tim44. In fact, an 18-aa deletion completely removing this region resulted in synthesis of a stable mutant protein that was inserted correctly in the membrane but was defective in interaction with Ssc1. These results suggested that this region of Tim44 interacts directly with Ssc1 (17). However, this scenario of Tim44 having evolved a J-like domain that interacts with Ssc1 at the J domain binding site is unlikely because no competition for functional interaction between Tim44 and the true J proteins Mdj1 and Pam18 were observed. Thus, the interaction sites of J proteins and Tim44 are likely distinct; more detailed structural information will be required to clearly delineate the exact sites of interactions among these proteins.
Our results indicate that Pam18 is the prime candidate for the J protein partner for Ssc1 in mitochondrial import. As is the case for other components of the import machinery, such as Tim44, Ssc1, and Mge1, Pam18 is essential. It is located in the inner membrane, with its J domain extending into the matrix, thus well positioned to interact with Ssc1 when it is tethered to the import channel. As expected of a bona fide J protein partner, it can stimulate the ATPase activity of the Hsp70 Ssc1, with which it functions, and it does not stimulate the ATPase activity of another matrix Hsp70, Ssq1. This stimulation is not affected by the presence of Tim44. Thus, Pam18 would be able to stimulate the ATPase activity of Ssc1 while it is tethered to the import channel by Tim44.
However, Tim44 should not be considered simply a membrane anchor for Ssc1. The interaction between the two proteins seems to have evolved to be exquisitely sensitive to the conformation of Ssc1. For example, a conformational change of Ssc1[ATP] induced upon interaction with a peptide substrate rapidly destabilizes the Ssc1:Tim44 interaction (20). Likewise, binding of Mge1 to Ssc1[ADP] destabilizes interaction with Tim44 (20, 37). Thus, Tim44 has evolved as a conformational-sensitive switch governing association of Ssc1 with the import channel.
The fact that Tim44 is not a J protein and the identification of a new J protein partner of Ssc1 allow us to put forward a revised model of protein import. In contrast to the endoplasmic reticulum, where tethering to the import channel and stimulation of ATPase activity are executed by one protein, the J protein Sec63 (38), mitochondria separate these critical functions into two, or perhaps more, proteins. Tim44 regulates Ssc1's interaction with the import channel, and Pam18 stimulates Ssc1's ATPase activity. Occupancy of the biologically relevant form of Ssc1, Ssc1[ATP], predominates at the import channel (20). Interaction with an incoming polypeptide stimulates dissociation of Ssc1 from Tim44 and, in combination with Pam18, stimulates ATP hydrolysis, stabilizing the Ssc1:polypeptide substrate interaction. Interestingly, Ssc1's release from Tim44 upon substrate binding is independent of ATP hydrolysis. Because Pam18's ability to stimulate the ATPase activity of Ssc1 is not affected by Tim44, determining the relative rates of release from Tim44 and ATP hydrolysis will be needed before a more definitive model of import can be put forth. However, it should be noted that mechanistically it is important that ATP hydrolysis be temporally coupled to release, because failure to hydrolyze ATP will result in dissociation of the polypeptide chain from Ssc1 due to the extreme instability of the Ssc1:substrate interaction when Ssc1 is bound to ATP. Pam18 surely plays a critical role in stimulating the hydrolysis rate. It may also play a role in coordinating the timing of hydrolysis, allowing efficient coupling of release from Tim44 and ATP hydrolysis.
Determination of the exact mechanistic details of protein import and the coordination of action of Ssc1 cofactors awaits further analysis. However, there is a precedent for additional proteins mediating interactions among Hsp70s/J proteins/substrate polypeptides. In Thermus thermophilus a stable complex between Hsp70 and a J protein is mediated by a third protein, DafA (39). Upon binding of a substrate polypeptide, DafA is displaced from the complex. Interactions of adapter proteins such as Tim44 and DafA may be more common than is presently appreciated and serve important roles in regulating interactions among Hsp70s, J proteins, and their substrate polypeptides.
Acknowledgments
We thank Jarek Marszalek for helpful discussions and thoughtful comments on the manuscript. This work was supported by National Institutes of Health Grants GM278709 (to E.A.C.) and 5T326M08349 (to A.A.).
Abbreviation: HA, hemagglutinin.
References
- 1.Neupert, W. & Brunner, M. (2002) Nat. Rev. Mol. Cell Biol. 8, 555–565. [DOI] [PubMed] [Google Scholar]
- 2.Pfanner, N. & Geissler, A. (2001) Nat. Rev. Mol. Cell Biol. 5, 339–349. [DOI] [PubMed] [Google Scholar]
- 3.Matouschek, A., Pfanner, N. & Voos, W. (2000) EMBO Rep. 5, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukau, B. & Horwich, A. L. (1998) Cell 92, 351–366. [DOI] [PubMed] [Google Scholar]
- 5.Hartl, F. & Hayer-Hartl, M. (2002) Science 295, 1852–1858. [DOI] [PubMed] [Google Scholar]
- 6.Cheetham, M. E. & Caplan, A. J. (1998) Cell Stress Chaperones 3, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelley, W. (1999) Curr. Biol. 9, R305–R308. [DOI] [PubMed] [Google Scholar]
- 8.Genevaux, P., Schwager, F., Georgopoulos, C. & Kelley, W. L. (2002) Genetics 162, 1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodsky, J. L. & Schekman, R. (1993) J. Cell Biol. 123, 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matlack, K. E. S., Plath, K., Misselwitz, B. & Rapoport, T. A. (1997) Science 277, 938–941. [DOI] [PubMed] [Google Scholar]
- 11.Corsi, A. K. & Schekman, R. (1997) J. Cell Biol. 137, 1483–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermann, J., Stuart, R., Craig, E. & Neupert, W. (1994) J. Cell Biol. 127, 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowley, N., Prip-Buus, C., Westermann, B., Brown, C., Schwarz, E., Barrell, B. & Neupert, W. (1994) Cell 77, 249–259. [DOI] [PubMed] [Google Scholar]
- 14.Lutz, T., Westermann, B., Neupert, W. & Herrmann, J. (2001) J. Mol. Biol. 307, 815–825. [DOI] [PubMed] [Google Scholar]
- 15.Voisine, C., Cheng, Y. C., Ohlson, M., Schilke, B., Hoff, K., Beinert, H., Marszalek, J. & Craig, E. (2001) Proc. Natl. Acad. Sci. USA 98, 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westermann, B. & Neupert, W. (1997) J. Mol. Biol. 272, 477–483. [DOI] [PubMed] [Google Scholar]
- 17.Merlin, A., Voos, W., Maarse, A., Meijer, M., Pfanner, N. & Rassow, J. (1999) J. Cell Biol. 145, 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krimmer, T., Rassow, J., Kunau, W. H., Voos, W. & Pfanner, N. (2000) Mol. Cell. Biol. 20, 5879–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strub, A., Rottgers, K. & Voos, W. (2002) EMBO J. 21, 2626–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, Q., D'Silva, P., Walter, W., Marszalek, J. & Craig, E. A. (2003) Science 300, 139–141. [DOI] [PubMed] [Google Scholar]
- 21.Sikorski, R. S. & Hieter, P. (1989) Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James, P., Pfund, C. & Craig, E. (1997) Science 275, 387–389. [DOI] [PubMed] [Google Scholar]
- 23.Roof, D. M., Meluh, P. B. & Rose, M. D. (1992) J. Cell Biol. 118, 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan, K. L. & Dixon, J. E. (1991) Anal. Biochem. 192, 262–267. [DOI] [PubMed] [Google Scholar]
- 25.Gari, E., Piedrafita, L., Aldea, M. & Herrero, E. (1997) Yeast 13, 837–848. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski, R. S. & Boeke, J. D. (1991) in Metholds in Enzymology: Guide to Yeast Genetics and Molecular Biology, eds. Guthrie, C. & Fink, G. R., Vol. 194, pp. 302–318. [Google Scholar]
- 27.Ryan, M. T., Voos, W. & Pfanner, N. (2001) Methods Cell Biol. 65, 189–215. [DOI] [PubMed] [Google Scholar]
- 28.Kang, P. J., Ostermann, J., Shilling, J., Neupert, W., Craig, E. A. & Pfanner, N. (1990) Nature 348, 137–143. [DOI] [PubMed] [Google Scholar]
- 29.Miroux, B. & Walker, J. E. (1996) J. Mol. Biol. 260, 289–298. [DOI] [PubMed] [Google Scholar]
- 30.Horst, M., Oppliger, W., Rospert, S., Schonfeld, H.-J., Schatz, G. & Azem, A. (1997) EMBO J. 16, 1842–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutkiewicz, R., Schilke, B., Knieszner, H., Walter, W., Craig, E. A. & Marszalek, J. (2003) J. Biol. Chem. 278, 29719–29727. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Q., Krzewska, J., Liberek, K. & Craig, E. A. (2001) J. Biol. Chem. 276, 6112–6118. [DOI] [PubMed] [Google Scholar]
- 33.Montgomery, D., Morimoto, R. & Gierasch, L. (1999) J. Mol. Biol. 286, 915–932. [DOI] [PubMed] [Google Scholar]
- 34.Lim, J. H., Martin, F., Guiard, B., Pfanner, N. & Voos, W. (2001) EMBO J. 20, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng, M., Hartl, F., Martin, J., Pollock, R., Kalousek, F., Neupert, W., Hallberg, E., Hallberg, R. & Horwich, A. (1989) Nature 337, 620–625. [DOI] [PubMed] [Google Scholar]
- 36.Gambill, B. D., Voos, W., Kang, P. J., Miao, B., Langer, T., Craig, E. A. & Pfanner, N. (1993) J. Cell Biol. 123, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider, H. C., Westermann, B., Neupert, W. & Brunner, M. (1996) EMBO J. 15, 5796–5803. [PMC free article] [PubMed] [Google Scholar]
- 38.Rapoport, T., Matlack, K., Plath, K., Misselwitz, B. & Staeck, O. (1999) Biol. Chem. 380, 1143–1150. [DOI] [PubMed] [Google Scholar]
- 39.Klostermeier, D., Seidel, R. & Reinstein, J. (1999) J. Mol. Biol. 287, 511–525. [DOI] [PubMed] [Google Scholar]