Abstract
Duchenne muscular dystrophy (DMD) is caused by out-of-frame mutations of the human DMD gene. Antisense oligonucleotides (AOs) have previously been used to skip additional exons that border the deletions such that the reading frame is restored and internally truncated, but functional, dystrophin expressed. We have designed phosphorodiamidate morpholino oligomer (PMO) AOs to various exons of the human dystrophin gene. PMOs were designed to have their target sites overlapping areas of open RNA structure, as defined by hybridization-array analysis, and likely exonic splicing enhancer (ESE)/silencer sites on the target RNA. The ability of each PMO to produce exon skipping was tested in vitro in normal human skeletal muscle cells. Retrospective analysis of design parameters used and PMO variables revealed that active PMOs were longer, bound to their targets more strongly, had their target sites closer to the acceptor splice site of the exon, overlapped areas of open conformation (as defined by the hybridization or the RNA secondary structure prediction software), and could interfere with the binding of certain SR proteins. No other parameter appeared to show significant association to PMO-skipping efficacy. No design tool is strong enough in isolation; however, if used in conjunction with other significant parameters it can aid AO design.
Introduction
Duchenne muscular dystrophy (DMD) is a severe X-linked muscle-wasting disease, affecting 1 of 3,500 boys. Prognosis is poor: loss of mobility by the age of 12, compromised respiratory and cardiac function by late teens, and probable death by the age of 30. The disease is caused by mutations within the large DMD gene such that the reading frame is disrupted, leading to lack of dystrophin protein expression and breakdown of muscle fiber integrity.1 The DMD gene is large, with 79 exons. The most common DMD mutation is genomic deletion of one or more exons, generally centered around hotspots involving exons 44–55 and the 5′end of the gene.2 Mutations of the DMD gene that preserve the reading frame result in the milder, non-life-threatening Becker muscular dystrophy.
Exon skipping induced by antisense oligonucleotides (AOs), generally based on an RNA backbone, is a future hope as a therapy for DMD. Indeed, by skipping out-of-frame mutations of the DMD gene, the reading frame can be restored and a truncated, yet functional, Becker muscular dystrophy–like dystrophin protein is expressed. Studies in human cells in vitro3,4 and in animal models of the disease in vivo5,6,7,8,9 have proven the principle of exon skipping as a potential therapy for DMD (reviewed in ref. 10). Initial clinical trials using two different AO chemistries [phosphorodiamidate morpholino oligomer (PMO) (F. Muntoni, personal communication) and phosphorothioate-linked 2′-O-methyl RNA (2′OMePS)]11 have recently been performed, with encouraging results. Indisputably impressive restoration of dystrophin expression in the TA muscle of four DMD patients injected with a 2′OMePS AO to exon 51 has been reported by van Deutekom et al.11
However, it should be noted that, relative to 2′OMePS AOs, PMOs have been shown to produce more consistent and sustained exon skipping in the mdx mouse model of DMD (refs. 12,13,14 and A. Malerba et al., manuscript submitted), in human muscle explants,15 and in dystrophic canine cells in vitro.16 Most important, PMOs have excellent safety profiles from clinical and preclinical data.17
The first step to a clinical trial is the choice of the optimal AO target site for skipping of those DMD exons most commonly deleted in DMD. In-depth analysis of arrays of 2′OMePS AOs have been reported,18,19 and relationships between skipping bioactivity and AO variables examined. Here, we report the first detailed study of the role that AO target-site variables have on the bioactivity of PMOs to induce skipping. The results reported here should have an impact on the initial planning and design of antisense PMOs for future potential clinical trials.
Results
PMO design and analysis of bioactivity
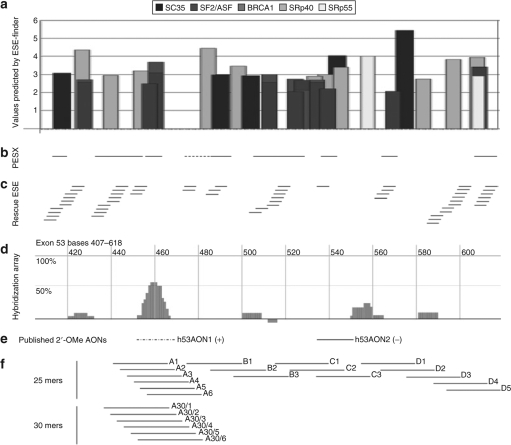
A unique set of 66 PMOs has been designed to target exons 44, 45, 46, 51, and 53 of the human gene for dystrophin. The design process for exon 53 is depicted in Figure 1, and has also been performed for the other four exons (data not shown). The exon sequence was analyzed for the presence of exonic splicing enhancers (ESEs) and exonic splicing suppressors or silencers and the outputs aligned for the three available algorithms, ESEfinder (Figure 1a),20,21 PESX (Figure 1b),22,23 and Rescue ESE (Figure 1c).24 Hybridization-array analysis was also performed for each exon in vitro, as described in Materials and Methods section. The peaks shown in Figure 1d indicate areas of the exon that are in a conformation able to hybridize to the array, and which may consequently prove more accessible to antisense AOs. The coincidence of ESEs, as predicted by two or more algorithms and hybridization peaks determined experimentally, was used to design arrays of 25 mer and, subsequently, 30 mer PMOs, the positions of which are shown in Figure 1f. The binding sites for 2′OMePS AOs described previously18 are shown for comparison (Figure 1e).
Figure 1.
Scheme summarizing the tools used in the design of PMOs to exon 53. (a) Results of ESEfinder analysis, showing the location and values above threshold for SF2/ASF, SF2/ASF (BRCA1), SC35, SRp40, and SRp55, shown as gray and black bars, as indicated in the legend above. (b) Output of PESX analysis, showing the location of exonic splicing enhancers as solid lines, and exonic splicing silencer as a dashed line. (c) Rescue ESE analysis for exon 53, showing predicted ESEs by lines, and where they overlap, by a ladder of lines. (d) AccessMapper analysis of in vitro hybridization. Synthetic pre-mRNA containing exon 53 and surrounding introns subjected to a hybridization screen against a random hexamer oligonucleotide array, as described in Materials and Methods section. Areas of hybridization, suggestive of areas of open conformation, are indicated by peaks on the graph. (e) The position of the target sites of two 2′OMePS AOs studied previously18 are shown for comparison. (f) The location of the target sites for all the 25 mer and 30 mer PMOs to exon 53 used in this study are indicated by lines, and numbered according to the scheme used in Supplementary Table S1, except for exclusion of the prefix “h53.”
Each PMO was tested in primary cultures of human skeletal muscle, in triplicate, in at least two experiments, and over a range of concentrations from 50 to 500 nmol/l. Their bioactivity was determined by reverse transcription–PCR analysis, which showed a wide variation in the level of exon skipping induced (Supplementary Figure S1, and data not shown), ranging, for example, for the targeted skipping of exon 53, from 0% for h53C1 (Figure 1f and Supplementary Figure S1, lane 2) to 80% for h53A30/3 (Figure 1f and Supplementary Figure S1, lane 6). Often when efficient skipping of the targeted exons was achieved, an additional high molecular–weight product that is slightly shorter than the full-length product was seen (Supplementary Figure S1, lanes 4 and 5). This has been reported previously and is attributable to heteroduplex formation.4,25 Sequencing of the PCR products verified accurate skipping of the targeted exon (data not shown). The activity of each PMO at the stated optimal concentration is summarized in Supplementary Table S1. Bioactivity is expressed as a percentage of the skipped amplicon relative to total PCR product, as assessed by densitometry. Specific, consistent, and sustained exon skipping was evident for 44 of the 66 PMOs tested.
In silico analysis of PMOs
We then performed a retrospective in silico analysis of the characteristics of all 66 PMOs tested in this study, with respect to PMO length, the distance of the PMO-target site from the splice donor and acceptor sites, PMO-to-target binding energy and PMO-to-PMO binding energy, as calculated using RNAstructure2.2 software for the equivalent RNA–RNA interaction, and percentage GC content of the PMO, the results of which are summarized in Supplementary Table S1. Also shown in Supplementary Table S1 is the percentage overlap of each PMO-target site with sequences shown to be accessible to binding as determined experimentally by the hexamer hybridization–array analysis. The relationship between PMO-target site and RNA secondary structure was also examined using the program MFOLD26 (Figure 2 and data not shown), with the percentage overlap of PMO-target site with sequence predicted to be in open conformation by MFOLD analysis given in Supplementary Table S1. ESEfinder and SSF (http://www.umd.be/SSF/) software analysis of exon sequences revealed the positions of putative SR protein–binding motifs (SF2/ASF (by two algorithms), SC35, SRp40, SRp55, Tra2β, and 9G8). The highest score over threshold for each SR protein is given for each PMO in the columns on the right of Supplementary Table S1. Also shown is the degree of overlap of each PMO-target site with the ESE and exonic splicing suppressor or silencer regions predicted by Rescue ESE and PESX.
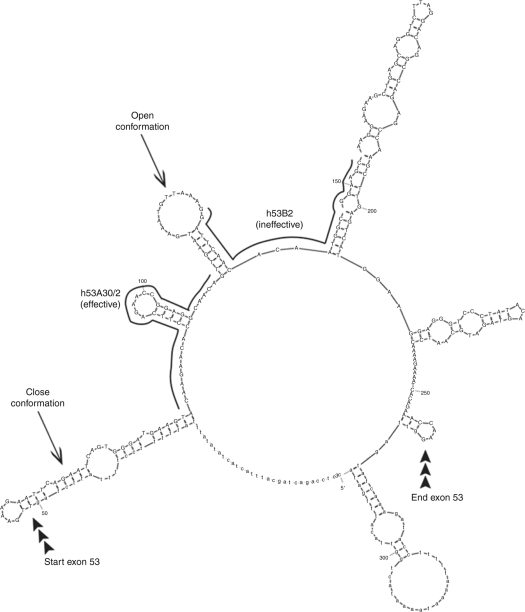
Figure 2.
Mfold secondary structure prediction for exon 53 of the human DMD gene. MFOLD analysis26 was performed using exon 53 plus 50 nt of the upstream and downstream introns, and with a maximum base-pairing distance of 100 nt. The intron and exon boundaries are indicated, as are the positions of the target sites of the bioactive PMO h53A30/2, and an inactive PMO (h53B2). Examples of open and closed RNA secondary structure are arrowed.
Statistical analysis of design parameters in relation to PMO bioactivity
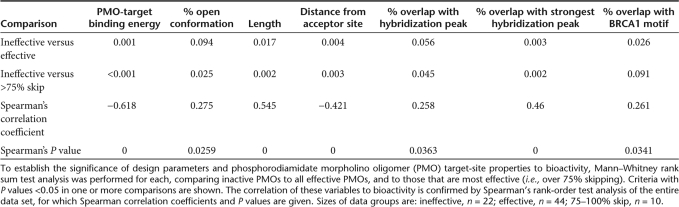
For this statistical analysis, bioactive PMOs are considered to be those that produce over 5% skipping, while those that produce <5% skipping are considered inactive. For each of the parameters listed in Supplementary Table S1, comparison was made between bioactive and inactive PMOs using the nonparametric Mann–Whitney rank sum test, or, when it was statistically valid to do so, the parametric Student's t-test (two-tailed). The P values obtained from such analyses are listed in Table 1 for the parameters that show varying levels of significance to bioactivity. Considering the data as a whole, the variable that showed the highest significance to PMO bioactivity was the binding energy of the PMO to the exon (P = 0.001); the most bioactive PMOs are predicted to bind better to their target sites. Those PMOs that overlap with peaks identified by the experimental hybridization-array analysis are not significantly more active than those that do not (P = 0.056), but when only the strongest peak for each exon is considered, this parameter becomes highly significant (P = 0.003). Distance of the PMO-target site to the splice acceptor site of the exon was also significant (P = 0.004), with PMOs whose target sites were closer to the acceptor site being more active. PMOs whose target sites showed coincidence with binding motifs for the SR protein SF2/ASF (as defined by the BRCA1 algorithm of Smith et al.21) produced significantly greater skipping (P = 0.026), but only when the data is considered as a whole. PMO length is also a significant parameter (P = 0.017), with longer PMOs being more effective at inducing skipping. Boxplots of the significant variables identified here are shown in Supplementary Figure S2. None of the other variables considered in this study were shown to have any significance to AO bioactivity.
Table 1.
The correlation of significant design parameters and PMO-target site properties to skipping efficacy
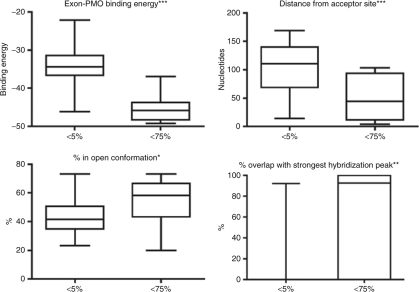
To ascertain which parameters/design tools are the most powerful, we also used the Mann–Whitney rank sum test to compare the most active PMOs (i.e., those that induce >75% skipping of the target exon) to those that were inactive (i.e., those that produce <5% skipping), and the P values obtained from such analyses are listed in Table 1. Boxplots of the significant variables for this comparison are shown in Figure 3. There is strong significance of overlap of the PMO-target site with the strongest hybridization peak for each exon (P = 0.002); more of the most bioactive PMOs had their target sites coincident with sequences accessible to binding than those that were inactive. This is reinforced by the observation that the target sites of PMOs that produced over 75% skipping overlapped significantly more RNA that was in open conformation, relative to inactive PMOs (P = 0.025). Stronger binding between the PMO and its target exon, PMO length, and proximity of the target to the acceptor site are also significant parameters when comparing the most and least effective reagents.
Figure 3.
Boxplots of parameters significant to strong PMO bioactivity. Comparisons were made between inactive PMOs and those inducing skipping at levels in excess of 75%. Boxplots are shown for parameters which are significant on a Mann–Whitney rank sum test: PMO-to-target binding energy, distance of the target site from the splice acceptor site, the percentage overlap with areas of open conformation, as predicted by MFOLD software, and the percentage overlap of the target site with the strongest area accessible to binding, as revealed by hexamer hybridization–array analysis,. Degrees of significance are indicated by asterisks. *P < 0.05. **P < 0.01. ***P < 0.001.
Spearman's rank-order correlation was used to establish potential relationships between design parameters and skipping bioactivity using the entire set of PMOs (see Table 1). This shows statistically significant correlations between skipping bioactivity and PMO-target-binding energy (rs = −0.618, P = 0), percentage open conformation (rs = 0.275, P = 0.0259), PMO length (rs = 0.545, P = 0), distance from the splice acceptor site (rs = –0.421, P = 0), percentage overlap with the strongest hybridization peak (rs = 0.46, P = 0), and overlap with an exonic splicing suppressor or silencer sequence, as predicted by PESX (rs = 0.261, P = 0.0348).
Linear discriminant analysis
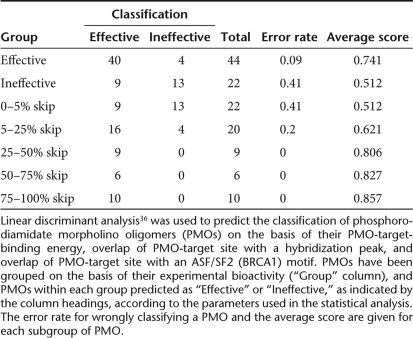
This analysis was performed on all possible combinations of length, overlap with the SF2/ASF (BRCA1) motif, percentage overlap with areas of open conformation, percentage overlap with hybridization peak and PMO-target-binding energy, that is, PMO parameters and design tools that showed significance or borderline significance. Using length, SF2/ASF (BRCA1) motif and hybridization peak data, nine of the inactive PMOs were classified as bioactive and four bioactive PMOs were classified as inactive (Table 2). These four misclassified PMOs were 25 mers to exon 46, three of which have borderline bioactivity, that is, they produce just 10% skipping, while the fourth produces about 20% skipping. Taken overall, this equates to 80% of the PMOs being predicted correctly when assessed according to their length, SF2/ASF (BRCA1) overlap, and hybridization peak overlap. This would suggest that these parameters have the potential to be effective design tools, with four out of every five PMOs designed to have these three properties likely to be bioactive. In line with this, there was a distinct trend for PMOs being correctly assigned as bioactive with increased skipping bioactivity (see Table 2). Indeed, the PMOs with greatest bioactivity were all 30 mers (10/10), bound to their target with a high binding energy of below −43.0 kDa (9/10), overlapped by over 50% with areas of open conformation (7/10), overlapped with SF2/ASF (BRCA1) peak (8/10), and overlapped with a hybridization peak (7/10).
Table 2.
Linear discriminant analysis of bioactive and inactive PMOs
Discussion
Clinical studies using AOs to skip exon 51 to correct DMD deletions are progressing well (ref. 11 and F. Muntoni, Principal Investigator of MDEX Consortium, personal communication). However, because the mutations that cause DMD are so diverse, skipping of exon 51 would have the potential to treat just 24.6% of DMD patients on the Leiden DMD database.27 It is, therefore, imperative that preclinical optimization of AO target sequence and chemistry is continually studied and improved. This study has examined the significance of design parameters for PMO-induced skipping of exons 44, 45, 46, 51, and 53, which would have the potential to treat, respectively, 11.5, 15.8, 8.4, 24.6 and 13.5% of DMD patients in the Leiden database (ref. 27 and A. Aartsma-Rus, personal communication).
Specific skipping was observed for the five DMD exons studied here, with two thirds of the PMOs tested being bioactive. This proportion of bioactive AOs within a cohort has been reported previously,18,19 but we have induced high-level (i.e., >75%) skipping in four of the five exons tested, some of which are achievable at relatively low doses of oligomer. The exception is exon 51, published previously,4 achieving a maximal skipping of 26%. The work of Wilton et al.19 demonstrated that only exons 51 and 53 can be skipped with high efficiency (>30% by their definition) and that exons 44, 45, and 46 are less “skippable” (>30% skipping). Furthermore, Aartsma-Rus et al.18 showed oligomers capable of high-level skipping (greater than a mere 25%) for only exons 44, 46, and 51.
Ease of skipping of certain DMD exons has been seen elsewhere18 and may be related to other factors affecting splicing, including strength of splice donor and acceptor sites and branchpoint, and the size of upstream and downstream introns, which may affect the order in which exons are spliced together. There is the potential of using a cocktail of AOs to induce greater skipping of the more difficult to skip exons.28,29
We provide here direct evidence that AO bioactivity shows a significant association with accessibility of its target site to binding. This is the first study to assess sequences practically within the pre-mRNA that are accessible to binding and then use them as an aid to AO design. The data we show underline the value of the hybridization analysis in determining what are likely to be the most bioactive oligomers (i.e., those that produce >75% skipping). In general, the 2′OMePS AOs displaying the highest bioactivity in the work of Aartsma-Rus et al.18 and Wilton et al.19 show some degree of overlap with the hybridization peaks that we have defined here for exons 45, 46, and 53. Previous studies have relied on in silico analysis of RNA structure to facilitate AO design. Although the hybridization data presented here may be considered merely a confirmation of the theoretical analyses used previously in AO design, it certainly provides an additional experimental tool, which others may wish to exploit in subsequent studies.
Accessibility of the AO to its target site depends directly on the secondary structure of the pre-mRNA, which has a major role in determining AO bioactivity in cells. A study in which the structure around an AO target site was changed revealed that AOs were unable to invade very stable stem-loop structures and their antisense activity was inhibited, but generally showed good activity when impeded by little local structure.30 Overlap of PMO-target sites with open conformations in the folded RNA showed a weak association with PMO bioactivity, which was more obvious when only the stronger PMOs were considered in the statistical analysis. It is also possible that there is selective pressure for SR-binding sites to be located preferentially on these open secondary structures. The presumption is that binding of bioactive PMOs to their target sites sterically block the binding of important factors involved in RNA processing, resulting in exon skipping.
One of the PMO parameters with high significance was length; 30 mer PMOs were far superior to their 25 mer counterparts. The influence of 2′OMePS AO length on bioactivity has been reported elsewhere31 and such an observation for PMO-induced skipping of exon 51 has been reported previously by us.4 The more persistent action of longer PMOs would have important cost and dose implications in the choice of AO for clinical trials. Longer AOs are likely to sterically block more of the regions that interact with the splicing machinery, but in general terms, the energy of binding of the longer PMO to its target would be increased, which we showed to be the most significant parameter in AO design. The strong significance of the binding energy of PMO-target complexes (i.e., free energy of AO target compared to free energy of the target) and PMO length to bioactivity suggests that PMO bioactivity depends on stability of the PMO-target complex, and implies that bioactive PMOs act by interference of target RNA folding. Computational analysis revealed that the thermodynamics of binding of active PMOs to their target site had a dramatic effect on the secondary folded structure of the RNA (data not shown). It is likely that these changes in secondary structure would have a profound effect on the binding of SR proteins to the RNA, thereby disrupting splicing, and exon skipping would ensue.
Overlap of a PMO-target site with a binding site motif for the SR protein SF2/ASF (BRCA1), as predicted by ESEfinder, showed a significant association to PMO bioactivity. This partly confirms the work of Aartsma-Rus et al.,18 who observed marginally significantly higher ESEfinder values for SF2/ASF and SC35 motifs for effective AOs when compared to inactive AOs. SC35 and SF2/ASF motifs are the two most abundant proteins assessed by ESEfinder. The reason why we do not see any significance of overlap with SC35 motif to PMO bioactivity may be due to the difference in AO chemistry used, and the number of AOs assessed. However, Aartsma-Rus et al.18 did note that not every bioactive AO has a high value for any of the SR protein–binding motifs, and some inactive AOs have high values. The apparent weakness and unreliability of SR protein–binding motifs as design tools for AOs may be a reflection of the lack of precision of the predictive software used. Overlap of PMO-target site with exonic splicing silencers appears to show a correlation with bioactivity in Spearman's rank-order test analysis. Such a correlation would be counterintuitive and the true significance questionable. Again the strength of the predictive software used may be in doubt. It should be noted that the software programs used predict SR-binding motifs on the linear exon sequence. The availability of these predicted motifs to bind SR proteins, or for binding PMOs to disrupt the binding of these proteins, is directly related to the folding of the pre-mRNA. The discrepancy in the relative significance of secondary RNA structure and SR protein–binding motifs may be due to active PMOs disrupting SR protein–binding, not sterically but indirectly, by altering the secondary pre-mRNA structure. A very recent study has shown the importance of co-transcriptional pre-mRNA folding in determining the accessibility of AO target sites and their effective bioactivity, and showed a direct correlation between previously reported AO bioactivity and potential interaction with pre-mRNA.32
It has been previously reported that ESE sites located within 70 nucleotides of a splice site are more active than ESE sites beyond this distance.33 Our results partially support this; PMOs with their target site closer to the splice acceptor site are significantly more bioactive. However, distance of the PMO-target site to the splice donor site showed no statistical significance to bioactivity. This bias has been previously reported for the analyses of 2′OMePS AOs,18,19 and may be related to the demonstration by Patzel et al.34 of the importance of an unstructured 5′ end of RNA in the initiation of hybridization of oligonucleotide binding. This would suggest that targeting any significant parameters located in the 5′ part of an exon may increase the probability of designing a bioactive AO.
In conclusion, our findings show that no single design tool is likely to be sufficient in isolation to allow the design of a bioactive AO, and empirical analysis is still required. However, this study has highlighted the potential of using a combination of significant PMO parameters/design tools as a powerful aid in the design of bioactive PMOs. Linear discriminant analysis revealed that using the parameters of PMO length, overlap with SF2/ASF (BRCA1) motif, and hexamer-array hybridization data in combination would have an 80% chance of designing a bioactive PMO, which is an exciting finding and should be exploited in further studies.
Materials and Methods
Hybridization analyses. Templates for the production of synthetic pre-mRNAs for exons 44, 45, 46, 51, and 53 of the human DMD gene were generated by PCR amplification from genomic clones of the exons, together with ~500 nt of upstream and downstream introns. PCR primers incorporated T7 RNA polymerase promoter sequences such that pre-mRNAs could be produced by in vitro transcription. Pre-mRNAs were then subjected to a hybridization screen against a spotted array of all 4,096 possible hexanucleotide sequences (Access Array 4000; Nyrion, Edinburgh, UK). Binding of the pre-mRNA to specific spots on the array was detected by reverse transcriptase-mediated incorporation of biotinylated nucleotides by primer extension, followed by fluorescent labeling. Scanning of the arrays followed by software analysis enabled sequences within the exons that were accessible to binding to the hexamer array to be identified. Using a hybridization assay, binding accessibility of each exons were analyzed, and hybridization peak was identified by AccessMapper software (Nyrion) (see Figure 1d).
AO design. Overlapping AOs were designed to exons 44, 45, 46, 51, and 53 of the human DMD gene using the following information: putative SR protein–binding domains as predicted by ESEfinder,20,21 Rescue ESE,24 and PESX22,23 analyses of exon sequence, sequences accessible to binding as determined by hybridization analyses (Nyrion), previously published work.18,19
All AOs were synthesized as PMOs by Gene Tools (Philomath, OR). To facilitate transfection of these uncharged oligonucleotides into cultured cells, the PMOs were hybridized to phosphorothioate-capped oligodeoxynucleotide leashes, as described by Gebski et al.,12 and stored at 4 °C. Sequences of leashes are available on request.
Cell culture and AO transfection. Normal human primary skeletal muscle cells (TCS Cellworks, Buckingham, UK) were seeded in 6-well plates coated with 0.1 mg/ml ECM Gel (Sigma-Aldrich, Poole, UK), and grown in supplemented muscle cell growth medium (Promocell, Heidelberg, Germany). Cultures were switched to supplemented muscle cell differentiation medium (Promocell) when myoblasts fused to form visible myotubes (elongated cells containing multiple nuclei and myofibrils). Transfection of PMOs was then performed using the transfection reagent Lipofectin (Invitrogen, Paisley, UK) at a ratio of 4 µl of Lipofectin per microgram of PMO (with a range of PMO concentrations tested from 50 to 500 nmol/l, equivalent to ~0.5–5 µg) for 4 hours, according to the manufacturer's instructions. All transfections were performed in triplicate in at least two different experiments.
RNA isolation and reverse transcriptase–PCR analysis. Typically 24 hours after transfection, RNA was extracted from the cells using the QIAshredder/RNeasy system (Qiagen, Crawley, UK) and ~200 ng RNA subjected to reverse transcription–PCR with DMD exon-specific primers using the GeneScript kit (Genesys, Camberley, UK). From this 20-cycle reaction, an aliquot was used as a template for a second nested PCR consisting of 25 cycles. PCR products were analyzed on 1.5% agarose gels in Tris-borate/EDTA buffer. Skipping efficiencies were determined by quantification of the PCR products by densitometry using GeneTools software (Syngene, Cambridge, UK). Primer sequences and detailed PCR protocols used are available on request.
Statistical analysis. The nonparametric Mann–Whitney rank sum test was used to identify whether parameters for effective PMOs were significantly different to those for ineffective PMOs. Where data was calculated to fit a normal distribution, the more powerful two-tailed Student's t-test was performed instead. According to accepted convention,35 outcomes of the tests were considered to be significant if P < 0.05. Correlations were generated using the Spearman's rank-order test. To determine the strength of the combined significant parameters/design tools to design effective PMOs, linear discriminant analysis was used,36 with the Ida function from the MASS package, using “effective” or “ineffective” as the two prior probabilities. The Ida function produces posterior probabilities for the two classes (effective and ineffective) for each PMO by leave-one-out classification.
Supplementary MaterialFigure S1. Comparison of active and inactive PMOs.Figure S2. Boxplots of parameters significantly different between bioactive and inactive PMOs.Table S1. Table summarizing the characteristics of PMOs used.
Supplementary Material
Comparison of active and inactive PMOs.
Boxplots of parameters significantly different between bioactive and inactive PMOs.
Table summarizing the characteristics of PMOs used.
Acknowledgments
We thank Dr Aartsma-Rus for her invaluable advice on the analysis of PMO-target sites, and for performing the linear discriminant analysis, and for critical reading of the manuscript, and we thank Dr Credland for advice on the statistical analyses used. The work is funded by the Department of Health (UK), Muscular Dystrophy Campaign (UK), and Muscular Dystrophy (Ireland). We are indebted to all members of the MDEX Consortium (Imperial College, London; Newcastle University; University of Oxford; Royal Holloway, University of London; University College, London).
REFERENCES
- Hoffmann EP, Brown RH., and , Kunkel LM. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Den Dunnen JT, Grootsscholten PM, Bakker E, Blonden LA, Ginjaar HB, Wapenaar MC, et al. Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet. 1989;45:835–847. [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Bremmer-Bout M, Janson AA, Ginjaar IB, Baas F, den Dunnen JT, et al. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum Mol Genet. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- Arechavala-Gomeza V, Graham IR, Popplewell LJ, Adams AM, Aartsma-Rus A, Kinali M, et al. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during pre-mRNA splicing in human muscle. Hum Gene Ther. 2007;18:798–810. doi: 10.1089/hum.2006.061. [DOI] [PubMed] [Google Scholar]
- Mann CJ, Honeyman K, Cheng AJ, Ly T, Lloyd F, Fletcher S, et al. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc Natl Acad Sci USA. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QL, Mann CJ, Lou F, Bou-Gharios G, Morris GE, Xue SA, et al. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med. 2003;9:1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- Graham IR, Hill VJ, Manoharan M, Inamati GB., and , Dickson G. Towards a therapeutic inhibition of dystrophin exon 23 splicing in mdx mouse muscle induced by antisense oligonucleotides (splicomers): target sequence optimisation using oligonucleotide arrays. J Gene Med. 2004;6:1149–1158. doi: 10.1002/jgm.603. [DOI] [PubMed] [Google Scholar]
- Bremmer-Bout M, Aartsma-Rus A, de Meijer EJ, Kaman WE, Janson AA, Vossen RH, et al. Targeted exon skipping in transgenic hDMD mice: A model for direct preclinical screening of human-specific antisense oligonucleotides. Mol Ther. 2004;10:232–240. doi: 10.1016/j.ymthe.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Jearawiriyapaisarn N, Moulton HM, Buckley B, Roberts J, Sazani P, Fucharoen S, et al. Sustained dystrophin expression induced by peptide-conjugated morpholino oligomers in the muscles of mdx mice. Mol Ther. 2008;16:1624–1629. doi: 10.1038/mt.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni C. Clinical approaches in the treatment of Duchenne muscular dystrophy (DMD) using oligonucleotides. Front Biosci. 2008;13:517–527. doi: 10.2741/2697. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Franzhuzen WS, Aartsma-Rus A, Bremmer-Bout M, et al. Local antisense dystrophin restoration with antisense oligonucleotide PRO051. N Eng J Med. 2007;357:2677–2687. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- Gebski BL, Mann CJ, Fletcher S., and , Wilton SD. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet. 2003;12:1801–1811. doi: 10.1093/hmg/ddg196. [DOI] [PubMed] [Google Scholar]
- Alter J, Lou F, Rabinowitz A, Yin H, Rosenfeld J, Wilton SD, et al. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- Fletcher S, Honeyman K, Fall AM, Harding PL, Johnsen RD., and , Wilton SD. Dystrophin expression in the mdx mouse after localized and systemic administration of a morpholino antisense oligonucleotide. J Gene Med. 2006;8:207–216. doi: 10.1002/jgm.838. [DOI] [PubMed] [Google Scholar]
- McClorey G, Fall AM, Moulton HM, Iversen PL, Rasko JE, Ryan M, et al. Induced dystrophin exon skipping in human muscle explants. Neuromus Disord. 2006;16:583–590. doi: 10.1016/j.nmd.2006.05.017. [DOI] [PubMed] [Google Scholar]
- McClorey G, Moulton HM, Iversen PL, Fletcher S., and , Wilton SD. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13:1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- Arora V, Devi GR., and , Iversen PL. Neutrally charged phosphorodiamidate morpholino antisense oligomers: uptake, efficacy and pharmacokinetics. Curr Pharm Biotechnol. 2004;5:431–439. doi: 10.2174/1389201043376706. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, De Winter CL, Janson AAM, Kaman WE, van Ommen G-JB, Den Dunnen JT, et al. Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides. 2005;15:284–297. doi: 10.1089/oli.2005.15.284. [DOI] [PubMed] [Google Scholar]
- Wilton SD, Fall AM, Harding PL, McClorey G, Coleman C., and , Fletcher S. Antisense oligonucleotide-induced exon skipping across the human dystrophin gene transcript. Mol Ther. 2007;15:1288–1296. doi: 10.1038/sj.mt.6300095. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ., and , Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MO., and , Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Zhang XH., and , Chasin LH. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Leslie CS., and , Chasin LA. Computational searches for splicing signals. Methods. 2005;37:292–305. doi: 10.1016/j.ymeth.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA., and , Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kamen WE, Bremmer-Bout M, Den Dunnen JT, Baas F, et al. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M., and , Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Bremmer-Bout M, Janson AAM, den Dunnen JT, van Ommen GJ., and , van Deutekom JCT. Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromus Disord. 2002;12:871–877. doi: 10.1016/s0960-8966(02)00086-x. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Kaman WE, Weij R, den Dunnen JT, van Ommen GJ., and , van Deutekom JC. Exploring the frontiers of therapeutic exon skipping for Duchenne muscular dystrophy by double targeting within one or multiple exons. Mol Ther. 2006;14:401–407. doi: 10.1016/j.ymthe.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Adams AM, Harding PL, Iversen PL, Coleman C, Fletcher S., and , Wilton SD. Antisense oligonucleotide induced exon skipping and the dystrophin gene transcript: cocktails and chemistries. BMC Mol Biol. 2007;8:57. doi: 10.1186/1471-2199-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers TA, Wyatt JR., and , Freier SM. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 2000;28:1340–1347. doi: 10.1093/nar/28.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding PL, Fall AM, Honeyman K, Fletcher S., and , Wilton SD. The influence of antisense oligonucleotide length on dystrophin exon skipping. Mol Ther. 2007;15:157–166. doi: 10.1038/sj.mt.6300006. [DOI] [PubMed] [Google Scholar]
- Wee KB, Pramono ZAD, Wang JL, MacDorman KF, Lai PS., and , Yee WC. Dynamics of co-translational pre-mRNA folding influences the induction of dystrophin exon skipping by antisense oligonucleotides. Plos one. 2008;3:e1844. doi: 10.1371/journal.pone.0001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, et al. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzel V, Steidl R, Kronenwell R, Haas R., and , Sczakiel G. A theoretical approach to select effective antisense oligodeoxyribonucleotides at high statistical probability. Nucleic Acids Res. 1999;27:4328–4334. doi: 10.1093/nar/27.22.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar JH. 4th edn. Prentice Hall: New Jersey. Biostatistical Analysis. 1999. pp. 80–83.
- Ihaka R., and , Gentleman RC. R: A language for data analysis and graphics. J Comput Graphic Stat. 1996;15:999–1013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of active and inactive PMOs.
Boxplots of parameters significantly different between bioactive and inactive PMOs.
Table summarizing the characteristics of PMOs used.