Abstract
Antisense oligonucleotides (AONs) can interfere with mRNA processing through RNase H–mediated degradation, translational arrest, or modulation of splicing. The antisense approach relies on AONs to efficiently bind to target sequences and depends on AON length, sequence content, secondary structure, thermodynamic properties, and target accessibility. We here performed a retrospective analysis of a series of 156 AONs (104 effective, 52 ineffective) previously designed and evaluated for splice modulation of the dystrophin transcript. This showed that the guanine-cytosine content and the binding energies of AON-target and AON–AON complexes were significantly higher for effective AONs. Effective AONs were also located significantly closer to the acceptor splice site (SS). All analyzed AONs are exon-internal and may act through steric hindrance of Ser-Arg-rich (SR) proteins to exonic splicing enhancer (ESE) sites. Indeed, effective AONs were significantly enriched for ESEs predicted by ESE software programs, except for predicted binding sites of SR protein Tra2β, which were significantly enriched in ineffective AONs. These findings compile guidelines for development of AONs and provide more insight into the mechanism of antisense-mediated exon skipping. On the basis of only four parameters, we could correctly classify 79% of all AONs as effective or ineffective, suggesting these parameters can be used to more optimally design splice-modulating AONs.
Introduction
Antisense oligonucleotides (AONs) are useful tools to modulate gene expression in a sequence-specific manner (reviewed in ref. 1). Generally, AONs are used to induce gene knockdown through RNase H cleavage of DNA:RNA hybrids of an mRNA. In addition, mRNA translation can be arrested by steric hindrance of the ribosomal complex by the AON. Finally, AONs can interfere with the splicing process to induce nonfunctional mRNAs that are subjected to the nonsense-mediated RNA decay pathway. Using the latter approach, it is also feasible to modulate alternative splicing, or to block aberrant, disease-causing splice sites (SSs).2 These mechanisms can be used for studies on developmental processes by allowing knockdown of genes at specific time points,3 or for therapeutic purposes. In fact, an RNase H–inducing AON is registered under the name Vitravene to treat cytomegaloviral-induced retinitis, and many AONs aiming at targeted gene downregulation are in late stage clinical trials mainly as putative anticancer drugs.1 Splice-modulating AONs are in early phase clinical trials for Duchenne muscular dystrophy (DMD).4 Here, the modulation of splicing (in this case the skipping of an exon) aims to restore the disrupted dystrophin-reading frame, allowing the generation of partly functional proteins and slowing down the severe, progressive muscle wasting phenotype.
Each antisense mechanism requires stable and efficient binding of the AON to its target sequence. One obvious determinant of AON efficacy is the accessibility of the target (Supplementary Figure S1). Several software programs are available to predict the secondary structure of RNA, of which the m-fold server is the most widely used.5 This server also provides a so-called SS-count for the target sequence, indicating the propensity of a nucleotide to be single stranded in a number of potential secondary structure predictions. This approach probably reflects the actual in vivo situation more closely than focusing only on the most energetically stable structure. In addition, the stability and binding energy of the AON to the target sequence influence AON efficiency. This depends on e.g., AON length and sequence constitution and the free energy of local structures.1 To efficiently bind a target sequence, the free energy of the AON-target complex must be higher than that of the target complex and that of the AON. As AONs are generally only 17–25-nucleotides long, they are unlikely to form stable secondary structures. However, most AONs can form AON–AON complexes with other AONs of the same sequence (Supplementary Figure S2). The software program RNAstructure 4.5 has a tool that provides the free energy of AON–AON complexes and AON-target complexes, in addition to the free energy of individual AONs and the target sequence.6 The aforementioned software programs (as well as others) can be used to facilitate AON design (reviewed in ref. 1). Nonetheless, none of them is 100% conclusive or predictive and in general a trial and error procedure is still involved to identify potent AONs.
In addition to more general AON requirements, the different applications may introduce further requirements as well. For instance, translation-blocking AONs often target the ATG initiation site, whereas splice-modulating AONs target sites involved in exon recognition and inclusion. The latter consist of the donor and acceptor SSs and branch point sequence, but also include exon-internal sequences [exonic splicing enhancers (ESEs) sites] that facilitate splicing by binding of so-called Ser-Arg-rich (SR) proteins, which in their turn recruit U1 snRNP and U2AF to the donor and acceptor SSs, respectively.7 Both AONs targeting (aberrant) SSs and ESEs have been shown to be efficient modulators of splicing.8,9 An advantage of exon-internal over SS AONs may be that they do not target consensus sequences as ESEs are weakly defined motifs, and that there is a wider range of target choice. Numerous software programs predict ESEs, such as ESEfinder that predicts binding sites for the four most abundant SR proteins (SF2/ASF, SC35, SRp40, and SRp55),10,11 RESCUE-ESE,12 and the PESX server13 that provide hexamers and octamers, respectively, significantly enriched in exons over introns. These programs are implemented in the human splicing finder (http://www.umd.be/HSF/), which also provides algorithms to predict binding sites for two other SR proteins (9G8 and Tra2β). In addition, the HSF program includes hexamers, octamers, and decamers associated with exonic splicing silencers (ESSs).13,14,15 These splicing silencers are the counterparts of ESEs and prevent exon inclusion. As exon-internal AONs are hypothesized to work through steric hindrance of SR proteins, it is expected that AONs targeting predicted ESEs are more likely to be effective. We have previously analyzed a series of 114 exon-internal AONs targeting dystrophin exons using ESEfinder and RESCUE-ESE in retrospect, and indeed found that effective AONs were significantly enriched in SR-binding sites and RESCUE-ESE hexamers.8,16 We have now extended our series of exon-internal dystrophin AONs to 156, of which 104 (67%) are effective and 52 (33%) are ineffective. In this study, we have analyzed our larger set for previously analyzed and new parameters to improve AON design guidelines and gain insight into the mechanism of AON-mediated exon skipping.
Results
AON constitution and thermodynamic properties
We here compared a series of 156 exon-internal AONs targeting exons of the DMD gene transcript. AONs were assigned to either the effective (n = 104) or ineffective (n = 52) group, depending on their ability to reproducibly induce targeted exon skipping at levels of at least 5% as assessed by RT-PCR analysis. Different parameters such as predicted Tm, length, AON constitution, and thermodynamic properties were determined for each AON (see Materials and Methods and Supplementary Table S1). The values of each parameter were compared between the effective and ineffective AONs (Supplementary Table S2), and a Wilcoxon signed–rank sum test was used to detect significant differences. Boxplots of the parameters that differed significantly between the two groups are shown in Figure 1.
Figure 1.
Boxplots of thermodynamic antisense oligonucleotide (AON) parameters that differed significantly between the effective and ineffective group. *P < 0.05, **P < 0.005. GC, guanine-cytosine.
First, we compared AON length in both groups. In our series AON length varies from 15 to 25 nucleotides and did not significantly differ between the two groups. Second, we evaluated AON constitution, as C and G content has been shown to correlate with AON-mRNA duplex stability.17 Indeed, the total number of Gs and Cs was significantly higher in effective AONs (P < 0.05). In contrast, no significant difference was observed for the percentage and total number of A, C, G and U, GA, and GT nucleotides. Focusing on the predicted basic melting temperature (Tm) and Tms using the nearest neighbor model, Tms were significantly higher for the effective group using both calculation methods.
As a third parameter, we used the m-fold program to predict the secondary structure of the local target sequence and the AONs, and the RNAstructure software version 4.5 to predict thermodynamic properties of AONs and AON-target binding. It is expected that the ability of AONs to form stable secondary structures with themselves or other AONs of the same sequence (see Supplementary Figure S2) will negatively influence their efficacy. As AONs are relatively short, predicted secondary structures are often not very stable (δG ~ 0 kcal/mol). No significant difference was observed for the free energy of individual AONs. Unexpectedly, for AON–AON complexes we observed that complexes formed by effective AONs had significantly lower free energy values (and where thus more stable) than those of ineffective AONs (P = 0.03). We also used the RNAstructure software to predict the free energy of the local target structure and the binding energy of AON-target complexes. We anticipated that it would be more difficult to find efficient AONs for exons with a stable secondary structure, as these are harder to disrupt. The free energies of the target exons seem slightly higher (less stable) for effective AONs, but this difference was not statistically significant (P = 0.11). No difference was observed between the free energy of the AON-target complexes. In contrast the binding energy of AON-target complexes (free energy of AON-target compared to free energy of target) was significantly higher for effective AONs (P = 0.003). This indicates that for splice-modulating AONs the efficacy of an AON depends not on the stability of the AON-target complex, but rather on the amount of energy that is released upon AON binding.
Finally, using SS-count (m-fold server) we determined the local accessibility of the target pre-mRNA. We defined the local accessibility as the average propensity of target nucleotides to be single stranded in the predicted secondary structures. We anticipated that effective AONs would be enriched in single-stranded target sequences, as we hypothesized SR protein binding to be more likely to predicted open secondary structures. Nevertheless, we observed that the accessibility of ineffective AONs was significantly higher (P = 0.021), although both groups targeted sequences that were at least partly accessible.
Splicing enhancer and silencer motifs in AON-target sequences
Exon-internal splice-modulating AONs are hypothesized to act through blocking of SR protein binding to ESE motifs. Thus, effective AONs are expected to be enriched in ESEs, while the targeting of splicing silencer motifs is expected to be counterproductive. In addition, we compared the location of the AON-target sequence to the SSs, as ESE motifs have been reported to be more active when located within 70 nucleotides from either splice side.18 An overview of parameters that differed significantly between effective and ineffective AONs is given in Figure 2, while all results are summarized in Supplementary Table S2B. Similar to our previous analysis of a smaller set of AONs,8 we observed that effective AONs are located significantly closer to the acceptor but not to the donor SS. In our current, larger set, the difference became more significant (P = 0.006 versus 0.008). For the donor site there was a trend toward effective AONs being located further away to the donor site (and thus closer to the acceptor SS, P = 0.09).
Figure 2.
Boxplots of exonic splicing enhancer (ESE) parameters of antisense oligonucleotide (AON)-target sequences that differed significantly between the effective and ineffective group. *P < 0.05. SS, splice site; PESE, putative ESE.
A variety of software programs predicting ESE and ESS motifs is available and most of these are implemented in the human splicing finder software (http://www.umd.be/HSF/). We analyzed the target sequence of each AON for the presence of ESE and ESS motifs and compared the effective and ineffective group as described above. No significant difference was observed when we analyzed the presence of any predicted ESE in the two groups. However, for the vast majority of AONs at least one ESE motif was predicted, which might explain the lack of significance. We then proceeded to compare individual ESE predicting programs starting with ESEfinder and RESCUE-ESE, which previously had shown significantly higher values for SC35 and the highest predicted value for any SR protein (ESEfinder)8 and significantly more hexamers (RESCUE-ESE)16 in our smaller series of AON. We now confirm these findings and for SC35 and the number of RESCUE-ESE sites the difference became more significant (SC35: 0.01 versus 0.05, RESCUE-ESE: 0.03 versus 0.04), while it became slightly less significant for the highest SR value (0.045 versus 0.037). Notably, we did not observe the previously identified trend for SF2/ASF values to be higher in the effective group, which bordered on statistical significance (P = 0.053).8 This may have been caused by the fact that in the previous analysis predicted values without a threshold were compared, while here a threshold was applied. Therefore, we also determined the values relative to the threshold (either over or below) for each of the SR proteins. This still did not result in a significant increase for SF2/ASF values in the effective group, and the difference became less significant for the predicted SC35 values (0.02 versus 0.01). However, it did lead to the finding that SRp55 values relative to the threshold are significantly higher for the effective AONs. The suggestion that AONs indeed act through blocking ESEs was further underlined by the finding that the number of ESEfinder sites over the threshold was significantly higher for effective AONs (P = 0.03), effective AONs were significantly enriched in the number of PESE octamers (P = 0.02) and targeted significantly higher PESE values (P = 0.02). The difference between the number and values for the 9G8 protein was not significant (0.054 and 0.09, respectively). Unexpectedly, for Tra2β, we observed that the ineffective AONs targeted significantly higher and more predicted motifs (P = 0.01 and 0.04, respectively).
Finally, we compared predicted ESS motifs between the two groups. We did not observe a significant difference in the presence of any ESS motif, the number of Sironi and Wang ESS motifs, and the number and values of PESS and hnRNP A1 (the ESS-binding counterpart of SR proteins) motifs between the groups. There was an unexpected trend for effective AONs to target more Sironi motifs (P = 0.09) and an expected trend for ineffective AONs to target more Wang motifs (P = 0.08).
Classification of effective and ineffective AONs
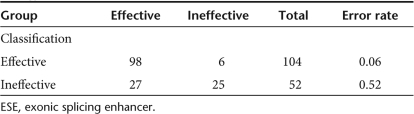
Dimension reduction using principal component analysis was used to extract those parameters that are largely independent and would yield optimal classification of effective and ineffective AONs. The first three principal components appeared most important for explaining the variance (data not shown). The contributions of each of the 54 evaluated variables to these three principal components are reported in Supplementary Table S3. The parameters that contribute most to these principal components were generally those for which significant differences had been observed between effective and ineffective AONs (Supplementary Table S2). Principal component 1 was dominated by the total amount of Gs and Cs, which was highly correlated with Tm and the number of ESE sites. Principal component 2 was dominated by the total number of Cs and the number of RESCUE-ESE hexamers, while percentage GT and Tra2β value contributed most to the third principal component. To assess the predictive value of these parameters, we selected one of the sequence composition parameters (total amount of Gs and Cs), the number of RESCUE-ESE hexamers, and the Tra2β value for linear discriminant analysis and classified the AONs in the effective and ineffective groups by leave-one-out analysis. The linear discriminant analysis showed a very good performance for the class of effective AONs (97 of 104 were correctly classified, Table 1). However, only 17 of 52 ineffective AONs were correctly classified (Table 1). Better classification was obtained with the following parameters: number of RESCUE-ESE hexamers, the Tra2β value, AON-target exon binding energy, and SC35 values over the threshold. Now 98 effective and 25 ineffective AONs are correctly classified [classification rate: 79% (Table 2)]. Classification based on other combinations of parameters that were found significantly different between the two classes gave lower classification rates, demonstrating that principal component analysis in combination with manual optimization resulted in the selection of an optimal set of parameters for AON design.
Table 1.
Classification results for effective and ineffective antisense oligonucleotides based on the parameters: total guanine-cytosine content, number of RESCUE-ESE hexamers, and the Tra2β value
Table 2.
Classification results for effective and ineffective antisense oligonucleotides (AONs) based on the parameters: number of RESCUE-ESE hexamers, the Tra2β value, AON-target exon binding energy, and SC35 values over the threshold
Discussion
We compared our set of effective and ineffective AONs for a variety of thermodynamic parameters as well as the presence of splice enhancer or silencer sequence motifs in the target sequence. For the thermodynamic parameters, effective AONs generally had a higher affinity to the target sequence than ineffective AONs. As Tms, the amount of Gs and Cs, and binding energy correlate, this is not surprising. We note that Tms were calculated using algorithms for unmodified RNA, as accurate software to predict 2′-O-methyl phosphorothioate AONs is not available. However, we expect that the increase in Tm caused by the 2′-O-methyl modification is negated by the phosphorothioate backbone, and that the actual Tms of our AONs may in fact be close to those of normal RNA. Along the same line, increasing the length of AONs should also increase the affinity to the target sequence. However, we did not find a difference in length between effective and ineffective AONs. This probably reflects the fact that a length of ~20 nucleotides is optimal for this chemistry, as shorter AONs may form AON-target complexes with suboptimal stability, while longer AONs are more likely to form stable secondary structures that interfere with target binding. Others and we have found that for some AONs increasing the length can improve their efficiency and the data from Wee et al. support this19,20 (Heemskerk et al., manuscript submitted). However, this rule does not apply to all AONs and for some increasing the length even decreases efficiency19 (Heemskerk et al., manuscript submitted).
An interesting finding was that the free energy of AON–AON complexes was higher for effective AONs. This is in contrast to what has been previously published for RNase H–dependent AONs.21 A possible explanation for this discrepancy could be the inherent differences of the two approaches. RNase H–mediated knockdown generally takes place in the cytoplasm and the AONs target mature mRNAs, while splice modulation occurs in the nucleus and targets pre-mRNA. Thus the formation of stable AON–AON complexes may favor splice-modulating AONs, because it could increase the chance that the AON reaches the nucleus intact and complex formation may even enhance uptake by the nucleus. Alternatively, the formation of AON–AON complexes might prevent the AON from binding aspecifically and thus increase the amount of AON left to bind specifically to its target. To underline this finding, the energy of AON–AON binding (free energy of AON–AON complexes compared to the free energy of structures formed by individual AONs) was even more significantly increased for the effective AONs (P = 0.0029).
When comparing accessibility, we observed that ineffective AONs targeted significantly more accessible targets, though both effective and ineffective AONs always targeted at least partially accessible regions. This indicates that a small accessible region is sufficient for AON binding. In addition, it is likely that targeting a highly accessible target will disrupt the secondary structure to a lesser extent than targeting a partially closed structure (e.g., a stem loop). This suggests that the mechanism of action perhaps does not depend solely on steric hindrance of SR protein binding to an ESE, but also on the manner in which AON binding disrupts the secondary structure. As SR proteins recognize only weakly defined sequence motifs, it is conceivable that they rely on secondary structure as well as on sequence, and that blocking these motifs or disrupting the secondary structure results in the inability of a certain SR protein to bind its target. Recently, a very thorough analysis of local accessibility during transcription was performed for a subset of these AONs.20 Here, no difference was observed between the accessibility of effective and ineffective AONs. The authors did find that the presence of multiple inaccessible nucleotides in the 3′ or 5′ region of the target attenuated efficacy more than inaccessible nucleotides elsewhere in the target. This effect was most significant for inaccessible nucleotides in the 3′ target region. When we compared the local accessibility of the 3′ and 5′ ends of the target sequences, we did not observe significant differences. The discrepancies with the report of Wee et al. are likely due to the different method of analysis (we considered several structures for a single target exon with 50–100 nucleotides flanking intron sequence, whereas they considered all possible structures for 1,500 different partly overlapping targets).
As all our AONs target exon-internal sequences, we also compared the presence of predicted ESE and ESS sites in the target sequences, and the location of the AON within the exon. We observed that effective AONs located closer to the start of the exon (acceptor SSs) than ineffective AONs, while there was a nonsignificant trend for effective AONs to be further away from the end of the exon (donor SS). The higher P value for the distance to the donor SS suggests that the highly significant effect of the distance to the acceptor SS may mask a possible beneficial effect of a target being in close proximity to the donor SS. Another explanation could be that targeting an AON close to the 3′-SS (i.e., the beginning of the exon) works better, as it increases the chance that the AON binds the target before the 5′-SS is transcribed and it thus decreases the chance of proper exon definition.20
In general we found an enrichment of predicted ESE sites in target sites of effective AONs, as anticipated. The exception is Tra2β, where we found an enrichment of target sites in ineffective AONs. Tra2β is an SR-like protein and has been shown to be involved in ESE binding and enhancement of exon inclusion for different genes, including the DMD gene.22 It is possible that prevention of Tra2β binding to ESEs is less likely to be successful, because Tra2β binding occurs rapidly after transcription or binding is very stable. Alternatively, Tra2β could also be involved in exon skipping with binding sites acting as both ESEs and ESSs. Regardless of the underlying mechanism, this finding does provide an additional design guideline as apparently AONs not targeting predicted Tra2β sites are more likely to be effective.
For ESS sites, no significant difference was observed between effective and ineffective AONs, though there was a trend for enrichment of Wang motifs in ineffective AONs and an unexpected trend of enrichment of Sironi motifs in effective AONs. Notably, the Wang motif occurred only once in our set of AONs, while by contrast, in the majority of AONs at least one Sironi motif was present. Some of these motifs overlap with ESE motifs as predicted by other programs, which most likely explains the observed discrepancy.
After principal component analysis, we could select four parameters to correctly classify 94% of effective AONs and 48% of ineffective AONs (Table 2). The observation that in particular the ineffective AONs are difficult to classify is likely due to the underrepresentation of ineffective AONs in our training set. We investigated whether ineffective AONs that were classified as effective AONs were more likely to demonstrate some extent of exon skipping (skip levels lower than 5%). However, the misclassified and correctly classified groups of ineffective AONs did not show significant differences in skip levels and the proportion of AONs with no exon skip activity was equal between the two classes. Future, prospective analysis will have to demonstrate how efficient the selected combination of parameters is for the prediction of exon skip activity. However, based on the current study we would advise to design AONs with a high number of RESCUE-ESE and SC35 sites, a low number of Tra2β sites, and a high AON-target binding energy. Using previously defined design rules, our success rate was ~65–70%. We expect from the classification results with these parameters that applying these new design rules will improve the success rate considerably.
In conclusion, our results are a further indication that exon-internal AONs work through prevention of SR binding to ESEs and that ESE predicting software packages are a valuable tool for AON design. In addition, they suggest that the mechanism of action and binding may differ for splice-modulating AONs and RNase H–dependent AONs as previously identified rules for the latter did not always apply to splice-modulating AONs, especially regarding the thermodynamic parameters. Finally, the selected parameters for optimal classification of effective and ineffective AONs provide guidelines for design of splice modulation AONs.
Materials And Methods
AON design. The majority of AONs have been previously published (see Supplementary Table S1).8 For each AON, design was primarily based targeting an open region in the secondary structure of the target exon as predicted by m-fold. In addition, for a subset of AONs (see Supplementary Table S1) design was in addition based on the presence of SF2/ASF and SC35 sites in the target sequence as predicted by ESEfinder.
AON analysis. AON efficacy was determined as described previously.8 An AON was considered effective when it was able to reproducibly induce skipping of the targeted exon at a concentration of 500 nmol/l and at levels of at least 5% as assessed by RT-PCR analysis. For each AON the following characteristics were compared: Tm (calculated with the oligonucleotide properties calculator; http://www.unc.edu/~cail/biotool/oligo/index.html) for single-stranded RNA using the basic Tm and the nearest neighbor model, AON length, number, and percentage of A, C, G, U, G and C, G and A and G and T nucleotides, the free energy of the predicted secondary structure of the AON (using the quickfold server from m-fold5), the free energy of the target exon [using RNAstructure version 4.5 (ref. 6)], the free energy of the AON-target exon complex (using RNAstructure version 4.5), the free energy of AON–AON complexes (using RNAstructure version 4.5), and the relative accessibility of the total target sequence as well as the accessibility of the most 3′ and the most 5′ target nucleotide (using the exon and 50–100 nucleotides of each flanking intron as input in the m-fold server). Free energy values were used to calculate the binding energy of AON-target complexes and AON–AON complexes (free energy AON-target complex minus free energy target; free energy AON–AON complex minus free energy AON and binding energy AON-target minus binding energy AON–AON).
For each target sequence several characteristics were determined: the number of nucleotides at the beginning and end of the AONs from the donor and acceptor SS. In addition, the human splicing finder software (http://www.umd.be/HSF/) was used to predict the presence of any ESE in the target sequence, the number of ESEfinder motifs over the threshold, predicted values for SF2/ASF, SC35, SRp40, and SRp55 binding sites with threshold and predicted values relative to the threshold, the highest ESEfinder value above the threshold, the number of RESCUE-ESE hexamers, the number of PESE octamers and the highest predicted PESE value, the number and highest predicted values of 9G8 and Tra2β binding sites, the presence of any ESS site, the number of ESS Sironi motifs, the number of ESS Wang motifs, the number and values of PESS sites, and finally the number and values of predicted hnRNP A–binding sites. Only predicted motifs completely covering the target sequence were included in the analysis.
The Wilcoxon signed–rank sum test (one and two tailed) was used to identify parameters that differed for effective (n = 104) and ineffective (n = 52) AONs as described previously.8
Principle component analysis was performed on all 54 variables using the princomp function from the stats package in R.23 Linear discriminant analysis was performed with the lda function from the MASS package, using “effective” or “ineffective” as the two prior probabilities. The lda function produces posterior probabilities for the two classes (effective and ineffective) for each AON by leave-one-out classification.
Supplementary Material Figure S1. Example of a secondary structure prediction for DMD target exon 50 by the m-fold server. H50AON1 and h50AON2 were primarily designed to target open (accessible) structures, rather than closed (less accessible) structures. Figure S2. Examples of secondary structures formed by one AON (A) and structures formed by two AONs of the same sequence (B). Table S1. Overview of AON parameters. Table S2. Comparison of AON parameters for the effective and ineffective group. Table S3. Results of the principal component analysis.
Supplementary Material
Example of a secondary structure prediction for DMD target exon 50 by the m-fold server. H50AON1 and h50AON2 were primarily designed to target open (accessible) structures, rather than closed (less accessible) structures.
Examples of secondary structures formed by one AON (A) and structures formed by two AONs of the same sequence (B).
Overview of AON parameters.
Comparison of AON parameters for the effective and ineffective group.
Results of the principal component analysis.
Acknowledgments
This work was supported by grants from the Dutch Duchenne parent project, the Center for Biomedical Genetics (the Netherlands), and ZonMw (the Netherlands). The Leiden University Medical Center is partner of the TREAT-NMD FP6 network of excellence.
REFERENCES
- Chan JH, Lim S., and , Wong WS. Antisense oligonucleotides: from design to therapeutic application. Clin Exp Pharmacol Physiol. 2006;33:533–540. doi: 10.1111/j.1440-1681.2006.04403.x. [DOI] [PubMed] [Google Scholar]
- Kole R, Williams T., and , Cohen L. RNA modulation, repair and remodeling by splice switching oligonucleotides. Acta Biochim Pol. 2004;51:373–378. [PubMed] [Google Scholar]
- Nasevicius A., and , Ekker SC. Effective targeted gene “knockdown” in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M., and , Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL., and , Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, De Winter CL, Janson AA, Kaman WE, van Ommen GJ, den Dunnen JT, et al. Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides. 2005;15:284–297. doi: 10.1089/oli.2005.15.284. [DOI] [PubMed] [Google Scholar]
- Wilton SD, Fall AM, Harding PL, McClorey G, Coleman C., and , Fletcher S. Antisense oligonucleotide-induced exon skipping across the human dystrophin gene transcript. Mol Ther. 2007;15:1288–1296. doi: 10.1038/sj.mt.6300095. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ., and , Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ., and , Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, et al. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH., and , Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M, Menozzi G, Riva L, Cagliani R, Comi GP, Bresolin N, et al. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 2004;32:1783–1791. doi: 10.1093/nar/gkh341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M., and , Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Heemskerk JA, De Winter CL, Van Ommen GJ., and , Van Deutekom JC. Therapeutic modulation of DMD splicing by blocking exonic splicing enhancer sites with antisense oligonucleotides. Ann NY Acad Sci. 2006;1082:74–76. doi: 10.1196/annals.1348.058. [DOI] [PubMed] [Google Scholar]
- Ho SP, Britton DH, Stone BA, Behrens DL, Leffet LM, Hobbs FW, et al. Potent antisense oligonucleotides to the human multidrug resistance-1 mRNA are rationally selected by mapping RNA-accessible sites with oligonucleotide libraries. Nucleic Acids Res. 1996;24:1901–1907. doi: 10.1093/nar/24.10.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Holste D, Burge CB., and , Sharp PA. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:E268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding PL, Fall AM, Honeyman K, Fletcher S., and , Wilton SD. The influence of antisense oligonucleotide length on dystrophin exon skipping. Mol Ther. 2007;15:157–166. doi: 10.1038/sj.mt.6300006. [DOI] [PubMed] [Google Scholar]
- Wee KB, Pramono ZA, Wang JL, MacDorman KF, Lai PS., and , Yee WC. Dynamics of co-transcriptional pre-mRNA folding influences the induction of dystrophin exon skipping by antisense oligonucleotides. PLoS ONE. 2008;3:e1844. doi: 10.1371/journal.pone.0001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva OV, Tsodikov AD, Giddings M, Freier SM, Wyatt JR, Spiridonov AN, et al. Identification of sequence motifs in oligonucleotides whose presence is correlated with antisense activity. Nucleic Acids Res. 2000;28:2862–2865. doi: 10.1093/nar/28.15.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disset A, Bourgeois CF, Benmalek N, Claustres M, Stevenin J., and , Tuffery- Giraud S. An exon skipping-associated nonsense mutation in the dystrophin gene uncovers a complex interplay between multiple antagonistic splicing elements. Hum Mol Genet. 2006;15:999–1013. doi: 10.1093/hmg/ddl015. [DOI] [PubMed] [Google Scholar]
- Ihaka R., and , Gentleman RC. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example of a secondary structure prediction for DMD target exon 50 by the m-fold server. H50AON1 and h50AON2 were primarily designed to target open (accessible) structures, rather than closed (less accessible) structures.
Examples of secondary structures formed by one AON (A) and structures formed by two AONs of the same sequence (B).
Overview of AON parameters.
Comparison of AON parameters for the effective and ineffective group.
Results of the principal component analysis.