Abstract
Aldehyde dehydrogenase class 3 (ALDH3) constitutes 20–40% of the total water-soluble proteins in the mammalian cornea. Here, we show by Northern blot analysis that ALDH3 expression in the mouse is at least 500-fold higher in the cornea than in any other tissue examined, with very low levels of expression detected in the stomach, urinary bladder, ocular lens, and lung. Histochemical localization reveals that this exceptional level of expression in the mouse cornea occurs in the anterior epithelial cells and that little ALDH3 is present in the keratocytes or corneal endothelial cells. A 13-kbp mouse ALDH3 promoter fragment containing >12 kbp of the 5′ flanking sequence, the 40-bp untranslated first exon, and 29 bp of intron 1 directed cat reporter gene expression to tissues that express the endogenous ALDH3 gene, except that transgene promoter activity was higher in the stomach and bladder than in the cornea. By contrast, when driven by a 4.4-kbp mouse ALDH3 promoter fragment [1,050-bp 5′ flanking region, exon 1, intron 1 (3.4 kbp), and 7 bp of exon 2] expression of the cat reporter gene was confined to the corneal epithelial cells, except for very low levels in the liver, effectively reproducing the corneal expression pattern of the endogenous ALDH3 gene. These results indicate that tissue-specific expression of ALDH3 is determined by positive and negative elements in the 5′ flanking region of the gene and suggests putative silencers located in intron 1. We demonstrate regulatory sequences capable of directing cornea-specific gene expression, affording the opportunity for genetic engineering in this transparent tissue.
The isolation of tissue-specific promoters and enhancers has allowed developmental mechanisms and functions of individual tissues to be examined experimentally at the molecular level in transgenic mice. Additionally, the ability of these promoters and enhancers to target the expression of foreign genes to a specific tissue affords researchers the opportunity to pursue potential gene-based therapeutic strategies. The eye in particular has a number of highly specialized tissues that have long served as models for development and cellular specializations. In the ocular lens, the development and use of lens-specific promoters derived from crystallin genes has led to significant insights into the regulation of gene expression and cellular processes (1, 2). Similarly, use of retina-specific promoters has led to a greater understanding of gene expression and differentiation in this complex ocular tissue (3–5). The expression of several proteins at crystallin-like levels in the cornea (6–8) offers the potential for development of tissue-specific promoters in this ocular tissue as well. The development of such a tissue-specific promoter for the cornea is of special interest considering its direct importance to vision and, because of its surface location, its accessibility for receiving foreign genes.
The avascular cornea is structurally and functionally specialized to provide a protective barrier between sensitive ocular tissues and the external environment. An anterior continuation of the opaque sclera, the transparent cornea, is covered by a nonkeratinized, stratified squamous epithelium with a slow cycling stem cell population localized in the peripheral limbal region. Examination of mammalian corneas reveals that a single 54-kDa protein accounts for 5–40% of the total water-soluble protein in the epithelium (6, 7, 9–12). Enzymatic (6, 13, 14) and molecular (9, 15, 16) characterization identified this 54-kDa corneal protein, originally described in bovine cornea and designated BCP 54 (17), as a class 3 aldehyde dehydrogenase (ALDH3: aldehyde:NAD(P)+ oxidoreductase, EC 1.2.1.3). Low levels of constitutively expressed ALDH3 also have been observed in several nonocular tissues (stomach and bladder), and induced expression of ALDH3 has been reported in several carcinomas and in response to xenobiotic activating agents (18).
As the anterior-most layer of the cornea, the epithelium is faced with continual impingement by environmental stressors, such as UV radiation, that can have deleterious effects on cellular function. The abundant expression of ALDH3 in the corneal epithelium is believed to play a role in protecting this vital tissue, as well as the rest of the eye, from damage generated by UV exposure (6, 19). With a demonstrated substrate preference for the medium-length aliphatic aldehyde products of UV-induced lipid peroxidation, ALDH3 is thought to be responsible for preventing the accumulation of these cytotoxic products in the cornea (6, 19). In addition to its detoxification function, ALDH3 also may protect the eye by the direct absorption of UV radiation. The high UV-absorbing capacity of ALDH3, which has been attributed both to its high tryptophan content and to its ability to bind NAD, has led some investigators to ascribe the term “absorbin” to this major corneal protein (10). In the present study, we show that a promoter fragment derived from the mouse ALDH3 gene is capable of targeting expression of the chloramphenicol acetyltransferase (cat) reporter gene specifically to the epithelial layer of the cornea in transgenic mice, mimicking the expression pattern of the endogenous ALDH3 gene.
MATERIALS AND METHODS
Identification and Cloning of the Mouse Corneal ALDH3 Transcript.
5′ rapid amplication of cDNA ends (RACE) (Life Technologies, Grand Island, NY) was used to identify the ALDH3 transcriptional start site. First strand cDNA synthesis involved reverse transcribing 1 μg of total RNA from mouse and human cornea with gene-specific synthetic oligonucleotide primers (5′-GATGGTGAGGTTGAAGGGGTAGTT-3′, GSP1) corresponding to the coding sequence in exon 3 of the rat and human ALDH3 genes. On completion of the first strand synthesis, the mRNA templates were degraded by treatment with RNase H. A poly(dCTP) tail was added to the 3′ end of the cDNAs with terminal D transferase. Amplification of the cDNAs was accomplished with a second gene-specific oligonucleotide primer corresponding to a region of exon 3 located 24 bp upstream of GSP1 (5′-CAGGGGCTCCGAGTGGATGTAGAG-3′, GSP2) in conjunction with the anchor primer provided by the manufacturer. The resulting 5′ RACE products were inserted into the unique Srf I site of the pCRScript (Stratagene) cloning vector. The sequence of the mouse and human ALDH3 cDNAs was determined by the dideoxy method. The 3′ end of the mouse cornea ALDH3 transcript, consisting of exon 3 through the polyadenylation signal, was determined with 3′ RACE. A poly(dTTP) primer was used to reverse transcribe DNA copies from cornea RNA. mRNA templates were removed by treatment with RNase H, and PCR amplification was performed with a gene-specific oligonucleotide primer (5′-GATGACCTCTACATCCACTCGGAG-3′) and the poly(dTTP) primer. The resulting 3′ RACE product was inserted into the unique Srf I site of the pCRScript cloning vector and sequenced by the dideoxy method.
Northern Blot Analysis of ALDH3 Expression in Adult Mouse Tissues.
Total RNA was prepared from the indicated tissues with RNAzol (Tel-Test, Friendsworth, TX), and concentrations were calculated from A260 determinations. RNA integrity and loading amounts were assessed by examining 18 S and 28 S ribosomal RNA banding of samples electrophoresed under nondenaturing conditions and stained with ethidium bromide. For analysis of ALDH3 mRNA levels, total RNAs (1–20 μg) were resolved by formaldehyde-agarose gel electrophoresis, transferred to GeneScreen Plus membrane (DuPont), and UV-crosslinked. Blots were prehybridized at 65°C in a solution containing 6× standard saline citrate [1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7.0 (SSC)], 0.1% Ficoll™ 400, 0.1% polyvinylpyrrolidone, 0.1% BSA, 1% SDS, and 100 μg/ml sheared salmon sperm DNA. The prehybridization buffer was replaced with fresh buffer containing the [α-32P]dCTP-labeled cDNA probe(s) (≈109 cpm), and the blots were hybridized overnight at 65°C. Blots were washed sequentially in buffer A (6× SSC/0.1% SDS) and buffer B (2× SSC/0.1% SDS) at 65°C, rinsed briefly in 2× SSC at room temperature, and autoradiographed on XAR film (Kodak) for 1–3 days at −80°C.
Screening of a Mouse Genomic Library for ALDH3-Positive Clones.
A λ FIX II mouse genomic library (Stratagene) was screened with cDNA probes to mouse ALDH3 (described above). Secondary and tertiary screenings of positive clones were carried out with exon-specific [γ-32P]dATP end-labeled oligonucleotide probes, corresponding to sequences contained within exons 1, 3, 7, and 11 of the ALDH3 gene. Primary screening hybridizations with the cDNA probes were carried out at 65°C. Hybridized filters were sequentially washed in buffer A (6× SSC/0.1% SDS) and buffer B (2× SSC/0.1% SDS) at 65°C, rinsed briefly in 2× SSC at room temperature, and autoradiographed on XAR film for 1–3 days at −80°C. Secondary and tertiary screening hybridizations and washes were carried out at 55°C. A 16-kbp genomic clone, found to contain the 5′ end of the mouse ALDH3 gene, was subcloned into the NotI site of pBlueScript (Stratagene) and subjected to dideoxy sequencing with vector-specific and ALDH3-specific oligonucleotide primers.
Histochemical Localization of ALDH3.
Enucleated mouse eyes and corneas isolated from enucleated monkey eyes by circumcision just lateral to the limbus were placed in molds containing Tissue Tek (Miles Scientific) and quick frozen on dry ice. Human corneas obtained from Lions Eye Tissue Bank (St. Louis, MO) were removed from storage media, rinsed in PBS, and embedded in OCT as above. Cryostatic sections were collected on uncoated glass slides and stored at −80°C until use. For histochemical localization of ALDH3 activity, sections were allowed to warm to room temperature before being placed in a histochemical staining solution (20) containing benzaldehyde (25 mM), NADP (1.5 mM), phenazine methylsulfate (0.32 mM), nitro blue tetrazolium (1.5 mM), 10% polyvinyl alcohol, and 10 mM sodium azide. The reaction was allowed to proceed for 5 min, and the sections were rinsed thoroughly and fixed for 10 min in 4% glutaraldehyde. After rinsing, coverslips were mounted, and the sections were viewed and photographed with a Zeiss Axiophot microscope.
Construction of cat Reporter Transgene and Generation of Transgenic Mice.
A 12-kbp XbaI fragment (Xb12) of the mouse ALDH3 gene, which included exon 1, 29 bp of intron 1, and ≈12 kbp of the 5′ flanking sequence, was cloned into the polylinker region of the promoter-less reporter vector pCAT basic (Promega). A second promoter fragment (−1050INT), consisting of 1,050 bp of the 5′ flanking sequence, the 40-bp untranslated exon 1, the 3.4-kb of intron 1, and 7-bp of exon 2 was generated by PCR from a 16-kbp mouse ALDH3 genomic clone and inserted into the polylinker of pCAT basic. For the generation of transgenic mice, the promoter/reporter mini genes (XB12/cat and −1050INT/cat, respectively) were excised by restriction endonuclease digestions (Life Technologies) and purified from agarose gels with Geneclean II (Bio 101). Transgenic mice were generated by using standard strategies (21).
Analysis of CAT Expression in Transgenic Mice.
To assay for CAT activity, tissues of interest were obtained from transgenic animals at the time of killing and immediately placed on ice. After homogenization, cellular debris was pelleted by centrifugation at 14,000 × g for 10 min, and the water-soluble fraction was retained for analysis. Before assaying for CAT activity, samples were heated at 65°C for 15 min, followed by centrifugation for 10 min at 14,000 × g. The concentration of protein in the final fractions was determined with the BCA™ method (Pierce). CAT activity was assayed with a biphasic assay (22) with 3H–acetyl CoA (DuPont/NEN) as substrate. A direct histochemical assay for CAT activity (23) was used to localize reporter gene expression in the cornea. Eyes were excised from 10-week-old transgenic mice, fixed in 5% paraformaldehyde in PBS, washed twice in PBS, placed in Tissue Tek, frozen on dry ice, and stored at −80°C until used. Cryosections (8 μm) were collected on uncoated glass slides, allowed to air dry for 5 min, and washed in PBS for 5 min. The slides were then removed from PBS, excess buffer was eliminated, the tissue sections were overlain with the staining solution (8 mM sodium citrate, 3 mM copper sulfate, 5 mM potassium ferricyanide, 4 mM chloramphenicol, 0.3 mM acetyl CoA, and 63 mM sodium maleate at pH 6.0), and the slides were placed in a humidified chamber at room temperature for 6–12 h. The reactions were terminated by washing three times in PBS, and the sections were dehydrated in ethanol, cleared in xylene, and coverslips were affixed with Permount (Fisher Scientific). Reacted sections were viewed and photographed with a Ziess Axiophot microscope.
RESULTS AND DISCUSSION
Mouse ALDH3 cDNA.
In the present investigation, 5′ and 3′ RACE techniques were used to determine the transcriptional start site and sequence of ALDH3 transcripts expressed in the mouse cornea. Analysis of the resulting cDNA products revealed a +1 transcriptional start site 45 bp upstream of the ATG translational initiation codon, in agreement with that identified for both constitutive and induced ALDH3 mRNAs in rats (24, 25) and humans (26, 27). Additionally, the 5′ untranslated region of the mouse corneal ALDH3 transcript is 87% and 64% identical with published rat (24, 25) and human (26, 27) sequences, respectively. Surprisingly, comparison of the mouse cornea ALDH3 transcript with a previously reported mouse ALDH3 cDNA (28) revealed no homology within the 5′ noncoding regions. Analysis of the mouse ALDH3 gene (see below) showed that our 5′ noncoding sequence, not reported earlier (28), is present in a distinct upstream exon. Significant interspecies sequence conservation is seen in the coding region with the mouse ALDH3 transcript sharing identities of 90% and 80% with published rat (24, 29) and human (26) ALDH3 cDNAs, respectively. When the deduced amino acid sequence of mouse ALDH3 is compared with that of rat and human ALDH3, the identities rise to 96% and 89%, indicating the highly conserved nature of the ALDH3 protein.
Cornea-Preferred ALDH3 Expression.
Northern blot analysis, utilizing total RNA isolated from mouse tissues, showed that the ALDH3 transcript is highly expressed in the cornea, with little or no expression detected in the other examined tissues (Fig. 1A). Scanning densitometry of Northern blot autoradiograms indicated that ALDH3 expression in the cornea is at least 500-fold higher than in the next highest expressing tissue (stomach), clearly establishing ALDH3 as a cornea-preferred transcript although not strictly a cornea-specific transcript. Significant levels of ALDH3 transcript also were found in human and bovine corneas (Fig. 1B), consistent with protein expression (7, 30). In Northern blot analysis to compare transcript levels in human corneal epithelial and endothelial cells, abundant expression of ALDH3 was detected only in the epithelial layer (Fig. 1B). An additional Northern blot experiment, in which the autoradiogram was purposely overexposed, revealed trace amounts of ALDH3 mRNA in the bladder, lens, and lung (Fig. 1C) but not in any other adult mouse tissues (data not shown).
Figure 1.
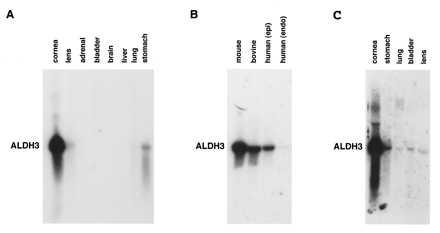
Northern blot analysis of ALDH3 expression in mouse, bovine, and human tissues. (A) Hybridization of a mouse ALDH3 cDNA probe to a Northern blot of total RNA from adult mouse cornea (2 μg), lens (20 μg), adrenal gland (20 μg), urinary bladder (20 μg), brain (20 μg), liver (20 μg), lung (20 μg), and stomach (20 μg) revealed ALDH3 transcript levels in the cornea to be at least 500-fold higher than those observed in any other tissue examined. (B) Significant levels of ALDH3 transcript also were detected in total RNA from bovine cornea (10 μg) and human corneal epithelium (10 μg) but not in human corneal endothelium (5 μg). (C) A Northern blot analysis in which the autoradiogram purposely has been overdeveloped to demonstrate hybridization of a mouse ALDH3 cDNA probe to total RNA from mouse cornea (2 μg), stomach (20 μg), urinary bladder (20 μg), lung (20 μg), and lens (20 μg), indicating very low but detectable levels of ALDH3 expression in these tissues.
ALDH3 Localization in the Mouse Cornea.
Hematoxylin/eosin staining of the central region of the adult mouse cornea (Fig. 2A) demonstrates the presence of three distinct cell types: (i) the anterior epithelial cells, (ii) keratocytes located within the collagenous stromal layer, and (iii) a layer of endothelial cells forming the posterior extent of cornea. Histochemical staining shows ALDH3 activity to be primarily within the epithelial layer of the mouse (Fig. 2B), monkey (not shown), and human (not shown) corneas. Immunohistochemical localization (W.T.K. and I. Gipson, unpublished work) confirmed restriction of ALDH3 protein expression to the corneal epithelium. Northern blot analysis (see Fig. 1B) and in situ hybridizations (W.T.K., F. X. Yu, J. Guo, and J.P., unpublished work) also indicated that the abundant expression of ALDH3 mRNA in the cornea is localized to the anterior epithelial cells. These findings differ from earlier studies that reported, in addition to high levels in the epithelia, ALDH3 immunoreactivity in the stroma and endothelium of bovine (30, 31), porcine (31), ovine (31), and baboon (31) corneas. Within the epithelial layer, ALDH3 activity was highest in the cells of the central cornea with expression diminishing near the transition to the stem cell-containing limbus (Fig. 2D).
Figure 2.
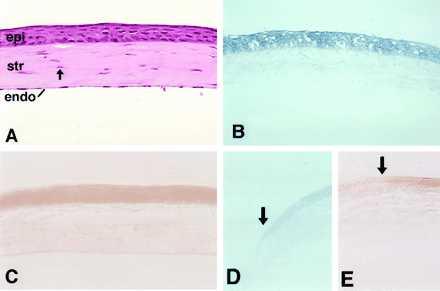
Histochemical examination of ALDH3 and cat reporter gene expression in the mouse cornea. (A) The structure of the adult mouse cornea is seen in a hematoxylin/eosin-stained section. The stratified corneal epithelium (epi) overlies a stromal layer (str) maintained by numerous corneal keratocytes (arrow). The one-cell-thick endothelium (endo) forms the posterior extent of the cornea. (B) Histochemical localization demonstrates ALDH3 activity primarily in the cells of the central epithelium of the normal mouse cornea. (C) Histochemical localization of CAT activity in the cornea of a −1050INT transgenic mouse demonstrates the ability of the mouse ALDH3 promoter fragment to target reporter gene expression to the corneal epithelium, faithfully reproducing the epithelial-specific pattern demonstrated for the endogenous mouse ALDH3 gene. In the peripheral cornea, localization of both ALDH3 (D) and CAT (E) activities diminishes near the limbal border (arrows), with little activity detected in limbal epithelial cells themselves.
ALDH3 Promoter Isolation.
We next attempted to identify a mouse ALDH3 promoter fragment that is active specifically in the cornea of transgenic mice. RACE-generated cDNAs were used as probes to obtain a 16-kbp ALDH3 genomic fragment with its 3′ terminus within exon 2, 111 bp downstream from the translation start site. The transcription initiation site and 40-bp noncoding first exon also were identified, residing 3.4 kbp upstream from the translational initiation ATG codon located in the second exon (Fig. 3). A TATA box and numerous potential regulatory sequences were present within the first kilobase pair of the 5′ flanking sequence of the mouse ALDH3 gene (Fig. 3). Examination of 5′ flanking regions of other genes found to be highly expressed in the cornea (24, 27, 32) revealed a similar array of candidate binding sites for transcription factors.
Figure 3.
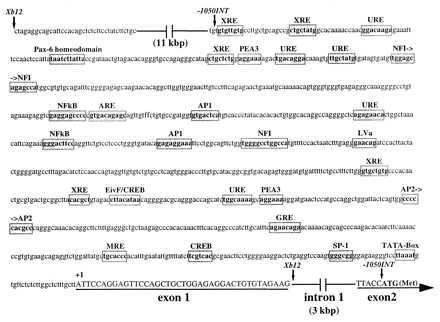
Structure of the 5′ end of the mouse ALDH3 gene. A 40-bp untranslated exon (exon 1) was identified ≈3 kbp upstream of the translation initiation ATG codon in exon 2. A sequence analysis, employing Wisconsin Sequence Analysis Package software (Genetics Computer Group, Madison, WI), identified putative binding sites for a number of transcription factors within the initial 1,050 bp of sequence 5′ to the +1 transcription initiation site. Of particular interest are the antioxidant responsive element (ARE) and UV-responsive elements (URE), which suggest a potential to up-regulate expression in response to oxidative and UV-induced stress in the cornea. The promoter fragment utilized to generate transgenic mice is delineated by arrows (↓). Exon sequence is indicated by underlined uppercase characters.
Cornea-Preferred Promoter Activity in Transgenic Mice.
Although cultured hepatoma cells have been used to study induced expression of the rat ALDH3 gene (33–35), examination of available primary and simian virus 40-transformed corneal epithelial cells showed that the endogenous ALDH3 gene is not expressed in cultured corneal cells (data not shown). Similarly, ALDH3 activity declines in intact rat corneas placed in culture (25). We thus resorted to transgenic mice to examine the ability of mouse ALDH3 promoter fragments to direct reporter gene expression in the cornea. Five independent lines of transgenic mice were generated with a 12-kbp XbaI fragment of the mouse ALDH3 gene, encompassing ≈12 kbp of the 5′ flanking sequence, the untranslated first exon, and 29 bp of intron 1 (see Fig. 3) fused to the cat reporter gene. Of the five Xb12/cat-positive lines obtained, significant CAT activity was detected in the tissues of F1 mice from four independently derived lines, establishing that this large promoter fragment is capable of directing reporter gene expression in transgenic mice. Analysis of CAT activity indicates that Xb12-driven expression is highest the stomach, followed by bladder, cornea, lens, and lung (trace) (Fig. 4A). Although this is similar qualitatively to the pattern of ALDH3 expression in the mouse (see Fig. 1C), it differs quantitatively from the expression pattern demonstrated for the endogenous mouse ALDH3 gene, in which corneal ALDH3 transcript levels are at least 500-fold higher than those detected in stomach and other tissues (see Fig. 1A). It is unlikely that this expression pattern was caused by the integration site of the transgene because similar results were obtained in four separate founder mice. The detection of significant CAT expression in noncorneal tissues indicates that the Xb12-promoter fragment lacks the regulatory elements required to reproduce the highly cornea-preferred expression pattern of the endogenous ALDH3 gene.
Figure 4.
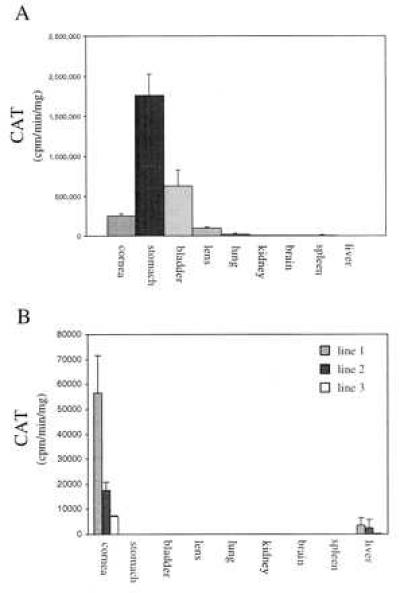
ALDH3 promoter-driven reporter gene expression in the tissues of adult transgenic mice. (A) In Xb12/cat transgenic animals (n = 4), significant expression of the reporter gene, expressed as CAT activity, is seen in stomach, bladder, cornea, lens, lung, and spleen (trace) although not at relative levels consistent with expression of the endogenous ALDH3 gene (see Fig. 1 A and C). (B) By contrast, transgene expression in −1050INT/cat transgenic mice is restricted primarily to the corneas of mice from three independent transgenic lines (n = 4 animals/transgenic line). Very low levels of transgene expression are seen in the livers of animals from each transgenic line, with CAT activity ranging from 5 to 12% of corneal activity in the respective transgenic line.
The higher than expected level of XP12/cat transgene expression detected in a few noncorneal tissues that normally express ALDH3 at low levels suggests the possible involvement of silencing elements in producing the cornea-preferred expression of ALDH3. Indeed, the extreme lens specialization of crystallin genes has been shown to depend on an array of positive and negative control elements (36–41). In the case of chicken δ1–crystallin, repression of gene expression in nonlens cells involves binding of the negative δEF1 transcription factor within the third intron (36, 39). In the case of rat βB2–crystallin, a complex interaction between multiple silencing elements situated within intron 1 and the 5′ flanking region of the gene appear critical for the developmentally controlled expression of this gene in the lens (40). Consequently, to examine the possible role of intronic sequences in regulating ALDH3 expression, a second promoter construct (−1050INT/cat) comprising 1,050 bp of the 5′ flanking sequence, exon 1, intron 1, and 7 bp of exon 2 of the mouse ALDH3 gene (see Fig. 3) fused to the cat reporter gene was utilized to generate transgenic mice. Five independent lines of transgenic animals that displayed integration of the transgene were obtained, with three of these lines exhibiting significant cornea-preferred CAT activity (Fig. 4B). Corneal CAT activity, although lower than that seen in Xb12/cat lines, was consistent within each −1050INT/cat transgenic line, with high, moderate, and low expressing lines corresponding to the relative transgene copy number observed. Histochemical localization reveals that ocular CAT activity in −1050INT/cat transgenic mice is restricted to the epithelial layer of the cornea (Fig. 2 C and E), reproducing the distribution of the endogenous ALDH3 protein (Figs. 2B and 3D). A low level of extra-ocular cat expression was detected only in the liver of −1050INT/cat mice (Fig. 4B), a tissue in which uninduced ALDH3 expression was not detected by Northern blot analysis (see Fig. 1A). It is likely that this weak expression in the liver is due to the absence of upstream negative regulatory elements, recently demonstrated to block uninduced ALDH3 expression in rat hepatoma cells (34), from the −1050INT/cat promoter fragment. The lack of detectable cat expression in the livers of Xb12/cat transgenic mice (see Fig. 4A) supports this explanation. These results indicate that the regulatory sequences included in the −1050INT promoter fragment are capable of targeting reporter gene expression to the cornea, reproducing the corneal expression pattern of the endogenous mouse ALDH3 gene.
The ability of these ALDH3 promoter fragments to direct reporter gene expression in transgenic mice indicates that the high expression of ALDH3 in corneal epithelial cells is controlled, at least in part, at the transcriptional level, similar to the expression of crystallin genes in the lens (42). The lack of significant extra-corneal cat expression in −1050INT/cat transgenic mice suggests the presence of silencing elements within the first intron of the mouse ALDH3 gene. Comparison of reporter gene expression in Xb12/cat and −1050INT/cat mice suggests that these elements act, possibly in conjunction with elements in the 5′ flanking region, to block or repress ALDH3 expression outside the cornea, suggesting similarities with the proposed roles of the intronic sequences for the highly lens-preferred expression of the δ–crystallin (36) and βB2–crystallin (40) genes. Additional experiments are required to identify and characterize the putative ALDH3 intronic silencer elements.
The transcription factors responsible for the high ALDH3 promoter activity in the corneal epithelium have not yet been identified. DNase I footprinting experiments indicate that AP1 and Sp1 are involved in expression of the rat ALDH3 gene in hepatoma cells (35). AP2 and Sp1 are involved in the regulation of keratin gene expression in the cornea. K3 keratin expression in the differentiated suprabasal cells of the corneal epithelium is associated with an elevated Sp1/Ap2 ratio (32, 43), although AP2 activates basal cell expression of the K12 gene (44). AP2 also has been implicated in the activation of gelatinase B expression in the corneal epithelium (45). Another transcription factor that may be involved in regulating gene expression in the cornea is Pax-6. This transcription factor is expressed at significant levels in the corneal epithelium (46), and consensus Pax-6 binding sites are found in the 5′ flanking regions of the mouse ALDH3 gene (see Fig. 3) and other genes that exhibit significant corneal expression.
Because ALDH3 is believed to function in protecting the eye from UV and UV-induced oxidative damage, the ability of these stresses to influence corneal gene expression is of particular interest. Within the first kilobase pair of the 5′ flanking sequence in both the mouse (see Fig. 3) and rat ALDH3 (34) genes are consensus binding sites for proteins known to mediate cellular responses to UV and oxidative damage (47–49). Evidence that these UV- and antioxidant-responsive elements function in regulating ALDH3 expression is provided by a recent study in which ALDH3 levels were found to decline in rat corneal epithelia cultured in the absence of light although elevated ALDH3 levels were maintained in epithelia cultured under cycling light conditions (25). Furthermore, ocular ALDH3 activity in mice raised in the dark was found to be significantly lower than that found in animals raised in a 12-h light/dark cycle (50). Although expression of ALDH3 in the mammalian cornea is generally referred to as “constitutive,” the likely involvement of these stress-responsive regulatory sequences in generating the profound levels of expression found in this tissue suggests that “modulated constitutive expression” would be a more accurate term to describe this phenomenon. The availability of a cornea-specific promoter that includes regulatory elements potentially responsive to UV and UV-induced oxidative damage will allow this question to be addressed in vivo.
By analogy with the studies on the lens and retina, the availability of a tissue-specific promoter provides a critical tool with which analysis of corneal development, function, and wound healing can be studied at the molecular level. The surface accessibility of the corneal epithelium also makes this tissue an attractive candidate for gene therapy, adding greatly to the usefulness of the −1050INT ALDH3 promoter fragment. We thus anticipate that use of this ALDH3 promoter fragment will provide the groundwork for the alleviation of corneal disorders.
Acknowledgments
We thank Drs. S. Tomarev, M. Duncan, and C. Sax for critical reading of the manuscript. The transgenic mice used in this study were generated by the NEI Transgenic Service Facility, headed by Dr. Eric Waurousek.
ABBREVIATIONS
- ALDH3
aldehyde dehydrogenase class 3
- cat
chloramphenicol acetyltransferase
- RACE
rapid amplication of cDNA ends
- SSC standard saline citrate.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF033034).
References
- 1.Overbeek P A, Chepelinsky A B, Khillan J S, Piatigorsky J, Westphal H. Proc Natl Acad Sci USA. 1985;82:7815–7819. doi: 10.1073/pnas.82.23.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cvekl A, Piatigorsky J. Bioessays. 1996;18:621–630. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 3.Liou G I, Geng L, al-Ubaidi M R, Matragoon S, Hanten G, Baehr W, Overbeek P A. J Biol Chem. 1990;265:8373–8376. [PubMed] [Google Scholar]
- 4.Lem J, Applebury M L, Falk J D, Flannery J G, Simon M I. Neuron. 1991;6:201–210. doi: 10.1016/0896-6273(91)90356-5. [DOI] [PubMed] [Google Scholar]
- 5.Kikuchi T, Raju K, Breitman M L, Shinohara T. Mol Cell Biol. 1993;13:4400–4408. doi: 10.1128/mcb.13.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abedinia M, Pain T, Algar E M, Holmes R S. Exp Eye Res. 1990;51:419–426. doi: 10.1016/0014-4835(90)90154-m. [DOI] [PubMed] [Google Scholar]
- 7.Cuthbertson R A, Tomarev S I, Piatigorsky J. Proc Natl Acad Sci USA. 1992;89:4004–4008. doi: 10.1073/pnas.89.9.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sax C M, Salamon C, Kays W T, Guo J, Yu F X, Cuthbertson R A, Piatigorsky J. J Biol Chem. 1996;271:33568–33574. doi: 10.1074/jbc.271.52.33568. [DOI] [PubMed] [Google Scholar]
- 9.Cooper D L, Baptist E W, Enghild J J, Isola N R, Klintworth G K. Gene. 1991;98:201–207. doi: 10.1016/0378-1119(91)90174-a. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell J, Cenedella R J. Cornea. 1995;14:266–272. doi: 10.1097/00003226-199505000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Evces S, Lindahl R. Arch Biochem Biophys. 1989;274:518–524. doi: 10.1016/0003-9861(89)90465-7. [DOI] [PubMed] [Google Scholar]
- 12.Holmes R S, Cheung B, VandeBerg J L. Comp Biochem Physiol B. 1989;93:271–277. doi: 10.1016/0305-0491(89)90081-3. [DOI] [PubMed] [Google Scholar]
- 13.Alexander R J, Silverman B, Henley W L. Exp Eye Res. 1981;32:205–216. doi: 10.1016/0014-4835(81)90009-9. [DOI] [PubMed] [Google Scholar]
- 14.Verhagen C, Hoekzema R, Verjans G M, Kijlstra A. Exp Eye Res. 1991;53:283–284. doi: 10.1016/0014-4835(91)90085-s. [DOI] [PubMed] [Google Scholar]
- 15.Verjans G M, Verhagen C, Hoekzema R, Kijlstra A. Curr Eye Res. 1990;9:1217–1220. doi: 10.3109/02713689009003478. [DOI] [PubMed] [Google Scholar]
- 16.Algar E M, Cheung B, Hayes J, Holmes R S, Beacham I R. Adv Exp Med Biol. 1993;328:153–157. doi: 10.1007/978-1-4615-2904-0_17. [DOI] [PubMed] [Google Scholar]
- 17.Holt W S, Kinoshita J H. Invest Ophthalmol. 1973;12:114–126. [PubMed] [Google Scholar]
- 18.Lindahl R. Crit Rev Biochem Mol Biol. 1992;27:283–335. doi: 10.3109/10409239209082565. [DOI] [PubMed] [Google Scholar]
- 19.Lindahl R, Petersen D R. Biochem Pharmacol. 1991;41:1583–1587. doi: 10.1016/0006-2952(91)90157-z. [DOI] [PubMed] [Google Scholar]
- 20.Chieco P, Stecca B, Bolondi L, Melchiorri C, Gaiani S, Barbara L. Liver. 1995;15:87–92. doi: 10.1111/j.1600-0676.1995.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 21.Hogan B, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 22.Neumann J R, Morency C A, Russian K O. Biotechniques. 1987;5:444–447. [Google Scholar]
- 23.Donoghue M J, Alvarez J D, Merlie J P, Sanes J R. J Cell Biol. 1991;115:423–434. doi: 10.1083/jcb.115.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asman D C, Takimoto K, Pitot H C, Dunn T J, Lindahl R. J Biol Chem. 1993;268:12530–12536. [PubMed] [Google Scholar]
- 25.Boesch J S, Lee C, Lindahl R G. J Biol Chem. 1996;271:5150–5157. doi: 10.1074/jbc.271.9.5150. [DOI] [PubMed] [Google Scholar]
- 26.Hsu L C, Chang W C, Shibuya A, Yoshida A. J Biol Chem. 1992;267:3030–3037. [PubMed] [Google Scholar]
- 27.Hsu L C, Chang W C, Chang C, Tsukamoto N, Yoshida A. Gene Expr. 1996;6:87–99. [PMC free article] [PubMed] [Google Scholar]
- 28.Vasiliou V, Reuter S F, Kozak C A, Nebert D W. Pharmacogenetics. 1993;3:281–290. doi: 10.1097/00008571-199312000-00002. [DOI] [PubMed] [Google Scholar]
- 29.Jones D E, Jr, Brennan M D, Hempel J, Lindahl R. Proc Natl Acad Sci USA. 1988;85:1782–1786. doi: 10.1073/pnas.85.6.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silverman B, Alexander R J, Henley W L. Exp Eye Res. 1981;33:19–29. doi: 10.1016/s0014-4835(81)80078-4. [DOI] [PubMed] [Google Scholar]
- 31.Downes J E, VandeBerg J L, Hubbard G B, Holmes R S. Cornea. 1992;11:560–566. doi: 10.1097/00003226-199211000-00013. [DOI] [PubMed] [Google Scholar]
- 32.Wu R L, Chen T T, Sun T T. J Biol Chem. 1994;269:28450–28459. [PubMed] [Google Scholar]
- 33.Takimoto K, Lindahl R, Pitot H C. Arch Biochem Biophys. 1992;298:493–497. doi: 10.1016/0003-9861(92)90440-8. [DOI] [PubMed] [Google Scholar]
- 34.Takimoto K, Lindahl R, Dunn T J, Pitot H C. Arch Biochem Biophys. 1994;312:539–546. doi: 10.1006/abbi.1994.1343. [DOI] [PubMed] [Google Scholar]
- 35.Xie Y Q, Takimoto K, Pitot H C, Miskimins W K, Lindahl R. Nucleic Acids Res. 1996;24:4185–4191. doi: 10.1093/nar/24.21.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamachi Y, Kondoh H. Mol Cell Biol. 1993;13:5206–5215. doi: 10.1128/mcb.13.9.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cvekl A, Sax C M, Bresnick E H, Piatigorsky J. Mol Cell Biol. 1994;14:7363–7376. doi: 10.1128/mcb.14.11.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sax C M, Cvekl A, Kantorow M, Sommer B, Chepelinsky A B, Piatigorsky J. Gene. 1994;144:163–169. doi: 10.1016/0378-1119(94)90374-3. [DOI] [PubMed] [Google Scholar]
- 39.Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa–Sehara A, Nabeshima Y, Kondoh H. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dirks R P, Kraft H J, Van Genesen S T, Klok E J, Pfundt R, Schoenmakers J G, Lubsen N H. Eur J Biochem. 1996;239:23–32. doi: 10.1111/j.1432-1033.1996.0023u.x. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Cvekl A, Bassnett S, Piatigorsky J. Dev Genet. 1997;20:258–266. doi: 10.1002/(SICI)1520-6408(1997)20:3<258::AID-DVG8>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 42.Piatigorsky J, Zelenka P S. Adv Dev Biochem. 1992;1:211–256. [Google Scholar]
- 43.Chen T T, Wu R L, Castro-Munozledo F, Sun T T. Mol Cell Biol. 1997;17:3056–3064. doi: 10.1128/mcb.17.6.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuchs E, Weber K. Ann Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 45.Fini M E, Bartlett J D, Matsubara M, Rinehart W B, Mody M K, Girard M T, Rainville M. J Biol Chem. 1994;269:28620–28628. [PubMed] [Google Scholar]
- 46.Koroma B M, Yang J M, Sundin O H. Invest Ophthalmol Vis Sci. 1997;38:108–120. [PubMed] [Google Scholar]
- 47.Stein B, Kramer M, Rahmsdorf H J, Ponta H, Herrlich P. J Virol. 1989;63:4540–4544. doi: 10.1128/jvi.63.11.4540-4544.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rutberg S E, Yang Y M, Ronai Z. Nucleic Acids Res. 1992;20:4305–4310. doi: 10.1093/nar/20.16.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wasserman W W, Fahl W E. Proc Natl Acad Sci USA. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Downes J, Holmes R. Biol Neonate. 1992;61:118–123. doi: 10.1159/000243539. [DOI] [PubMed] [Google Scholar]


