Abstract
Interest in the use of recombinant retroviral vectors for clinical gene therapy has been tempered by evidence of vector-mediated genotoxicity involving the activation of cellular oncogenes flanking sites of vector integration. We report here that the rate of gammaretroviral vector genotoxicity can be significantly reduced by addition of the cHS4 chromatin insulator, based on two complementary approaches for assessing vector-mediated genotoxicity. One approach involves the direct, genomewide assessment of cellular gene dysregulation using panels of transduced cell clones and genomic microarrays, whereas the other involves the functional assessment of malignant transformation using a factor-dependent cell line. Both assays are robust and quantitative, and indicate the cHS4 chromatin insulator can reduce vector-mediated genotoxicity approximately sixfold (ranged three to eight fold). These approaches also provide a means for assessing various aspects of vector-mediated genotoxicity, including the overall rate of cellular gene dysregulation, the potential influence of vector provirus over large genomic distances, and the involvement of oncogenic pathways in vector-mediated malignant transformation.
Introduction
Gene transfer vectors based on recombinant retroviruses are proving increasingly useful in biomedical research and as therapeutic agents in clinical trials. However, with this success has come the revelation of toxicity related to the ability of these vectors to activate cellular oncogenes flanking sites of vector integration.1,2,3 This “genotoxicity” has typically been attributed to the potent enhancers or, to a lesser extent, promoters located within these vectors. Examples include clinical gene therapy trials,4,5 studies in nonhuman primates,6 and both normal and disease-specific mouse models.3,7,8,9 Several advances have been made toward the development of methods for both assessing and reducing the rate of functional vector-mediated genotoxicity.10,11,12 However, the methods described to date for assessing the rate of vector-mediated genotoxicity are limited by compounding factors such as the potential influence of in vivo reconstitution and expansion,10 low sensitivity due to high background rates of tumor formation,11 or lack of direct quantification.12,13 In addition, the use of self-inactivating vector design, as described in these studies, is not always adaptable to vector designs currently in use for clinical and research applications that rely on vector long terminal repeat (LTR) promoters and enhancers.
We and others have proposed that a prototypical chromatin insulator mapped to DNase hypersensitive site 4 of the chicken β-globin loci (cHS4) could potentially reduce the rate of vector-mediated genotoxicity.2,14,15,16 Chromatin insulators are a naturally occurring DNA element that functionally separate differentially expressed genetic loci through their ability to block the repressive effects of heterochromatin, the activating effects of enhancers, or both.17,18 The cHS4 insulator has been shown to reduce the rate of transgene silencing in both gammaretroviral and lentiretroviral vectors,14,19,20,21,22,23 reducing the vector dose and associated risks necessary to reach therapeutic expression levels. More important, insulators such as the cHS4 element can also efficiently block the interaction between enhancers and promoters when placed between these elements.24,25,26,27 It is this directional enhancer-blocking property that holds the most promise for reducing vector-mediated genotoxicity.
Here, we describe the use of two complementary methods to demonstrate the ability of the cHS4 to reduce the rate of gammaretorival vector-mediated genotoxicity. The first involves measuring the rate of cellular gene dysregulation using panels of transduced cell clones and genomic expression microarrays, and the second involves measuring the rate of malignant transformation in the interleukin-3 (IL-3)-dependent cell line 32D. Both systems are robust and quantitative, and revealed an approximately six-fold reduction in the rate of genotoxicity associated with flanking the vector with the cHS4 chromatin insulator, with a range of three to eight fold depending on particular parameters. These studies also support previous findings for a remarkably high rate of cellular gene dysregulation,10,28 suggesting that vector provirus may influence expression of cellular genes at remote locations, and implicate specific oncogenes in 32D cell transformation.
Results
Assessing vector-mediated genotoxicity by genomewide expression analysis
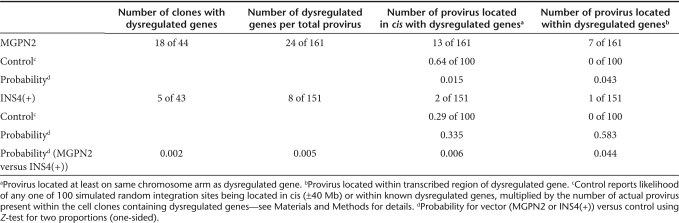
As a first approach for assessing the effects of the cHS4 element on vector genotoxicity, we sought to directly measure the frequency of dysregulated cellular genes in panels of vector-transduced cell lines using genomic expression microarrays. The genetically stable human fibrosarcoma cell line HT1080 was transduced with gammaretroviral vectors either unflanked (MGPN2) or flanked (INS4(+)) with the cHS4 chromatin insulator (Figure 1a), and panels of 44 and 43 independent clones were generated for each vector, respectively, in the absence of drug selection. RNA from each individual clone was then analyzed using Codelink UniSet Human 20K I Bioarrays. A gene was deemed dysregulated if its expression level within an individual clone was at least five-fold higher or lower than the mean from the other 86 clones and two untransduced controls. We used this five-fold threshold because it was the minimum level necessary to achieve no background when comparing independent isolates of the untransduced controls. Validation by independent real-time reverse transcriptase-PCR confirmed gene dysregulation in 80% of these cases, supporting the specificity of the microarray analysis. As diagramed in Figure 1b and summarized in Table 1, we validated 24 dysregulated genes in 18 of 44 cell clones from the uninsulated vector panel, and 8 dysregulated genes in 5 of 43 cell clones from the insulated vector panel. This represents a statistically significant three-fold reduction in the overall frequency of dysregulated genes associated with inclusion of the cHS4 insulator (P = 0.002). As detailed in Supplementary Table S2, the majority of these genes (21 of 32) were upregulated, with absolute changes ranging from 5.1 to 91.7 fold, and a median of 7.9 fold.
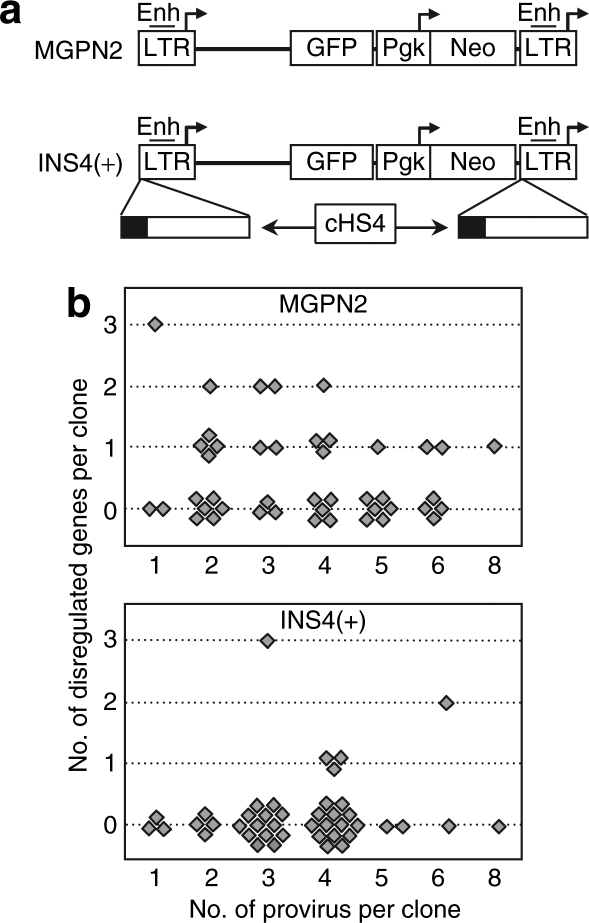
Figure 1.
The cHS4 insulator reduces the frequency of dysregulated cellular genes in transduced HT1080 cell clones. (a) Vector design. The control vector MGPN2 (top) is based on the gammaretroviral vector murine stem cell virus (MSCV) and has two reporter expression cassettes: GFP transcribed from the 5′-LTR promoter and Neo transcribed from the a Pgk promoter. The insulated vector INS4(+) was generated by flanking this vector with a 1.2 kb cHS4 fragment (bottom) using a double-copy arrangement, wherein fragments inserted in the 3′-LTR are copied into the 5′-LTR during proviral integration.14 This results in an arrangement such that the 5′-LTR and internal vector sequences are insulated from genomic sequences on both side of the vector and the 3′-LTR is insulated from genomic sequences upstream of the vector. The DNase-I hypersensitive site core located at the proximal end of the cHS4 fragment is indicated as a solid block. Enh, enhancer; arrows, transcription start sites; GFP, green-fluorescent protein; LTR, long terminal repeat. (b) Frequency of dysregulated cellular genes in transduced HT1080 cell clones. Graphs report the number of dysregulated genes, as well as the number of vector provirus, for individual clones from the panels of HT1080 cells transduced with the uninsulated vector MGPN2 (top) and insulated vector INS4(+) (bottom). See Supplementary Tables S1 and S2 for details on individual clones. Genes were considered dysregulated if expressed at five-fold higher or lower levels within a single clone compared to the mean of the remaining clones and two untransduced controls as determined by expression microarray analysis, and this difference in expression was confirmed by independent real-time PCR. Linear regression analysis indicated there was no significant correlation between the number of provirus and the number of dysregulated genes on a per-clone bases for either dataset (R2 = 0.032, P = 0.24 for vector MGPN2 and R2 = 0.023, P = 0.24 for vector INS4(+)). Although difficult to explain, this result is similar to that found in a tumor-prone mouse model of vector genotoxicity, where no correlation was observed between vector copy number and the time to vector-mediated tumor onset.11
Table 1.
Summary of dysregulated genes in HT1080 cell clones
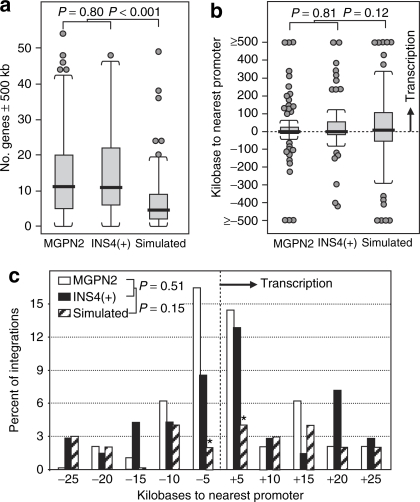
Subsequent molecular and cellular analysis ruled out several trivial explanations for this difference. Southern analysis demonstrated the two panels had equivalent numbers of vector provirus, with a total of 161 provirus for the uninsulated vector and 151 provirus for the insulated vector (Supplementary Table S1, Supplementary Figure S1a). Dividing the number of dysregulated genes by the number of actual provirus confirmed an overall three-fold reduction in frequency of dysregulated genes associated with inclusion of the cHS4 insulator (Table 1). Although flow cytometric analysis revealed a slightly higher level of vector green-fluorescent protein (GFP) expression for the insulated vector panel (with a median of 478 versus 359 mean fluorescent units per vector copy for the uninsulated control, Supplementary Table S1, Supplementary Figure S2a), this difference is consistent with the ability of the cHS4 insulator to reduce silencing chromosomal position effects in this cell line.29 In order to assure that the differences observed between the two panels of transduced cell clones was not simply due to the influence of the cHS4 element on the general pattern of vector integration, we cloned and mapped 97 of the estimated 161 integration sites from the uninsulated MGPN2 vector panel and 70 of the estimated 151 integration sites from the insulated INS4(+) vector panel. As seen in Figure 2, both vectors have a clear preference for integration in gene-dense regions, and a modest preference for integration near transcription start sites, compared to a simulated control dataset, as seen in other settings.30,31 However, there were no statistically significant differences between the uninsulated and insulated vector datasets involving these parameters, indicating the cHS4 does not impact overall vector integration site selection.
Figure 2.
Vector integration patterns in transduced HT1080 cell clones. The sites of vector integration for 97 MGPN2 provirus and 70 INS4(+) provirus from the panels of transduced HT1080 cells clones were mapped, and the patterns of integration compared to the pattern for 100 simulated random integration sites as previously described.30 (a) Boxplots showing the density of genes within a 1 Mb window (500 kb on either side) of integration sites. Heavy horizontal bars, median; open box ends, first and third quartiles; whiskers, 1.5 times the interquartile range; circles, individual outlier data points. P values were determined by the nonparametric Kolmogorov–Smirnov test. (b) Boxplots showing the distance between integration sites and the nearest transcription start site, in kilobases. Negative numbers, integrations located 5′ (upstream) of nearest transcription start sites; positive numbers: integrations 3′ (downstream) of nearest transcription start sites. (c) Histogram plot showing the frequency of vector integrations near transcription start sites. The percentage of integration sites within the indicated discrete windows around known transcription start sites are shown for both the experimental and simulated random datasets. Negative numbers, integrations located 5′ (upstream) of nearest transcription start sites; positive numbers: integrations 3′ (downstream) of nearest transcription start sites. P values were determined by the nonparametric Kolmogorov–Smirnov test. *P = 0.002 for the –5 kb window and P = 0.009 for the +5 kb window, comparing the combined vector datasets to the simulated mock dataset by one-sided Z-test for two proportions. Note that both vectors exhibited the expected preference for gene-rich regions and transcriptional start sites seen in other settings;30,31 however, there was no difference between the two vectors with regards to the general pattern of vector integration sites.
Correlating dysregulated genes with specific provirus
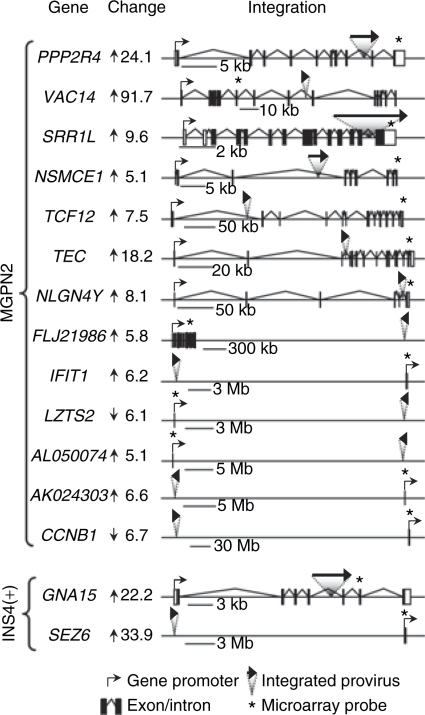
We mapped the integration sites for all of the provirus present in the cell clones exhibiting dysregulated genes (as determined by Southern blot analysis and confirmed by one or more of three independent cloning methods) and compared these sites to the locations of the dysregulated genes. As summarized in Table 1, this analysis revealed that about half of the dysregulated genes were located in cis with specific vector provirus (see Supplementary Table S2 for details). As diagramed in Figure 3, this included seven examples of intragenic and six examples of intergenic integration for the uninsulated vector and one case each of intragenic and intergenic integration for the insulated vector. All cases of intragenic integration involved provirus located within introns in a sense orientation. Cases of intergenic integration involved provirus located at distances of 9.1–111 Mb from gene promoters, with no clear preference in orientation. The difference between the uninsulated and insulated vector datasets was significant, with a clear reduction associated with inclusion of the cHS4 chromatin insulator when considering both the total number of provirus located in cis with dysregulated genes (P = 0.006) and the number of provirus physically located within dysregulated genes (P = 0.044) (Table 1). Furthermore, the frequency of dysregulated genes associated with the uninsulated provirus was significantly higher than that observed for a simulated random control dataset (P = 0.015), indicating a causal relationship (Table 1), whereas the frequency of dysregulated genes associated with the insulated provirus was indistinguishable from background. These differences argue that the high rate of dysregulated genes directly associated with the uninsulated vector cannot be explained by chance, but rather must reflect functional significance. Taken together, these studies demonstrate a significant decrease in the frequency of dysregulated genes associated with flanking the vector with the cHS4 insulator, ranging from three fold (taking into account all dysregulated genes identified) to six fold (taking into account the dysregulated genes located in cis with specific provirus).
Figure 3.
Dysregulated genes associated with specific provirus in transduced HT1080 cell clones. The location and orientation of vector provirus relative to dysregulated genes are diagrammed for every case where a specific provirus was found to be located in cis with a specific dysregulated gene (see Supplementary Table S2 for additional information). A provirus was considered to be located in cis relative to a dysregulated gene if it was mapped to the same chromosome arm as the gene. To the left of each diagram is listed the gene name and the degree and direction of expression dysregulation. The location and orientation of promoters, exons/introns, integrated provirus, and probes used in the expression microarray analysis, are indicated.
Assessing vector-mediated genotoxicity by functional transformation
As an alternative means of assessing the ability of the cHS4 insulator to reduce vector-mediated genotoxicity, we turned to functional transformation studies using the factor-dependent cell line 32D. This is a myeloid cell line derived from mouse bone marrow that is diploid, nonleukemic, and dependent on supplemental IL-3 for growth.32 Previous studies demonstrated that 32D cells can be transformed to IL-3 independence by transfection with oncogenes or cytokine receptor genes.33,34 Transformation can also lead to the ability of this cell line to form tumors on transplantation into congenic C3H/HeJ mice.32,33
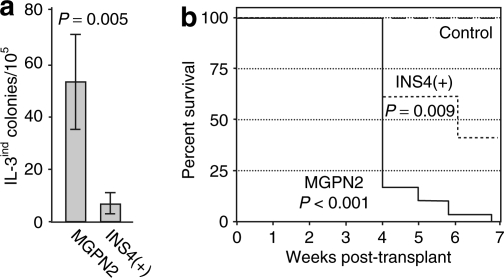
We started by transducing 32D cells with the uninsulated and insulated vectors used in the previous studies (Figure 1a) at a low multiplicity of infection, and then scoring for colony formation in semisolid methylcellulose cultures with and without exogenous IL-3 (see Supplementary Table S3 for details). As summarized in Figure 4a, inclusion of the cHS4 chromatin insulator reduced the rate of IL-3-independent colony formation 8.4 fold, from an average of 53.0 ± 18.5 colonies to an average of 6.3 ± 4.0 colonies per 105 transduced (vector-containing) cells. In order to control for gene transfer rates, we typically used flow cytometry to measure the fraction of GFP-positive cells within a few days of transduction (before silencing position effects could become established; for example, see Supplementary Figure S2b). In some cases, we also used quantitative real-time PCR to determine directly the amount of vector provirus in transduced pools. In one case, we also confirmed the results seen by flow cytometry by using drug selection to assess the fraction of cells expressing the vector-derived Neo gene that we have previously shown to be relatively resistant to silencing position effects.14 As detailed in Supplementary Table S3, all three approaches gave similar results.
Figure 4.
The cHS4 insulator reduces the frequency of vector-mediated transformation of 32D cells assessed by IL-3 independence. (a) Frequency with which 32D cells transduced with the uninsulated vector MGPN2 and the insulated vector INS4(+) formed colonies in the absence of exogenous IL-3. Results represent the average ± SD for the transformation rates from five independent experiments. See Supplementary Table S3 for details of individual experiments. P value based on two-sided, paired Student's t-test. (b) Kaplan–Meier tumor-free survival curves for mice transplanted with clones of 32D cells derived from IL-3-independent colonies. Uninsulated arm: six independent clones transduced with vector MGPN2 and transplanted into 5 mice each. Insulated arm: one independent clone transplanted into 5 mice. Control arm: untransduced 32D cells transplanted into 13 mice. Each mouse received 107 cells administered via tail vein injection. P values based on Logrank test versus control.
The colonies that formed in the absence of IL-3 were notably smaller than the typical colonies that formed in the presence of IL-3. In addition, most of these clones could not be propagated long term in liquid culture without IL-3 supplementation (for example, see Supplementary Table S4). After subsequent expansion in liquid culture with IL-3, most clones were still capable of forming colonies in methylcellulose in the absence of IL-3, but only at a low plating efficiency (ranging around 1% or less). However, Southern blot analysis confirmed the presence of vector provirus in every clone tested (Supplementary Figure S1b). In addition, as shown in Figure 4b, every clone tested was capable of forming tumors upon transplantation into congenic C3H/HeJ mice, with a median latency of 29 days. Tumor-baring mice typically presented with palpable splenomegaly. Cursory histopathology indicated involvement of spleen, liver, and lymph nodes. The apparently stochastic nature of this IL-3-independent phenotype may reflect the relative inefficiency of the murine stem cell virus LTR promoter/enhancer in 32D cells (Supplementary Figure S2), or promoter competition/transcriptional interference between the LTR and flanking genes involved in the oncogenic transformation. The latter hypothesis in particular is consistent with studies were vector expression was found to be lost specifically in transformed or dominant clones.11,35
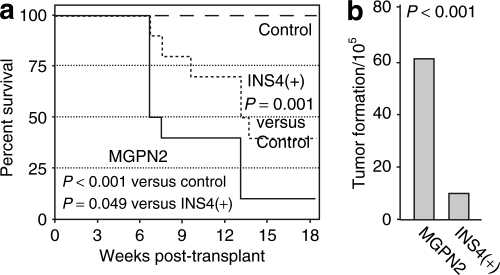
Given the apparent robustness of the tumor formation phenotype, we next measured the rate of vector-mediated transformation using this assay directly. 32D cells were transduced as above, immediately split into independent pools, expanded with IL-3 to generate the requisite number of cells (107 cells per mouse), and then transplanted into congenic mice. As seen in Figure 5a, 9 of 10 mice in the uninsulated vector arm developed tumors, with a median latency of 47 days. In contrast, only 6 of 10 mice in the insulated vector arm developed tumors, with a median latency of 92 days. Taking into account the frequencies of vector transduction and tumor formation, we estimate that inclusion of the cHS4 reduced the frequency of vector-mediated tumor formation six fold, from 61 to 10 per 105 transduced cells (Figure 5b; see legend and Materials and Methods section for details).
Figure 5.
The cHS4 insulator reduces the frequency of vector-mediated transformation of 32D cells assessed by in vivo tumor formation. (a) Kaplan–Meier tumor-free survival curves for mice transplanted with pools of vector-transduced 32D cells. Uninsulated arm: 10 mice from two independent experiments (5 mice each), where each mouse was transplanted with cells from independent pools separated immediately following transduction with vector MGPN2. Insulated arm: 10 mice from two independent experiments (5 mice each) treated the same as the mice in the uninsulated arm, except with the insulated vector INS4(+). Control arm: 13 mice transplanted with untransduced 32D cells. Each mice received 107 cells administered via tail vein injection. P values based on Logrank test. (b) Estimated transformation rate. The frequency of vector-mediated tumor formation was calculated by first estimating the number of transformation events based on the fraction of tumor-free animals at 130 days and the Poisson distribution (23 for the MGPN2 vector arm and 9 for the INS4(+) vector arm), and then dividing these by the estimated number of cells that were transduced during the original transduction culture (found in Supplemental Table S3). See the Statistics section in Supplementary Materials and Methods for details. P values based on one-sided Z-test for two proportions.
Several transformed 32D cell lines were chosen for further characterization, including 12 lines transduced with the uninsulated vector MGPN2 and 9 lines transduced with the insulated vector INS4(+) (Supplementary Table S4). Flow cytometric analysis demonstrated that all clones expressed vector GFP, but at low levels and with a high degree of variation between clones (e.g., see Supplementary Figure S2c). All of the clones also expressed sufficient levels of vector Neo to allow for G418 selection in culture. However, only a total of six clones could be readily cultured in the complete absence of IL-3, and most of these were derived as tumors from mice transplanted with vector-transduced cells. Immunophenotyping indicated that all 21 lines were positive for the panhematopoietic marker CD45 (Ly-5), and were negative for the myeloid differentiation antigen Gr1 (Ly-6G and Ly-6C), the T lymphocyte differentiation antigen CD3, the B lymphocyte differentiation antigen CD19, and the erythroid antigen Ter119 (Ly-76) (data not shown). This is the same profile as the parental 32D cell line and suggests that the transformation events did not result in differentiation down these specific lineages.
Candidate genes involved in vector-mediated transformation
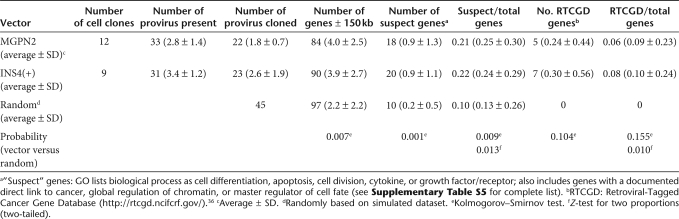
As a first step toward understanding the underlying genetic lesions giving rise to transformed cells in this model, we mapped vector integration sites in a subset of transformed 32D clones using linear amplification–mediated PCR and considered the genes with promoters located within a 150 kb window of each site (Table 2). As expected, there was a nearly two-fold enrichment in the total number of genes surrounding the authentic vector integration sites compared to simulated random controls, reflecting the well-described integration bias for gene-rich regions.30,31 However, among the genes flanking the authentic vector integration sites, there was a further two-fold enrichment for genes that could be considered likely candidates in the 32D transformation process (Table 2; Supplementary Table S5). Of these, 5 of the 84 genes flanking the uninsulated vector provirus and 7 of the 90 genes flanking the insulated vector provirus were listed in the mouse Retroviral Tagged Cancer Gene Database (RTCGD; http://rtcgd.ncifcrf.gov/),36 whereas none of the 97 genes flanking the set of simulated random integration sites were listed in this database. In total, 11 of 12 clones transduced with the uninsulated vector and 8 of 9 clones transduced with the insulated vector contained at least one “suspect” gene within a 150 kb window of a mapped integration site, and the two clones where no “suspect” genes were identified contained additional provirus that could not be readily cloned. We noted two common integration sites (sites of integration near the same gene(s) in two or more independent clones): Aatk, which has previously been shown to play a role in the response of 32D cells to IL-3;37 and Gata2, which has been implicated in oncogenic transformation mediated through Evi1, another gene commonly associated with gammaretroviral vector genotoxicity.2,5,38
Table 2.
Summary of integration site analysis in transformed 32D cells
In order to confirm the functional significance of the correlation between specific provirus–oncogene combinations in the transformed 32D cells clones, we used quantitative real-time reverse transcriptase-PCR to analyze the expression level of a subset of these genes. Based on a previously described algorithm,10 we compared the level of expression for candidate genes within the clones containing the linked provirus to the levels of expression for the same genes in 8–13 other transformed clones that did not contain provirus near the candidate genes. As detailed in Supplementary Table S4, we found a significant level of dysregulation for a total of 14 genes in 14 different clones, out of a total of 22 genes and 19 clones analyzed. The level of dysregulation ranged from 1.6 to over 671 fold, with P values typically <0.0001. The gene ontogeny terms for these genes were varied and included genes involved in cytokine pathways, transcription factors, cell fate and differentiation, and cell cycle control. For those clones where the candidate genes were not found to be dysregulated, there were other candidate genes that were not tested and other sites of vector integration that were not identified, which could serve to explain the source of transformation. Of note, four of five clones containing provirus at the Gata2 and Aatk common integration sites exhibited dysregulation of these genes, emphasizing the functional importance of these sites. Taken together, these results serve to further validate this assay for assessing vector-mediated genotoxicity by providing potential mechanistic explanations for the observed transformed phenotype.
Discussion
Reduced vector-mediated genotoxicity associated with the cHS4 insulator
Because most, if not all, transformation events associated with recombinant retroviral vectors arise from the activation of cellular oncogenes by vector enhancers,4,6,7,8,39 it is the ability of chromatin insulators such as cHS4 to block the activating effects of such enhancers that hold the most promise for reducing the rate of vector-mediated genotoxicity. This potential is bolstered by several recent studies demonstrating the ability of the cHS4 insulator to block a gammaretroviral vector enhancer27 or to block the activation of a promoter trap reporter gene in a lentiviral vector.40 Results of one study also suggested that flanking a lentiviral vector with the cHS4 insulator could also reduce the rate of vector-mediated genotoxicity based on the clonal complexity of a transduced Jurkat cell pool.16 Unfortunately, this study failed to provide a mechanistic rational for the differences observed, such as differences in clonal growth or death rates. In addition, the lack of experimental (culture) replicates in the study made it difficult to assess the robustness of the assay itself.
The demonstrated ability of insulators to interfere with normal genomic interactions, as demonstrated in drosophila models,41 suggests that inclusion of insulators in retroviral vectors could actually increase vector-mediated genotoxicity. Although the studies presented here do not rule out these possibilities, they do serve to demonstrate, on the whole, that these risks are far outweighed by the benefits afforded by the cHS4 insulator. The expression microarray studies demonstrated a three- to six-fold reduction in the frequency of dysregulated cellular genes associated with inclusion of the cHS4 insulator, depending on whether one considers all dysregulated genes or only those genes associated in cis with mapped vector provirus, respectively. The studies in the factor-dependent cell line 32D also demonstrated a six- to eight-fold reduction in the frequency of malignant transformation associated with inclusion of the cHS4 insulator, depending on whether one considers the in vivo tumor formation assay or the in vitro colony assay, respectively. This is the first example, to our knowledge, that inclusion of the cHS4 insulator can reduce the rate of functional, malignant transformation associated with gammaretroviral vector transduction.
These results are in excess of the two- to three-fold reduction expected from strictly topological considerations because the deployment of the cHS4 insulator using a double-copy approach should, at best, only restrict three of four possible interactions between the vector LTRs and flanking sequences: interactions of the 5′-LTR with sequences located upstream and downstream of the vector, and interactions of the 3′-LTR with sequences located upstream of the vector (see Figure 1a). Others have shown that the degree of vector-mediated genotoxicity is determined by the overall transcriptional activity of a particular vector, rather than the number or nature of vector-specific enhancers11 and that reducing the number of LTR enhancers in a gammaretroviral vector from two to one can reduce vector-mediated genotoxicity well in excess of two-fold.12 Thus, it appears that inclusion of the cHS4 element may reduce the transcriptional activity profile of retroviral vectors beyond the level predicted solely by topological considerations, in a manner similar to that achieved using a SIN vector design.
Approaches for assessing vector-mediated genotoxicity
We report here the development of two complementary approaches for assessing vector-mediated genotoxicity. The expression microarray approach provides an unbiased and genomewide assessment of cellular gene dysregulation associated with gammaretroviral vector transduction. By using individual, unselected cell clones, this approach avoids the potential skewing of integration site repertoires introduced by selection,30 competitive growth as pools,12,42 or in vivo reconstitution.6,7,8 By using genomic microarrays, this approach also avoids the use of discrete windows of analysis.10 These studies provided three insights into the nature of retroviral vector-mediated genotoxicity. First, we found that the frequency of dysregulated cellular genes associated with transduction by the uninsulated vector is remarkably high, averaging approximately one cellular gene for every seven provirus. This frequency is very similar to the one in five to one in nine rates reported from primary tissue samples.10,28 Second, our studies suggest that vector provirus may affect cellular gene expression over very large distances. Such observations could arise from indirect effects on transcription factors that escaped detection by the microarray analysis, or by authentic long-range enhancer–promoter interactions of the kind recently reported for the human H19 imprinting control region.43 Third, we found several examples of cellular genes downregulated by greater than five fold, an unexpected result given a diploid genome. However, recent studies suggest that monoallelic expression on human autosomes is surprisingly widespread.44 It is also possible these cases may arise from the indirect effects of provirus at distant locations. Regardless of mechanism, these findings imply that screening methods based on assessing gene expression within a limited window around individual vector provirus, the approach of choice used in most genotoxicity studies, may underestimate the impact of vector transduction on the rate of cellular gene dysregulation.
The studies in the factor-dependent cell line 32D served to confirm and extend the expression microarray studies by providing a functional assay for vector-mediated transformation. This approach is quicker and more cost-effective than previously described methods that rely on bone-marrow transduction and transplantation studies in mice and large animals,6,8,11 yet provides a functional in vivo readout (tumor formation), which is lacking in strictly ex vivo culture models.12 This approach also provides a ready means of quantifying transformation rates, either through the enumeration of discrete transformed colonies in vitro, or the formation of tumors in mice transplanted with discrete cultures. The low background rate of malignant transformation renders this assay more sensitive than tumor-prone mouse models,11 and increases the ability to correlate specific integration events with specific transformation events. Indeed, we were able to identify oncogenes or other genes likely to be involved in the transformation process associated with specific provirus in almost all of the transformed 32D cell clones analyzed. In addition to serving as a means of validating this assay, these correlations also provided modest new insights into one specific pathway by which retroviral vectors can induce malignant transformation, namely, that the Evi1 pathway of vector-mediated transformation can be accessed through the downstream mediator Gata-2.
Implications for gene therapy vector design
It is important to point out that the degree of protection associated with inclusion of the cHS4 element is far from complete and that we demonstrated this protection in the setting of one particular gammaretroviral vector containing two intact LTRs. Several other approaches have been proposed for improving the safety of retroviral vectors, including the use of vectors based on lentiviruses,11 the use of self-inactivating LTR's,12 and the use of tissue-specific promoters.45 As such, chromatin insulators are only one of many possible tools for reducing vector-mediated genotoxicity. However, only the cHS4 element can be easily and directly applied to essentially any vector design.14,20,22 In our experience and that of some others, the cHS4 element has little to no impact on vector titers, although the addition of a 1.2 kb fragment can push large vectors beyond an efficient packaging size or create unintended incompatibilities with other vector sequences.14,20,21,22,30 Although the enhancer-blocking activity of the cHS4 insulator has been mapped to a single DNA footprint on a 42 bp fragment25 and most of the repressor-blocking activity has been mapped to a 250 bp fragment,24 recent studies from our laboratory indicate that a slightly larger 400 bp fragment is needed to obtain full insulating barrier activity, at least in the setting of gammaretroviral and lentiviral vectors.46 Studies with nonretroviral vector delivery systems indicate that a high level of insulation can also be achieved with two copies of the 250 bp cHS4 core fragment. However, this doublet, like all duplicated sequences, tends to recombine when incorporated into gammaretroviral and lentiviral vectors.47
In conclusion, our studies describe the use of two robust and quantitative systems to demonstrate the ability of the cHS4 chromatin insulator to reduce the rate of gammaretroviral vector-mediated genotoxicity. Together with the demonstrated ability of this element to reduce the incidence of vector silencing, we believe these studies provide a strong impetus for the inclusion of this or similar chromatin insulators in integrating gene transfer vectors used for research and clinical applications.
Materials and Methods
Cell culture. All cell lines were maintained in Dulbecco's Modified Eagle's Medium with 10% heat-inactivated fetal bovine serum. Media for the murine myeloid bone marrow cell line 32D32 also contained 5% murine IL-3 culture supplement (BD Biosciences, Bedford, MA), except where noted. Colony assays were performed by plating 32D cells in methylcellulose base medium (catalog H4230; Stemcell Technologies, Vancouver, BC) at a dose of 1 × 105 to 4 × 105 cell/ml (without IL-3), and scoring colonies after 1–2 weeks. Mock-transduced cultures were always performed in parallel to control for background reversion rates.
Retroviral vectors. The gammaretroviral reporter vectors MGPN2 and INS4(+) (Figure 1a) have been previously described.14 Producer lines were generated using the amphotropic packaging line PA317 and the ecotropic packaging line GP+E86. Vector titers for matched producer clones were determined by the transfer of G418 resistance to naive NIH3T3 cells and were ~1 × 06 transducing units per ml for the amphotropic producer clones and ~2 × 105 for the ecotropic producer clones.
Retroviral vector transductions. The human fibrosarcoma cell line HT108048 was transduced by culturing 50,000 cells for 48 hours in amphotropic–pseudotyped virus supernatant containing 4 µg/ml polybrene at a multiplicity of infection (moi) of 10. The cells were then washed and replated at limiting dilution in the absence of selection. Discrete clones were subsequently isolated and expanded for further analysis. In order to reduce variability, all clones were generated and analyzed in parallel. Vector transduction had no measurable effect on the plating efficiency or expansion capacity of the HT1080 cell clones. The cell line 32D was transduced by culturing ~106 cells for 24 hours in ecotropic–pseudotyped virus supernatant containing 4 µg/ml polybrene at an moi of ~2-4. The cells were then washed extensively before further analysis.
Southern analysis. Genomic DNA prepared by column purification (DNA Midi kit, Qiagen, Valencia, CA) was digested with EcoRI, separated on 0.8% agarose gels, blotted onto nylon filters, probed with a radiolabeled 632 bp PstI fragment for Neo, and analyzed by PhosphorImager (Molecular Dynamics, Sunnyvale, CA). For studies in 32D cells, EcoRI cuts outside of the provirus for vector INS4(+) and cuts once inside of the provirus for vector MGPN2. For studies in HT1080 cells, the EcoRI site inside vector MGPN2 was disabled by site-directed mutagenesis so that EcoRI cuts outside both vectors. This was done strictly for cloning purposes and does not effect vector expression, titer, or genetic stability.46 In all cases, the number of provirus was determined by assessing both the number and intensity of independent bands for each clone.
Flow cytometry analysis. Transduced cell were analyzed for vector GFP expression by flow cytometry on a FACScan (Beckton Dickinson, Franklin Lakes, NJ) using CellQuest software. The fraction of GFP-positive cells was determined using specific gates and subtracting any background observed from untransduced controls (typically 0.0–2.0%). For the HT1080 cell-clone studies, the level (intensity) of GFP expression was determined as mean fluorescent units (mfu) for the cells within the specific gates used to determine the fraction of GFP-positive cells.
Integration site analysis. Vector integration sites in transduced HT1080 cell clones were identified by a variety of methods, including inverse PCR, linear amplification–mediated PCR, and, in some cases, with a directed DNA library screening approach. Vector integration sites in transduced 32D cell clones were identified by linear amplification–mediated PCR. Sequences were BLAST searched against either the human genome (March 2006 assembly) or the mouse genome (February 2006 assembly) using the UCSC Genome Browser (http://genome.ucsc.edu/) as previously described.30 See Supplementary Materials and Methods for details.
Expression microarray analysis. Codelink UniSet Human 20K I Bioarrays and gene expression system (Amersham/GE Healthcare Bio-Sciences, Piscataway, NJ) were used following the manufacturer's directions. A gene was considered to be dysregulated if the intensity of that gene's signal within any one cell clone was either five-fold higher or five-fold lower than the mean signal intensity for the remaining cell clones and untransduced controls, if the signal was considered reliable by the manufacturer's criteria, and if the signal was not found to be dysregulated in more than one clone. See Supplementary Materials and Methods for details.
Quantitative PCR analysis. Column-purified total RNA was reverse transcribed using the QuantiTec Reverse Transcription kit (Qiagen). Real-time PCR was performed (in triplicate) using the Fast Start DNA Master Mix SYBR Green I kit and LightCycler thermocycler (Roche, Indianapolis, IN), according to the manufacture's directions. For the HT1080 cell studies, primer pairs consisted of one oligonucleotide derived from the microarray trap sequence and a second oligonucleotide chosen to generate a 150–200 bp gene-specific amplicon. For the 32D cell studies, primer pairs were chosen from the 3′ portion of the candidate genes to generate 150–200 bp gene-specific amplicons. Signal intensities were normalized to an internal control (glyceraldehydephosphate dehydrogenase) and were compared to the normalized signal intensity from untransduced parental cells. For vector copy number analysis, column-purified DNA was analyzed by real-time PCR (in triplicate) essentially as described above using primer pairs specific to vector–GFP sequences. The level of vector provirus was determined by comparing normalized signals from the transduced pools to a single copy reference clone.
Mouse transplantation studies. 32D cells were split into independent pools immediately after transduction, expanded ~2 weeks with IL-3 in order to generate the requisite number of cells, and transplanted into congenic female C3H/HeJ mice by tail vein injection at doses of 107 cells per mouse (one independent pool per mouse, no myeloablation). Transplant recipients were monitored routinely and were euthanized following criteria established by our Institutional Animal Care and Use Committee. In essentially every case tumor-baring mice presented with palpable splenomegaly. Studies in mice were carried out in compliance with federal guidelines and institutional policies.
Statistical analysis. Supplementary Materials and Methods for details.
Website URLs. UCSC Genome Browser, http://genome.ucsc.edu. Mouse retroviral tagged cancer gene database, http://RTCGD.ncifcrf.gov. VizX Labs LLC GeneSifter microarray analysis software, http://www.genesifter.net/web.
Supplementary MaterialFigure S1. Southern copy number analysis. (A) Genomic DNA from transduced HT1080 cell clones was analyzed by digestion with Eco RI (which does not cut in either vector used in these studies), and Southern blot analysis using a probe for Neo. The clone numbers are shown at the top, and the estimated number of vector copies, reflecting the number of individual bands per lane on the Southern blot, are shown at the bottom. Clone numbers 74-101, vector MGPN2; clone numbers 224-239, vector INS4(+); n.a., clones not analyzed; Marker, size marker (shown in kb). See Supplementary Table 1 for results of analysis for complete panel of HT1080 cell clones. (B) Genomic DNA from transduced 32D cell clones was analyzed by digestion with Eco RI and Southern blot analysis using a probe for Neo. Eco RI cuts once in the version of the uninsulated MGPN2 vector used in these studies and does not cut in the insulated vector INS4(+). Left panel, uninsulated vector MGPN2; right panel, vector INS4(+). The clone numbers are shown at the top. Mock, untransduced control; G418pool, pools of clones selected with G418 but grown with IL-3; (sp), lines derived from enlarged spleens of mice transplanted with pools of transduced cells; Marker, size marker (shown in kb). The estimated numbers of vector copies are shown at the bottom. Southern copies, copy number based on the number of major bands and both major and minor bands (total) for each lane; LAM-PCR copies, number of copies cloned and mapped by 3-arm LAM-PCR.Figure S2. Vector GFP expression analysis. (A) Flow cytometric analysis of pools and single-copy clones of HT1080 cells transduced with either the uninsulated vector MGPN2 or the insulated vector INS4(+). The percentage of GFP-positive cells (pools only) and the level of GFP expression (all samples, in mean fluorescence units, mfu) are shown above the gates used for the analysis. Gray histograms, untransduced controls; open histograms with dark lines, experimental samples. (B) Flow cytometric analysis of 32D pools shortly after transduction (Experiment D, Supplementary Table 3). Scatter dot plots (forward light scatter versus GFP fluorescence) are shown for mock-transduced control, and pools transduced with either the uninsulated vector MGPN2 or the insulated vector INS4(+). The percentage of GFP-positive cells for the experimental samples was determined after subtracting background from mock-transduced control (0.1% in this case) using the indicated gates. (c) Flow cytometric analysis of transformed 32D clones (MGPN2 clones 9 and 25 and INS4(+) clones 30 and 45 from Supplementary Table 4). The level of GFP expression (in mean fluorescence units, mfu) are shown above the gates used for the analysis. Gray histograms, untransduced controls; open histograms with dark lines, experimental samples.Table S1. Basic characterization of transduced HT1080 clone panels.Table S2. Dysregulated genes in transduced HT1080 clone panels.Table S3. Details of 32D transduction studies.Table S3. Details of 32D transduction studies.Table S4. Characterization of transformed 32D clones.Table S5. Association between specific provirus and “suspect” genes in transformed 32D cell clones.Materials and Methods.
Supplementary Material
Southern copy number analysis. (A) Genomic DNA from transduced HT1080 cell clones was analyzed by digestion with Eco RI (which does not cut in either vector used in these studies), and Southern blot analysis using a probe for Neo. The clone numbers are shown at the top, and the estimated number of vector copies, reflecting the number of individual bands per lane on the Southern blot, are shown at the bottom. Clone numbers 74-101, vector MGPN2; clone numbers 224-239, vector INS4(+); n.a., clones not analyzed; Marker, size marker (shown in kb). See Supplementary Table 1 for results of analysis for complete panel of HT1080 cell clones. (B) Genomic DNA from transduced 32D cell clones was analyzed by digestion with Eco RI and Southern blot analysis using a probe for Neo. Eco RI cuts once in the version of the uninsulated MGPN2 vector used in these studies and does not cut in the insulated vector INS4(+). Left panel, uninsulated vector MGPN2; right panel, vector INS4(+). The clone numbers are shown at the top. Mock, untransduced control; G418pool, pools of clones selected with G418 but grown with IL-3; (sp), lines derived from enlarged spleens of mice transplanted with pools of transduced cells; Marker, size marker (shown in kb). The estimated numbers of vector copies are shown at the bottom. Southern copies, copy number based on the number of major bands and both major and minor bands (total) for each lane; LAM-PCR copies, number of copies cloned and mapped by 3-arm LAM-PCR.
Vector GFP expression analysis. (A) Flow cytometric analysis of pools and single-copy clones of HT1080 cells transduced with either the uninsulated vector MGPN2 or the insulated vector INS4(+). The percentage of GFP-positive cells (pools only) and the level of GFP expression (all samples, in mean fluorescence units, mfu) are shown above the gates used for the analysis. Gray histograms, untransduced controls; open histograms with dark lines, experimental samples. (B) Flow cytometric analysis of 32D pools shortly after transduction (Experiment D, Supplementary Table 3). Scatter dot plots (forward light scatter versus GFP fluorescence) are shown for mock-transduced control, and pools transduced with either the uninsulated vector MGPN2 or the insulated vector INS4(+). The percentage of GFP-positive cells for the experimental samples was determined after subtracting background from mock-transduced control (0.1% in this case) using the indicated gates. (c) Flow cytometric analysis of transformed 32D clones (MGPN2 clones 9 and 25 and INS4(+) clones 30 and 45 from Supplementary Table 4). The level of GFP expression (in mean fluorescence units, mfu) are shown above the gates used for the analysis. Gray histograms, untransduced controls; open histograms with dark lines, experimental samples.
Basic characterization of transduced HT1080 clone panels.
Dysregulated genes in transduced HT1080 clone panels.
Details of 32D transduction studies.
Characterization of transformed 32D clones.
Association between specific provirus and “suspect” genes in transformed 32D cell clones.
Acknowledgments
We thank G. Felsenfeld for providing the cHS4 insulator, R.G. Hawley for providing the MGPN2 vector, M.A. Harkey for providing the linear amplification-mediated PCR analysis service, T. Papayannopoulou for providing the 32D cells and for technical advice, and D.W. Russell for helpful discussions. This work was supported by grants from the National Heart, Lung, and Blood Institute, National Institutes of Health (D.W.E., RO1 HL75713; G.S., PO1 HL53750). All authors designed the research and wrote the paper. C.L.L., D.X., and D.W.E. performed the research and analyzed the data. None of the authors have a conflict of interest to declare.
REFERENCES
- Kohn DB, Sadelain M, Dunbar C, Bodine D, Kiem HP, Candotti F, et al. American Society of Gene Therapy (ASGT) ad hoc subcommittee on retroviral-mediated gene transfer to hematopoietic stem cells. Molec Ther. 2003;8:180–187. doi: 10.1016/s1525-0016(03)00212-0. [DOI] [PubMed] [Google Scholar]
- Baum C, von Kalle C, Staal FJ, Li Z, Fehse B, Schmidt M, et al. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol Ther. 2004;9:5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Nienhuis AW. Assays to evaluate the genotoxicity of retroviral vectors. Mol Ther. 2006;14:459–460. doi: 10.1016/j.ymthe.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dullmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Dullmann J, Kamino K, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Shou Y, Ma X, Lu T., and , Sorrentino BP. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc Natl Acad Sci USA. 2006;103:11730–11735. doi: 10.1073/pnas.0603635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia A, Bonini C, Magnani Z, Urbinati F, Sartori D, Muraro S, et al. Retroviral vector integration deregulates gene expression but has no consequences on the biology and function of transplanted T cells. Proc Natl Acad Sci USA. 2006;103:1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knoss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery DW. The SINister side of retrovirus vectors. Blood. 2006;108:2495. [Google Scholar]
- Emery DW, Yannaki E, Tubb J., and , Stamatoyannopoulos G. A chromatin insulator protects retrovirus vectors from position effects. Proc Natl Acad Sci USA. 2000;97:9150–9155. doi: 10.1073/pnas.160159597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nienhuis AW, Dunbar CE., and , Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Evans-Galea MV, Wielgosz MM, Hanawa H, Srivastava DK., and , Nienhuis AW. Suppression of clonal dominance in cultured human lymphoid cells by addition of the cHS4 insulator to a lentiviral vector. Mol Ther. 2007;15:801–809. doi: 10.1038/sj.mt.6300103. [DOI] [PubMed] [Google Scholar]
- West AG, Gaszner M., and , Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- Emery DW, Aker M., and , Stamatoyannopoulos G. Makrides SC. Gene Transfer and Expression in Mammalian Cells. EIC Laboratories: Norwood, MA; 2003. Chromatin insulators and position effects; pp. 381–395. [Google Scholar]
- Yannaki E, Tubb J, Aker M, Stamatoyannopoulos G., and , Emery DW. Topological constraints governing the use of the chicken HS4 chromatin insulator in oncoretrovirus vectors. Mol Ther. 2002;5:589–598. doi: 10.1006/mthe.2002.0582. [DOI] [PubMed] [Google Scholar]
- Emery DW, Yannaki E, Tubb J, Nishino T, Li Q., and , Stamatoyannopoulos G. Development of virus vectors for gene therapy of beta chain hemoglobinopathies: flanking with a chromatin insulator reduces gamma-globin gene silencing in vivo. Blood. 2002;100:2012–2021. doi: 10.1182/blood-2002-01-0219. [DOI] [PubMed] [Google Scholar]
- Rivella S, Callegari JA, May C, Tan CW., and , Sadelain M. The cHS4 insulator increases the probability of retroviral expression at random chromosomal integration sites. J Virol. 2000;74:4679–4687. doi: 10.1128/jvi.74.10.4679-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani A, Hawley TS., and , Hawley RG. Performance- and safety-enhanced lentiviral vectors containing the human interferon-beta scaffold attachment region and the chicken beta-globin insulator. Blood. 2003;101:4717–4724. doi: 10.1182/blood-2002-09-2991. [DOI] [PubMed] [Google Scholar]
- Yao S, Osborne CS, Bharadwaj RR, Pasceri P, Sukonnik T, Pannell D, et al. Retrovirus silencer blocking by the cHS4 insulator is CTCF independent. Nucleic Acids Res. 2003;31:5317–5323. doi: 10.1093/nar/gkg742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Bell AC., and , Felsenfeld G. Characterization of the chicken beta-globin insulator. Proc Natl Acad Sci USA. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, West AG., and , Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, et al. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu BY, Evans-Galea MV, Gray JT, Bodine DM, Persons DA., and , Nienhuis AW. An experimental system for the evaluation of retroviral vector design to diminish the risk for proto-oncogene activation. Blood. 2008;111:1866–1875. doi: 10.1182/blood-2007-04-085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove PW, Kepes S, Hanawa H, Obenauer JC, Pei D, Cheng C, et al. Globin lentiviral vector insertions can perturb the expression of endogenous genes in beta-thalassemic hematopoietic cells. Mol Ther. 2008;16:525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- Li CL., and , Emery DW. The cHS4 chromatin insulator reduces gammaretroviral vector silencing by epigenetic modifications of integrated provirus. Gene Ther. 2008;15:49–53. doi: 10.1038/sj.gt.3303009. [DOI] [PubMed] [Google Scholar]
- Aker M, Tubb J, Miller DG, Stamatoyannopoulos G., and , Emery DW. Integration bias of gammaretrovirus vectors following transduction and growth of primary mouse hematopoietic progenitor cells with and without selection. Mol Ther. 2006;4:226–235. doi: 10.1016/j.ymthe.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J., and , Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberger JS, Sakakeeny MA, Humphries RK, Eaves CJ., and , Eckner RJ. Demonstration of permanent factor-dependent multipotential (erythroid/neutrophil/basophil) hematopoietic progenitor cell lines. Proc Natl Acad Sci USA. 1983;80:2931–2935. doi: 10.1073/pnas.80.10.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald TJ, Henault S, Sakakeeny M, Santucci MA, Pierce JH, Anklesaria P, et al. Expression of transfected recombinant oncogenes increases radiation resistance of clonal hematopoietic and fibroblast cell lines selectively at clinical low dose rate. Radiat Res. 1990;122:44–52. [PubMed] [Google Scholar]
- Guo XY, Cuillerot JM, Wang T, Wang T, Wu Y, Arlinghaus R, et al. Peptide containing the BCR oligomerization domain (AA 1-160) reverses the transformed phenotype of p210bcr-abl positive 32D myeloid leukemia cells. Oncogene. 1998;17:825–833. doi: 10.1038/sj.onc.1201999. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Stein S, Schwarzwaelder K, Siler U, Koehl U, et al. Long-term clinical follow-up and safety/toxicity analysis in 3 X-CGD patients treated by gene therapy and non-myeloablative conditioning. Blood. 2006;108:63a. [Google Scholar]
- Akagi K, Suzuki T, Stephens RM, Jenkins NA., and , Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaozza E, Baker SJ, Vora RK., and , Reddy EP. AATYK: a novel tyrosine kinase induced during growth arrest and apoptosis of myeloid cells. Oncogene. 1997;15:3127–3135. doi: 10.1038/sj.onc.1201575. [DOI] [PubMed] [Google Scholar]
- Yuasa H, Oike Y, Iwama A, Nishikata I, Sugiyama D, Perkins A, et al. Oncogenic transcription factor Evi1 regulates hematopoietic stem cell proliferation through GATA-2 expression. EMBO J. 2005;24:1976–1987. doi: 10.1038/sj.emboj.7600679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkers H, Allen J, Knipscheer P, Knipscheer P, Romeijn L, Hart A, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32:153–159. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- Ryu BY, Persons DA, Evans-Galea MV, Gray JT., and , Nienhuis AW. A chromatin insulator blocks interactions between globin regulatory elements and cellular promoters in erythroid cells. Blood Cells Mol Dis. 2007;39:221–228. doi: 10.1016/j.bcmd.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Savitskaya E, Melnikova L, Kostuchenko M, Kravchenko E, Pomerantseva E, Boikova T, et al. Study of long-distance functional interactions between Su(Hw) insulators that can regulate enhancer-promoter communications in Drosophila melanogaster. Mol Cell Biol. 2006;26:754–761. doi: 10.1128/MCB.26.3.754-761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Jenkins NA., and , Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR., and , Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Chang AH, Stephan MT., and , Sadelain M. Stem cell-derived erythroid cells mediate long-term systemic protein delivery. Nat Biotechnol. 2006;24:1017–1021. doi: 10.1038/nbt1227. [DOI] [PubMed] [Google Scholar]
- Aker M, Tubb J, Groth AC, Bukovsky AA, Bell AC, Felsenfeld G, et al. Extended core sequences from the cHS4 insulator are necessary for protecting retroviral vectors from silencing position effects. Hum Gene Ther. 2007;18:333–343. doi: 10.1089/hum.2007.021. [DOI] [PubMed] [Google Scholar]
- Ramezani A, Hawley TS., and , Hawley RG. Stable gammaretroviral vector expression during embryonic stem cell-derived in vitro hematopoietic development. Mol Ther. 2006;14:245–254. doi: 10.1016/j.ymthe.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed S, Nelson-Rees WA, Toth EM, Arnstein P., and , Gardner MB. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Southern copy number analysis. (A) Genomic DNA from transduced HT1080 cell clones was analyzed by digestion with Eco RI (which does not cut in either vector used in these studies), and Southern blot analysis using a probe for Neo. The clone numbers are shown at the top, and the estimated number of vector copies, reflecting the number of individual bands per lane on the Southern blot, are shown at the bottom. Clone numbers 74-101, vector MGPN2; clone numbers 224-239, vector INS4(+); n.a., clones not analyzed; Marker, size marker (shown in kb). See Supplementary Table 1 for results of analysis for complete panel of HT1080 cell clones. (B) Genomic DNA from transduced 32D cell clones was analyzed by digestion with Eco RI and Southern blot analysis using a probe for Neo. Eco RI cuts once in the version of the uninsulated MGPN2 vector used in these studies and does not cut in the insulated vector INS4(+). Left panel, uninsulated vector MGPN2; right panel, vector INS4(+). The clone numbers are shown at the top. Mock, untransduced control; G418pool, pools of clones selected with G418 but grown with IL-3; (sp), lines derived from enlarged spleens of mice transplanted with pools of transduced cells; Marker, size marker (shown in kb). The estimated numbers of vector copies are shown at the bottom. Southern copies, copy number based on the number of major bands and both major and minor bands (total) for each lane; LAM-PCR copies, number of copies cloned and mapped by 3-arm LAM-PCR.
Vector GFP expression analysis. (A) Flow cytometric analysis of pools and single-copy clones of HT1080 cells transduced with either the uninsulated vector MGPN2 or the insulated vector INS4(+). The percentage of GFP-positive cells (pools only) and the level of GFP expression (all samples, in mean fluorescence units, mfu) are shown above the gates used for the analysis. Gray histograms, untransduced controls; open histograms with dark lines, experimental samples. (B) Flow cytometric analysis of 32D pools shortly after transduction (Experiment D, Supplementary Table 3). Scatter dot plots (forward light scatter versus GFP fluorescence) are shown for mock-transduced control, and pools transduced with either the uninsulated vector MGPN2 or the insulated vector INS4(+). The percentage of GFP-positive cells for the experimental samples was determined after subtracting background from mock-transduced control (0.1% in this case) using the indicated gates. (c) Flow cytometric analysis of transformed 32D clones (MGPN2 clones 9 and 25 and INS4(+) clones 30 and 45 from Supplementary Table 4). The level of GFP expression (in mean fluorescence units, mfu) are shown above the gates used for the analysis. Gray histograms, untransduced controls; open histograms with dark lines, experimental samples.
Basic characterization of transduced HT1080 clone panels.
Dysregulated genes in transduced HT1080 clone panels.
Details of 32D transduction studies.
Characterization of transformed 32D clones.
Association between specific provirus and “suspect” genes in transformed 32D cell clones.