Abstract
The herpes simplex virus (HSV) amplicon vector produces an initial host response that limits transgene expression. In this study, we hypothesized that restoration of the HSV gene infected cell protein (ICP0) into the amplicon could circumvent this host response and thus overcome silencing of encoded transgenes. To test this, we constructed an amplicon vector that encodes the ICP0 under control of its native promoter (ICP0+ amplicon). Expression of ICP0 was transient and, at a multiplicity of infection (MOI) of 1, did not significantly alter interferon (IFN)-based responses against the vector or cell kinetics/apoptosis of infected cells. Chromatin immunoprecipitation (ChIP) PCR analysis revealed that conventional amplicon DNA became associated with histone deacetylase 1 (HDAC1) immediately after infection, whereas ICP0+ amplicon DNA remained relatively unbound by HDAC1 for at least 72 hours after infection. Mice administered systemic ICP0+ amplicon exhibited significantly greater and more sustained transgene expression in their livers than did those receiving conventional amplicon, likely due to increased transcriptional or post-transcriptional activity rather than increased copy numbers of vector DNA. These findings indicate that restoration of ICP0 expression may be employed within HSV amplicon constructs to decrease transgene silencing in vitro and in vivo.
Introduction
The herpes simplex virus (HSV) amplicon vector possesses unique features for gene therapy studies, including (i) ability to accommodate up to 150 kbp of exogenous DNA;1,2,3 (ii) capacity for transduction of a wide variety of cell types across a broad range of species; (iii) relative ease and flexibility of vector construction;4 (iv) the availability of several hybrid amplicon vectors to achieve extended transgene expression;5,6,7,8,9,10 and (v) the limited cytotoxicity, which is due to the lack of viral coding sequences and availability of helper virus–free packaging systems.4,11,12 However, stability and long-term expression with this episomal construct can remain a challenge.
Mammalian hosts have evolved a variety of sensors, both cytoplasmic and membrane-associated, for viral infection. Activation of these “sensors” ultimately leads to the production of type I interferons (IFNs), such as IFN-α and IFN-β.13 As secreted factors, type I IFNs regulate a range of immune responses through their specific type I IFN receptor complex. Type I IFN stimulation leads to the transcription of >100 IFN-stimulated genes (ISGs), whose concerted action leads to the generation of an “antiviral state.”14 Recently, we investigated the stability of transgene expression and early host responses in mice after systemic delivery of an HSV amplicon.15 Immediately after vector administration, transgene expression was readily detected, primarily in the liver. However, transgene expression declined rapidly and was undetectable within 2 weeks. Type I IFNs signal transducers and activators of transcription 1 signaling were found to be a critical pathway, responsible for early silencing of vector-encoded transgene expression. This induction of type I IFNs initiated a cascade of immune responses and suppressed vector-encoded transgene expression at the transcriptional level via the activation of signal transducers and activators of transcription 1 and possibly through recruitment of histone deacetylases (HDACs) to vector DNA. Recent discoveries employing mutant HSV1 viruses and expression plasmids have shown that the HSV gene product, infected cell protein (ICP0), can dissociate HDAC from viral genes, leading to increased gene expression particularly at low multiplicities of infection.16,17 In addition, ICP0 interacts with several proteins and appears to perform multiple functions.18 ICP0 blocks IFN-dependent host responses designed to block viral replication.19,20,21,22 ICP0 is also a promiscuous transactivator of genes introduced by infection or transfection.16,17,23 In addition, ICP0 has been shown to possess E3 ubiquitin ligase activity24,25 and to block cell cycle progress.26,27,28,29
In the current study, we thus hypothesized that restoration of ICP0 into the HSV1 amplicon vector would overcome the cellular antiviral response leading to transcriptional silencing of a transgene. In fact, ICP0 expression did lead to significantly greater and more sustained transgene expression than that observed with a conventional amplicon in vitro and in vivo. In vitro, this was found to correlate with ICP0-mediated displacement of HDAC1 from vector DNA, thus inhibiting the rapid cellular-mediated transcriptional silencing of transgenes transduced by vector.
Results
ICP0 gene expression mediated by HSV amplicon
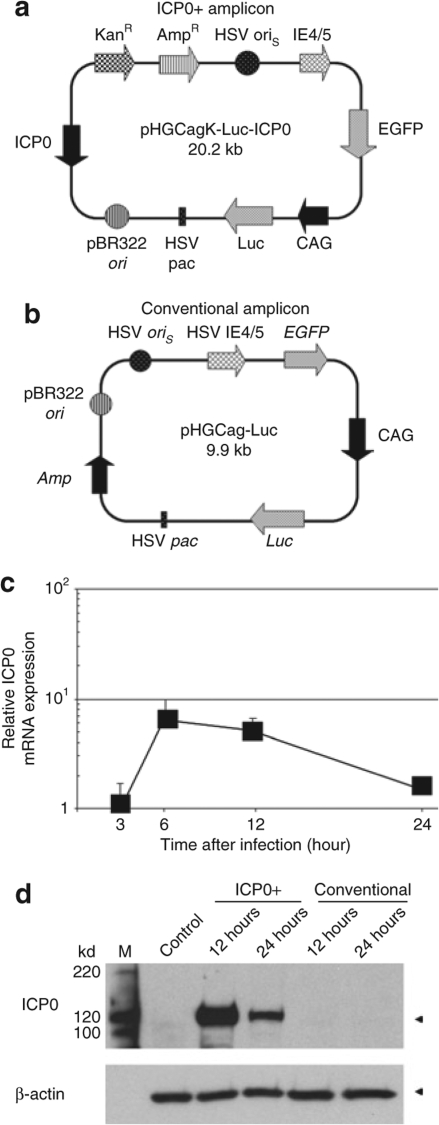
For the current study, an ICP0-encoding HSV amplicon plasmid, pHGCakgICP0-Luc (Figure 1a), was constructed and packaged into infectious HSV virions using a helper-free system (ICP0+ amplicon).11 A pBR322 backbone-containing conventional amplicon plasmid pHGCag-Luc (Figure 1b) was used as a control. To determine whether the native promoter of ICP0 regulated the expression of ICP0 in the context of the HSV amplicon vector system, the temporal kinetics of its mRNA were assayed in mouse embryonic fibroblast cells (MEFs) infected with ICP0+ amplicon (Figure 1c). Expression of ICP0 mRNA peaked at 6 hours after infection and gradually decreased to basal level by 24 hours. The temporal kinetics of ICP0 protein were also significantly increased in MEFs infected with ICP0+ amplicon 12 hours (and less so 24 hours) after infection of MEFs, whereas no ICP0 was detected in MEFs infected with conventional amplicon and control (Figure 1d). These results thus indicated that the ICP0 promoter derived from wild-type HSV was capable of regulating the expression of an ICP0 gene in the context of an amplicon vector with an initial peak of ICP0 expression after infection followed by gradual shutoff. Therefore, the effect of ICP0 in this construct would be expected to be transient.
Figure 1.
Expression of infected cell protein 0 (ICP0) in infected mouse embryonic fibroblast cells (MEFs). (a) Schematic representation of the ICP0+ amplicon vector plasmid pHGCakg-Luc-ICP0. The plasmid contains two transgene cassettes, the enhanced green fluorescent protein (EGFP) gene driven by the herpes simplex virus (HSV) IE4/5 promoter and firefly luciferase (Luc) gene under the control of the CMV-IE/chicken β-actin/rabbit β-globin (CAG) chimeric promoter. (b) Map of pHGCag-Luc amplicon vector plasmid used for control in this study. (c) Time course induction of ICP0 mRNA in MEFs infected with ICP0+ amplicon vector was determined by real-time reverse transcriptional PCR analysis. Each value was calculated relative to that of infected MEFs after 3 hours. Data are presented as mean ± SD (n = 3). (d) Western blot analysis of ICP0. MEFs infected with ICP0+ amplicon or conventional amplicon vector were harvested at 12 and 24 hours after infection and subjected to Western blotting. M, protein size marker; Control, untreated control MEFs sample. AmpR, ampicillin resistance gene; HSV oriS, HSV1 origin of replication; HSV pac, HSV1 packaging signal; KanR, kanamycin resistance gene; pBR322 ori, Plasmid pBR322 origin of replication.
Differential transgene expression in MEFs infected with the ICP0+ amplicon
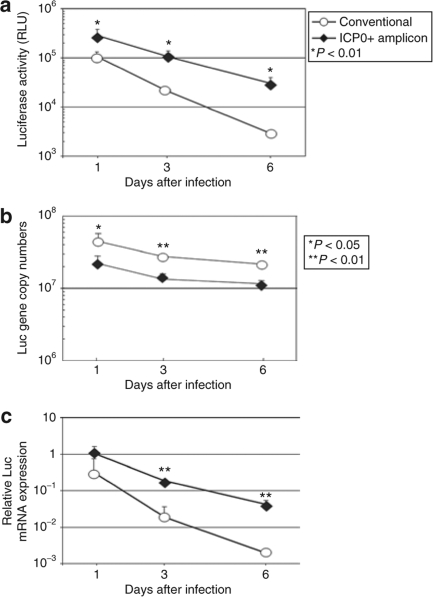
We first evaluated the time course of expression of a firefly luciferase (Luc) gene product in MEFs delivered by the two HSV amplicons. The temporal kinetics of Luc activity in MEFs infected with ICP0+ versus conventional amplicon was assayed. Luc activity of ICP0+ amplicon–infected MEFs was consistently and significantly higher than that of conventional amplicon-infected MEFs (Figure 2a). Luc activity in conventional amplicon-infected cells was only ~1/30 its original level by day 6, whereas that in cells infected with ICP0+ amplicon decreased more gradually to ~1/8 its original level by day 6. Similar findings were obtained using different doses of vector and also using nondividing human fibroblasts (data not shown), to show that the ICP0 effect was not dose dependent or species specific. The copy number of vector DNA for cells infected with conventional amplicon was approximately twofold that of cells infected with the ICP0+ amplicon, as a result of the size difference of their monomer DNA (9.9 kbp versus 20.2 kbp, respectively). Because each amplicon is packaged into the infectious virion particle (vp) as a concatamer of ~150–160 kb, more copies of the smaller conventional amplicon will be packaged into each infectious vp compared to the larger ICP0+ amplicon. In both cases, at least 50% of the originally introduced vector DNA was maintained at least until 6 days after infection (Figure 2b). At all time points, Luc mRNA expression was significantly higher in the ICP0+ amplicon-infected cells than in the conventional amplicon-infected cells (Figure 2c). These results suggested that vector DNA loss was not the only factor to account for the observed decline of Luc expression and that ICP0 expression provided increased retention of expression of the delivered Luc gene product.
Figure 2.
Time course of transgene expression in mouse embryonic fibroblast cells (MEFs). Primary cultured MEFs were infected with ICP0+ amplicon or conventional amplicon vector (multiplicity of infection = 1). Cells were harvested at various time points and subjected to (a) Luc activity assay and real-time PCR analysis of (b) Luc-encoding vector DNA as well as (c) messenger RNA transcripts. The data for cells infected with the conventional and ICP0+ amplicons are plotted as circles and diamonds, respectively (RLU, relative light units). Data are presented as mean ± SD (n = 4). ICP0, infected cell protein 0.
The expression of GFP mRNA in MEFs infected with ICP0+ amplicon also exhibited significant higher expression compared with that of MEFs infected with conventional amplicon (Supplementary Figure S1). There was a gradual decrease of GFP mRNA from day 1 to 6 day after infection, similar to what was observed with Luc mRNA. This suggested that transient expression of ICP0 led to enhanced transgene expression regardless of promoter activity (IE4/5 versus CAG).
The expression of IFN and ISG is altered after high, but not low doses of ICP0
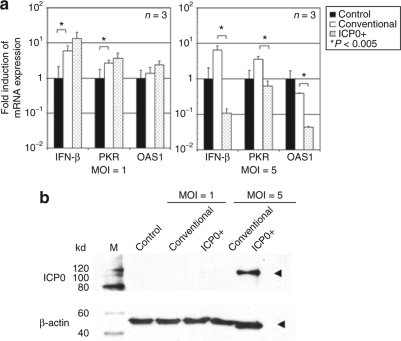
We sought to determine whether the observed increase in Luc gene expression by ICP0+ versus conventional amplicon could be explained by a differential response of IFN and ISGs in infected MEFs. MEFs were treated at an multiplicity of infection (MOI) of 1 or 5. Surprisingly, an MOI of 1 (MOI employed for the experiments of Figures 1 and 2) did not result in a change in the induction of IFN-β, IFN-inducible double stranded RNA protein kinase (PKR), or 2′, 5′-oligoadenylate synthetase 1 (OAS1) between ICP0+ versus conventional amplicons (Figure 3a). Only when the MOI was increased to 5, did we observe a significant reduction in the levels of IFN and ISG mRNAs (Figure 3a). In fact, 6 hours after infection, accumulation of ICP0 in cells was observed only in cells infected with an MOI of 5, but not with an MOI of 1 (Figure 3b). These results thus indicate that only at high MOI (and with ICP0 present at 6 hours) was there suppression of the ISG after amplicon infection. There was no evidence of DNA fragmentation of MEFs infected at an MOI of 5 compared with untreated control cells, 24 hours postinfection (Supplementary Figure S2). Cell cycle analysis failed to show changes induced by ICP0 expression at an MOI of 1. However, at an MOI of 5, there was a significant increase in apoptotic cells as well as a change in cell cycle kinetics (Supplementary Table S1). This result suggested that, at high MOI, transient and rapid expression of ICP0 suppressed ISGs, with an increase in apoptotic cells and changes in cell cycle kinetics, albeit without gross DNA fragmentation. However, at low MOI, more gradual onset of ICP0 expression did not lead to a change in IFN and ISGs in infected MEFs or to a change in cell cyle kinetics/apoptosis, although it did result in an increase in transgene expression.
Figure 3.
Interferon (IFN) responses to infection of ICP0+ amplicon or conventional amplicon vector in mouse embryonic fibroblast cells (MEFs). (a) Induction of IFN-β, IFN-inducible double stranded RNA protein kinase (PKR), and 2′,5′-oligoadenylate synthetase (OAS) mRNA 6 hours postinfection with ICP0+ amplicon or conventional amplicon vector (MOI = 1 or 5) were determined in MEFs by real-time RT-PCR analysis. Each value was calculated relative to that of untreated MEFs. Data are presented as mean ± SD (n = 3). Control, untreated control MEFs sample. *P < 0.005 comparing control to conventional amplicon and conventional amplicon to ICP0+ amplicon. (b) Western blot analysis of ICP0. MEFs infected with MOI = 1 or 5 of ICP0+ amplicon or conventional amplicon vector were harvested at 6 hours after infection and subjected to Western blotting. M, protein size marker; Control, untreated control MEFs sample. ICP0, infected cell protein 0; MOI, multiplicity of infection.
Presence of ICP0 leads to differential chromatin modification of amplicon DNA
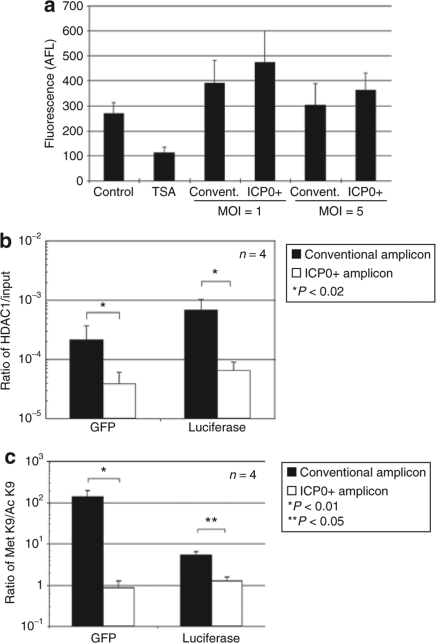
In order to determine whether ICP0 altered HDAC activity, we infected MEFs in vitro with ICP0+ or conventional amplicon. Figure 4a shows that MEFs infected with either amplicon vector exhibited similar levels of upregulation of HDAC activity, thus indicating that ICP0 did not suppress HDAC activity. ICP0 has recently been shown to interact with CoREST and to displace HDAC1 from the CoREST–REST–HDAC1/2 complex.17 In order to determine whether expression of ICP0 influences the chromatin status of amplicon vector DNA, chromatin immunoprecipitation (ChIP) PCR analysis was performed. DNA samples were harvested 24 hours after infection of MEFs with ICP0+ or conventional amplicon vector and immunoprecipitated with an antibody specific to HDAC1. The immunoprecipitated DNA–chromatin complexes were subjected to real-time PCR analysis using the enhanced green fluorescent protein (EGFP) and Luc-specific primers. The ratio of HDAC1-associated DNA to input DNA was calculated for each sample. Figure 4b shows that the HDAC1-associated DNA/input DNA ratio was significantly higher for both assayed GFP and Luc transgenes in control (Luc) amplicon–infected MEFs compared to ICP0+ (ICP0-Luc) amplicon-infected MEFs. DNA samples were also immunoprecipitated with an antibody specific to histone H3, methylated at lysine 9 (Met-K9-H3), or acetylated at lysine 9 (Ac-K9-H3). Met-K9-H3 is known to associate with transcriptionally inactive heterochromatic domains, whereas Ac-K9-H3 is known to associate with transcriptionally active euchromatic domains.30 The ratio of Met-K9-H3-associated DNA to Ac-K9-H3-asociated DNA was thus calculated (Figure 4c). Vector DNA in conventional amplicon-infected cells showed a significantly higher ratio of transcriptionally inactive chromatin compared with vector DNA in ICP0+ amplicon-infected cells at 24 hours. Taken in conjunction, this data thus suggest that ICP0 expression in the context of an amplicon vector leads to reduced transcriptionally inactive vector DNA chromatin by decreasing the association of HDAC1 to amplicon DNA
Figure 4.
Histone deacetylase (HDAC) activity and chromatin modification of transgenes after amplicon infection. (a) HDAC activity of mouse embryonic fibroblast cells (MEFs) infected with conventional amplicon or ICP0+ amplicon vector (MOI = 1 or 5) were measured, 24 hours after infection. Data are presented as mean ± SD (n = 3). Control, untreated control MEFs sample. The experiments were repeated twice with similar results. (b) Ratios of vector DNA to that with input DNA associated with HDAC1 were calculated 24 hours postinfection of MEFs with conventional amplicon or ICP0+ amplicon vectors. Data are presented as mean ± SD (n = 4). *P < 0.02 compared with the conventional amplicon vector samples. (c) Ratios of vector DNA associated with Met-K9-H3 to that associated with Ac-K9-H3 were calculated 24 hours postinfection. Data are presented as mean ± SD (n = 3). **P < 0.01 and *P < 0.05 compared with conventional amplicon-infected cells. GFP, green fluorescent protein; ICP0, infected cell protein 0; MOI, multiplicity of infection; TSA, trichstatin A.
Differential transgene expression in mice injected with ICP0+ or conventional amplicon
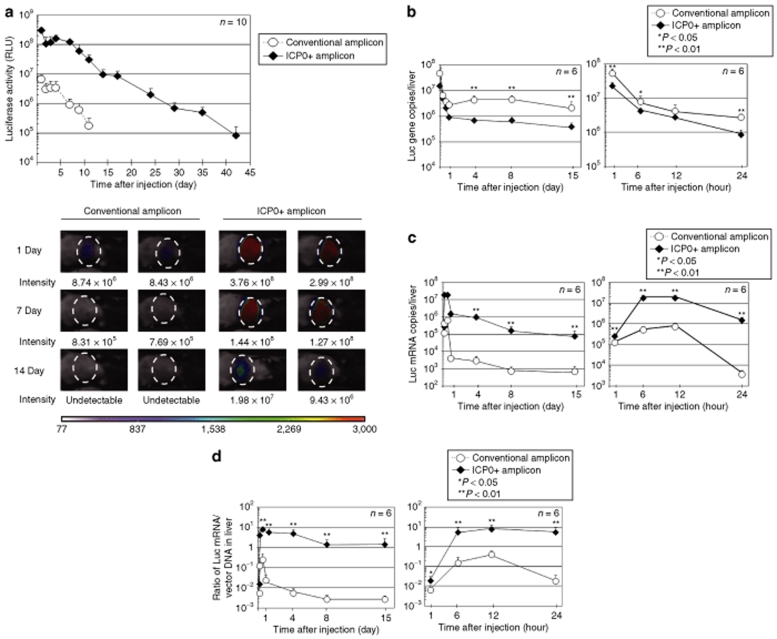
The above results were pertinent to in vitro cultured MEFs. We next evaluated ICP0+ and conventional amplicon vectors for their transgene expression in vivo. An injection of 1 × 107 TU of each amplicon was made into the tail veins of adult C57Bl/6 mice (day 0) and in vivo Luc activity was measured. Notably, at day 1, an ~50-fold higher level of Luc activity was detected in ICP0+ amplicon-injected mice than in conventional amplicon-injected mice (Figure 5a). Most of the expression was detected almost exclusively in liver for either amplicon with very little expression in other organs (lower panel of Figure 5a). Luc expression in mice injected with conventional amplicon started to decline by day 6 and became undetectable by day 14, whereas that of mice injected with ICP0+ amplicon remained detectable at least until day 42. As expected, there was an initial surge of expression of ICP0 mRNA levels, peaking at 12 hours and then gradually declining to almost basal levels by day 4 (data not shown). These results indicate that ICP0 plays a significant role in enhancing both initial and long-term transgene expression in the mouse model of systemic amplicon delivery. In order to delineate the effects of ICP0 on the amplicon vector–mediated transgene expression in vivo, we evaluated the time course of vector copies and transgene mRNA copies in the liver of mice. Whole cellular DNA as well as total RNA were isolated from the livers of the ICP0+ or conventional amplicon–injected mice at several time points (1, 6, 12, 24 hours, and day 4, day 8, and day 15 after injection). The copy number of Luc-encoding vector DNA was determined in each DNA sample by real-time PCR analysis (Figure 5b). Conventional amplicon vector DNA number was significantly greater than ICP0+ amplicon DNA copy number. This was consistent with the in vitro finding and was due to the larger number of delivered conventional amplicon DNA per vp. These results indicate that temporal changes in vector DNA copy number in liver were not the likely culprit for the differences in Luc activities between ICP0+ and conventional amplicon-injected mice. We then measured the amounts of Luc mRNA in the same liver samples by real-time RT-PCR. As shown in Figure 5c, mice injected with ICP0+ amplicon revealed increased Luc mRNA, at least by two orders of magnitude, than mice injected with conventional amplicon vector. After an initial increase in Luc mRNA in mice injected with ICP0+ amplicon, levels decreased gradually from 12 hours to day 15 to ~105 copies/liver, whereas in mice injected with conventional amplicon, Luc transcription declined by 99% by 24 hours to <104 copies/liver after injection and were <103 copies/liver by day 15 (Figure 5c). The ratio of Luc mRNA/vector DNA in the liver of amplicon-treated mice was upregulated from 1 to 24 hours and then remained relatively stable over the remaining 14 days of the assay for both ICP0+ and conventional amplicon-treated (Figure 5d).
Figure 5.
In vivo time course of transgene expression in C57Bl/6 mice. (a) Wild-type C57Bl/6 mice were systemically injected via teil vein with 1 × 107 TU of ICP0+ amplicon or conventional amplicon vector and their Luc expression was monitored using a NightOWL LB981 bioluminescence imaging system. Total Luc activity in each animal was evaluated by calculating the integration of photon counts obtained from the liver over the course of 5 minutes. Total photon counts were calculated from the acquired images using the CMIR-Image program. The data for animals injected with the conventional and ICP0+ amplicons are plotted as circles and diamonds, respectively (RLU: relative light units). Data are presented as mean ± SD (n = 10). The animals were sacrificed at various time points, and the copy numbers of (b) Luc-encoding vector DNA and (c) mRNA transcripts present in their livers were determined by real-time PCR analyses. (d) The ratio of Luc mRNA copies to vector DNA copies was calculated at each time points and plotted. The data for animals injected with the conventional and ICP0+ amplicons are plotted as circles and diamonds, respectively. Data are presented as mean ± SD (n = 6). ICP0, infected cell protein 0. CMIR, Center for Molecular Imaging Research.
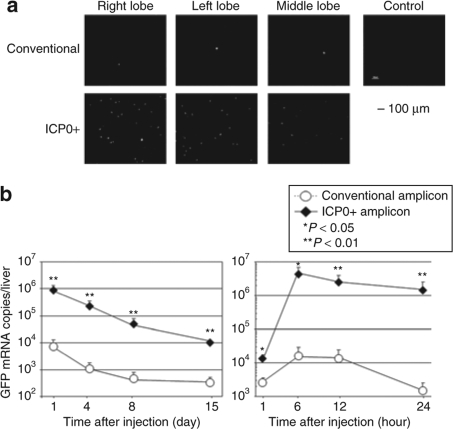
A similar result was obtained when GFP expression (under control of the IE4/5 promoter, see Figure 1) was visualized in the liver of mice injected with the ICP0+ versus conventional amplicon, 24 hours postinjection (Figure 6a). GFP mRNA in the liver of ICP0+ amplicon-injected mice was one to two orders of magnitude higher than that of conventional amplicon-injected mice over the 15 days of the experiment (Figure 6b). These results indicated that ICP0 increased expression of both delivered transgenes in vivo.
Figure 6.
GFP transgene expression under the control of IE4/5 promoter in C57Bl/6 mice. (a) Wild-type C57Bl/6 mice were systemically injected with 1 × 107 TU of ICP0+ amplicon or conventional amplicon vector. Twenty-four hours later, animals were sacrificed and their livers were harvested to evaluate GFP expression. Bar = 100 µm. Control, untreated control liver sample. (b) The animals were sacrificed at various time points and the copy number of GFP mRNA present in their liver was determined by real-time PCR analyses. The data for animals injected with the conventional and ICP0+ amplicons are plotted as circles and diamonds, respectively. Data are presented as mean ± SD (n = 6). GFP, green fluorescent protein; ICP0, infected cell protein 0.
Finally, we performed a dose–response experiment where increasing doses of the ICP0+-amplicon (including the dose of 1 × 107 TU employed in Figures 5 and 6 to detect prolonged gene expression) was injected systemically in mice, and IFN, PKR, and OAS responses were evaluated. Supplementary Figure S3 shows that ICP0 did not block the induction of these antiviral responses, and, in fact, these antiviral IFN responses were significantly increased. Thus, we concluded that ICP0's action in vivo did not appear to block the usual antiviral response of the host, suggesting that other mechanisms (such as decreased transgene silencing) may be operative in the increased expression of transgenes shown in Figures 5 and 6.
Discussion
In this study, we hypothesized that ICP0 would increase the duration and levels of expression of transgenes delivered by an HSV1 amplicon vector. In fact, we were able to show that (i) ICP0 in the context of an amplicon did mediate a significant increase in the duration and levels of transgene expression in vitro and in vivo, primarily by retarding gradual loss of transgene mRNA, when compared to conventional amplicon; (ii) in vitro, although amplicon treatment led to the expected increase in IFN and ISG gene responses, the addition of ICP0 did not lead to a reduction of these responses in a significant fashion at low MOI (of 1), and in vivo systemic ICPO+-amplicon (at 1 × 107 TU) did not block the induction of ISG genes in liver in spite of prolonged transgene expression; (iii) however, ICP0 did provide dissociation of HDAC from the transgenes (Luc and GFP), providing the most likely reason for the observed increase in transgene expression.
In vitro, the experiments of Figure 2 showed that the time-course profiles of decrease in Luc activity and Luc mRNA were similar, indicating that ICP0 was unlikely to be affecting Luc activity in MEFs at the translational level. In fact, the relative stability of Luc DNA copy number coupled with the decrease in Luc mRNA suggested that ICP0-mediated regulation occurred at the transcriptional or post-transcriptional level. In addition, the relative difference in the slopes of the lines for Luc mRNA in ICP0+ versus conventional amplicon-infected cells suggested that one of the actions of ICP0 was to maintain transcription of the Luc transgene and that this effect persisted for some time even after ICP0 expression had returned to baseline consistent with a mechanism that involved dissociation of HDAC-mediated silencing of the transgene (as shown in Figure 4). In vivo, the situation appeared to be similar: The experiments of Figures 5 and 6 showed that there was increased maintenance of both Luc and GFP mRNA levels in ICP0+ amplicon-injected mice (in spite of less gene copy numbers), which could have been due to a combination of both transcriptional and post-transcriptional mechanisms. This led to significantly higher and prolonged expression of the reporter transgene in livers.
ICP0 is a multifunctional protein and affects multiple cellular and viral processes.18,19,20,24,26,27,28,29,31,32,33 Interestingly, in our experiments, ICP0 expression in vitro reduced the expected increase in levels of type 1 IFN, PKR, or OAS upon cellular infection only at an MOI of 5 but not at an MOI of 1. In addition, changes in cell cycle kinetics occurred only at an MOI of 5 and not at an MOI of 1, thus indicating that this was not a reason for the observed increase in transgene expression. Because the in vitro experiments of Figures 2 and 4 were performed at an MOI of 1, this may suggest that, at low dose, ICP0 functioned by relieving transcriptional silencing of the transgene, while higher doses of ICP0 were required to affect the cellular antiviral IFN response. Similarly, in vivo, systemic injection of the ICP0+ amplicon at a dose of 1 × 107 TU led to increased transgene expression in liver without abrogating the expected increase in IFN and ISGs.
Our data are in agreement with the recent discovery that ICP0 leads to dissociation of HDAC1 from DNA by binding to CoREST thus displacing it from the REST/HDAC1 transcriptional repressor.16,17 Although the study showed that ICP0 can provide an avenue for securing longer-term transgene expression from the amplicon, transgene expression still gradually became reduced with time. Our data suggest that the lack of ICP0 after 24 hours (due to the use of the endogenous ICP0 promoter) probably led to the reassociation of HDAC with transgenes causing the continued decline in expression. This data thus suggest that future constructs may want to employ promoters that would allow for continued expression of ICP0, although there may be a possible concern for cytotoxicity if expression of ICP0 was elevated.34 In fact, studies have shown that ICP0 expression within recombinant virus may be cytotoxic and that deletion of ICP0 allowed for prolonged expression in cells.35,36 The differences may be due to vector type (amplicon versus recombinant vector). The product of the viral gene Us3 has also been shown to mediate increased expression of transgenes by phosphorylation of HDACs and thus future constructs may require this gene as well for more prolonged transgene expression.23,37
Achieving long-term gene expression in an organ outside the central nervous system using extrachromosomal vectors has been elusive.38 Episomal vectors possess a reduced risk of causing secondary oncogenic events through random integration in the host chromosome. The in vivo studies of this report where systemic administration of the vector was employed, provide relatively significant evidence of ICP0's action on reducing the rate of loss of transgene expression in liver. This provides a significant advance over current technology. Previously, we showed that transient transgene expression occurred after systemic administration of a 17.1 kb HSV amplicon vector encoding β-galactosidase under the control of an IE4/5 promoter and Luciferase under the control of the CAG promoter.15 This amplicon vector did not encode ICP0 sequences. Luciferase activity in mice injected with this amplicon vector became undetectable within 2 weeks. It is noteworthy that this time frame is similar to what was observed with the conventional 9.9 kb in this study, but significantly less in duration than what was observed with the 20.2 kb ICP0+ amplicon. Therefore, comparing the experiments reported in this study with our previously published study lead us to the conclusion that ICP0 function, rather than plasmid size or ICP0 sequences, was the critical variable for increasing duration of transgene expression.
Another consideration related to transgene expression is the recent discovery that microRNAs are produced in an antisense configuration to ICP0.39 In fact, miR-H2-3p is the mature form of the latency associated transcript known to downregulate ICP0 expression during neuronal latency. However, the ICP0 transcriptional cassette in the ICP0+ amplicon does not contain the entire sequence that allows expression of this latency associated transcript.
The reasons for ICP0 action in vivo may be more complex than in vitro. In fact, 1 and 6 hours after injection at the dose tested for transgene expression (1 × 107 TU) (Supplementary Figure S3), there actually was an increase in IFN or ISG responses when compared to animals injected with lower doses. In addition, the ICP0+ amplicon did not change the magnitude or time frame of neutralizing humoral immunity to the vector (data not shown). Because ICP0 in this context did not decrease IFN/ISG and because these increased IFN/ISG responses would be expected to diminish transgene expression, these findings suggest that ICP0 in vivo may also act primarily through dissociation of HDAC1 from transgene transcription sites, though the sheer complexity of performing in vivo ChIP assays have rendered it difficult to provide this evidence. However, we cannot exclude at present that in vivo loss of transgene expression may also occur by immune-mediated clearance of transgene-expressing cells or vector DNA loss. In addition, it is possible that HSV amplicon infection of phagocytic and/or antigen-presenting cells may also be a contributor to immunogenicity and transgene loss.40 The higher expression of transgene products in ICP0+ amplicon-injected mice versus conventional amplicon may induce a stronger innate and adaptive immune response. The transgenes used in this study are reporter transgenes (Firefly Luciferase and Green Fluorescent Protein) that unfortunately are immunogenic and thus unlikely to be expressed long-term,41,42 though there is a report that GFP is poorly immunogenic in C57/Bl6 mice.43 Nevertheless, decays in long-term transgene expression would be better studied using a nonimmunogenic mammalian (murine) gene.44 Future experiments will hopefully enable us to answer issues related to long-term maintenance of transgene expression.
In summary, the studies in this report provide evidence that inclusion of ICP0 mediates improved expression of foreign transgenes in vitro and in vivo, thus providing a framework for future advances in the area.
Materials and Methods
The following topics are discussed in Supplementary Materials and Methods section: plasmid construction, cell culture, and vector production, preparation of MEFs, tail-vein injection of HSV amplicon vectors into mice, in vivo bioluminescence imaging studies, histological analyses, and statistical analyses.
Luciferase activity assay in vitro. MEF cells were seeded at 1 × 105/well in 24-well plates. After 24 hours, MEF were infected with 1 × 105 TU of the HSV amplicon vectors in 250 µl Dulbecco's modified Eagle's medium (DMEM) supplemented with 2% fetal bovine serum (FBS). After 1 hour of incubation, the cells were washed with HBSS and fed with 500 µl of fresh DMEM supplemented with 2% FBS. At various time points (day 1, day 3, and day 6 after infection), each well of cells were washed with Dulbecco's phosphate-buffered saline (PBS), and 500 µl of Passive Lysis Buffer (Promega) was added. After a freeze-thaw cycle, the cell extracts were transferred into 1.5-ml Eppendorf tubes and centrifuged at 7,000g for 3 minutes at 4 °C. The resulting supernatant was measured for luciferase activity using the Firefly Luciferase Assay Reagent (Promega, Madison, WI) and an Auto Lumat LB953 luminometer (Berthold Technologies, Oak Ridge, TN).
Quantitative PCR analysis of transgene-encoding vector DNA and mRNA expression in vitro. For DNA extraction MEF cells were seeded at 1 × 105 cells/well in 12-well plates. After 24 hours, MEF were infected with 1 × 105 TU of the HSV amplicon vectors (MOI = 1) in 1500 µl DMEM supplemented with 2% FBS. For RNA extraction MEF cells were seeded at 2 × 105/well in 6-well plates. After 24 hours, MEF were infected with 2 × 105 TU of the HSV amplicon in 1 ml DMEM supplemented with 2% FBS. After 3 hours of incubation, the cells were washed with HBSS and fed with 1 ml of fresh DMEM supplemented with 2% FBS. At various time points (3, 6, 12 hours, and day 1, day 3, and day 6 after infection), each well of cells were washed with PBS. Total DNA was extracted using QIAamp DNA mini kit (Qiagen, Valencia, CA), and total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). Nucleotide concentration of each sample was measured using a DU530 spectrophotometer (Beckman Coulter, Fullerton, CA). RNA samples were treated with TURBO DNA-free Kit (Ambion, Austin, TX) and then reverse transcribed into complementary DNA (cDNA) using the Superscript II first-strand cDNA synthesis system (Invitrogen, Carlsbad, CA). DNA and cDNA samples were subjected to quantitative real-time PCR analysis (10 minutes at 95 °C and then 40 cycles of 15 seconds at 94 °C, 30 seconds at 60 °C, and 1 minute at 72 °C) using an ABI PRISM 7500 sequence detection system, Power SYBR Green PCR Master Mix (Applied Biosystems, Foster, CA), and the Luciferase-specific primers (5′-GGAATCCATCTTGCTCCAAC-3′ and 5′-TCTCTC TGATTTTTCTTGCGTC-3′). GFP-specific primers were same as that used for in vivo experiments. Total Luciferase copy numbers were calculated from the standard curve generated with the pHGCag–Luc DNA.
Quantitative real-time RT-PCR analysis of IFN, PKR, and OAS. MEFs were infected with MOI = 1, 5 of ICP0+ or conventional amplicon vector as described above. After 6 hours, cells were washed with PBS, and total RNA was extracted from each well using the TRIzol reagent. First-strand cDNA was synthesized from RNA samples and subjected to quantitative real-time PCR analysis as described above. The following sequences of PCR primers were used for the analysis:
5′-CAGCTCCAAGAAAGGACGAACATTC-3′ and 5′-AGCTCTTC AAGTGGAGAGCAGTTG A-3′ for IFN-β;
5′-GTGATACAAGTCGATACAAAACCCG-3′ and 5′-AATAACTG TTCTGGACTCATGTATT-3′ for PKR;
5′-CATGAGAGACGTTTTAGAGTCAAGTTTG-3′ and 5′-GCAAT GGCTTCCCCAGCTTCTCCTT-3′ for OAS1.
All sets of the primers were verified for specific amplification of their target DNA fragments. As an internal control, the mRNA level of glyceraldehyde-3-phosphate dehydrogenase was determined using specific primers (5′-AGCCTCGTCCCGTAGACAAAAT-3′ and 5′-GAAG ACACCAGTAGACTCCAC GAC AT-3′).
Immunoblot analysis of ICP0. MEF cells were seeded at 2 × 105/well in 6-well plates. After 24 hours, MEF were infected with 1 × 106 TU (MOI = 5) of the HSV amplicon vectors in 1500 µl DMEM supplemented with 2% FBS. After 6, 12, and 24 hours of incubation, each well of cells were washed with PBS, and 500 µl of Sample Buffer containing 62.5 mmol/l Tris-HCl (pH 6.8), 1% sodium dodecyl sulfate and 25% Glycerol was added. The cell extracts were transferred into 1.5-ml Eppendorf tubes, homogenized through 19G 1 1/2 needle, and centrifuged at 20,000g for 30 minutes at 4 °C. The concentrations of protein samples were measured with BCA Protein Assay Kit (PIERCE, Rockford, IL). The protein extracts were boiled in Laemmli sample buffer containing 0.01% Bromophenol Blue and 5% mercaptoethanol, loaded onto 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, and transferred to polyvinylidene difluoride membranes (Millipore, Billeria, MA). The blotted membranes were then incubated with a mouse monoclonal anti-HSV1 ICP0 antibody (Santa Cruz Biotechnology, Santa Cruz, CA). HSV1-ICP0 protein was detected using the ECL Plus detection kit (GE Healthcare, Waukesha, WI) as recommended by the manufacturer. After removing anti-HSV1 ICP0 antibody with Restore Western Blotting Stripping Buffer (PIERCE), the same membrane was incubated with a mouse monoclonal anti-β-actin antibody (Sigma, Saint Louis, MI).
In vitro HDAC activity assay. HDAC activity in infected cells was measured using the HDAC assay kit (Upstate, Charlottesville, VA). MEFs were seeded at subconfluency in 6-well plates and infected with the packaged pHGCakg-Luc-ICP0 amplicon vector (ICP0+ amplicon) or pHGCag-Luc amplicon vector (conventional amplicon) as described above. After 24 hours, HDAC activity was measured using the HDAC assay kit (Upstate) as recommended by the manufacturer. Briefly, infected cells were harvested with RIPA buffer. The supernatant of samples were mixed with HDAC assay substrate and incubated at 37 °C for 40 minutes. The samples were added the activator solution and incubated room temperature for 15 minutes. The resulting sample was measured the excitation at 355 nm and emission at 440 nm with FLUOstar OPTIMA microplate fluorometer (BMG labtech, Durham, NC). The HDAC activity of each sample (AFL) were calculated from the standard curve.
In vitro ChIP assay. ChIP PCR assay was performed using the ChIP assay kit (Upstate, Charlottesville, VA) as described previously.15,45 MEF cells were seeded at 100% confluence in 10-cm dishes (3 × 106 cells/plate) and infected with HSV amplicon vectors (1.5 × 107 TU/plate). After 24 hours postinfection, the ChIP assay was performed using the ChIP Assay Kit (Upstate, Charlottesville, VA) with anti-K9-dimethylated H3 and anti-K4–dimethylated H3 antibodies (Upstate) as recommended by the manufacturer. Briefly, chromatin was cross-linked with 1% formaldehyde for 10 minutes at room temperature. After quenching with 100 mmol/l glycine, the cells were washed and resuspended in sodium dodecyl sulfate lysis buffer and then sonicated. The soluble chromatin in the supernatant was diluted in ChIP dilution buffer and precleared by incubating with protein A-agarose/salmon sperm DNA slurry. Antibodies were added to the precleared supernatant and immunoprecipitated overnight at 4 °C. After washing, antibody-bound histone–DNA complex was eluted from the agarose beads and histone–DNA crosslinks were reversed by heating to 65 °C for 4 hours. The immunoprecipitated DNA from each sample was subjected to quantitative PCR analyses using a 7500 Real-Time PCR System and the POWER SYBR GREEN PCR Master Mix (Applied Biosystems). PCR amplification was performed in the following conditions: 15 minutes at 95 °C, 40 cycles of 1 minute at 94 °C, 30 seconds at 60 °C and 1 minute at 72 °C. The following primer sets were used for the assay:
5′-GGAATCCATCTTGCTCCAAC-3′ and 5′-TCTCTCTGATTTTT CTTGCGTC-3′ for Luciferase;
5′-GCGACGTAAACGGCCACAAGTTCAG-3′ and 5′-GTCGTCCT TGAAGAAGATGGTGCGC-3′ for GFP.
The DNA of amplicon plasmid pHGCag-Luc was used as control standard.
DNA fragmentation and cell cycle analysis. MEF cells were seeded at 1 × 106 cells/well in 6-well plates. After 24 hours, MEF were infected with 5 × 106 TU of the HSV amplicon vectors in 1 ml DMEM supplemented with 2% FBS. After 3 hours of incubation, cells were washed with PBS and 1 ml of fresh DMEM with 2% FBS was added. Twenty-four hours after infection, each well was washed with PBS and cells were harvested with 20 µl of cell lysis buffer containing 50 mmol/l Tris (pH 7.8), 10 mmol/l EDTA (pH 8) and 0.5% sodium dodecyl sulfate. Cell extracts were loaded onto 0.6% agarose gels and electrophoresed. DNA was detected with Ethidium Bromide staining. For cell cycle analyses, murine embryonic fibroblasts (106 cells per 6-cm dish) were incubated overnight in DMEM + 10F medium. The following day, the ICP0+ or conventional amplicons were added at MOIs of 1 or 5. The following day, cells were fixed with 70% ethanol, after washing and resuspension in PBS. After low-speed centrifugation, the cell pellet was resuspended again in PBS and recentrifuged at low speed once more, before suspension in PBS (0.25 ml). To extract low molecular weight DNA, 0.2 ml of DNA extraction buffer (0.2 M Na2HPO4; 0.1 M citric acid, pH 7.8) was added. After low-speed centrifugation, the pellet was resuspended in propidium iodide staining solution before FACS.
Quantitative PCR analysis of transgene-encoding vector DNA and mRNA expression in vivo. Mice were injected with 1 × 107 TU (or other indicated doses) of the packaged pHGCakg-Luc-ICP0 amplicon vector (ICP0+ amplicon) or pHGCag-Luc amplicon vector (conventional amplicon) as described above. At defined time points (1, 6, 12, 24 hours, and day 4, day 8, day 15 day postinjection), mice were euthanized by CO2 inhalation, and liver was harvested and homogenized. Total DNA was extracted from each liver using QIAamp DNA mini kit (Qiagen, Valencia, CA), and total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). Nucleotide concentration of each sample was measured using a DU530 spectrophotometer (Beckman Coulter, Fullerton, CA). RNA samples were treated with TURBO DNA-free Kit (Ambion, Austin, TX) and then reverse transcribed into cDNA using the Superscript II first-strand cDNA synthesis system (Invitrogen). DNA and cDNA samples prepared from each liver were subjected to quantitative real-time PCR analysis (10 minutes at 95 °C and then 40 cycles of 15 seconds at 94 °C, 30 seconds at 60 °C, and 1 minute at 72 °C) using an ABI PRISM 7500 sequence detection system, Power SYBR Green PCR Master Mix (Applied Biosystems, Foster, CA). The following sequences of PCR primers were used for the analysis:
5′-GGAATCCATCTTGCTCCAAC-3′ and 5′-TCTCTCTGATTTTT CTTGCGTC-3′ for Luc;
5′-GCGACGTAAACGGCCACAAGTTCAG-3′ and 5′-GTCGTCCT TGAAGAAGATGGTGCGC-3′ for EGFP.
Total Luc and EGFP copy numbers were calculated from the standard curve generated with the Luc, EGFP-encoding pHGCag-Luc plasmid DNA.
Supplementary MaterialFigure S1. Time course of GFP mRNA expression in MEFs. Primary cultured MEFs were infected with ICP0+ amplicon or conventional amplicon vector (MOI=1). Cells were harvested at various time points and subjected to real-time polymerase chain reaction analysis of GFP messenger RNA transcripts. The data for cells infected with the conventional and ICP0+ amplicons are plotted as ○ and ♦, respectively. Data are presented as mean ± SD (n=4). *P <0.01 compared with conventional amplicon-infected cells.Figure S2. DNA fragmentation of MEFs, 24 hrs post-infection. Primary cultured MEFs were infected with ICP0+ amplicon or conventional amplicon vector (MOI=5). After 24 hours, cells were harvested and electrophoresed. DNA was detected with Ethidium Bromide staining. M, DNA size marker; Control, untreated control MEFs sample.Figure S3. Interferon and ISG responses to infection of ICP0+ amplicon in liver of C57Bl/6 mice. Wild-type C57Bl/6 mice were intravenosuly injected with the indicated escalating dose of ICP0+ amplicon vector. After 6 h, induction of IFN and ISG mRNA was determined in the liver of C57Bl/6 mice by real-time RT-PCR analysis. Each value was calculated relative to that of un-injected mice. Data are presented as mean ± SD (n=6). *P<0.0001 compared with mice injected with 1 x 106 TU. **P <0.0001 compared with mice injected with 5 x 106 TU.Table S1.Materials and Methods.
Supplementary Material
Time course of GFP mRNA expression in MEFs. Primary cultured MEFs were infected with ICP0+ amplicon or conventional amplicon vector (MOI=1). Cells were harvested at various time points and subjected to real-time polymerase chain reaction analysis of GFP messenger RNA transcripts. The data for cells infected with the conventional and ICP0+ amplicons are plotted as ○ and ♦, respectively. Data are presented as mean ± SD (n=4). *P <0.01 compared with conventional amplicon-infected cells.
DNA fragmentation of MEFs, 24 hrs post-infection. Primary cultured MEFs were infected with ICP0+ amplicon or conventional amplicon vector (MOI=5). After 24 hours, cells were harvested and electrophoresed. DNA was detected with Ethidium Bromide staining. M, DNA size marker; Control, untreated control MEFs sample.
Interferon and ISG responses to infection of ICP0+ amplicon in liver of C57Bl/6 mice. Wild-type C57Bl/6 mice were intravenosuly injected with the indicated escalating dose of ICP0+ amplicon vector. After 6 h, induction of IFN and ISG mRNA was determined in the liver of C57Bl/6 mice by real-time RT-PCR analysis. Each value was calculated relative to that of un-injected mice. Data are presented as mean ± SD (n=6). *P<0.0001 compared with mice injected with 1 x 106 TU. **P <0.0001 compared with mice injected with 5 x 106 TU.
Cell cycle analysis.
Acknowledgments
This project was supported in part by National Institutes of Health grant R21 NS44614 to Y.S. and E.A.C. and by the Dardinger Center Fund for Neuro-Oncology Research at the Arthur G. James Cancer Hospital, The Ohio State University Medical Center. We thank Drs Sandri-Goldin and Miyazaki for the Vero 2-2 cells, and pCAGGSneo, respectively.
REFERENCES
- Wade-Martins R, Smith ER, Tyminski E, Chiocca EA., and , Saeki Y. An infectious transfer and expression system for genomic DNA loci in human and mouse cells. Nat Biotechnol. 2001;19:1067–1070. doi: 10.1038/nbt1101-1067. [DOI] [PubMed] [Google Scholar]
- Wade-Martins R, Saeki Y., and , Chiocca EA.Infectious delivery of a 135-kb LDLR genomic locus leads to regulated complementation of low-density lipoprotein receptor deficiency in human cells Mol Ther 20037604–612.5 Pt 1 [DOI] [PubMed] [Google Scholar]
- Inoue R, Moghaddam KA, Ranasinghe M, Saeki Y, Chiocca EA., and , Wade-Martins R. Infectious delivery of the 132 kb CDKN2A/CDKN2B genomic DNA region results in correctly spliced gene expression and growth suppression in glioma cells. Gene Ther. 2004;11:1195–1204. doi: 10.1038/sj.gt.3302284. [DOI] [PubMed] [Google Scholar]
- Kasai K., and , Saeki Y. DNA-based methods to prepare helper virus-free herpes amplicon vectors and versatile design of amplicon vector plasmids. Curr Gene Ther. 2006;6:303–314. doi: 10.2174/156652306777592081. [DOI] [PubMed] [Google Scholar]
- Wang Y, Camp SM, Niwano M, Shen X, Bakowska JC, Breakefield XO, et al. Herpes simplex virus type 1/adeno-associated virus rep(+) hybrid amplicon vector improves the stability of transgene expression in human cells by site-specific integration. J Virol. 2002;76:7150–7162. doi: 10.1128/JVI.76.14.7150-7162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., and , Vos JM. A hybrid herpesvirus infectious vector based on Epstein-Barr virus and herpes simplex virus type 1 for gene transfer into human cells in vitro and in vivo. J Virol. 1996;70:8422–8430. doi: 10.1128/jvi.70.12.8422-8430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena-Esteves M, Saeki Y, Camp SM, Chiocca EA., and , Breakefield XO. Single-step conversion of cells to retrovirus vector producers with herpes simplex virus-Epstein-Barr virus hybrid amplicons. J Virol. 1999;73:10426–10439. doi: 10.1128/jvi.73.12.10426-10439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard N, Cosset FL., and , Epstein AL. Defective herpes simplex virus type 1 vectors harboring gag, pol, and env genes can be used to rescue defective retrovirus vectors. J Virol. 1997;71:4111–4117. doi: 10.1128/jvi.71.5.4111-4117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heister T, Heid I, Ackermann M., and , Fraefel C. Herpes simplex virus type 1/adeno-associated virus hybrid vectors mediate site-specific integration at the adeno-associated virus preintegration site, AAVS1, on human chromosome 19. J Virol. 2002;76:7163–7173. doi: 10.1128/JVI.76.14.7163-7173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers WJ, Mastrangelo MA, Howard DF, Southerland HA, Maguire-Zeiss KA., and , Federoff HJ. Neuronal precursor-restricted transduction via in utero CNS gene delivery of a novel bipartite HSV amplicon/transposase hybrid vector. Mol Ther. 2006;13:580–588. doi: 10.1016/j.ymthe.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Breakefield XO., and , Chiocca EA. Improved HSV-1 amplicon packaging system using ICP27-deleted, oversized HSV-1 BAC DNA. Methods Mol Med. 2003;76:51–60. doi: 10.1385/1-59259-304-6:51. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Fraefel C, Ichikawa T, Breakefield XO., and , Chiocca EA. Improved helper virus-free packaging system for HSV amplicon vectors using an ICP27-deleted, oversized HSV-1 DNA in a bacterial artificial chromosome. Mol Ther. 2001;3:591–601. doi: 10.1006/mthe.2001.0294. [DOI] [PubMed] [Google Scholar]
- Akira S., and , Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A., and , Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Chiocca EA., and , Saeki Y. Early STAT1 Activation After Systemic delivery of HSV amplicon vectors suppresses Transcription of The Vector-encoded Transgene. Mol Ther. 2007;15:2017–2026. doi: 10.1038/sj.mt.6300273. [DOI] [PubMed] [Google Scholar]
- Gu H, Liang Y, Mandel G., and , Roizman B. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci USA. 2005;102:7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., and , Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci USA. 2007;104:17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Gu H., and , Mandel G. The first 30 minutes in the life of a virus: unREST in the nucleus. Cell Cycle. 2005;4:1019–1021. doi: 10.4161/cc.4.8.1902. [DOI] [PubMed] [Google Scholar]
- Melroe GT, Silva L, Schaffer PA., and , Knipe DM. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: Potential role in blocking IFN-beta induction. Virology. 2007;360:305–321. doi: 10.1016/j.virol.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melroe GT, DeLuca NA., and , Knipe DM. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J Virol. 2004;78:8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Noyce RS, Collins SE, Everett RD., and , Mossman KL. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J Virol. 2004;78:1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harle P, Sainz B Jr, Carr DJ., and , Halford WP. The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-alpha/beta. Virology. 2002;293:295–304. doi: 10.1006/viro.2001.1280. [DOI] [PubMed] [Google Scholar]
- Poon AP, Gu H., and , Roizman B. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc Natl Acad Sci USA. 2006;103:9993–9998. doi: 10.1073/pnas.0604142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J Virol. 2000;74:9994–10005. doi: 10.1128/jvi.74.21.9994-10005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sant C, Hagglund R, Lopez P., and , Roizman B. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2001;98:8815–8820. doi: 10.1073/pnas.161283098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchet D, Ferrera R, Lomonte P., and , Epstein AL. Characterization of antiproliferative and cytotoxic properties of the HSV-1 immediate-early ICPo protein. J Gene Med. 2005;7:1187–1199. doi: 10.1002/jgm.761. [DOI] [PubMed] [Google Scholar]
- Hadjipanayis CG., and , DeLuca NA. Inhibition of DNA repair by a herpes simplex virus vector enhances the radiosensitivity of human glioblastoma cells. Cancer Res. 2005;65:5310–5316. doi: 10.1158/0008-5472.CAN-04-3793. [DOI] [PubMed] [Google Scholar]
- Hobbs WE., 2nd, and , DeLuca NA. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J Virol. 1999;73:8245–8255. doi: 10.1128/jvi.73.10.8245-8255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Baskaran R, Krisky DM, Bein K, Grandi P, Cohen JB, et al. Chk2 is required for HSV-1 ICP0-mediated G2/M arrest and enhancement of virus growth. Virology. 2008;375:13–23. doi: 10.1016/j.virol.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., and , Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Hagglund R., and , Roizman B. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J Virol. 2004;78:2169–2178. doi: 10.1128/JVI.78.5.2169-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman HM, Connor V, Cheng ZS, Grey F, Preston CM., and , Efstathiou S.Histone modifications associated with herpes simplex virus type 1 genomes during quiescence and following ICP0-mediated de-repression J Gen Virol 20088968–77.Pt 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidson KM, Hobbs WE, Manning BJ, Carlson P., and , DeLuca NA. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J Virol. 2002;76:2180–2191. doi: 10.1128/jvi.76.5.2180-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs WE, Brough DE, Kovesdi I., and , DeLuca NA. Efficient activation of viral genomes by levels of herpes simplex virus ICP0 insufficient to affect cellular gene expression or cell survival. J Virol. 2001;75:3391–3403. doi: 10.1128/JVI.75.7.3391-3403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaniego LA, Wu N., and , DeLuca NA. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J Virol. 1997;71:4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Watkins SC, Schaffer PA., and , DeLuca NA. Prolonged gene expression and cell survival after infection by a herpes simplex virus mutant defective in the immediate-early genes encoding ICP4, ICP27, and ICP22. J Virol. 1996;70:6358–6369. doi: 10.1128/jvi.70.9.6358-6369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon AP, Liang Y., and , Roizman B. Herpes simplex virus 1 gene expression is accelerated by inhibitors of histone deacetylases in rabbit skin cells infected with a mutant carrying a cDNA copy of the infected-cell protein no. 0. J Virol. 2003;77:12671–12678. doi: 10.1128/JVI.77.23.12671-12678.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt A, Haase R, Schepers A, Deutsch MJ, Lipps HJ., and , Baiker A. Episomal vectors for gene therapy. Curr Gene Ther. 2008;8:147–161. doi: 10.2174/156652308784746440. [DOI] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM., and , Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos K, Simon DA, Conway E, Bowers WJ, Mitra S, Foster TH, et al. Spatial and temporal expression of herpes simplex virus type 1 amplicon-encoded genes: implications for their use as immunization vectors. Hum Gene Ther. 2007;18:93–105. doi: 10.1089/hum.2006.082. [DOI] [PubMed] [Google Scholar]
- Ratanamart J, Huggins CG, Kirby JA., and , Shaw JA. In vitro and in vivo evaluation of intrinsic immunogenicity of reporter and insulin gene therapy plasmids. J Gene Med. 2007;9:703–714. doi: 10.1002/jgm.1066. [DOI] [PubMed] [Google Scholar]
- Gambotto A, Dworacki G, Cicinnati V, Kenniston T, Steitz J, Tuting T, et al. Immunogenicity of enhanced green fluorescent protein (EGFP) in BALB/c mice: identification of an H2-Kd-restricted CTL epitope. Gene Ther. 2000;7:2036–2040. doi: 10.1038/sj.gt.3301335. [DOI] [PubMed] [Google Scholar]
- Skelton D, Satake N., and , Kohn DB. The enhanced green fluorescent protein (eGFP) is minimally immunogenic in C57BL/6 mice. Gene Ther. 2001;8:1813–1814. doi: 10.1038/sj.gt.3301586. [DOI] [PubMed] [Google Scholar]
- Cerullo V, Seiler MP, Mane V, Cela R, Clarke C, Kaufman RJ, et al. Correction of murine hemophilia A and immunological differences of factor VIII variants delivered by helper-dependent adenoviral vectors. Mol Ther. 2007;15:2080–2087. doi: 10.1038/sj.mt.6300308. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kasai K., and , Saeki Y. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J Virol. 2006;80:3293–3300. doi: 10.1128/JVI.80.7.3293-3300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time course of GFP mRNA expression in MEFs. Primary cultured MEFs were infected with ICP0+ amplicon or conventional amplicon vector (MOI=1). Cells were harvested at various time points and subjected to real-time polymerase chain reaction analysis of GFP messenger RNA transcripts. The data for cells infected with the conventional and ICP0+ amplicons are plotted as ○ and ♦, respectively. Data are presented as mean ± SD (n=4). *P <0.01 compared with conventional amplicon-infected cells.
DNA fragmentation of MEFs, 24 hrs post-infection. Primary cultured MEFs were infected with ICP0+ amplicon or conventional amplicon vector (MOI=5). After 24 hours, cells were harvested and electrophoresed. DNA was detected with Ethidium Bromide staining. M, DNA size marker; Control, untreated control MEFs sample.
Interferon and ISG responses to infection of ICP0+ amplicon in liver of C57Bl/6 mice. Wild-type C57Bl/6 mice were intravenosuly injected with the indicated escalating dose of ICP0+ amplicon vector. After 6 h, induction of IFN and ISG mRNA was determined in the liver of C57Bl/6 mice by real-time RT-PCR analysis. Each value was calculated relative to that of un-injected mice. Data are presented as mean ± SD (n=6). *P<0.0001 compared with mice injected with 1 x 106 TU. **P <0.0001 compared with mice injected with 5 x 106 TU.
Cell cycle analysis.