Abstract
Stable addition of U2 small nuclear ribonucleoprotein (snRNP) to form the prespliceosome is the first ATP-dependent step in splicing, and it requires the DEXD/H box ATPase Prp5p. However, prespliceosome formation occurs without ATP in extracts lacking the U2 snRNP protein Cus2p. Here we show that Prp5p is required for the ATP-independent prespliceosome assembly that occurs in the absence of Cus2p. Addition of recombinant Cus2p can restore the ATP dependence of prespliceosome assembly, but only if it is added before Prp5p. Prp5p with an altered ATP-binding domain (Prp5-GNTp) can support growth in vivo, but only in a cus2 deletion strain, mirroring the in vitro results. Other Prp5 ATP-binding domain substitutions are lethal, even in the cus2 deletion strain, but can be suppressed by U2 small nuclear RNA mutations that hyperstabilize U2 stem IIa. We infer that the presence of Cus2p and stem IIa-destabilized forms of U2 small nuclear RNA places high demands on the ATP-driven function of Prp5p. Because Prp5p is not dispensable in vitro even in the absence of ATP, we propose that the core Prp5p function in bringing U2 to the branchpoint is not directly ATP-dependent. The positive role of Cus2p in rescuing mutant U2 can be reconciled with its antagonistic effect on Prp5 function in a model whereby Cus2p first helps Prp5p to activate the U2 snRNP for prespliceosome formation but then is displaced by Prp5p before or during the stabilization of U2 at the branchpoint.
Keywords: pre-mRNA splicing, branch site, RNA helicase, commitment complex, mRNA
Pre-mRNA splicing is a dynamic process, occurring within a large ribonucleoprotein complex called the spliceosome. Multiple ATP-dependent RNA and protein rearrangements take place before, between, and after the two transesterifications required to produce mature mRNAs (for reviews, see refs. 1–5). Important unsolved questions in splicing concern how these rearrangements are catalyzed at each ATP-dependent step. A total of eight members of the DEXD/H family of ATP-dependent RNA helicases have been assigned roles in splicing, each apparently responsible for catalyzing a specific transition in a particular splicing complex in conjunction with ATP hydrolysis (4, 5). In yeast, Prp5p and Sub2p have been implicated in U2 small nuclear ribonucleoprotein (snRNP) recruitment (6–11); Prp28p has been implicated in the addition of the U4/U6.U5 tri-snRNP (12) and destabilization of U1 snRNP (13); Brr2p has been implicated in the destabilization of U4 snRNP before catalysis (14); Prp2p, Prp16p, and Prp22p have been implicated in the activation of the complex for the cleavage-ligation reactions of the pre-mRNA substrate (15–18); and Prp22p and Prp43p have been implicated in the release of mRNA and disassembly of the splicing complex (17–19). RNA mutations consistent with hyperstabilization or destabilization of specific spliceosomal RNA duplexes cause consistent changes in the demand for DEXD/H protein function in genetic tests (4), but the molecular nature of these spliceosomal transitions and how these proteins accelerate them is largely unclear. A common theme is the activation, release, or stabilization of a distinct RNA–protein complex.
Some members of the DEXD/H protein family can carry out ATP-dependent RNA helicase reactions using model nucleic acid substrates (20–22). However, in some cases these may bear little resemblance to the natural substrate. Removal of proteins from RNA is also a possible function for this class of proteins (20, 23). Because the core conserved structural elements of this family are known to be important for RNA binding and ATP-dependent RNA unwinding (24), it has been assumed that these activities must reflect a core biochemical function of the family, with substrate specificity and timing restricted by particular characteristics of each family member. However, work by Schwer and Gross (18) shows that the ATP-binding activity of Prp22p is not required for its role in the second catalytic step of splicing but is required for mRNA release, suggesting that some key functions of the DEXD/H family may not be strictly ATP-dependent.
In the case of Prp5p, genetic and biochemical data indicate that it acts to stabilize the association of U2 snRNP with pre-mRNA during prespliceosome formation (6, 7, 11, 25). Heat treatment of splicing extracts from temperature-sensitive prp5-1 yeast prevents prespliceosome formation and splicing in vitro (6), and this block can be overcome with recombinant Prp5p (rPrp5p) (7). Annealing of oligonucleotides complementary to the branchpoint interaction region of U2 small nuclear RNA (snRNA), a model reaction thought to reflect U2-branchpoint interaction, is stimulated by Prp5p and ATP in vitro and is significantly reduced in heat-treated prp5-1 splicing extracts (7, 11). Immunodepletion of hPrp5p from HeLa cell extracts blocks splicing during or before prespliceosome assembly (25). Thus, Prp5p has been implicated as the mediator of the ATP requirement at this step (6, 7, 11, 27). Recently, splicing extracts lacking Cus2p have been shown to be relieved of the requirement for ATP in prespliceosome formation (27). Cus2p, isolated as a dominant suppressor of U2 snRNA folding mutants in yeast (26), acts both in support of and in opposition to Prp5p (27). Prp5p and Cus2p share genetic interactions with SF3a and SF3b subunit genes, indicative of shared function in prespliceosome assembly (6, 26, 28, 29). In splicing extracts, Cus2p enforces the ATP dependence of prespliceosome formation, because prespliceosomes form without ATP in extracts from a cus2 deletion strain. Despite this, cus2Δ does not bypass the need for Prp5p, because temperature-sensitive prp5-1 splicing extracts lacking Cus2p are not functional (27).
Here we present in vitro and in vivo evidence that Prp5p has multiple roles in prespliceosome formation, including an ATP-independent function. Order of addition experiments shows that, in the absence of Cus2p, the ATP-dependent activity of Prp5p is unnecessary for prespliceosome assembly in vitro, suggesting it may have to do with displacing Cus2p. An ATP-binding domain mutant of Prp5p (GKT to GNT) rescues a prp5 deletion in vivo, but only if Cus2p is absent. Other similar mutants (GKT to GAT or GHT) do not survive even without Cus2p but can be suppressed by mutant U2 snRNAs in which U2-stem IIa is hyperstabilized. Taken together, these results demonstrate that the normally stringent requirement for the ATP-dependent activity of Prp5p is relaxed when Cus2p is absent, and U2 snRNA can readily form stem IIa. Furthermore, there exists an underlying ATP-independent activity of Prp5p that is essential for prespliceosome formation. We suggest a model in which Cus2p helps Prp5p activate the U2 snRNP but then must be removed from the U2 snRNP before stable association with the pre-mRNA. In this model, the ATP-requiring activity of Prp5p helps ensure that competing RNA structures are disrupted to allow formation of U2-stem IIa and removal of Cus2p. This step could help the U2 snRNA to base-pair with the pre-mRNA branchpoint.
Methods
In Vitro Splicing and Native Gel Analysis. Splicing extracts were isolated as described (27) from yeast strain RP01, which is PRP5+;cus2Δ (Fig. 1 and ref. 27), or RP51–62, which is prp5-1;cus2Δ (Figs. 1, 2, 3 and ref. 27). Splicing reactions and native gel analysis were done by using in vitro transcribed RP51A pre-mRNA (Figs. 1 and 2) or actin pre-mRNA (Fig. 3 and ref. 27). Unless otherwise noted, reactions and preincubations were at 25°C. ATP depletions were during preincubation with 0.2 mM glucose added to splicing extract plus buffer.
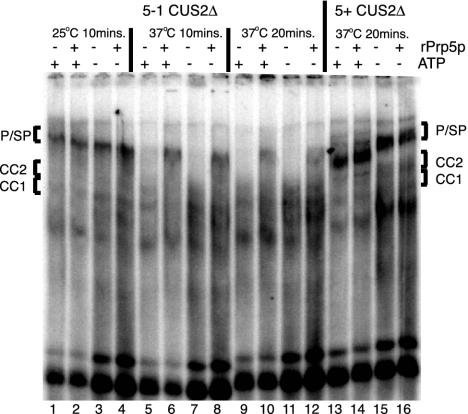
Fig. 1.
Prp5p has ATP-dependent and ATP-independent roles in prespliceosome assembly. Shown is native gel analysis of spliceosome assembly in ATP-depleted splicing extracts. Data are derived from a strain carrying prp5-1 and cus2Δ (5-1 CUS2Δ) preincubated at 25°C (lanes 1–4) or preincubated at 37°C (lanes 5–12) for 10 min (lanes 5–8) or 20 min (lanes 9 –12). Data in lanes 13–16 are from a control extract carrying PRP5+ and cus2Δ (5+CUS2Δ) preincubated at 37°C. Spliceosome assembly is analyzed in the presence (+; lanes 1, 2, 5, 6, 9, 10, 13, and 14) or absence (–; lanes 3, 4, 7, 8, 11, and 12) of exogenous ATP and the presence (+; lanes 2, 4, 6, 8, 10, 12, 14, and 16) or absence (–; lanes 1, 3, 5, 7, 9, 11, 13, and 15) of rPrp5 protein. CC1 and CC2, commitment complexes; P/SP, prespliceosomes/spliceosomes.
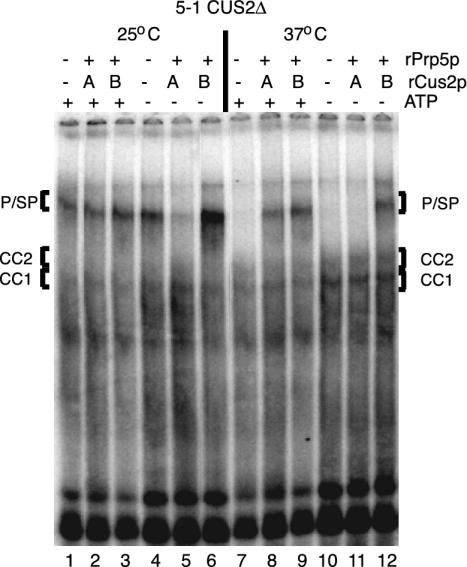
Fig. 2.
Cus2p acts before Prp5p to control the ATP dependence of prespliceosome assembly in vitro. Shown is native gel analysis of spliceosome formation in the presence (+; lanes 1–3 and 7–9) or absence (–; lanes 4–6 and 10 –12) of ATP in experiments in which rCus2p is added before (A; lanes 2, 5, 8, and 11) or together with (B; lanes 3, 6, 9, and 12) rPrp5p to prp5-1, cus2Δ splicing extracts preincubated at 25°C (lanes 1–6) or 37°C (lanes 7–12). In lanes 1, 4, 7, and 10, no recombinant protein was added. Bracketed species are as in Fig. 1.
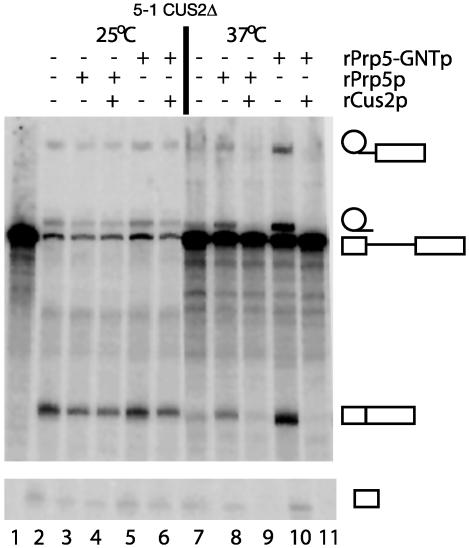
Fig. 3.
rPrp5-GNT protein can rescue splicing in the absence of Cus2p in vitro. Shown is denaturing gel analysis of splicing in prp5-1;cus2Δ splicing extracts preincubated at 25°C (lanes 2–6) or 37°C (lanes 7–11) for 30 min with buffer D (lanes 2, 3, 5, 7, 8, and 10) or rCus2p (lanes 4, 6, 9, and 11). Subsequent addition of buffer D (lanes 2 and 7), rPrp5p (lanes 3, 4, 8, and 9), or rPrp5-GNTp (lanes 5, 6, 10, and 11) was 2 min before pre-mRNA, and the reactions were incubated at 25°C for 15 min. Lanes 1, 2, and 7 have no recombinant protein added, with lane 1 also lacking splicing extract. Labeled products (from top to bottom) are lariat 3′ exon intermediate, lariat product, pre-mRNA, splice exon product, and free 5′ exon intermediate.
Recombinant Proteins. rPrp5 and rPrp5-GNT protein were isolated from Escherichia coli by successive binding to anti-FLAG and nickel-nitriloacetic acid columns to 6HIS and FLAG tags at the N terminus of both proteins. These were added to splicing reactions to a final reaction concentration of ≈4 nM. Recombinant Cus2p (rCus2p) purification was described in ref. 26, and this protein was supplemented to a final concentration of ≈200 nM. For treatment A in Fig. 2, rCus2p or buffer D were added to splicing extract plus splicing buffer plus 0.2 mM glucose and preincubated for 10 min at 25°C or 37°C. For treatment B, preincubation proceeded without rCus2p, which was added with rPrp5p and pre-mRNA plus ATP. In Fig. 3, rCus2p or buffer D was added to splicing extract plus splicing buffer and preincubated for 30 min at the indicated temperature. Note that ATP was not depleted from reactions in Fig. 3. rPrp5p or rPrp5-GNT was added 2 min before pre-mRNA plus ATP. After pre-mRNA addition, splicing reactions in Figs. 2 and 3 were incubated for 15 min at 25°C.
Construction of Strain DS4D. Strain DS4D; MATa, prp5::KanR, cus2::KanR, SNR20::HIS3, trp1, ura3, leu2, lys2, pIP45 (PRP5 plus SNR20 on URA) was made by crossing YIP90; MATa, prp5::KanR, SNR20::HIS3, trp1, ura3, leu2, pIP45 (PRP5 plus SNR20 on URA) with BY4742cus2Δ. Tetrad dissection and spore analysis were done by using standard genetic techniques. SNR20 is the standard name for the yeast U2 gene.
Analysis of Prp5 and U2 snRNA Mutants. Mutant U2 alleles are cloned on centromeric LEU2 plasmids. U2-WT and U2-G100A are as described in ref. 30. U2-A52G;U63C, U2-U54C;A61G, U50G;A65C, U2-ΔCC, and U2-A52G;U63C plus U50G;A65C were made by site-directed mutagenesis by using the pRS315-U2 template. CUS2 is on pRS317 (centromeric plasmid with LYS2), and PRP5, PRP5-GAT, PRP5-GHT, and PRP5-GNT are on pRS314 (centromeric plasmid with TRP1). Yeast strain DS4D was transformed with pRS317 or pRS317CUS2+ selecting for LYS+, then cotransformed with U2 and Prp5 mutant plasmids selecting on dextrose plates lacking lysine, tryptophan, and leucine. The phenotype of combinations of mutant Prp5, Cus2p, and U2 were determined by shuffling out the pIP45 plasmid encoding wild-type U2 and Prp5p on plates containing 5-fluoroorotic acid and lacking lysine, tryptophan, and leucine at 30°C for 3–5 days.
Results
Prp5p Is Required for Prespliceosome Formation in the Absence of ATP. Prespliceosome formation can occur in vitro without ATP if Cus2p is absent or U2 RNA is altered, but it cannot occur in heat-treated temperature-sensitive prp5-1 extracts lacking Cus2p (27). Thus, although the ATP requirement is relieved by the absence of Cus2p, the requirement for Prp5p in splicing appears not to be bypassed by the absence of Cus2p. This suggested, but did not prove, that a function of Prp5p is required in the absence of ATP. To address this, we asked whether addition of rPrp5p (7) can reconstitute prespliceosome formation in heat-treated prp5-1, cus2Δ splicing extracts in the absence of ATP (Fig. 1). Treatment of these extracts at 37°C inactivates assembly of prespliceosomes on rp51A pre-mRNA whether ATP is present or not (Fig. 1, lanes 5, 7, 9, and 11) (27). Addition of rPrp5p restores formation of prespliceosomes (Fig. 1, lanes 6 and 10), even in the absence of ATP (lanes 8 and 12). Control reactions in which prp5-1;cus2Δ extracts are preincubated at 25°C (lanes 1–4) or in which PRP5;cus2Δ extracts are preincubated at 37°C (lanes 13–16) lead to efficient prespliceosome formation under all treatment conditions. We conclude that the function of Prp5p in prespliceosome formation can be uncoupled from ATP hydrolysis in vitro if Cus2p is absent. Thus, Prp5p has two activities: an ATP-independent activity essential for prespliceosome formation, and an ATP-dependent activity that is essential in vitro only when Cus2p is present. This indicates that the prespliceosome formation observed in the absence of ATP and Cus2p (27) is due to the ATP-independent activity of Prp5p.
Ordered Addition of rCus2p Blocks the Function of rPrp5p in the Absence of ATP. ATP dependence of prespliceosome formation can be restored in cus2Δ extracts by the addition of rCus2p, and this activity requires residues in the first Cus2p RNA recognition motif (27). To analyze how Cus2p influences Prp5p function, we performed order-of-addition experiments (Fig. 2) in which we added rCus2p before (treatment A) or together with (treatment B) rPrp5p to heat-treated or mock-treated prp5-1;cus2Δ splicing extracts, and we assessed prespliceosome formation with (Fig. 2, lanes 1–3 and 7–9) or without (lanes 4–6 and 10–12) ATP. If ATP is present, both treatments reconstitute prespliceosome formation (Fig. 2, compare lane 7 to lanes 8 and 9). In the absence of ATP, treatment A fails to rescue prespliceosome formation (Fig. 2, compare lanes 10 and 11), although treatment B (both proteins together) reconstitutes prespliceosome formation (lane 12). In control reactions to which rCus2-Y48Dp is added (a mutant Cus2p that cannot bind RNA) (26), the ATP dependence of prespliceosome is not enforced (data not shown and ref. 27). Thus, Cus2p blocks the ATP-independent activity of Prp5p necessary for prespliceosome formation. To impose this block, Cus2p must be present in the extract before Prp5p and requires its RNA binding activity. This finding suggests that, rather than directly binding to Prp5p, Cus2p binds or modifies a substrate of Prp5p, either the U2 snRNP or the commitment complex. Furthermore, this finding shows that an ATP-dependent function of Prp5p is required to overcome the Cus2p block of the Prp5p ATP-independent function and suggests that antagonism of Prp5p by Cus2p represents the ATP requirement for prespliceosome assembly observed in yeast splicing extracts.
A Prp5p ATP-Binding Domain Mutant Rescues Splicing in Vitro, but Only When Cus2p Is Absent. If the presence of Cus2p enforces the requirement for an ATP-dependent function of Prp5p, then mutant Prp5 protein unable to bind ATP might rescue prespliceosome formation and splicing, as long as Cus2p is absent. The invariant lysine of the conserved GKT, the Walker A motif (31) of the DEXD/H box protein family, contacts the β and γ phosphates of bound nucleotides in the crystal structures of several NTPases (32–34). Lysine (K) to alanine (A), asparagine (N), or glutamine (Q) substitution in this motif of various DEXD/H family members results in loss of function and significant reduction in ATP-binding and ATP-hydrolysis activities (32–38). Thus, we made rPrp5p containing the GKT lysine to asparagine substitution, rPrp5-GNTp, and asked whether this protein could rescue splicing of actin pre-mRNA in heat-treated prp5-1;cus2Δ splicing extracts (Fig. 3).
Preincubation of prp5-1;cus2Δ extracts for 30 min at 37°C results in a drastic reduction in splicing as measured by the increase in unspliced pre-mRNA and the reduction in splicing intermediates and products (Fig. 3, lane 7). Addition of rPrp5p (Fig. 3, lane 8) or rPrp5-GNTp (lane 10) during the heat treatment can rescue this splicing, but the addition of rCus2p reduces rescue by rPrp5p (lane 9) and completely inhibits rescue by rPrp5-GNTp (lane 11). From this we conclude that rPrp5-GNT can rescue splicing in heat-inactivated prp5-1;cus2Δ splicing extracts, but only when Cus2p is absent. Preincubation of prp5-1;cus2Δ extract at 25°C with no protein (Fig. 3, lane 2), rPrp5p (lane 3), rCus2p plus rPrp5p (lane 4), rPrp5-GNT (lane 5), or rCus2p plus rPrp5-GNT (lane 6) demonstrates that the addition of these recombinant proteins does not generally affect splicing activity. Although we have not determined that rPrp5p-GNT is inhibited for ATP binding and hydrolysis, this experiment supports the observation that wild-type rPrp5p rescues prespliceosome formation in the absence of ATP (Fig. 2). The above experiment also suggests that the requirement for ATP hydrolysis by Prp5p is limited to prespliceosome assembly and is not necessary for other ATP-requiring steps in splicing.
The prp5-GNT Gene Can Rescue a PRP5 Deletion in Vivo, but Only When CUS2 Is Deleted. If the in vitro results above represent an accurate picture of Prp5p function in vivo, then Prp5p mutants lacking ATP-binding residues might support growth if Cus2p is absent from the cell. To test this, we constructed a yeast strain deleted for CUS2 and carrying the prp5-GNT allele. Cells carrying this PRP5 mutation are able to grow only if they also carry the CUS2 deletion (Fig. 4), demonstrating that the conserved ATP binding residues are not critical when Cus2p is absent in vivo. These observations show that, in vivo as well as in vitro, efficient ATP binding by Prp5p is not required for splicing if Cus2p is absent.
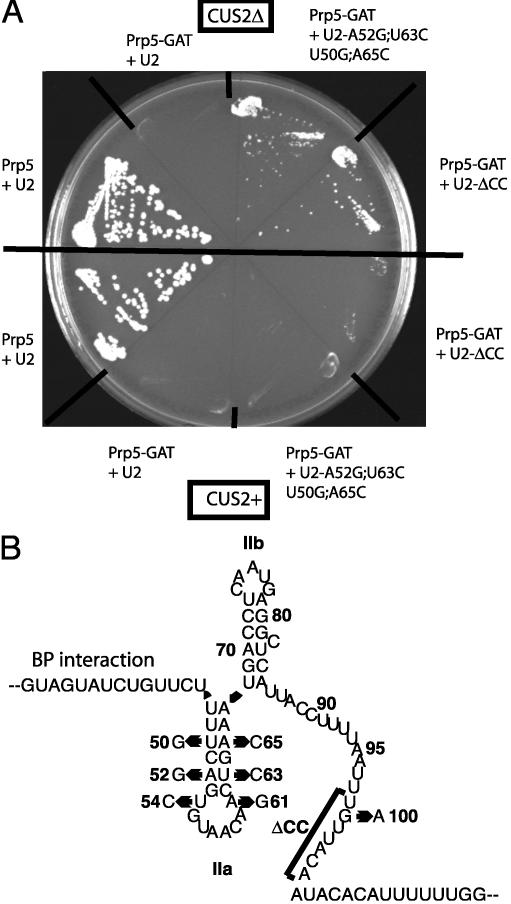
Fig. 4.
Prp5-GNT can rescue growth in the absence of Cus2p in vivo. Strain DS4D (prp5Δ;cus2Δ;u2Δ) carrying pRS315U2 and either pRS317 (right half) or pRS317CUS2+ (left half) plus one of pRS314, pRS314Prp5+, or pRS314Prp5-GNT plasmids plated on 5-fluoroorotic acid-Lys-Trp-Leu at 30°C for 5 days.
U2 snRNA Mutations Suppress the Lethal Effects of More Severe PRP5 ATP-Binding Domain Mutations. If Prp5p unwinds RNA, U2 snRNA is a likely substrate. Of several RNAs tested, U2 RNA fragments proved best at activating the ATP-hydrolyzing activity of rPrp5p (7). Oligonucleotide-based probing methods suggest that Prp5p modifies U2 snRNP structure in an ATP-dependent fashion (7, 11). We reasoned that, if U2 RNA is the target of Prp5p ATP-dependent activity, mutations in U2 RNA that are less dependent on these functions might be able to suppress defects caused by ATP-binding-deficient Prp5p proteins in vivo. In addition to the prp5-GNT allele, we made two other ATP-binding domain alterations, prp5-GAT and prp5-GHT. These alleles are more severely affected, because neither can complement a PRP5 deletion in a wild-type U2 background, even when CUS2 is deleted (Table 1). We screened a pool composed of available U2 snRNA mutations (30, 35), including a complex library of U2 mutations (29) and several mutants that hyperstabilize stem IIa directly or disrupt structures that compete with stem IIa (35). None of the mutant U2 alleles tested display any growth defects in wild-type (CUS2, PRP5), or cus2Δ, PRP5 backgrounds (refs. 29, 33, and 35 and data not shown).
Table 1. Growth of PRP5 and U2 mutants with and without CUS2.
| U2 | CUS2 |
PRP5
|
|||
|---|---|---|---|---|---|
| wt | GAT | GNT | GHT | ||
| wt | wt | ++++ | - | - | - |
| Δ | ++++ | - | ++ | - | |
| G100A | wt | ++++ | - | ++ | - |
| Δ | ++++ | + | +++ | - | |
| ΔCC | wt | ++++ | - | +++ | - |
| Δ | ++++ | +++ | +++ | ++ | |
| A52G;U63C | wt | ++++ | - | + | - |
| Δ | ++++ | ++ | +++ | + | |
| U50G;A65C | wt | ++++ | - | + | - |
| Δ | ++++ | + | +++ | - | |
| U50G;A65C + | wt | ++++ | - | +++ | - |
| A52G;U63C | Δ | ++++ | +++ | +++ | + |
| U54C;A61G | wt | ++++ | - | + | - |
| Δ | ++++ | ++ | +++ | - | |
Strain DS4D deleted of the PRP5, CUS2, and U2 genes and carrying PRP5 and U2 on a URA3 plasmid were transformed with the indicated U2, PRP5, and CUS2 plasmids, and then wild-type PRP5 and U2 were removed by plasmid shuffling on plates containing 5-fluoroorotic acid for 5 days at 30°C. The number of plus signs indicates the level of growth.
Strikingly, a subset of U2 snRNA mutants suppresses the lethal effects of prp5-GAT or prp5-GHT (Table 1 and Fig. 5A), suggesting that they further relieve the requirement for Prp5p function. Among these are U2 alleles in which stem IIa stability seems increased by one of several mechanisms: substituting A-U base pairs in stem IIa with G-C pairs (Fig. 5B), reducing the potential pairing between the loop of stem loop IIa and the conserved complementary region (G100A), or completely deleting the conserved complementary region (ΔCC). In the cus2Δ background, the prp5-GAT allele is rescued by all six mutant U2 snRNA alleles, whereas prp5-GHT is rescued by a subset of these (Table 1). None of the mutant U2 snRNA alleles is able to suppress prp5-GAT or prp5-GHT in the presence of CUS2; however, all of the U2 mutants suppress the lethal effects of prp5-GNT in the presence of CUS2 (Table 1). These data indicate that increasing the stability of U2-stem IIa in the absence of Cus2p allows cell viability even with severe Prp5-GKT mutant alleles.
Fig. 5.
U2 snRNA mutations that hyperstabilize U2-stem IIa suppress prp5-GAT. (A) Growth of strains. Strain DS4D plus pRS317 (upper half) or pRS317CUS2+ (lower half) was transformed with pRS314Prp5 or pRS314Prp5-GAT plus one of wild-type U2, U2-A52G;U63C+, U50G;A65C, or U2-ΔCC and plated on 5-fluoroorotic acid-Lys-Leu-Trp at 30°C for 5 days. (B) Secondary structure of nucleotides 29 –120 of yeast U2 snRNA. Nucleotide substitutions for U2 snRNA mutant alleles A52G;U63C, U50G;A65C, G100A, and ΔCC are shown.
Discussion
By reconstituting splicing extracts lacking Prp5p and Cus2p activity with recombinant proteins, we demonstrate that the DEXD/H box family member Prp5p has a role in U2 snRNP recruitment that is independent of ATP hydrolysis (Figs. 1, 2, 3). We find a strict requirement for Cus2p to be present before Prp5p to enforce the ATP dependence of U2 snRNP addition observed in wild-type extracts (Fig. 2). The conclusion that Cus2p enforces an ATP-dependent activity of Prp5p is supported by the following observations. First, mutations in the conserved GKT ATP-binding motif of Prp5p rescue splicing in vitro (Fig. 3), only if Cus2p is absent. Second, the same mutant PRP5 allele can support growth in vivo (Figs. 4 and 5 and Table 1), but, again, only when CUS2 is deleted. Some GKT ATP-binding motif mutants are more severe, and, in addition to cus2Δ, require hyperstabilized U2-stem IIa for rescue in vivo (Fig. 5 and Table 1), suggesting that U2-stem IIa formation may also be aided by ATP-dependent Prp5p activity. We conclude that the ATP-dependent functions for Prp5p involve removing Cus2p from the U2 snRNP and helping stabilize U2 stem IIa before prespliceosome formation. Because Prp5p is essential even when the ATP-dependent demands are removed, we also propose an ATP-independent activity for Prp5p in stabilizing the binding of U2 to the pre-mRNA branchpoint.
A Model for Cus2p and Prp5p Control of U2 snRNA and snRNP Function. Previous work has demonstrated complex functional links among Cus2p, Prp5p, and U2 snRNA. First, a set of genetic interactions indicates that each gene product contributes to splicing function and that each supports the function of the other two (6, 7, 11, 25–28). Second, biochemical evidence supports the idea that these gene products function together. Both Cus2p and Prp5p coimmunoprecipitate U2 snRNA in yeast (11, 25), and human Prp5 is a component of the 17S U2 snRNP (25). Furthermore, U2 RNA fragments stimulate rPrp5p ATPase activity in vitro (7). Finally, there is a negative effect of Cus2p on the ATP-dependent addition of the U2 snRNP, because, in the absence of Cus2p, this step no longer requires ATP in vitro (27). In this article we show that this negative effect of Cus2p is specific for the ATP-dependent activity of Prp5p itself, because (i) Prp5p but not ATP is required for prespliceosome assembly in vitro if Cus2p is absent (Figs. 1 and 2), and (ii) Prp5 ATP-binding site mutants are functional only in the absence of Cus2p in vitro (Fig. 3) and in vivo (Figs. 4 and 5 and Table 1).
Taken together, these data suggest a model for how Cus2p and Prp5p work with the U2 snRNP during prespliceosome assembly (Fig. 6). The positive role of Cus2p is to help refold U2 snRNA to make the U2 snRNP active for prespliceosome assembly (26). In this view, Cus2p helps create a form of the U2 snRNP (the “activated U2 snRNP”) that is an efficient substrate for Prp5p. This is not an essential function, so activation of the U2 snRNP can occur without Cus2p. However, mutant U2 RNAs with folding defects are dependent on Cus2p (26), suggesting that activation of the U2 snRNP requires correct U2 snRNA folding. When Prp5p function is compromised, as in the temperature-sensitive alleles, the positive function of Cus2p becomes essential (27). It is possible that Cus2p contributes to Prp5p function indirectly by increasing the levels of activated U2 snRNP in vivo.
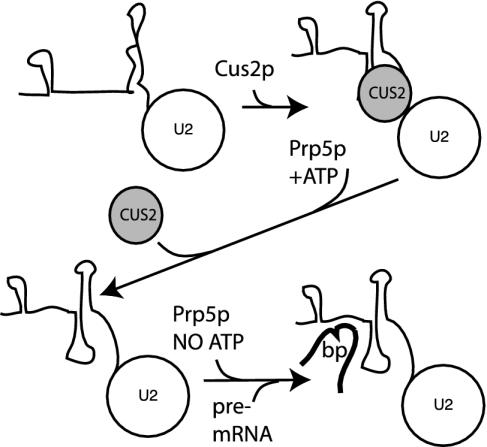
Fig. 6.
A model for the roles of Cus2p and the ATP-dependent activity of Prp5p in U2 snRNP recruitment and prespliceosome formation. Cus2p binds U2 snRNA and helps form stem IIa during formation of the activated U2 snRNP and engagement with pre-mRNA. The ATP-dependent functions of Prp5p include removing Cus2p, and stabilizing U2-stem IIa. This may include unwinding competing RNA structures to form stem IIa. The ATP-independent function is required for stable U2 snRNP-pre-mRNA binding.
To incorporate the negative effect of Cus2p on the ATP-dependent activity of Prp5p (ref. 27 and Fig. 2), we suggest that Cus2p occupies a site on the U2 snRNP that Prp5p must access to promote stable association of the U2 snRNP with pre-mRNA. In this view, Cus2p is displaced from the U2 snRNP by an ATP-dependent function of Prp5p before Prp5p can stabilize U2 association with pre-mRNA. In the absence of Cus2p, Prp5p has full access to the U2 snRNP and does not need ATP to perform the latter function (Fig. 6).
How does folding of U2 snRNA fit into this process? Hyperstabilizing stem IIa rescues GKT mutants (Fig. 5 and Table 1). Because aiding stem IIa formation would seem to reduce the demand for ATP binding, this finding suggests that stem IIa could be the end product of a Prp5p unwinding activity that acts on competing structures (for an example, see ref. 35). A Prp5p role in modifying U2 snRNA structure has been suggested (6, 7, 11, 27, 28, 44). Wiest et al. (44) and O'Day et al. (7) proposed alternate U2 snRNA states by assaying U2 branchpoint interaction region accessibility to oligonucleotide binding and RNaseH cleavage in inactivated prp5-1 yeast splicing extracts. When no Prp5p is present U2 RNA is less accessible and a closed state is proposed, but when active Prp5p and ATP are present U2 RNA becomes accessible and an open state is proposed. If so, the ATP-dependent functions of Prp5p may be involved in two distinct activities: (i) to remove Cus2p and (ii) to unwind helices that oppose stem IIa formation. Alternatively, a more stable form of stem IIa could contribute indirectly to Prp5p general function by increasing the levels of activated U2 snRNP. This possibility is consistent with the interpretation that Cus2p helps form the activated the U2 snRNP in part by stabilizing stem IIa (26).
Whether or not Prp5p unwinds U2 snRNA, the model describes a plausible pathway for wild-type prespliceosome assembly inferred from the phenotype of mutants and from ordered reactions in extracts depleted of ATP and supplemented with recombinant proteins. The two main features of the model are that Cus2p contributes early to U2 snRNP activation but then must be removed, and that Prp5p has an ATP-dependent function that removes Cus2p and an ATP-independent function that stabilizes binding of the U2 snRNP to the pre-mRNA branchpoint.
Multiple Functions for Prp5p. Our experiments have resolved ATP-dependent and ATP-independent roles for Prp5p in U2 snRNP recruitment to the pre-mRNA branchpoint. An ATP-independent function for Prp5p was first suspected when cus2Δ extracts able to support prespliceosome assembly in the absence of ATP failed to do so when prp5-1 was heat-inactivated (27). Using recombinant protein we now show that the missing activity required for prespliceosome assembly in the absence of ATP is Prp5 (Figs. 1 and 2). Supporting this is the observation that ATP-binding site mutants (GKT mutants) complement splicing in vitro (Fig. 3) and growth in vivo (Fig. 4 and Table 1) when Cus2p is absent, arguing that core Prp5 function does not require ATP or efficient binding of ATP to Prp5p. Although we have not assayed the ATP binding of the Prp5-GKT mutants, we are confident that they are significantly reduced, because equivalent lysine substitutions in other DEXD/H box proteins show negligible ATP-dependent protein activity (36–40). However, we cannot discount the possibility that the three PRP5 alleles may have subtle differences in residual ATP binding, and this could explain variations in viability we observe in the presence of different U2 snRNA alleles as well as their different responses to the presence of Cus2p (Table 1). Clearly however, if any PRP5 ATP-binding activity remains, it is not enough to support growth in wild-type cells.
The ATP-dependent functions of Prp5p are uncovered only when they are bypassed and an ATP-requirement is relieved. One ATP-dependent function for Prp5p is to overcome the presence of Cus2p (ref. 27 and Figs. 1 and 2). Similarly, when stem IIa is hyperstabilized, the requirement for ATP binding site residues becomes less stringent in vivo (Fig. 5 and Table 1), suggesting that another ATP-dependent function is to stabilize stem IIa. We do not know to what extent these two ATP-dependent functions are related, but it seems possible to both remove Cus2p and stabilize a defined structure of U2 snRNA (as well as making other changes) by a single structural rearrangement that remodels the U2 snRNP. The ATP-independent step could involve a conformational change in Prp5p that can occur without ATP hydrolysis. Alternatively, because Prp5p extends 290 aa N-terminally and 230 aa C-terminally from the conserved helicase core, these regions may contain elements directly required for the ATP-independent functions.
Prp5p is the second DEXD/H box protein family member for which ATP-dependent and ATP-independent roles have been demonstrated. Elegant work by Schwer and colleagues (18, 41) has resolved multiple roles for Prp22p before and after the second catalytic step of splicing. Prp22 mutants incapable of ATP hydrolysis can perform a Prp22p function required for the second step of splicing but cannot supply function for mRNA release from the spliceosome (18, 41). Prp5p appears different in that its ATP-dependent function is required to act before its ATP-independent function. Yeast cells with prp5 alleles altered in critical ATP-binding residues are inviable in an otherwise wild-type background. Requirement for these Prp5p residues is bypassed by CUS2 deletion (Figs. 4 and 5 and Table 1). This is the third such example of bypass of an essential DEXD/H box family protein in the splicing pathway. Deletion of MUD2 removes the requirement for the DEXD/H protein Sub2p (8), whereas specific alterations in U1-C protein eliminate the need for the DEXD/H box protein Prp28p (42). These findings demonstrate that removal of a specific protein or protein activity renders the ATP hydrolysis function of the corresponding DEXD/H protein dispensable in vivo and suggests that their primary ATP-driven function is to overcome the antagonizing effects of specific target proteins. DEXD/H box protein family members hydrolyze ATP to alter RNA structures (for a review, see ref. 24); however, these proteins could also disrupt RNA–protein complexes (4, 23, 43).
Acknowledgments
We thank Andrey Balakin for making U2-ΔCC, Carrie Davis and Stephanie Ruby for critical reading of the manuscript, and the reviewers for constructive comments. This work was supported by National Institutes of Health Grants GM47408 (to M.A.) and GM32637 (to J.A.) and an American Cancer Society Postdoctoral Fellowship (to R.P.).
Abbreviations: snRNA, small nuclear RNA; snRNP, small nuclear ribonucleoprotein; rPrp5p, recombinant Prp5p; rCus2p, recombinant Cus2p.
References
- 1.Moore, M., Query, C. & Sharp, P. A. (1993) in The RNA World, eds. Gesteland, R. & Atkins, J. (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY), pp. 303–357.
- 2.Ares, M., Jr., & Weiser, B. (1995) Prog. Nucleic Acid Res. Mol. Biol. 50, 131–159. [DOI] [PubMed] [Google Scholar]
- 3.Kramer, A. (1996) Annu. Rev. Biochem. 65, 367–409. [DOI] [PubMed] [Google Scholar]
- 4.Staley, J. P. & Guthrie, C. (1998) Cell 92, 315–326. [DOI] [PubMed] [Google Scholar]
- 5.Brow, D. A. (2002) Annu. Rev. Genet. 36, 333–360. [DOI] [PubMed] [Google Scholar]
- 6.Ruby, S. W., Chang, T. H. & Abelson, J. (1993) Genes Dev. 7, 1909–1925. [DOI] [PubMed] [Google Scholar]
- 7.O'Day, C. L., Dalbadie-McFarland, G. & Abelson, J. (1996) J. Biol. Chem. 271, 33261–33267. [DOI] [PubMed] [Google Scholar]
- 8.Kistler, A. L. & Guthrie, C. (2001) Genes Dev. 15, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Libri, D., Graziani, N., Saguez, C. & Boulay, J. (2001) Genes Dev. 15, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, M. & Green, M. R. (2001) Genes Dev. 15, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abu Dayyeh, B. K., Quan, T. K., Castro, M. & Ruby, S. W. (2002) J. Biol. Chem. 277, 20221–20233. [DOI] [PubMed] [Google Scholar]
- 12.Strauss, E. J. & Guthrie, C. (1994) Nucleic Acids Res. 22, 3187–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staley, J. P. & Guthrie, C. (1999) Mol. Cell 3, 55–64. [DOI] [PubMed] [Google Scholar]
- 14.Raghunathan, P. L. & Guthrie, C. (1998) Curr. Biol. 8, 847–855. [DOI] [PubMed] [Google Scholar]
- 15.Kim, S. H. & Lin, R. J. (1996) Mol. Cell. Biol. 16, 6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwer, B. & Guthrie, C. (1992) EMBO J. 11, 5033–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner, J. D., Jankowsky, E., Company, M., Pyle, A. M. & Abelson, J. N. (1998) EMBO J. 17, 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwer, B. & Gross, C. H. (1998) EMBO J. 17, 2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arenas, J. E. & Abelson, J. N. (1997) Proc. Natl. Acad. Sci. USA 94, 11798–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwer, B. (2001) Nat. Struct. Biol. 8, 113–116. [DOI] [PubMed] [Google Scholar]
- 21.Wang, Y., Wagner, J. D. & Guthrie, C. (1998) Curr. Biol. 8, 441–451. [DOI] [PubMed] [Google Scholar]
- 22.Jankowsky, E., Gross, C. H., Shuman, S. & Pyle, A. M. (2000) Nature 403, 447–451. [DOI] [PubMed] [Google Scholar]
- 23.Jankowsky, E., Gross, C. H., Shuman, S. & Pyle, A. M. (2001) Science 291, 121–125. [DOI] [PubMed] [Google Scholar]
- 24.Tanner, N. K. & Linder, P. (2001) Mol. Cell 8, 251–262. [DOI] [PubMed] [Google Scholar]
- 25.Will, C. L., Urlaub, H., Achsel, T., Gentzel, M., Wilm, M. & Luhrmann, R. (2002) EMBO J. 21, 4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan, D., Perriman, R., Igel, H., Howe, K. J., Neville, M. & Ares, M., Jr. (1998) Mol. Cell. Biol. 18, 5000–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perriman, R. & Ares, M., Jr. (2000) Genes Dev. 14, 97–107. [PMC free article] [PubMed] [Google Scholar]
- 28.Wells, S. E. & Ares, M., Jr. (1994) Mol. Cell. Biol. 14, 6337–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan, D. & Ares, M., Jr. (1996) Mol. Cell. Biol. 16, 818–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ares, M., Jr., & Igel, A. H. (1990) Genes Dev. 4, 2132–2145. [DOI] [PubMed] [Google Scholar]
- 31.Walker, J. E., Saraste, M., Runswick, M. J. & Gay, N. J. (1982) EMBO J. 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fry, D. C., Kuby, S. A. & Mildvan, A. S. (1986) Proc. Natl. Acad. Sci. USA 83, 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Story, R. M. & Steitz, T. A. (1992) Nature 355, 374–376. [DOI] [PubMed] [Google Scholar]
- 34.Subramanya, H. S., Bird, L. E., Brannigan, J. A. & Wigley, D. B. (1996) Nature 384, 379–383. [DOI] [PubMed] [Google Scholar]
- 35.Zavanelli, M. I., Britton, J. S., Igel, A. H. & Ares, M., Jr. (1994) Mol. Cell. Biol. 14, 1689–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pause, A. & Sonenberg, N. (1992) EMBO J. 11, 2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, D. W., Kim, J., Gwack, Y., Han, J. H. & Choe, J. (1997) J. Virol. 71, 9400–9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tai, C. L., Pan, W. C., Liaw, S. H., Yang, U. C., Hwang, L. H. & Chen, D. S. (2001) J. Virol. 75, 8289–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider, S., Hotz, H. R. & Schwer, B. (2002) J. Biol. Chem. 277, 15452–15458. [DOI] [PubMed] [Google Scholar]
- 40.Martin, A., Schneider, S. & Schwer, B. (2002) J. Biol. Chem. 277, 17743–17750. [DOI] [PubMed] [Google Scholar]
- 41.Schwer, B. & Meszaros, T. (2000) EMBO J. 19, 6582–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen, J. Y., Stands, L., Staley, J. P., Jackups, R. R., Jr., Latus, L. J. & Chang, T. H. (2001) Mol. Cell 7, 227–232. [DOI] [PubMed] [Google Scholar]
- 43.Linder, P., Tanner, N. K. & Banroques, J. (2001) Trends Biochem. Sci. 26, 339–341. [DOI] [PubMed] [Google Scholar]
- 44.Wiest, D. K., O'Day, C. L. & Abelson, J. (1996) J. Biol. Chem. 271, 33268–33276. [DOI] [PubMed] [Google Scholar]