Abstract
This study demonstrates proof of concept for delivery and expression of compacted plasmid DNA in the central nervous system. Plasmid DNA was compacted with polyethylene glycol substituted lysine 30-mer peptides, forming rod-like nanoparticles with diameters between 8 and 11 nm. Here we show that an intracerebral injection of compacted DNA can transfect both neurons and glia, and can produce transgene expression in the striatum for up to 8 weeks, which was at least 100-fold greater than intracerebral injections of naked DNA plasmids. Bioluminescent imaging (BLI) of injected animals at the 11th postinjection week revealed significantly higher transgene activity in animals receiving compacted DNA plasmids when compared to animals receiving naked DNA. There was minimal evidence of brain inflammation. Intrastriatal injections of a compacted plasmid encoding for glial cell line–derived neurotrophic factor (pGDNF) resulted in a significant overexpression of GDNF protein in the striatum 1–3 weeks after injection.
Introduction
Nearly 70% of ongoing clinical trials use viral vectors as vehicles to shuttle DNA into cells as a means to repair faulty genes.1 While viral vectors can be effective and are widely used, there still remains considerable safety concerns when using viral particles in a therapeutic setting.2 Benefits for developing nonviral vectors for human gene therapy include relatively low costs for large-scale manufacturing, and lack of risk of vector replication and no preexisting vector immunity may translate to potential safety advantages when compared to viral vectors.
Compacted DNA nanoparticles represent a promising nonviral technology that is safe and effective in the airway3,4,5 and eye.6 Single molecules of plasmid DNA can be compacted by polycations to form colloidally stable nanoparticles that have the minimum possible theoretical size based on the partial, specific volumes of the constituent components.7 A preferred polycation is polyethylene glycol (PEG)-substituted lysine 30-mer peptides, which facilitate cell entry and nondegradative trafficking to the nuclei of postmitotic cells by associating with cell-surface nucleolin protein.4,7,8 These particles are completely synthetic, can be designed to diminish an adverse immune response, are stable in saline and nuclease-rich environments, and can be formulated as particles with minimum diameters of 8–11 nm.9 In contrast, naked DNA is quite susceptible to enzymatic degradation, particularly after direct injection into brain tissue.10 In postmitotic cells, strands of naked DNA that ultimately survive cell entry and the cytoplasmic environment must then pass through the nuclear membrane pore, which presents another obstacle given the extended size of hydrated DNA. Nanoparticle technology can circumvent these problems by (i) protecting DNA from degradation and (ii) facilitating transit of compacted DNA, if the nanoparticles are sufficiently small, across the 25 nm nuclear membrane pore of postmitotic cells.7 When dosed in the lung, compacted DNA generates several hundred fold increased transgene activity compared to naked DNA in postmitotic lung epithelial cells.4 In the mouse retina, local delivery of compacted DNA transfects >90% of postmitotic photoreceptor cells.6
In this study we evaluated compacted DNA nanoparticles following direct injection into the rat brain. We report successful transfection and long-term transgene expression in brain cells using DNA plasmids encoding for several reporter genes and glial cell line–derived neurotrophic factor (GDNF).
Results
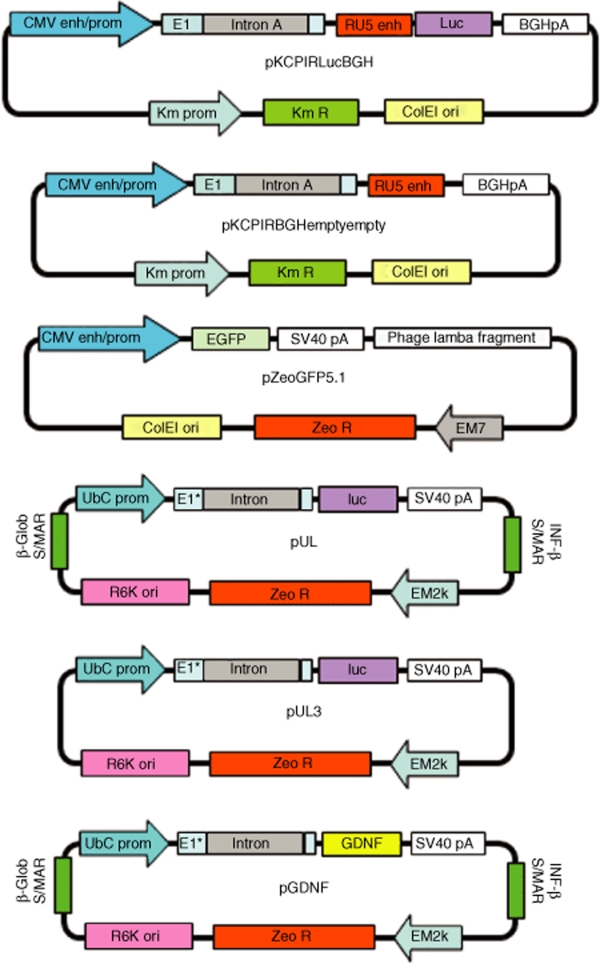
Figure 1 shows the plasmid maps for all plasmids used in this study.
Figure 1.
Plasmid maps.
Intracerebral injection of compacted DNA nanoparticles encoding for EGFP: in vivo transduction
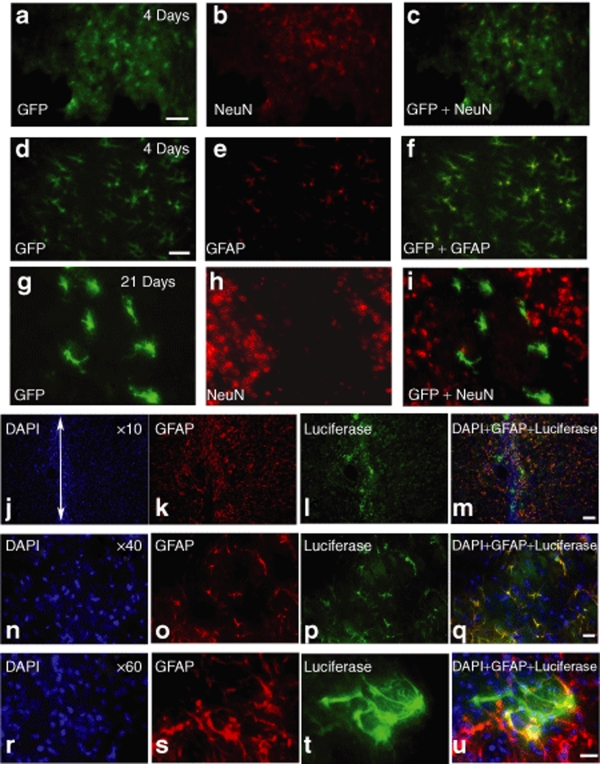
We injected naked DNA plasmids or compacted DNA plasmid nanoparticles directly into the brains of naive adult male Sprague-Dawley rats; the pZeoGFP5.1 plasmid [cytomegalovirus (CMV) promoter] was used in this experiment. Nanoparticles containing plasmids encoding for enhanced green flourescent protein (EGFP) (4.07 µg/µl; 2.0 µl) were stereotactically injected into the left striatum in a single injection tract. Histology was performed 4 or 21 days postinjection. Figure 2a–f shows at 4 days postinjection neurons (NeuN+) and glia (GFAP+) at the injection site coexpressed EGFP. However, at 21 days postinjection very few, if any, neurons showed EGFP colocalization (Figure 2g–i) while most EGFP+ cells were also GFAP+ (data not shown). At no time point did we observe detectable EGFP expression in brain tissue following an injection of naked pZeoGFP5.1 plasmid. No EGFP expression was observed in the substantia nigra.
Figure 2.
Histological analysis of in vivo transfection in neurons or glia by nanoparticles. Nanoparticles were injected into the left striatum using a single injection tract at the following concentrations: compacted pZeoGFP5.1 (4.07 µg/µl; 2.0 µl) or compacted pUL3 (4.1 µg/µl; 4.0 µl). (a–i) Enhanced green flourescent protein (EGFP) immunohistochemical (IHC) in the striatum at 4 or 21 days postinjection. (j–u) Luciferase IHC in the striatum at 28 days postinjection. Single-label IHC for NeuN+ (b,h), GFAP+ (e,k,o,s), EGFP (a,d,g), or luciferase+ (l,p,t) cells. Double-label IHC showing colocalization of EGFP to GFAP+ or NeuN+ cells at 4 days (c,f) or 21 days (i); yellow indicates cells that colabel for EGFP and NeuN or GFAP. Double-label IHC showing colocalization of luciferase to GFAP+ and DAPI+ cells at 28 days postinjection (m,q,u); yellow or reddish-yellow cells indicate colocalization of luciferase and GFAP. White arrow (j) indicates the site of injection. Scale bar (a,d,m) = 50 µm and (q,u) = 20 µm. DAPI, 4',6-diamidino-2-phenylindole; GFAP, glial fibrillary acidic protein.
Intracerebral injection of compacted DNA nanoparticles encoding for luciferase: chemiluminescent detection of transgene expression
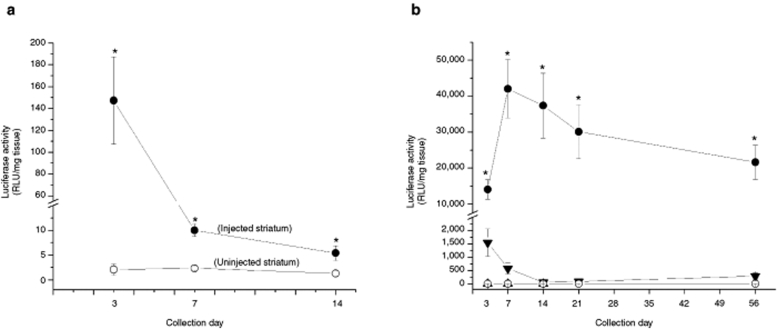
In this experiment, we injected naked DNA or compacted DNA nanoparticles directly into the brains of naive adult male Sprague-Dawley rats. The transferred plasmid, pKCPIRlucBGH, encodes for the reporter gene luciferase and contains the CMV promoter. Nanoparticles containing pKCPIRlucBGH (4.33 µg/µl; 4.0 µl) were stereotactically injected into the left striatum in a single injection tract; no injections were made into the contralateral (right) side of the brain. At 3, 7, or 14 days postinjection, animals were euthanized, brains were removed on ice, and the left and right striata were dissected and stored at −80 °C. Subsequently, frozen tissue was homogenized and luciferase activity in each sample was determined by a chemiluminescent assay. Statistical analysis revealed a significant side × day interaction [two-way ANOVA: F(2,18) = 59.7, P < 0.001] for luminescent values (Figure 3a); side is an independent variable and indicates the injected (left striatum) or noninjected (right striatum). No luminescence was detected in striatal tissue injected with naked pKCPIRlucBGH or in the ventral midbrain tissue samples from any treatment group. In Figure 3a, it can be seen that luciferase activity in the treated striatum remains significantly higher than luciferase expression in the untreated striatum for up to 2 weeks postinjection. While luciferase expression is significantly elevated in the treated striatum at 2 weeks, there was a precipitous drop in activity from the initial time point (3 days) until the 14th day.
Figure 3.
Luciferase activity in the striatum at various postinjection time points as determined by chemiluminescent analysis. All compacted plasmids used the acetate formulation of CK30PEG10k to form the nanoparticle. (a) Compacted pKCPIRlucBGH plasmid nanoparticles encoding for luciferase and containing the cytomegalovirus promoter was injected into the left striatum only (4.33 µg/µl; 4.0 µl); the right striatum served as a control (uninjected); n = 5 for each treatment. Tissue samples were collected at 3, 7, or 14 days postinjection. Filled circles represent mean luciferase activity (±SEM) on the injected side and measured in relative light units (RLU)/mg tissue while open circles represent mean luciferase activity (±SEM) on the uninjected side; *P < 0.05, compacted versus uninjected side. (b) The pUL3 plasmid encoded for luciferase and contains the polyubiquitin C promoter and pKCIRBGHemptyempty is a control plasmid lacking any promoter or transgene. Nanoparticles were injected into the left striatum only; naked pUL3 (filled triangles; 4.2 µg/µl, 4.0 µl), compacted pUL3 (filled circles; 4.1 µg/µl, 4.0 µl), and compacted pKCIRBGHemptyempty (open triangles; 3.9 µg/µl, 4.0 µl). The right striatum served as a control (open circles; no injection). Symbols indicate mean luciferase activity (±SEM) for each treatment group at each time point. *P < 0.05, compacted versus naked pUL3, compacted pKCIRBGHemptyempty, or no injection; n = 5 for each treatment group.
Because of the gene silencing we observed with the pKCPIRlucBGH plasmid in the previous experiment, we decided to try another luciferase expression plasmid that contained a polyubiquitin C promoter instead of CMV. This plasmid, pUL3, was compacted into nanoparticles and injected as naked DNA plasmid (4.2 µg/µl; 4.0 µl) or compacted DNA nanoparticles (4.1 µg/µl; 4.0 µl) into the left striatum of naive Sprague-Dawley rats; in addition, we also injected an “empty” nanoparticle (pKCIRBGHemptyempty; 3.9 µg/µl, 4.0 µl), which was a control plasmid lacking any promoter or transgene. At 3, 7, 14, 21, or 56 days postinjection, animals were euthanized, brains were removed on ice, and the left and right striata were dissected and stored at −80 °C. Subsequently, frozen tissue was homogenized and luciferase activity in each sample was determined by a chemiluminescent assay (Figure 3b). Statistical analysis of luminescent values obtained for each sample at each time point revealed a significant effect of treatment [three-way ANOVA: F(2,85) = 102.85, P < 0.001] and a significant treatment × side × day interaction [three-way ANOVA: F(8,85) = 3.60, P < 0.001]. As expected, no luminescence could be detected in tissue samples taken from animals that received injections of compacted plasmid nanoparticles containing the noncoding plasmid (pKCIRBGHemptyempty) or from the noninjected side of the brain (no injection). On the other hand, compacted pUL3 plasmid showed sustained luminescence in brain tissue from 3 to 56 days after an intracerebral injection; for our chemiluminescent analysis, 56 days was the longest postinjection time point that we examined. In addition, sustained and dose-related luminescence was observed in striatal tissue samples taken 4 weeks following an intrastriatal injection of 16.0 or 8.0 µg of compacted pUL3 nanoparticles: 21,352 ± 4,030 or 8,828 ± 850 relative light units (RLU)/mg tissue, respectively.
Immunohistochemical detection of luciferase enzyme in brain tissue
Figure 2j–u shows a typical example of cells immunohistochemically (IHC) labeled for luciferase following single injections of compacted pUL3 (4.1 µg/µl; 4.0 µl). The propensity of compacted pUL3 nanoparticles to transfect astrocytes was confirmed at 4 weeks postinjection using double-label IHC to identify the types of cells expressing luciferase. Figure 2j–u shows that at 4 weeks postinjection most cells around the injection site that are luciferase+ were also GFAP+. A separate NeuN analysis was performed at 4 weeks and we observed few, if any, NeuN+ cells that were also luciferase+ (data not shown); this analysis is similar to what we observed with the EGFP-expression plasmid at day 21 (Figure 2g–i). However, it is clear in the high power (×60) photomicrographs that not all luciferase+ cells are also GFAP+ and some luciferase+ cells may have a neuronal morphology.
Intracerebral injection of compacted DNA nanoparticles encoding for luciferase: in situ hybridization for luciferase mRNA
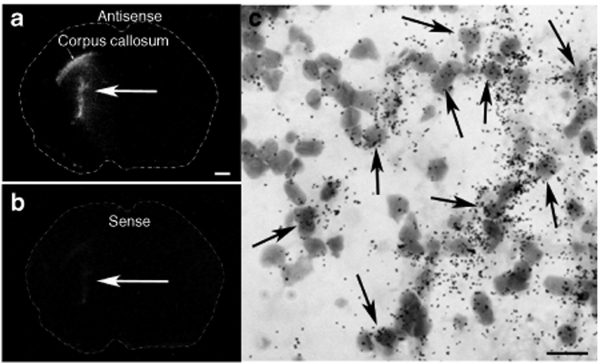
Two weeks following an intrastriatal injection of compacted pUL3 plasmid (4.1 µg/µl; 2 µl) into the left striatum of a naive rats, brain tissue was analyzed for the in situ hybridization localization of luciferase mRNA. Figure 4 shows film autoradiograms of antisense (Figure 4a,c) and sense (Figure 4b) cRNA riboprobe hybridization in adjacent brain sections. Antisense labeling shows hybridization to luciferase mRNA at a site coincident with the injection of the nanoparticles containing compacted pUL3 plasmid. Labeled cells appeared to have both glial and neuronal morphologies. This analysis also shows hybridization to luciferase mRNA occurring within the corpus callosum; this suggests nanoparticles may have a propensity to migrate along the fiber tracts of white matter. Control sense hybridization showed minimal activity.
Figure 4.
Film autoradiograms showing the spatial expression of luciferase mRNA in brain 2 weeks following an intrastriatal injection of compacted pUL3 nanoparticles. Dotted lines indicate the outline of the coronal brain section. Brain section hybridized with the antisense cRNA riboprobe (a,c) and adjacent brain section hybridized with the sense cRNA riboprobe (b). (c) Emulsion autoradiogram showing the cellular expression of luciferase mRNA (clustered dark grains) in striatal cells (gray; Nissl-stained); labeling is most likely occurring in neurons as well as glia. Black arrows point to cells showing cRNA riboprobe hybridization. a, scale bar = 1 mm; c, scale bar = 20 µm.
Intracerebral injection of compacted DNA nanoparticles encoding for luciferase: bioluminescent imaging studies
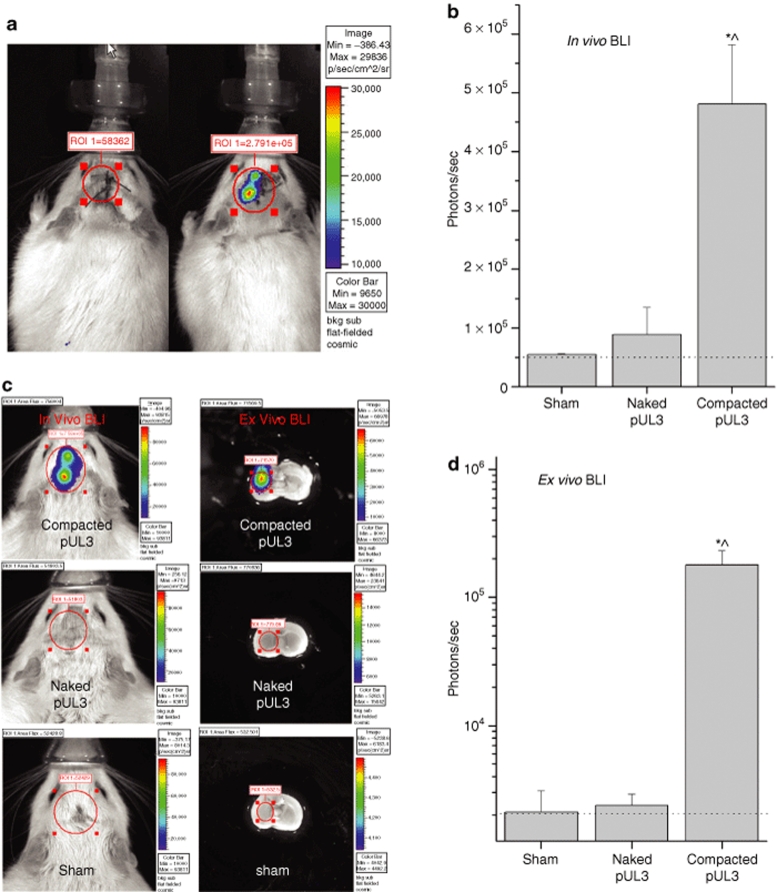
Animals received three injections of naked pUL3 (4.2 µg/µl; 4.0 µl) or compacted pUL3 (3.9 µg/µl; 4.0 µl) into the left hemisphere of the brain. Bioluminescent imaging (BLI) was performed 1, 8, and 11 weeks postinjection. During each imaging session, animals received an injection of luciferin (150 mg/kg, intraperitoneal) and were placed into the IVIS 200 imager. Figure 5a shows an example of in vivo BLI for rats receiving naked pUL3 or compacted pUL3 nanoparticles at 1 week postinjection. Between 8 and 11 weeks postinjection (Figure 5b), photon counts from animals receiving compacted pUL3 nanoparticles were stable and significantly greater than photon counts for animals receiving an equivalent injection of naked pUL3 plasmid or sham-injected animals [one-way ANOVA: F(2,11) = 5.45, P < 0.05]. At the 8th or 11th postinjection week, in vivo BLI was performed and then the animals were euthanized and the brains quickly dissected from the skull in order to image postmortem brain tissue (Figure 5c). Between 8 and 11 weeks postinjection (Figure 5d), statistical analysis of ex vivo BLI data revealed that photon counts in the striatum of an animal receiving compacted pUL3 nanoparticles were stable and significantly greater than the photon counts from animals receiving equivalent injections of naked pUL3 plasmid or sham treatment [one-way ANOVA: F(2,14) = 12.25, P < 0.001].
Figure 5.
In vivo and ex vivo bioluminescent imaging 1–11 weeks following an intracerebral injection of naked or compacted pUL3 plasmid. Rats were injected with luciferin (150 mg/kg, intraperitoneal) 10 minutes prior to image acquisition and maintained under 2.0% isofluorane anesthesia while in the IVIS 200 bioluminescent imager. The pseudocolor image superimposed on a gray-scale image of the rat body or coronal brain section represents photons emitted by live cells following the luciferin/luciferase light reaction that occurs within transfected brain cells and detected by the IVIS 200 imaging system; color scale bar on the right shows the photon counts (photon/s/cm2/sr). Red circle indicates region of interest (ROI) for bioluminescent imaging (BLI) analysis; ROI is centered over the left striatal region and values are reported as photons/s. (a) In vivo BLI 1 week following an intracerebral injection of naked or compacted pUL3. Animal on the left received naked pUL3 (4.2 µg/µl, 4.0 µl) and animal on the right received compacted pUL3 (3.9 µg/µl, 4.0 µl) into the left striatum; injections were targeted for a region 5–6 mm below the surface of the brain. (b) Graph summarizing in vivo BLI data for animals receiving intrastriatal injections of naked pUL3, compacted pUL3, or sham injections at 8–11 weeks postinjection. As shown in a, an approximate 2.5 cm2 circular ROI was centered over the striatal region and photon emissions were quantified within the ROI. Bars represent the average photon emission (photons/s + SEM) for compacted pUL3 (n = 4), naked pUL3 (n = 4), or sham (n = 4). *P < 0.05, compacted pUL3 versus naked pUL3; ^P < 0.05, compacted pUL3 versus sham. (c) In vivo and ex vivo bioluminescent imaging of live animals or postmortem brain 11 weeks following an intrastriatal injection of compacted pUL3 (top row), naked pUL3 (middle row), or sham injection (bottom row). BLI images of the animals receiving compacted or naked pUL3 were obtained from the same two animals shown in a but 10 weeks later. After the initial in vivo BLI session, the animals were euthanized, the brains removed, placed into an ice-cold brain matrix (ASI Instruments, Warren, MI) and cut into 3-mm sections so that the injection sites lie just below the surface facing the camera. While the coronal section for the brain treated with naked pUL3 (middle) is slightly more anterior than that shown for the brain treated with compacted pUL3 (top), the injection site was still fully contained within the sample. It is also important to note that imaging of the coronal sections was completed within 20 minutes following the injection of luciferin, and it is our experience that luciferase remains active 30 minutes postinjection. (d) Graph summarizing ex vivo BLI data for animals receiving intrastriatal injections of naked pUL3, compacted pUL3 or sham injections at 8–11 weeks postinjection. As shown in the ex vivo BLI images (c), an approximate 0.15 cm2 circular ROI was centered over the striatal region of the coronal brain section and photon emissions were quantified within the ROI. Bars represent the average photon emission (+SEM) for compacted pUL3 (n = 5), naked pUL3 (n = 5), or sham (n = 5). Scale for y-axis is log10. Dotted lines in graphs (b,d) indicate background levels of photon emissions. *P < 0.001, compacted pUL3 versus naked pUL3; ^P < 0.001, compacted pUL3 versus sham.
Intracerebral injection of nanoparticles encoding for luciferase: immune markers
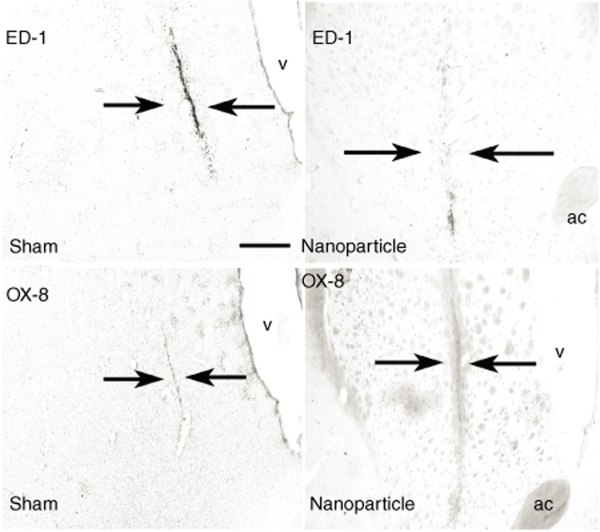
We also determined the extent of the immunogenic response within the brain following injection of compacted pUL3 nanoparticles. Figure 6 shows a minimal immunogenic response to the nanoparticle injection 3 weeks following the injection; most of the immune marker immunostaining for ED-1 (macrophage) or OX-8 (cytotoxic T cell) occurred directly in the cannula track and this is similar to what is seen in animals that receive a sham injection, i.e., made by the injector cannula alone.
Figure 6.
Immunostaining for immune markers in rat brain following an intrastriatal injection of nanoparticles or a sham injection. A single-tract injection of compacted pUL3 (4.1 µg/µl; 4.0 µl) was made into the left striatum; for the control, the injection cannula was lower into the striatum and an equal volume of sterile saline was injected into the site. Arrows point to the location of the injector tract. Low-level expression of the macrophage and microglia marker, ED-1, and the marker for cytotoxic T-cells, OX-8, were identified in brain sections 3 weeks after the injection of nanoparticles; this level of expression was equivalent to the sham-injected site. ac, anterior commissure and scale bar = 500 µm.
Characterization of the compacted pGDNF plasmid
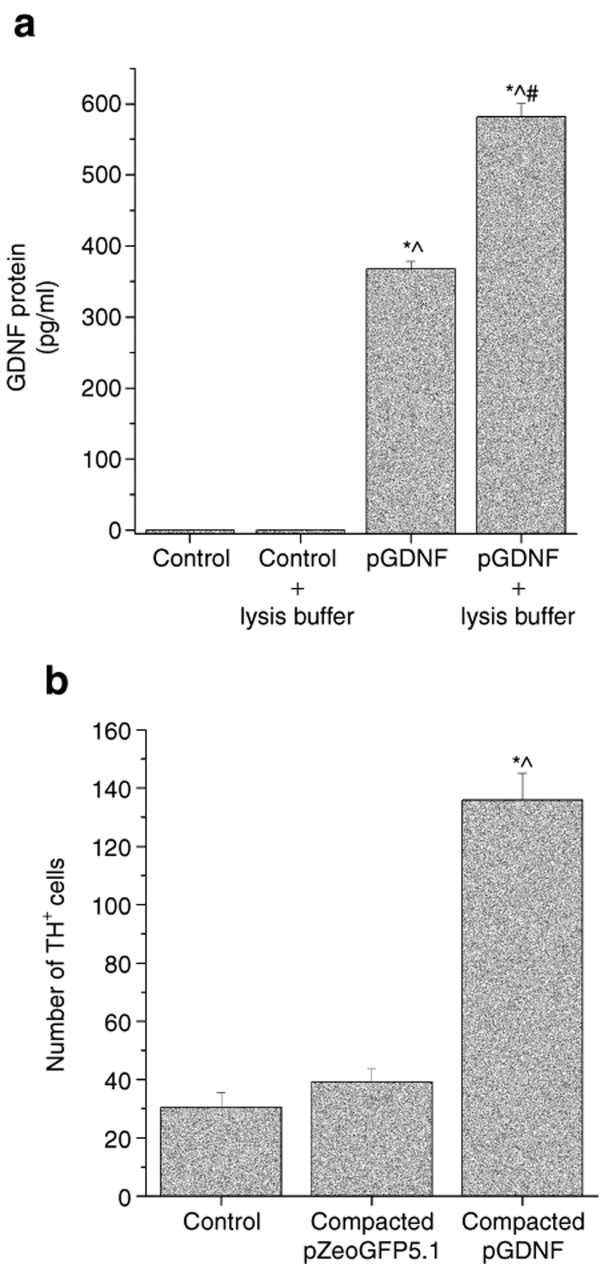
The preceding experiments demonstrated proof of concept that DNA compacted into nanoparticles can be injected into brain, transfect brain cells, and induce transgene expression of several reporter genes. Our next set of experiments was designed to determine whether a relevant neurotrophic factor could be overexpressed in brain cells. pGDNF was designed to overexpress GDNF. pGDNF is a modification of the pUL plasmid (Figure 1); This plasmid contains the polyubiquitin C promoter and the sequence for luciferase has been replaced with the sequence for rat GDNF. In order to determine the biological activity of pGDNF, we first tested pGDNF in cultures of HEK 293 cells. HEK 293 cells were transfected by naked pGDNF and these cells produced and released detectable GDNF protein into the culture media (Figure 7a). Next, we tested the ability of naked or compacted pGDNF to transfect primary cultures of ventral midbrain cells. Cultured ventral midbrain cells were treated with naked pGDNF or compacted pGDNF nanoparticles; the same amount of naked and compacted pGDNF nanoparticles (1.0 µg) were added to each culture well. Statistically higher levels of GDNF (1,771 ± 344 pg/ml) were measured in cultures treated with compacted pGDNF when compared to GDNF (980 ± 327 pg/ml) in cultures treated with naked pGDNF [t(6) = 10.74, P < 0.001]. Because GDNF is a potent neurotrophic factor for dopamine neurons, we dosed another set of cultures to determine whether ventral midbrain cells transfected with compacted pGDNF exerted a neurotrophic effect on the dopamine cells within primary ventral midbrain cultures. In this study, compacted pZeoGFP5.1 nanoparticles were used as a control. Primary cultures were established and then transfected with same amount (1.0 µg) of either compacted pGDNF nanoparticles or compacted pZeoGFP5.1. Histological analysis revealed a significantly higher number of TH+ cells in cultures treated with compacted pGDNF than in cultures treated with compacted pZeoGFP5.1 or untreated controls (Figure 7b). In addition, there was no statistically significant difference in the average number of NeuN+ cells between control (untreated), compacted pZeoGFP5.1, and compacted pGDNF treated cultures [one-way ANOVA: F(2,19) = 1.50; P ≥ 0.25]; this suggests there was no toxic effect of nanoparticles on cultured cells because there was no reduction in the number of neurons.
Figure 7.
In vitro transfection of HEK 293 cells or primary cultures of ventral midbrain cells using pGDNF. (a) HEK 293 cells were cultured and transfected with 1.0 µg/well pGDNF using Lipofectamine. Some of the transfected cells were treated with lysis buffer in order to release intracellular GDNF into the culture media; control cells were not transfected but received Lipofectamine. Media was collected at DIV 7 and GDNF content was measured in the culture media using enzyme-linked immunosorbent assay (ELISA). *P < 0.05, pGDNF or pGDNF + lysis buffer versus control; ^P < 0.05, pGDNF or pGDNF + lysis buffer versus control + lysis buffer; #P < 0.05, pGDNF + lysis buffer versus pGDNF. (b) Transfection of rat ventral midbrain cells and dopaminergic differentiation mediated by compacted pGDNF. Cells were plated at a density of 3.75 × 105 cells per well. One microgram of compacted pZeoGFP5.1 or compacted pGDNF plus Lipofectamine were added per well; control did not receive any plasmid treatment. Media was collected at DIV 7 and GDNF content was measured in the culture media using ELISA. Each bar represents the average number of TH+ cells (+SEM) counted per cell well (n = 8 for each treatment). Statistical analysis revealed significantly higher number of TH+ cells in cultures treated with compacted pGDNF when compared to control cultures or cultures treated with pGeoGFP5.1 [F (2,20) = 99.74, P < 0.001]. *P < 0.001, compacted pGDNF versus pZeoGFP5.1; ^P < 0.001, compacted pGDNF versus control. GDNF, glial cell line–derived neurotrophic factor.
Intracerebral injection of compacted pGDNF nanoparticles
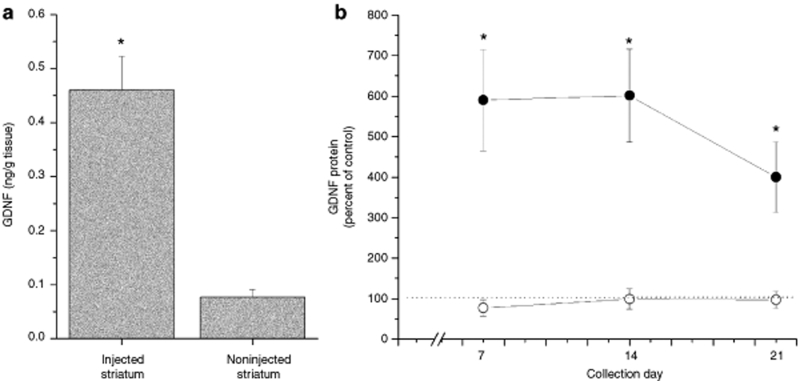
Nanoparticles containing pGDNF were stereotactically injected into two different sites within the left striatum as explained in the methods (3.5 µg/µl; 8.0 µl); no injections were made into the contralateral (right) side of the brain. At 1 week postinjection (Figure 8a), we observed a significant overexpression of GDNF in the injected striatum as compared to the noninjected striatum [t(10) = 5.44, P < 0.001]. Over a 3-week postinjection period, we observed stable overexpression of GDNF in the injected striatum of animals treated with compacted pGDNF nanoparticles that was statistically greater than GDNF levels in the injected striatum of animals treated with naked pGDNF (Figure 8b); the main effect of treatment was statistically significant [one-way ANOVA: F(1, 21) = 16.9, P < 0.001] but the main effect of time was not statistically significant [one-way ANOVA: F(2, 21) = 0.07, P = 0.93].
Figure 8.
Glial cell line–derived neurotrophic factor (GDNF) protein expression in the striatum 1–3 weeks following a dual injection of compacted pGDNF into the left striatum. (a) Animals were euthanized 1 week following the injection and both the left and right striata were dissected from the brain. Tissue was analyzed using enzyme-linked immunosorbent assay (ELISA). Bars represent the average (+SEM) GDNF protein values for the injected (left striatum) and the noninjected (right striatum). *P < 0.001, injected striatum versus noninjected striatum. (b) GDNF protein expression in the striatum 1–3 weeks following a dual injection of compacted or naked pGDNF into the left striatum. Animals were euthanized at 1, 2, or 3 weeks following the injection and both the left and right striata were dissected from the brain. Tissue was analyzed using ELISA. Circles represent the average value of GDNF protein (+SEM) calculated as a percentage of the control side (left/right) for compacted pGDNF (filled circles) or naked pGDNF (open circles). Dotted line indicates control levels of GDNF. *P < 0.001, compacted pGDNF versus naked pGDNF.
Discussion
Here we report that compaction of plasmid DNA into rod-like nanoparticles ranging in size from 8 to 11 nm in diameter can be injected directly, which effectively transfects cells in the brain so that transgene expression of reporter genes or neurotrophic factors is much greater than what is observed following an injection of naked plasmid DNA. Although effective treatments for numerous disorders are predicted based on introduction of normal copies of mutant genes (or antisense RNAi moieties) into disease sites, technical barriers such as cytoplasmic trafficking and nuclear uptake have limited the safe and effective delivery of therapeutic DNA. This nonviral nucleic acid delivery technology has largely overcome various physiologic barriers to introducing DNA into the nuclei of nondividing cells. This technology is based on condensation of single molecules of nucleic acids with PEG-substituted lysine peptides to formulate nanoparticles having the minimum possible theoretical size based on the partial specific volumes of lysine and DNA.7 These DNA nanoparticles are stable >3 years at 4 °C, nonimmunogenic (repeat dosing in the lungs of mice has been demonstrated), noninflammatory,5 and very effective in the airway,3,4,6 and eye.6 The results of this study have extended these findings to brain tissue.
Initial studies with naked DNA or nanoparticles suggested they were less efficient than viral vectors in cell transfection studies, particularly in postmitotic brain cells. Leone et al.11 reported limited success using a nonviral, lipid entrapped, polycation-condensed delivery system (LPD) to induce transgene expression in the brains of rodents, primates, and humans; these nanoparticles had an average particle size of 80–100 nm in diameter. Using polyethylenimine/DNA (PEI/DNA) complexes,12 Oh et al. reported that PEI/DNA nanoparticles with an average size of 100 nm could be injected into dog brain with minimal toxicity.13 However, in this same study it was also reported that intracerebral injection of plasmids encoding for luciferase as naked plasmids or PEI/DNA complexes elicited nearly equivalent levels of transgene activity at 3 days suggesting no real benefit for the PEI/DNA complexes. Efficiency problems in initial nanoparticle gene delivery studies were most likely because these complexes were still too large (≥25 nm) to diffuse efficiently in the extracellular brain matrix14 or cross the nuclear membrane pore. A more recent study demonstrated successful transfection of brain cells using smaller nanoparticles. For instance, Bharali et al. encapsulated DNA plasmids within silica nanoparticles (~30 nm diameter) and injected the nanoparticles directly into the brain and successfully demonstrated transgene expression of reporter genes and neurotrophic factor.15 Of note, polymeric small nanoparticles (<25 nm) but not larger nanoparticles (>42 nm) appear to enter cells and traffic through the cytoplasm of cells using a nondegradative pathway, which underscores the importance of using small particles to deliver plasmid DNA to the nucleus.16
Previous studies using immunoliposomes to deliver a luciferase expression plasmid to brain demonstrated only short-term transgene expression. For instance, intravenous administration of PEGylated brain–targeted immunoliposomes that used luciferase as the transgene demonstrated only a low level of luciferase expression which peaked at 48 hours and a return to baseline activity by 72 hours; the average particle size in this experiment was 72 nm.17 While the route of delivery of the particles used in this cited study and our present study differ, in the cited study particles did cross the blood brain barrier and successfully transfected brain cells but generated detectable luciferase activity that lasted only 2–3 days postadministration. In our present study, we observed sustained luciferase activity following an intracerebral injection that lasted up to the termination point of the study (77 days); therefore, transgene expression in brain may actually continue well beyond 77 days using this DNA nanoparticle formulation.
A recent viral vector study demonstrated the utility of BLI for measuring luciferase activity in the brain of mice or rats injected with a lentiviral vector encoding for luciferase.18 In this study, Deroose et al. used a lentiviral vector to transduce brain cells in various brain regions including the striatum and substantia nigra and then followed luciferase activity over several months. Compared to brain imaging data reported by Deroose et al., where lentiviral vectors encoding luciferase were used, we observed similar levels of luminescence in brains injected with pUL3; both studies used comparable instrumentation to quantify photon emissions. The stability of the in vivo BLI signal for animals receiving intrastriatal injections of compacted pUL3 can be seen in Figure 5a and c; the same animal is shown in both figures, and we observe a consistent pattern of activity from week 1 through week 11. Moreover, ex vivo BLI analysis performed at 8–11 weeks did not reveal photon emissions emanating from the injection target site (striatum) that were significantly greater than background for any of the animals treated with naked pUL3 while all animals treated with compacted pUL3 showed robust BLI signals emanating from the striatum. To date, no other nonviral gene therapy technique has shown the same intensity and duration of luminescence in the brain as what is reported in this study.
Our initial studies used a CMV promoter to drive the expression of the reporter genes EGFP or luciferase. However, we found that transgene expression in brain tissue dropped precipitously during the first week following an intracerebral injection of compacted plasmid DNA, which is not surprising in view of the well-described downregulation of the CMV promoter following in vivo gene transfer. On the other hand, nanoparticles containing plasmids encoding for luciferase and driven by a polyubiquitin C promoter provided a stable level of luminescence up to 11 weeks postinjection. Furthermore, our IHC analysis of EGFP and luciferase expression indicate that transgene expression was colocalized primarily to GFAP+ cells. Our initial IHC study used pZeoGFP5.1, which is an EGFP expression plasmid driven by the CMV promoter. While we observed that both neurons and astrocytes initially (3 days) expressed EGFP, by week 3 most EGFP+ cells were GFAP+ and NeuN−. This is not surprising because adenoviral vectors encoding for GFP and the CMV promoter tend to show stronger expression in astrocytes than neurons in both in vitro and in vivo studies.19 The sustained expression of luciferase in cells transfected with luciferase expression plasmids encoding for the polyubiquitin C promoter is somewhat novel in that this promoter is relatively untested in brain.
Our dissection technique was designed to obtain an area of the striatum that not only included the injection site of the nanoparticles but also a significant amount of striatal tissue that surrounded the injection site. Tissue samples most likely included a fair amount of the striatum that did not come into contact with nanoparticles; therefore, the RLU/mg tissue values that we report are diluted by the inclusion of nontransfected tissue in our assays. However, it is important to point out that there was no significant difference between tissue sample weights for each treatment group [mean values (mg): compacted = 14.8 ± 0.45, naked = 14.8 ± 0.40, empty = 14.1 ± 0.47, noninjected = 14.1 ± 0.36; F(2, 157) = 0.53, P = 0.59]; therefore, we are confident that the dilution factor was the same for each treatment group. Furthermore, our dissections were made ventral to the corpus callosum so it is also unlikely that our tissue analyses included transgene expression of luciferase within the corpus callosum that was observed in the in situ hybridization analysis.
Compacted DNA is an attractive alternative to viral vectors because of their noninflammatory and nonimmunogenic properties;5,6 immunogenicity is an inherent problem with many viral vectors,2 especially if viral vectors need to be redosed.20 In fact, compacted DNA can be repetitively dosed to murine airways without any decrement in transgene expression (Copernicus, unpublished results), suggesting that chronic administration is feasible. We report only a minimal amount of immune activity within the injection site based on macrophage (ED-1) and cytotoxic T cell (OX-8) infiltration into the injection site, which may be due to the injection process itself.
In addition to the reporter gene studies, we also report that this type of nonviral gene therapy may be useful for overexpressing genes that are relevant for the maintenance of cells in the brain. We designed a plasmid that encodes for the neurotrophic factor, GDNF, and its expression was driven by a polyubiquitin C promoter. Here we show that compacted DNA technology can be used to overexpress GDNF in the rat striatum to levels that are 400–600% greater than normal levels for at least 3 weeks, which happens to be the longest time point we have studied using compacted pGDNF nanoparticles. This level of GDNF overexpression is significant because it was recently reported that continuous delivery of GDNF to the striatum using an AAV vector which resulted in a threefold increase over baseline was sufficient to provide neuroprotection to dopamine neurons.21 Future studies will be designed to determine the duration of transgene expression and as well as determining whether these levels are therapeutically significant. A recent study from our laboratory indicates that this level of GDNF overexpression provided trophic support for dopamine neurons grafted into the denervated/transfected striatum.22 The significance of overexpressing GDNF within the striatum as a treatment for Parkinson's disease has been well established. Clinical trials have provided encouraging but controversial evidence that direct injection of GDNF protein into the putamen is an effective therapeutic approach toward counteracting the symptoms of Parkinson's disease. Open-label clinical trials have reported modest improvements in Parkinson's disease patients receiving direct, intracerebral injections of neurotrophic factor GDNF;23,24,25,26 however, a recently concluded phase II double-blind clinical trial was stopped because primary and secondary end points were not met and some patients developed antibodies against GDNF.27 The controversy surrounding these trials is centered on the pump and catheter system used to deliver GDNF protein to the brain and whether the delivery system used in the phase II trial was as efficacious as those used in the other trials28 and whether injection of GDNF into a single site results in poor diffusion throughout the target site.29 Gene delivery offers a means to achieve a continuous and selectively distributed supply of neurotrophic factors to degenerating neurons in specific brain regions. Indeed, several groups have established proof of principle for GDNF gene delivery directly to the nigrostriatal system in animal models of Parkinson's disease using viral vectors.30,31,32,33
In summary, we report successful long-term transgene expression in brain using PEG-substituted lysine peptide DNA nanoparticles as a vehicle for gene delivery. Reporter gene studies demonstrated long-term expression of transgene up to 11 weeks in brain following a direct intracerebral injection of DNA nanoparticles. Moreover, we have used these same nanoparticles to overexpress a relevant neurotrophic factor (GDNF) in brain tissue. More important, this significant level of GDNF expression was produced with minimal evidence of vector inflammation. This study demonstrates the utility of using a nonviral, nanoparticle-based technology for transferring genes to brain cells as a possible therapeutic approach for treating neurodegenerative diseases.
Materials and Methods
Plasmid construction. DNA vectors were constructed using standard molecular biology techniques, following by restriction analysis and sequencing of subcloned regions.34 pKCPIRlucBGH (5,352 bp) is described in ref. 4. pKCPIRBGHemptyempty (3,069 bp) was obtained by deletion of the luc gene and CMV promoter/enhancer sequences from pKCPIRlucBGH. pZeoGFP5.1 (5,147 bp) is described in ref. 7. pUL: the CpG-depleted luciferase gene was amplified from the plasmid pModLucSh (Invivogen) using 5′-ATACACCATGGAGGATGCCAAGAATATTAAG-3′ and 5′-ATACAACTAGTCTAGATTATTTGCCACCCTTCTTGGCCT-3′ primers. The stop codon (TAA) and two restriction sites (XbaI and SpeI) were added to the 3′ end during amplification. The obtained PCR fragment was digested with NcoI and SpeI and subcloned into the pCpG-mcs/NcoI/NheI vector (Invivogen). Then hEF1-α promoter and synthetic intron were deleted using SpeI and NcoI. The polyubiquitin C promoter, and first exon and intron sequences were amplified from the pUbLux plasmid35 using 5′-ACATATCTAGACTGCAGGCCTCC-GCGCCGGGTTTTG-3′ and 5′-GTCTTCCATGGTGGCTAGCTCGTCTAACA-3′ primers. Additional XbaI, PstI, and NcoI sites were added during amplification. The PCR fragment was digested with XbaI and NcoI and subcloned into the prepared promoterless SpeI/NcoI vector described above. The mCMV enhancer was deleted from the obtained vector using PstI. pUL3 (3,850 bp) is derivative of pUL. The S/MAR regions were deleted in the following way. A fragment containing the luciferase expression cassette was prepared by digestion of pUL with EcoRI and blunting with Klenow (~3.1 kb). This fragment was subcloned into pUL/PacI (blunted with T4 polymerase) vector. pGDNF (3,997 bp): The GDNF gene open-reading frame was cut from pLenti-GDNF-IRES-GFP/BamHI (blunted with Klenow)/NheI36,37 and subcloned into pUL/XbaI (blunted with Klenow)/NheI vector. Figure 1 shows plasmid maps for each plasmid used in these studies.
Preparation of condensing peptide. L-Cysteinyl-poly-L-lysine (UCB Bioproducts) was conjugated with 10 kd PEG (Nektar Therapeutics, San Carlos, CA) as described in Liu et al.7 except that trifluoroacetate counterion was replaced with acetate by size-exclusion chromatography on Sephadex G-25 before lyophilization of the PEGylated peptide.
Formulation of compacted DNA nanoparticles. Compacted DNA was manufactured by adding 20 ml of DNA solution (0.1 mg/ml in water) to 2.0 ml of PEGylated condensing peptide (3.2 mg/ml in water) at a rate of 4 ml/min by a syringe pump and through sterile tubing ended with a blunt cannula. During this addition, the tube with peptide was vortexed at a controlled rate so that the two materials mixed instantaneously. Peptide and DNA were formulated at a final amine-to-phosphate ratio of 2:1. The compacted DNA was then filtered through a vacuum-driven sterile filter with 0.2-µm polyethersulfone membrane. The filtered compacted DNA was then concentrated 20- to 30-fold using VIVASPIN centrifugal concentrators (MWCO 100 k). The concentrated DNA was then diluted 20- to 30-fold with sterile 0.9% NaCl for injection (Baxter, Deerfield, IL) and concentrated again 20- to 30-fold to remove excess peptide and exchange water with physiologic saline. The final concentration of compacted DNA was ~4 mg/ml. After formulation, the compacted DNA underwent several quality-control tests, including sedimentation, turbidity, gel electrophoresis, transmission electron microscopy, and fluorescamine assays, as described in Ziady et al.4 and Liu et al.7 Also, endotoxin levels were checked using an ENDOSAFE PTS (Portable Test System) manufactured by Charles River Laboratories (Boston, MA). Estimated number of transfecting nanoparticles: pZeoGFP5.1 = 7.2 × 1011 particles/µl; pKCPIRlucBGH = 7.4 × 1011 particles/µl; pUL3 = 9.2 × 1011 particles/µl; pGDNF = 8.0 × 1011 particles/µl; and pKCIRBGHemptyempty = 11.6 × 1011 particles/µl.
Stereotactic microinjection of compacted DNA nanoparticles or naked DNA into brain. Compacted DNA nanoparticles or naked DNA plasmids were suspended in sterile saline and loaded into a sterile 30-gauge Hamilton syringe needle. Injections were made stereotactically into the brain of isoflurane-anesthetized rats at a rate of 0.5 µl/min for 4 minutes per site. For single-tract injections, the injector was lowered into the striatum to the deepest DV coordinate (DV1) and a 2.0 µl deposit of nanoparticles or naked plasmids was made and then the injector was raised 0.5 mm and a second 2.0 µl deposit was made at this site (DV2); coordinates for injections: AP +0.5, ML +2.5, DV1 −6.5, DV2 −5.0. Injector was left in place for 4–5 minutes after the end of the injections and then slowly withdrawn from the brain at a rate of 1 mm/min. Double-tract injections (pGDNF) were performed similarly using the following coordinates: (i) tract 1, AP + 0.5, ML + 3.5, DV1 −6.5, DV2 −5.0; (ii) tract 2, AP +0.5, ML +2.5, DV1 −6.5, DV2 −5.0. For the BLI study, three-tract injections were performed using the following coordinates: (i) tract 1, AP +1.0, ML +2.8, DV1 −6.6, DV2 −5.0; (ii) tract 2, AP +0.3, ML +2.4, DV1 −6.6, DV2 −5.0; and (iii) tract 3, AP +0.3, ML +3.6, DV1 −6.5, DV2 −5.0.
In vivo bioluminescence imaging. Rats were dosed at the University of Kentucky and then transported to the Case Center for Imaging Research (Case Western Reserve University). Rats were imaged in an IVIS 200 system (Xenogen, Alameda, CA). Anesthesia was induced in an induction chamber with 2.5% isoflurane in 100% oxygen at a flow rate of 1 l/min and maintained in the IVIS with a 2.0% mixture at 0.5 l/min. The rats were injected with d-luciferin (150 mg/kg, intraperitoneal) dissolved in phosphate-buffered saline. Subsequently, the rat was placed in the prone position inside the IVIS imaging chamber. Photons emitted from live rats were acquired as photons/s/cm2/steradian (sr) and analyzed using LivingImage software (Xenogen). Data for the 8–11 week time points was averaged for this postinjection time period. For in vivo BLI analyses, photon emissions were integrated over 10 minutes. For ex vivo BLI analyses, photon emissions were integrated over 2 minutes.
Luciferase chemiluminescent assays. The striatum and substantia nigra from both the treated and untreated sides of the brain were dissected on ice, placed into microcentrifuge tubes, and flash frozen on dry ice. Next, 100 µl of cell culture lysis buffer (Promega, Madison, WI) was added to each sample. Sample was homogenized by pulsing with a Fisher Scientific sonic dismembrator model series 60 for 2–3 seconds and spun at 12,000g at 4 °C for 5 minutes. Furthermore, 100 µl of luciferase reagent was added to disposable luminometer cuvettes; to this, 10 µl of sample supernatant was added and vortexed. Samples shown in Figure 3a were read on a Turner 20/20 luminometer (Turner Biosystems, Sunnyvale, CA) and samples shown in Figure 3b were read on a Modulus luminometer (Turner Biosystems); both luminometers had the following settings: 2-second delay and 10-second read time.
Quantification of GDNF by enzyme-linked immunosorbent assay. Protein levels of GDNF were measured in tissue samples. Each tissue sample was weighed and immediately frozen on dry ice. Subsequently, each tissue sample was homogenized in 300 µl volumes of homogenate buffer [400 mmol/l NaCl, 0.1% Triton-X, 2.0 mmol/l EDTA, 0.1 mmol/l benzethonium chloride, 2.0 mmol/l benzamidine, 0.1 mmol/l phenylmethylsulphonyl fluoride, aprotinin (9.7 TIU/ml), 0.5% bovine serum albumin, 0.1 mol/l phosphate buffer, pH = 7.4]. The homogenate was centrifuged for 10 minutes at 10,000g at 4 °C. Tissue homogenates were assayed using a GDNF Emax ImmunoAssay System (Promega, Madison, WI).
IHC techniques. All rats were deeply anesthetized with sodium pentobarbital and perfused transcardially with ice-cold saline followed by 4% buffered paraformaldehyde (pH 7.4). The brains were postfixed overnight at 4 °C in 4% paraformaldehyde and placed in 30% sucrose. Brain sections (40 µm) were cut on a sliding microtome and stored in cryoprotectant at −20 °C. For IHC detection of markers, free-floating sections were first rinsed in 0.1 mol/l phosphate buffer (22 mmol/l NaH2PO4 and 80 mmol/l K2HPO4, pH = 7.2) followed by 3% H2O2 treatment to inhibit endogenous peroxidase activity. Sections were then rinsed in 0.1 mol/l PO4 and 0.1 mol/l PO4-Triton followed by an overnight incubation in primary antisera. On the next day, tissue was rinsed with buffers and incubated in secondary antisera for 1.5 hours. The following antisera combinations were used: mouse α-NeuN (1:100) (Bioscience Research Reagents, Temecula, CA) with affinity-purified biotinylated goat α-mouse IgG (1:500) or with affinity-purified Cy3-conjugated goat α-mouse (1:500) (Bioscience Research Reagents, Temecula, CA), mouse α-TH (1:4,000) (Bioscience Research Reagents, Temecula, CA) with affinity-purified biotinylated goat antimouse IgG (1:400) (Bioscience Research Reagents, Temecula, CA), rabbit α-GFP (1:1,000) (Abcam, Cambridge, MA) with affinity-purified fluorescein isothiocyanate–conjugated goat α-rabbit IgG (1:500) (Bioscience Research Reagents, Temecula, CA), rabbit α-GFAP (1:1,000) (Bioscience Research Reagents, Temecula, CA) with affinity purified fluorescein isothiocyanate–conjugated goat α-rabbit IgG (1:500) (Bioscience Research Reagents, Temecula, CA), rabbit α-luciferase (1:100) (Fitzgerald Industries, Concord, MA) with affinity-purified biotinylated goat antirabbit IgG (1:500) or with affinity-purified fluorescein isothiocyanate–conjugated goat α-rabbit IgG (1:500) (Bioscience Research Reagents, Temecula, CA), mouse α-OX-8 (1:200) (Bioscience Research Reagents, Temecula, CA) with affinity-purified biotinylated goat α-mouse IgG (1:400) (Bioscience Research Reagents, Temecula, CA), and mouse α-ED-1 (1:1,000) (Bioscience Research Reagents, Temecula, CA) with affinity-purified biotinylated goat α-mouse IgG (1:400) (Bioscience Research Reagents, Temecula, CA). The sections incubated in a biotinylated secondary antiserum were then incubated in an avidin–biotin–peroxidase complex (Vector Labs, Burlingame, CA). Staining was completed by placing the sections in 0.003% H2O2 that contained diaminobenzidine chromogen (1 mmol/l in Tris buffer, pH 7.6) to visualize the peroxidase-catalyzed reaction product. To control for nonspecific staining, some brain sections were incubated without primary antibody.
Statistical analyses. The α level of significance was set at P < 0.05. ANOVA or t-tests were used for statistical analyses; choice of test was dependent on the experimental design. Statistical tests are indicated for each analyses. For chemiluminescent data, a three-way ANOVA was used to analyze luminescent values (RLU/mg tissue); statistically significant interactions between the three independent variables treatment, side (injected or noninjected), and day. Where appropriate, all statistically significant interactions were followed up with the post hoc Tukey test for mean comparisons. Descriptive statistics: means are reported with their corresponding SEM.
For a description of animals, transfection of HEK293 and ventral midbrain culture, tissue dissection technique, immunocytochemical techniques for cultures, cell counts for ventral mesencephalic culture, and in situ hybridization for luciferase mRNA assay, see the Supplementary Material and Methods.
Supplementary MaterialMaterials and Methods.
Supplementary Material
Acknowledgments
This research was supported in part by NS50311 (D.M.Y.), NS040592 (G.M.S.), the Michael J. Fox Foundation for Parkinson's Research (D.M.Y., M.J.C.), the Jelm Foundation (D.M.Y.), and the State of Ohio Biomedical Research and Technology Transfer fund (A.G.Z., M.J.C.). The authors thank Kerstin Lundgren for her excellent technical assistance.
REFERENCES
- Kirchweger G. Nanoparticles—the next big thing. Mol Ther. 2002;6:301–302. doi: 10.1006/mthe.2002.0686. [DOI] [PubMed] [Google Scholar]
- Thomas CE, Ehrhardt A., and , Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- Konstan MW, Davis PB, Wagener JS, Hilliard KA, Stern RC, Milgram LJ, et al. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum Gene Ther. 2004;15:1255–1269. doi: 10.1089/hum.2004.15.1255. [DOI] [PubMed] [Google Scholar]
- Ziady AG, Gedeon CR, Miller T, Quan W, Payne JM, Hyatt SL, et al. Transfection of airway epithelium by stable PEGylated poly-L-lysine DNA nanoparticles in vivo. Mol Ther. 2003;8:936–947. doi: 10.1016/j.ymthe.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Ziady AG, Gedeon CR, Muhammad O, Stillwell V, Oette SM, Fink TL, et al. Minimal toxicity of stabilized compacted DNA nanoparticles in the murine lung. Mol Ther. 2003;8:948–956. doi: 10.1016/j.ymthe.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Farjo R, Skaggs J, Quiambao AB, Cooper MJ., and , Naash MI. Efficient non-viral ocular gene transfer with compacted DNA nanoparticles. PLoS ONE. 2006;1:e38. doi: 10.1371/journal.pone.0000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Li D, Pasumarthy MK, Kowalczyk TH, Gedeon CR, Hyatt SL, et al. Nanoparticles of compacted DNA transfect postmitotic cells. J Biol Chem. 2003;278:32578–32586. doi: 10.1074/jbc.M305776200. [DOI] [PubMed] [Google Scholar]
- Chen X, Kube DM, Cooper MJ., and , Davis PB. Cell surface nucleolin serves as receptor for DNA nanoparticles composed of pegylated polylysine and DNA. Mol Ther. 2008;16:333–342. doi: 10.1038/sj.mt.6300365. [DOI] [PubMed] [Google Scholar]
- Fink TL, Klepcyk PJ, Oette SM, Gedeon CR, Hyatt SL, Kowalczyk TH, et al. Plasmid size up to 20 kbp does not limit effective in vivo lung gene transfer using compacted DNA nanoparticles. Gene Ther. 2006;13:1048–1051. doi: 10.1038/sj.gt.3302761. [DOI] [PubMed] [Google Scholar]
- Schwartz B, Benoist C, Abdallah B, Rangara R, Hassan A, Scherman D, et al. Gene transfer by naked DNA into adult mouse brain. Gene Ther. 1996;3:405–411. [PubMed] [Google Scholar]
- Leone P, Janson CG, Bilaniuk L, Wang Z, Sorgi F, Huang L, et al. Aspartoacylase gene transfer to the mammalian central nervous system with therapeutic implications for Canavan disease. Ann Neurol. 2000;48:27–38. doi: 10.1002/1531-8249(200007)48:1<27::aid-ana6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Nimesh S, Goyal A, Pawar V, Jayaraman S, Kumar P, Chandra R, et al. Polyethylenimine nanoparticles as efficient transfecting agents for mammalian cells. J Control Release. 2006;110:457–468. doi: 10.1016/j.jconrel.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Oh S, Pluhar GE, McNeil EA, Kroeger KM, Liu C, Castro MG, et al. Efficacy of nonviral gene transfer in the canine brain. J Neurosurg. 2007;107:136–144. doi: 10.3171/JNS-07/07/0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne RG., and , Nicholson C. In vivo diffusion analysis with quantum dots and dextrans predicts the width of brain extracellular space. Proc Natl Acad Sci USA. 2006;103:5567–5572. doi: 10.1073/pnas.0509425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharali DJ, Klejbor I, Stachowiak EK, Dutta P, Roy I, Kaur N, et al. Organically modified silica nanoparticles: a nonviral vector for in vivo gene delivery and expression in the brain. Proc Natl Acad Sci USA. 2005;102:11539–11544. doi: 10.1073/pnas.0504926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SK, Hida K, Man ST, Chen C, Machamer C, Schroer TA, et al. Privileged delivery of polymer nanoparticles to the perinuclear region of live cells via a non-clathrin, non-degradative pathway. Biomaterials. 2007;28:2876–2884. doi: 10.1016/j.biomaterials.2007.02.021. [DOI] [PubMed] [Google Scholar]
- Shi N., and , Pardridge WM. Noninvasive gene targeting to the brain. Proc Natl Acad Sci USA. 2000;97:7567–7572. doi: 10.1073/pnas.130187497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroose CM, Reumers V, Gijsbers R, Bormans G, Debyser Z, Mortelmans L, et al. Noninvasive monitoring of long-term lentiviral vector-mediated gene expression in rodent brain with bioluminescence imaging. Mol Ther. 2006;14:423–431. doi: 10.1016/j.ymthe.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Kugler S, Meyn L, Holzmuller H, Gerhardt E, Isenmann S, Schulz JB, et al. Neuron-specific expression of therapeutic proteins: evaluation of different cellular promoters in recombinant adenoviral vectors. Mol Cell Neurosci. 2001;17:78–96. doi: 10.1006/mcne.2000.0929. [DOI] [PubMed] [Google Scholar]
- Lowenstein PR, Mandel RJ, Xiong WD, Kroeger K., and , Castro MG. Immune responses to adenovirus and adeno-associated vectors used for gene therapy of brain diseases: the role of immunological synapses in understanding the cell biology of neuroimmune interactions. Curr Gene Ther. 2007;7:347–360. doi: 10.2174/156652307782151498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslamboli A, Georgievska B, Ridley RM, Baker HF, Muzyczka N, Burger C, et al. Continuous low-level glial cell line-derived neurotrophic factor delivery using recombinant adeno-associated viral vectors provides neuroprotection and induces behavioral recovery in a primate model of Parkinson's disease. J Neurosci. 2005;25:769–777. doi: 10.1523/JNEUROSCI.4421-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurek DM, Fletcher AM, Smith GM, Padegimas L., and , Cooper MJ. Nano particle gene therapy in an animal model of Parkinson's disease: neuroprotection and expression of GDNF in the nigrostriatal pathway. Mol Ther. 2008;16 suppl. 1:S196. [Google Scholar]
- Gill SS, Patel NK, Hotton GR, O'Sullivan K, McCarter R, Bunnage M, et al. Direct brain infusion of glial cell line-derived neurotrophic factor in Parkinson disease. Nat Med. 2003;9:589–595. doi: 10.1038/nm850. [DOI] [PubMed] [Google Scholar]
- Slevin JT, Gerhardt GA, Smith CD, Gash DM, Kryscio R., and , Young B. Improvement of bilateral motor functions in patients with Parkinson disease through the unilateral intraputaminal infusion of glial cell line-derived neurotrophic factor. J Neurosurg. 2005;102:216–222. doi: 10.3171/jns.2005.102.2.0216. [DOI] [PubMed] [Google Scholar]
- Slevin JT, Gash DM, Smith CD, Gerhardt GA, Kryscio R, Chebrolu H, et al. Unilateral intraputamenal glial cell line-derived neurotrophic factor in patients with Parkinson disease: response to 1 year of treatment and 1 year of withdrawal. J Neurosurg. 2007;106:614–620. doi: 10.3171/jns.2007.106.4.614. [DOI] [PubMed] [Google Scholar]
- Love S, Plaha P, Patel NK, Hotton GR, Brooks DJ., and , Gill SS. Glial cell line-derived neurotrophic factor induces neuronal sprouting in human brain. Nat Med. 2005;11:703–704. doi: 10.1038/nm0705-703. [DOI] [PubMed] [Google Scholar]
- Lang AE, Gill S, Patel NK, Lozano A, Nutt JG, Penn R, et al. Randomized controlled trial of intraputamenal glial cell line-derived neurotrophic factor infusion in Parkinson disease. Ann Neurol. 2006;59:459–466. doi: 10.1002/ana.20737. [DOI] [PubMed] [Google Scholar]
- Morrison PF, Lonser RR., and , Oldfield EH. Convective delivery of glial cell line-derived neurotrophic factor in the human putamen. J Neurosurg. 2007;107:74–83. doi: 10.3171/JNS-07/07/0074. [DOI] [PubMed] [Google Scholar]
- Salvatore MF, Ai Y, Fischer B, Zhang AM, Grondin RC, Zhang Z, et al. Point source concentration of GDNF may explain failure of phase II clinical trial. Exp Neurol. 2006;202:497–505. doi: 10.1016/j.expneurol.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Choi-Lundberg DL, Lin Q, Schallert T, Crippens D, Davidson BL, Chang YN, et al. Behavioral and cellular protection of rat dopaminergic neurons by an adenoviral vector encoding glial cell line-derived neurotrophic factor. Exp Neurol. 1998;154:261–275. doi: 10.1006/exnr.1998.6887. [DOI] [PubMed] [Google Scholar]
- Mandel RJ, Snyder RO., and , Leff SE. Recombinant adeno-associated viral vector-mediated glial cell line-derived neurotrophic factor gene transfer protects nigral dopamine neurons after onset of progressive degeneration in a rat model of Parkinson's disease. Exp Neurol. 1999;160:205–214. doi: 10.1006/exnr.1999.7203. [DOI] [PubMed] [Google Scholar]
- Kordower JH, Emborg ME, Bloch J, Ma SY, Chu Y, Leventhal L, et al. Neurodegeneration prevented by lentiviral vector delivery of GDNF in primate models of Parkinson's disease. Science. 2000;290:767–773. doi: 10.1126/science.290.5492.767. [DOI] [PubMed] [Google Scholar]
- Kirik D, Rosenblad C, Bjorklund A., and , Mandel RJ. Long-term rAAV-mediated gene transfer of GDNF in the rat Parkinson's model: intrastriatal but not intranigral transduction promotes functional regeneration in the lesioned nigrostriatal system. J Neurosci. 2000;20:4686–4700. doi: 10.1523/JNEUROSCI.20-12-04686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF., and , Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY; 1989. [Google Scholar]
- Gill DR, Smyth SE, Goddard CA, Pringle IA, Higgins CF, Colledge WH, et al. Increased persistence of lung gene expression using plasmids containing the ubiquitin C or elongation factor 1alpha promoter. Gene Ther. 2001;8:1539–1546. doi: 10.1038/sj.gt.3301561. [DOI] [PubMed] [Google Scholar]
- Baumgartner BJ., and , Shine HD. Targeted transduction of CNS neurons with adenoviral vectors carrying neurotrophic factor genes confers neuroprotection that exceeds the transduced population. J Neurosci. 1997;17:6504–6511. doi: 10.1523/JNEUROSCI.17-17-06504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner BJ., and , Shine HD. Permanent rescue of lesioned neonatal motoneurons and enhanced axonal regeneration by adenovirus-mediated expression of glial cell-line-derived neurotrophic factor. J Neurosci Res. 1998;54:766–777. doi: 10.1002/(SICI)1097-4547(19981215)54:6<766::AID-JNR4>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.