Abstract
Small interfering RNAs (siRNAs) are short, double-stranded RNAs that mediate efficient gene silencing in a sequence-specific manner by utilizing the endogenous RNA interference (RNAi) pathway. The current standard synthetic siRNA structure harbors a 19–base-pair duplex region with 3′ overhangs of 2 nucleotides (the so-called 19+2 form). However, the synthetic 19+2 siRNA structure exhibits several sequence-independent, nonspecific effects, which has posed challenges to the development of RNAi therapeutics and specific silencing of genes in research. In this study, we report on the identification of truncated siRNA backbone structures with duplex regions shorter than 19 bp (referred to as asymmetric shorter-duplex siRNAs or asiRNAs) that can efficiently trigger gene silencing in human cell lines. Importantly, this asiRNA structure significantly reduces nonspecific effects triggered by conventional 19+2 siRNA scaffold, such as sense-strand–mediated off-target gene silencing and saturation of the cellular RNAi machinery. Our results suggest that this asiRNA structure is an important alternative to conventional siRNAs for both functional genomics studies and therapeutic applications.
Introduction
Small interfering RNAs (siRNAs) are short, double-stranded RNAs (dsRNAs) that mediate efficient gene silencing in a sequence-specific manner.1 The specific cleavage of mRNA molecules targeted by siRNAs is mediated by the endogenous RNA interference (RNAi) pathway, which is present in most eukaryotic cell types.2 Using Drosophila melanogaster embryo lysates, Elbashir et al.3 identified siRNAs with 19-base-pair (bp) duplex regions and 2-nucleotide (nt) 3′ overhangs (the so-called 19+2 structure) as the most efficient triggers of sequence-specific mRNA degradation. In contrast, blunt-ended siRNAs or siRNAs that harbor duplex regions shorter than 19 bp were shown to be inefficient silencing structures in Drosophila embryo lysates even at the highest concentration tested (100 nmol/l).3 The 19+2 siRNA structure has thus become the standard for designing gene-silencing RNA molecules for research and therapeutic applications.
Despite the significant promise of siRNA technology, several studies have demonstrated nonspecific effects triggered by conventional 19+2 siRNA structures. First, siRNA can silence nontarget genes either by imperfect pairing between mRNA molecules and the antisense strand of siRNA,4,5 or by incorporation of sense strand into RNA-induced silencing complex (RISC) that results in the cleavage of mRNAs complementary to the sense strand.6 Second, excess amounts of siRNAs can saturate the cellular RNAi machinery7 and competitively inhibit the activity of other siRNAs8,9,10 or microRNAs (miRNAs).11 Third, while siRNAs were originally designed to circumvent the dsRNA-induced innate immune response, several studies have reported that nonspecific innate immune response can be induced by siRNAs.12,13,14,15,16 These nonspecific effects triggered by siRNAs limit the development of siRNA as a specific tool for functional genomics studies and as a therapeutic modality. Although several studies have investigated chemical modifications to circumvent some of the nonspecific effects of 19+2 siRNAs,17,18,19 modification of the backbone structure of 19+2 siRNAs to alleviate nonspecific effects has not been reported.
In recent reports, researchers have attempted to introduce diverse structural variations into the siRNA backbone without the loss of gene-specific silencing activity.20,21,22 It appears that some flexibility in siRNA backbone structure exists and that deviation from the originally proposed 19+2 structure is possible. For example, we have recently shown that the 2-nt 3′ overhang is dispensable for efficient gene silencing by an siRNA that targets the gene encoding Lamin A/C in HeLa cells.20 siRNAs longer than 19 bp, that is, the 27-mer Dicer-substrate siRNA, have been tested and shown, in some cases, to display silencing efficiencies greater than those of the corresponding 19+2 siRNAs.23
In this study, we report an asymmetric shorter-duplex siRNA backbone structure with duplexes shorter than 19 bp for mediating efficient gene silencing. Importantly, we show that this RNA duplex structure significantly reduces nonspecific effects caused by conventional 19+2 siRNA scaffold, such as sense strand–mediated off-target gene silencing and saturation of the cellular RNAi machinery.
Results
Asymmetric shorter-duplex siRNAs with 3′-antisense overhang trigger efficient gene silencing comparable to conventional 19+2 siRNAs
While we were studying blunt-ended siRNA structural variants targeting TIG3 mRNA, we observed that siRNAs with duplex region shorter than 19 bp can silence their target gene expression significantly. TIG3 mRNA level was reduced to about 13% of original level by 15 bp-long blunt siRNA (Supplementary Figure S1), suggesting that duplex RNA as short as 15 bp can be successfully incorporated into RISC and trigger gene silencing via RNAi. However, these blunt-ended, shorter-duplex siRNAs showed somewhat reduced silencing activities at low concentration when directly compared with a conventional 19+2 siRNA (Supplementary Figure S1). The reduced activities of blunt-ended, shorter-duplex siRNAs could be due to reduced incorporation into RISC or insufficient length of antisense strand for optimal gene silencing.
We wondered whether we could increase the activity of these shorter-duplex siRNAs while maintaining the duplex length, by increasing the length of the antisense strand. To test this, we synthesized a series of siTIG3 structural variants (Figure 1a). The length of the guide (antisense; AS) strand of these siRNAs was fixed to 19 nt, but the passenger (sense)-strand length was varied. This process yielded several siRNA structures with duplex regions shorter than 19 bp and AS strands with 3′-overhangs of various lengths (Figure 1a). We named these as 17+2A, 16+3A, 15+4A, 14+5A, and 13+6A, which refer to a 17-bp duplex with a 2-nt 3′-overhang on the AS strand, a 16-bp duplex with 3-nt 3′-overhang on the AS strand, a 15-bp duplex with 4-nt 3′-overhang on the AS strand, a 14-bp with 5-nt 3′-overhang on the AS strand, and a 13-bp duplex with 6-nt 3′-overhang on the AS strand, respectively. These short RNA duplexes were transfected into HeLa cells using Lipofectamine 2000, and TIG3 gene-silencing efficiency was measured at two different concentrations (10 and 1 nmol/l) using a quantitative real-time reverse transcription–PCR (RT-PCR). To our surprise, two structures with duplexes shorter than 19 bp, 17+2A and 16+3A, showed silencing activity comparable to that of the conventional 19+2 siRNA at both tested concentrations (Figure 1b). At 1 nmol/l, the 15+4A and 14+5A siRNAs showed slightly reduced gene-silencing activity relative to the 19+2 siRNA, while the 13+6A siRNAs showed reduced silencing at both concentrations tested (Figure 1b). Taken together, these results demonstrate that siRNAs with duplexes shorter than 19 bp can trigger effective gene silencing in human cells. Similar results were observed when we transfected these siRNA structural variants into T98G cells (Supplementary Figure S2). We also tested the importance of the overhang position with fixed AS sequence. As shown in Supplementary Figure S3, siRNA structures that have a 3′-overhang on the AS strand (15+4A structure) showed silencing activities that were comparable to that of the 19+2 siRNA, while siRNA with 5′-overhang on the AS strand (15-4A structure) showed reduced silencing activity. We termed this siRNA backbone structure “asymmetric shorter-duplex siRNA (asiRNA).”
Figure 1.
Gene silencing triggered by asymmetric shorter-duplex siRNAs. (a) Structures of asiRNAs that target TIG3 mRNA. (b) Activities of asiRNAs (shown on the x-axis) that target TIG3 mRNA in HeLa cells. siRNAs were transfected into HeLa cells using Lipofectamine 2000, and TIG3 mRNA levels were analyzed by quantitative real-time reverse transcription–PCR 24 hours following transfection. The values plotted as “TIG3 mRNA levels” on the y-axis in the figure were calculated as the TIG3 mRNA level divided by the GAPDH (control) mRNA level. All data in the graph represent mean ± SD values of three independent experiments. (c) Activities of asiRNAs that target Survivin mRNA in HeLa cells. (d) Activities of asiRNAs that target LaminA/C mRNA in HeLa cells. (e) Activities of asiRNAs that target Integrin mRNA in HeLa cells. For c–e, see the legend of b for details. 0 nmol/l, control with no siRNA transfected. asiRNA, asymmetric shorter-duplex siRNA; siRNA, small interfering RNA.
We then tested whether active asiRNA structures can be designed for other mRNA targets. siLamin 19+2 (ref. 1), siSurvivin 19+2 (ref. 24), and siIntegrin 19+2 (ref. 25), which are conventional 19+2 siRNAs that target the LaminA/C, Survivin, and Integrin αv subunit, respectively, were converted to asiRNA structures (Supplementary Figure S4). The gene-silencing efficiencies of these asiRNA structures were then compared with those of their corresponding 19+2 structures in HeLa cells (Figure 1c–e). The 17+2A and 16+3A asiRNA structural variants efficiently reduced the amounts of LaminA/C, Survivin, and Integrin mRNAs at all concentrations tested, and silencing efficiencies were comparable to those of the 19+2 structures (Figure 1c–e). IC50 values of active 16+3A asiRNAs were either almost identical to those of the 19+2 siRNAs (in the case of siTIG3 and siSurvivin), or slightly increased, about twofold higher than those of the 19+2 siRNAs (in the case of siLamin and siIntegrin) (Supplementary Figure S5). We also converted 16+3A structures of siTIG3 and siSurvivin to 16+5A structures by adding dTdT overhangs at the 3′-end of antisense strands (Supplementary Figure S6a,b), and the resulting 16+5A siRNAs showed comparable activities with those of 19+2 structures (Supplementary Figure S6c,d). We then designed additional asiRNAs for seven different target genes, and all asiRNAs showed comparable activities with those of 19+2 siRNAs (Supplementary Figure S6e–k). We also performed western blot to confirm the efficient target protein knockdown by asiRNAs (Supplementary Figure S7).
Survivin, an inhibitor of apoptosis protein (IAP), is required for cell viability and proper cell cycle progression.26 A variety of cancer cells show abundant expression of Survivin, and several studies have shown that blocking Survivin function inhibits cell proliferation and induces polyploid phenotypes in diverse cell lines.27 These findings suggest that Survivin may be a prime drug target for cancer therapy. We transfected the Survivin-specific siRNAs, siSurvivin 19+2 and 16+3A, into HeLa cells and then used diamidinophenyl indole (DAPI) staining to visualize the phenotypic changes triggered by these siRNAs. HeLa cells transfected with siSurvivin 19+2 showed a larger cell size and an increase in the number of polyploid cells relative to control cells (Supplementary Figure S8a). siSurvivin 16+3A also induced the polyploid phenotype in HeLa cells (Supplementary Figure S8a). To quantify the percentage of polyploid cells in the cell populations, we performed propidium iodide staining of the siRNA-transfected cells followed by flow cytometry. Supplementary Figure S8b showed that the 19+2 and 16+3A siRNAs induced the production of a similar percentage of polyploid cells. Therefore, siSurvivin 16+3A not only silenced Survivin mRNAs to the same extent as did the 19+2 siRNA (Figure 1c), but also induced similar phenotypic changes.
AsiRNAs induce gene silencing through the RNAi pathway utilized by the conventional 19+2 siRNAs
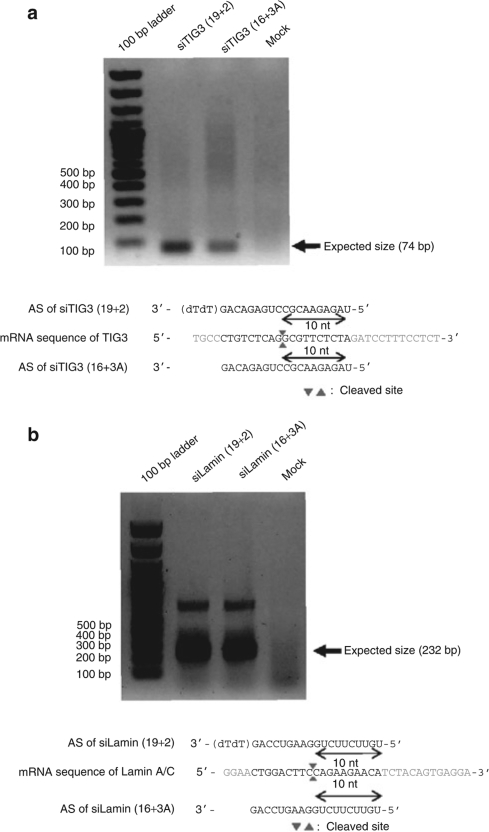
To test whether the asiRNAs silence genes via the same RNAi pathway used by conventional 19+2 siRNAs, we first knocked-down Argonaute-2 (Ago2) levels using an siRNA-targeting Ago2 (siAgo2) in HeLa cells. Both Ago2 mRNA and protein levels were reduced (Supplementary Figure S9a,b). We found that gene silencing by both 19+2 and 16+3A structures of siSurvivin, siLamin, and siIntegrin was markedly reduced following cellular Ago2 knockdown, confirming that gene silencing by asiRNA is also Ago2 dependent (Supplementary Figure S9d–f). We note that TIG3 gene silencing by 19+2 or 16+3A siTIG3 was unaffected by Ago2 reduction in our experiment (Supplementary Figure S9c). These findings suggest that both 19+2 and 16+3A siTIG3 can function normally at reduced Ago2 levels because we were unable to completely knock down Ago2 in HeLa cells. We further performed 5′-RACE analysis to analyze the cleavage site of target mRNAs by shorter-duplex siRNAs. Both TIG3 and LaminA/C mRNAs were cleaved at a position 10 nt from the 5′-end of the antisense strand of siTIG3 and siLamin structural variants (Figure 2a,b), confirming that asiRNAs induce gene silencing through the RNAi pathway utilized by the conventional 19+2 siRNAs.
Figure 2.
16+3A and 19+2 siRNAs cleave identical site within target mRNAs. (a) PCR products of 5′-RACE assay from siTIG3-transfected cells. TIG3 mRNA cleavage sites were analyzed by 5′-RACE assay and sequencing. The antisense strand of siTIG3(19+2) is shown in blue and the antisense strand of siTIG3(16+3A) is shown in red. Cleavage sites are marked with arrowheads. (b) PCR products of 5′-RACE assay using siLamin-transfected cells. LaminA/C mRNA cleavage sites were analyzed by 5′-RACE assay and sequencing. The antisense strand of siTIG3(19+2) is shown in blue and the antisense strand of siTIG3(16+3A) is shown in red. Cleavage sites are marked with arrowheads. siRNA, small interfering RNA.
Studies have shown that siRNA antisense strands alone can sometimes execute RNAi-mediated gene silencing.28 We tested the gene-silencing activity of antisense strands from each siRNA, and found no activity for three siRNA antisense strands (siTIG3, siLamin, and siIntegrin; data not shown) and measurable but severely reduced gene-silencing activity for siSurvivin antisense strand (Supplementary Figure S5b). These data suggest that the gene-silencing activity of asiRNA is due to dsRNA-mediated RNAi.
Because asiRNAs have a shortened duplex region and longer overhangs than 19+2 structures, we tested whether it might be more susceptible to degradation by serum nucleases than the latter. Supplementary Figure S10 shows that both 19+2 and 16+3A siRNAs have similar kinetics of degradation when incubated with serum, suggesting that the serum stability of asiRNAs is comparable to that of conventional 19+2 siRNAs.
AsiRNAs show reduced sense-strand–mediated off-target silencing activity
Off-target gene silencing is a key obstacle to the development of RNAi therapeutics. One mechanism of off-target gene silencing is the incorporation of sense strand into RISC.6 In the assembly and maturation of RISC, an ATP-dependent helicase activity initiates siRNA duplex unwinding at the thermodynamically unstable end of the siRNA, resulting in the preferential incorporation of one strand into RISC over the other.29 Therefore, in order to minimize incorporation of the sense strand into the RISC, synthetic siRNAs are designed such that the 5′-end of the AS strand is thermodynamically more unstable than that of the sense strand.29,30 Nonetheless, it has been observed that manipulating the thermodynamic stability of each end of a synthetic siRNA is not sufficient to eliminate sense-strand–mediated off-target gene silencing.6
We hypothesized that because asiRNA structures are highly asymmetric and have sense strands whose lengths are suboptimal for effective gene silencing, these asiRNAs should yield less sense-strand–mediated off-target gene silencing than 19+2 siRNAs. To test this hypothesis, we designed expression vector constructs that encoded luciferase mRNA bearing either a sense or an AS siRNA target sequence in the 3′ untranslated region. We then co-transfected HeLa cells with these luciferase expression vector constructs and siSurvivin or siTIG3 structural variants and measured the resulting luciferase activity. Although siSurvivin 19+2 and siTIG3 19+2 were designed with AS strands that had thermodynamically unstable 5′ ends, these two siRNAs showed strong sense-strand–mediated gene-silencing activity (Figure 3a,b). In contrast, 16+3A siTIG3 and siSurvivin yielded significantly reduced gene-silencing activity by their sense strands (Figure 3a,b). These data demonstrate that asiRNAs display less sense-strand–mediated off-target silencing than 19+2 siRNAs.
Figure 3.
AsiRNAs show reduced sense-strand–mediated off-target gene silencing. HeLa cells were transfected with a luciferase reporter plasmid that carried either a Survivin AS, Survivin sense, TIG3 AS, or TIG3 sense-target sequence, without (0 nmol/l) or with 10 nmol/l of Survivin or TIG3 19+2 or 16+3A siRNAs. Luciferase activity was measured 48 hours after transfection. (a) Gene-silencing activities of sense and AS strands of siSurvivin variants. (b) Gene-silencing activities of sense and AS strands of siTIG3 variants. asiRNA, asymmetric shorter-duplex siRNA.
To test whether the asymmetric nature of the asiRNA is responsible for the reduction in sense-strand–mediated off-target silencing, we synthesized symmetric shorter-duplex siRNAs with 3 nt overhang at the 3′ end of both strands (designated as 16+3) (Supplementary Figure S11a). We then compared the activities of sense and antisense strand of 16+3 siRNAs with those of 16+3A siRNA variants. As shown in Supplementary Figure S11b, both symmetric (16+3) siRNAs showed stronger sense-mediated off-target silencing activities than their corresponding asymmetric (16+3A) siRNAs. These results demonstrate that reduced sense off-target silencing is most efficiently achieved in asymmetric structures.
AsiRNAs have less or no inhibition of exogenous siRNA and endogenous miRNA activity
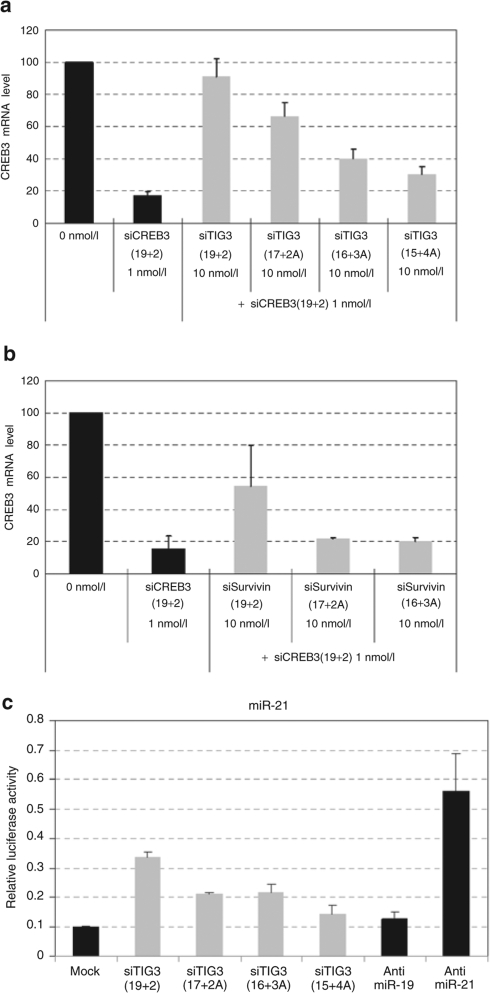
Recent reports have documented another unexpected complication of RNAi-mediated gene silencing: saturation of the RNAi machinery (e.g., Ago2) by exogenously introduced siRNAs,7 which can result in competition between two siRNAs.8,31 We assessed the ability of our asiRNA variants to saturate the RNAi machinery by measuring the competition potency of these siRNAs when co-transfected into HeLa cells along with another siRNA. When co-transfected with 19+2 siCREB3, 19+2 siTIG3 effectively reduced siCREB3-mediated gene silencing (Figure 4a). To our surprise, the ability of 17+2A and 16+3A siTIG3 to reduce 19+2 siCREB3–mediated gene silencing was significantly reduced, compared with that of its 19+2 counterpart, and 15+4A siTIG3 displayed almost no competition potency (Figure 4a). A similar trend in competition potency was observed for the asiRNA structural variants of siSurvivin (Figure 4b).
Figure 4.
AsiRNAs have less or no inhibition of the activities of other siRNAs or miRNAs. (a) siRNAs that target CREB3 mRNA [siCREB3 (19+2)] and competitor siRNA variants (shown on the x-axis) were co-transfected into HeLa cells at the indicated concentrations. CREB3 mRNA levels were analyzed by quantitative real-time reverse transcription–PCR 24 hours after transfection. The CREB3 mRNA levels plotted on the y-axis in the figure were calculated as the CREB3 mRNA level divided by the GAPDH (control) mRNA level. All data in the graph represent mean ± SD values of three independent experiments. The black bars labeled 0 nmol/l and siCREB3(19+2) 1 nmol/l represent transfections of Lipofectamine 2000 only and 1 nmol/l of siCREB3(19+2) only, respectively. For experiments represented by the gray bars, 10 nmol/l competitor siRNAs (siTIG3 19+2, siTIG3 17+2A, siTIG3 16+3A, or siTIG3 15+4A) were transfected with 1 nmol/l siCREB3(19+2). (b) HeLa cells were transfected with 1 nmol/l siCREB3(19+2) with or without 10 nmol/l Survivin siRNAs (Survivin 19+2, Survivin 17+2A, or Survivin 16+3A) into HeLa cells. See the legend of a for details. (c) Changes in the gene-silencing activity of miR-21 by transfection of exogenous siRNAs into HeLa cells. HeLa cells were transfected with a pMIR-luc–based firefly luciferase reporter plasmid (200 ng/ml) that contained a miR-21 binding site, the control pRL-SV40 Renilla luciferase expression vector (2 ng/ml), and one of several siTIG3 variants at a concentration of 10 nmol/l. The firefly luciferase activity was normalized by dividing it by the Renilla luciferase activity. Luciferase activity of the control luciferase reporter was set as one. All data in the graph represent means ± SD values of two independent experiments. Mock, mock-transfected control HeLa cells; anti-miR-19 and anti-miR-21, HeLa cells treated with an antagomir against miR-19 or miR-21, respectively.
It has been also shown that siRNAs compete with endogenous miRNAs to inhibit miRNA function.31 Our observation that asiRNAs have little or reduced competition with 19+2 structures (Figure 4a,b) suggests that cellular RNAi machinery saturation may be reduced or prevented with our asiRNA structures. Thus, we tested whether asiRNA structures have less inhibition of endogenous miRNA activity. We transfected HeLa cells with a luciferase reporter plasmid harboring target sequences for the endogenous miRNA miR-21 in the 3′ untranslated region and the ability of miR-21 to inhibit luciferase gene expression was measured (Figure 4c). Luciferase activity was dramatically reduced (approximately tenfold) in HeLa cells transfected with the miR-21 luciferase reporter construct, relative to that produced by cells transfected with the control luciferase reporter without miR-21 target sequence (Figure 4c). Consistent with a recent report,31 co-transfection of HeLa cells with the miR-21 luciferase reporter and the 19+2 siTIG3 yielded an increase in luciferase activity (Figure 4c). These results suggest that the exogenous 19+2 siTIG3 interferes with miR-21-mediated luciferase gene silencing. Importantly, the corresponding 17+2A and 16+3A structures showed less inhibition of miR-21 activity, with the 15+4A showing almost no inhibition of miR-21 activity in cells (Figure 4c). These findings demonstrate that asiRNAs have less or no inhibition of endogenous miRNA activity.
A recent study has shown that inhibition of phosphorylation of the 5′-end of the sense strand of siRNAs through chemical modifications results in reduced sense-strand–mediated off-target silencing.19 We tested whether the reduction of sense-strand activity is sufficient to alleviate siRNA-mediated RNAi saturation. 5′-end of the sense strand of 19+2 siTIG3 was amine-modified (Supplementary Figure S12a), and the resulting structural variant showed reduced sense-strand–mediated gene silencing as expected (Supplementary Figure S12b). However, this siRNA still maintained strong competition potency that is similar to the unmodified 19+2 siTIG3 (Supplementary Figure S12c). Therefore, the reduction of competition potency of asiRNA cannot be explained by the lack of sense-strand activity. Rather, we suggest that the unique backbone structure of asiRNA is responsible for the reduced competition potency.
Discussion
Chemical modifications to the conventional 19+2 siRNA duplex structure have been reported to improve the specificity of siRNA-mediated gene silencing at the cost of reduced gene-silencing efficiency. Despite these efforts, the nonspecific effect of 19+2 siRNA structure remains a major bottleneck in the application of RNAi. Unlike previous reported attempts at chemical modifications, we have introduced changes to the backbone structure of conventional 19+2 siRNAs. Using these modified asiRNA backbone structures, we were able to reduce multiple nonspecific effects triggered by conventional siRNAs, such as off-target effect mediated by the sense strand and saturation of the RNAi machinery. The asiRNAs used in this study only have modifications to the backbone structure and do not contain any chemical modifications. Further chemical modification of asiRNAs can be expected to add additional benefits.
One important feature of asiRNA structural variants is that they have a reduced ability to saturate the RNAi machinery. What is the mechanism by which asiRNAs alleviate saturation of the RNAi machinery? One possible mechanism is that the suboptimal length of the sense strands of asiRNAs renders them poor substrates for incorporation into the RISC. Because RNAi pathway components exist in limited amounts, it has been suggested that the critical point for saturation of the RNAi machinery is siRNA incorporation into the RISC.10 Therefore, lack of activity by the sense strand of the asiRNA relieves RNAi saturation by the sense strand. We think this hypothesis is unlikely, because we showed that the 5′-amine modification at the end of the sense strand, which eliminates sense-strand–mediated RNAi, did not affect the degree of RNAi saturation by the siRNA. Another possible mechanism is due to the shortened length of the duplex. It has been suggested that siRNA duplex unwinding is the rate-determining step of siRNA-mediated gene silencing.32 Because asiRNAs are likely to be unwound more easily than their 19-base–pair counterparts, these siRNAs might proceed relatively quickly through the rate-limiting step. This fast structural transition might impart upon the asiRNAs reduced ability to saturate the cellular RNAi pathway.
Recently, Chu and Rana33 reported a symmetric short siRNA structure with 16 bp-long duplex region. While our asiRNA structure is similar to theirs in terms of the duplex length, the asymmetric structure of asiRNAs, which is responsible for the reduced off-target silencing of sense strand (Supplementary Figure S11), is a unique feature of our asiRNAs that makes them more specific gene silencers than the symmetric siRNA structures. Our observation is also consistent with the report by Rose et al.,34 in which they showed that the siRNA strand with 3′-overhang is preferentially incorporated into RISC over the other strand with blunt-end or 5′-overhang.
The algorithm for optimizing 19+2 siRNAs are mainly to provide thermodynamic asymmetry to the symmetric siRNA duplex structure.29 In contrast, our asiRNA structures are highly asymmetric. Therefore, it is possible that shorter-duplex siRNAs might have different sequence preferences than the 19+2 structure. Similarly, it is well known that the sequence rules for siRNA and shRNA are different.35 Future work should focus on identifying sequence rules that confer optimal gene-silencing efficiency upon asiRNAs.
While siRNAs were originally selected to circumvent the innate immune surveillance system,1 several reports have demonstrated that synthetic siRNAs can trigger nonspecific innate immune responses.12 Strikingly, a recent report demonstrated that siRNA can nonspecifically suppress neovascularization when introduced into eye, via Toll-like receptor-3 activation.36 Further in this study, siRNAs with duplex regions shorter than 19 bp showed reduced or no immune activation, which suggests that our shorter-duplex siRNAs may not trigger nonspecific Toll-like receptor-3 activation. Therefore, we expect that shorter-duplex siRNA may circumvent or reduce the challenge of nonspecific immune responses observed with 19+2 siRNA, which is especially critical in therapeutic development of siRNA-based drugs.
In conclusion, the asymmetric shorter-duplex siRNA structures described in this report showed gene-silencing activities that are comparable to the conventional 19+2 siRNA structures. Importantly, our asiRNA strcuture provides several advantages, which includes reduced sense-strand–mediated off-target gene silencing, reduced saturation of the cellular RNAi machinery, and a reduction in production costs. Our findings provide a new structural scaffold for designing silencing RNA duplex that can potentially overcome nonspecific effects evoked by siRNAs, a significant challenge in broad RNAi applications in functional genomics and therapeutics.
Materials and Methods
siRNAs. Chemically synthesized RNAs were purchased from Bioneer and annealed according to the manufacturer's protocol. RNA sequences for siAgo2 and siCREB3 were:
siAgo2 antisense: 5′-AAUCUCUUCUUGCCGAUCGGG-3′
siAgo2 sense: 5′-CGAUCGGCAAGAAGAGAUUAG-3′
siCREB3 antisense: 5′-GGCUCAGACUGUGUACUCC(dTdT)-3′
siCREB3 sense: 5′-GGAGUACACAGUCUGAGCC(dTdT)-3′
Sequences and structures of other siRNAs used in experiments are shown in figures.
Cell culture and siRNA transfection. HeLa or T98G cells were cultured in Dulbecco's modified Eagle's medium (Hyclone) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin. Cells were plated on 12-well plates 24 hours before transfection at 30–50% confluence in complete medium without antibiotics. All transfections were performed using the Lipofectamine 2000 reagent at the indicated concentrations following the manufacturer's protocol (Invitrogen, Carlsbad, CA). Cells were harvested 48 hours (T98G cells) or 24 hours (HeLa cells) after transfection. In the case of Ago2 knockdown experiments, HeLa cells were transfected with 10 nmol/l siAgo2 or Lipofectamine 2000 alone. Twenty-four hours following siAgo2 or Lipofectamine 2000 treatment, siRNAs were transfected and the cells were harvested after further 24 hours.
Quantitative real-time RT-PCR. Total RNA was extracted from cell lysates using a Tri-reagent kit (Ambion, Austin, TX). Total RNA (1 µg) was used as a template for cDNA synthesis, which was performed using a ImProm-II Reverse Transcription System (Promega, Madison, WI) according to the manufacturer's protocol. Aliquots (1/20) of each cDNA reaction were analyzed by quantitative real-time RT-PCR using a Rotor-Gene 3000 (Corbett Research, Sydney, Australia). Gene-specific primers were mixed with Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) containing the cDNA to be analyzed and the mixture was placed into 0.1 ml tubes. Samples were assayed in duplicate and the data obtained were analyzed using Rotor-Gene 6 software (Corbett Research). The primer sequences for each gene were:
Ago2-forward 5′-GGA AGT ACC GTG TCT GCA ATG-3′
Ago2-reverse 5′-ACT GCT TGT TCC GCA TGT C-3′
ATF6-forward 5′-GCC TTT ATT GCT TCC AGC AG-3′
ATF6-reverse 5′-TGA GAC AGC AAA ACC GTC TG-3′
Calcineurin-forward 5′-GCA ACC ATG AAT GCA GAC AC-3′
Calcineurin -reverse 5′-TGG TGA AAG TCC ACC ATG AA-3′
CREB3-forward 5′-TTC TGA GGT ACC GAG CGA CT-3′
CREB3-reverse 5′-GGA GGG AGT AGG TGT GGT CA-3′
DBP-forward 5′-GTA GAC CTG GAC GCC TTC CT-3′
DBP-reverse 5′-CGG GTT CAA AGG TCA TCA AC-3′
GAPDH-forward 5′-GAG TCA ACG GAT TTG GTC GT-3′
GAPDH-reverse 5′-GAC AAG CTT CCC GTT CTC AG-3′
HIF1α-forward 5′-CCA GCA ACA GAA AGT CGT CA-3′
HIF1α-reverse 5′-GGC TAT ACT TGG GCA TGG AA-3′
Integrin-forward 5′-CGT ATC TGC GGG ATG AAT CT-3′
Integrin-reverse 5′ GGG TTG CAA GCC TGT TGT AT-3′
Lamin-forward 5′-CCG AGT CTG AAG AGG TGG TC-3′
Lamin-reverse 5′-AGG TCA CCC TCC TTC TTG GT-3′
NF-κB-forward 5′-CCT GGA GCA GGC TAT CAG TC-3′
NF-κB -reverse 5′-CAC TGT CAC CTG GAA GCA GA-3′
Survivin-forward 5′-GCA CCA CTT CCA GGG TTT AT-3′
Survivin-reverse 5′-CTC TGG TGC CAC TTT CAA GA-3′
TEF-forward 5′-CCC CAG CCT ATG ATC AAA AA-3′
TEF-reverse 5′-CCG GAT GGT GAT CTG ATT CT-3′
Plasmids. DNA oligonucleotides corresponding to the sense or antisense strands of TIG3 or Survivin siRNA (see below for the DNA sequences) were cloned into the Speі and Hindііі sites of the pMIR Report-luciferase vector (Ambion).
TIG3 sense target:
5′-CTAGTTAGAGAACGCCTGAGACAGA-3′
3′-A ATCTCTTGCGGACTCTGTCTTCGA-5′
TIG3 antisense target:
5′-AGCTTTAGAGAACGCCTGAGACAGA-3′
3′-A ATCTCTTGCGGACTCTGTCTGATC-5′
Survivin sense target:
5′-CTAGTTGAAAATGTTGATCTCCTTA-3′
3′-A ACTTTTACAACTAGAGGAATTCGA-5′
Survivin antisense target:
5′-AGCTTTGAAAATGTTGATCTCCTTA-3′
3′-AACTTTTACAACTAGAGGA ATGATC-5′
TIG3 sense target-2:
5′-AGCTTGCCCUGUCUCAGGCGUUCUCUAGAUA-3′
3′-ACGGGACAGAGUCCGCAAGAGAUCUATGATC-5′
TIG3 antisense target-2:
5′-AGCTTAUCUAGAGAAGCCCUGAGACAGGGCA-3′
3′-AUAGAUCUCUUGCGGACUCUGUCCCGTGATC-5′
Survivin sense target-2:
5′-AGCTTGGAAAGGAGAUCAACAUUUUCAAATA-3′
3′-ACCUUUCCUCUAGUUGUAAAAGUTTATGATC-5′
Survivin antisense target-2:
5′-AGCTTATTUGAAAAUGUUGAUCUCCUUUCCA-3′
3′-ATAAACUUUUACAACUAGAGGAAAGGTGATC-5′
Assay for RNAi saturation by siRNAs. HeLa cells were plated on 12-well plates in complete medium without antibiotics and incubated for 24 hours at 37 °C until they reached ~50% confluency. The cells were then transfected according to the manufacturer's instructions using Lipofectamine 2000 reagent and the target siRNA (siCREB3) (at a final concentration of 1 nmol/l), with or without competitor siRNA variants (at a final concentration of 10 nmol/l). Forty-eight hours following transfection the cells were harvested, total RNA was isolated and RT-PCR was performed. To measure miRNA-mediated luciferase gene silencing, HeLa cells were plated at 50% confluency on 24-well plates 24 hours before transfection. The cells were then transfected using Oligofectamine (Invitrogen) and siRNAs (at a final concentration 10 nmol/l). Forty-eight hours following the initial transfection, a second transfection was performed using Lipofectamine 2000 reagent, siRNAs (at a final concentration 10 nmol/l), 200 ng of either the pMIR-luc luciferase reporter DNA (Ambion) or the pMIR-luc reporter that contained various miRNA target sites37 in its 3′ untranslated region, and 2 ng of pRL-SV40 reporter DNA. 48 hours following the second transfection, cells were harvested and luciferase assays were carried out. Antagomirs against miR-19 and miR-21 were purchased from Ambion and transfected into HeLa cells as described.37
Luciferase reporter assay. Cells were lysed using Passive lysis buffer (Dual-luciferase Reporter Assay System; Promega). Firefly and Renilla luciferase activity was then measured in 20 µl of each cell extract using a Victor3 plate reader (PerkinElmer, Boston, MA).
Flow cytometric analysis. HeLa cells were transfected with siSurvivin variants using Lipofectamine 2000. Forty-eight hours following transfection the cells were harvested, washed twice with 2% fetal bovine serum in phosphate-buffered saline, and fixed with 70% ethanol overnight. Cells were then resuspended in 250 µl of phosphate-buffered saline containing 50 µg/ml RNase A, incubated for 30 minutes at 37 °C, and treated 500 µl of propidium iodide (25 µg/ml). The propidium iodide–stained cells were then analyzed using a FACSCalibur System (Becton Dickinson, Franklin Lakes, NJ). Data shown are representative of two independent experiments.
DAPI staining. HeLa cells were treated with siSurvivin variants, collected 48 hours after transfection and fixed in 3.7% formaldehyde. The fixed cells were then stained with DAPI solution (2 µg/ml), and visualized using a fluorescence microscope.
5′-RACE assay. Twenty-four hours after siRNA transfection into HeLa cells, total RNA was extracted using Tri-reagent kit (Ambion). Total RNA (2 µg) was then ligated to 0.25 µg of GeneRacer RNA oligo without prior treatment.38 The GeneRacer RNA oligo-ligated total RNA was then reverse transcribed using a GeneRacer oligo dT and a SuperScript III RT kit (Invitrogen). After 35 cycles of PCR using the GeneRacer 5′ primer and a gene-specific 3′ primer, 25 cycles of nested PCR was performed using the GeneRacer 5′ nested primer and a gene-specific 3′ nested primer. The resulting PCR product was then cloned into a T&A vector (RBC) and sequenced. Sequences of primers used are as follows:
TIG3 Gene specific 3′ primer: 5′-GGGGCAGATGGCTGTTTAT TGATCC-3′
TIG3 Gene specific 3′ nested primer: 5′-ACTTTTGCCAGCGAGA GAGGGAAAC-3′
Lamin Gene specific 3′ primer: 5′-CCAGTGAGTCCTCCAGGTCT CGAAG-3′
Lamin Gene specific 3′ nested primer: 5′-CCTGGCATTGTCCAGCT TGGCAGA-3′
Western blot analysis. HeLa cells were transfected with 10 nmol/l of siAgo2 using Lipofectamine 2000. Forty-eight hours after transfection, cells were lysed using RIPA buffer containing 150 mmol/l NaCl, Tris pH 7.5, 0.5% sodium dodecyl sulfate, 0.1% sodium deoxycholate, 0.02% sodium azide, 1 mmol/l EDTA, and protease inhibitors. Protein concentration was measured by BCL protein assay kit (Pierce, Rockford, IL). Equal amounts of protein were resolved on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel in Tris-glycine sodium dodecyl sulfate running buffer and transferred to a nitrocellulose membrane. The membranes were blocked for 1 hour in Tris-buffered saline buffer containing 5% milk powder. After overnight incubation at 4 °C with a mouse monoclonal antibody to hAgo2 (Abcam), membranes were washed in Tris-buffered saline containing 1% Tween-20. After 1-hour incubation with peroxidase-coupled secondary mouse antibody, membranes were developed using enhanced chemiluminescence detection system (Amersham Biosciences, Piscataway, NJ) and exposed to X-ray films (Kodak, Rochester, NY).
Serum stability of siRNAs. 0.1 nmole of siTIG3(19+2) or siTIG3(16+3A) was incubated in 40 µl of 10% fetal bovine serum solution. Seven microliter of each sample was taken at the indicated time points and immediately frozen at −80 °C. A 3-µl aliquot of each sample was then separated in a 15% (wt/vol) nondenaturing polyacrylamide gel, stained with EtBr, and visualized by UV transillumination.
Supplementary MaterialFigure S1. Structures and activities of blunt-ended, shorter-duplex siRNAs targeting TIG3 mRNA in HeLa cells. siRNAs were transfected into HeLa cells using Lipofectamine 2000, and TIG3 mRNA levels were analyzed by quantitative real-time RT-PCR 24 h after transfection. The “TIG3 mRNA levels” on the y-axis in the figure were calculated as the TIG3 mRNA level divided by the GAPDH (control) mRNA level. All data in the graph represent mean ± SD values of three independent experiments.Figure S2. Activities of asiRNAs targeting TIG3 mRNA in T98G cells. siRNAs were transfected into T98G cells using Lipofectamine 2000, and TIG3 mRNA levels were analyzed by quantitative real-time RT-PCR 48 h after transfection. The “TIG3 mRNA levels” on the y-axis in the figure were calculated as the TIG3 mRNA level divided by the GAPDH (control) mRNA level. All data in the graph represent mean ± SD values of three independent experiments.Figure S3. Structures and activities of siTIG3 variants with 15-bp duplex regions. (A) Structures of siRNAs that have 15-bp duplex region but different overhang structures. 15-4A refers to a 15 bp duplex with 4 nt 5'-overhang at the end of antisense strand. (B) Gene silencing activities of siTIG3 variants with different overhangs. See the legend of Fig. S1 for details. All data in the graph represent mean ± SD values of three independent experiments.Figure S4. Structures of other asiRNAs. (A) Structures of asiRNAs that target Survivin mRNA. (B) Structures of asiRNAs that target LaminA/C mRNA. (C) Structures of asiRNAs that target Integrin αv mRNA.Figure S5. IC50 values of 19+2 and 16+3A siRNAs. (A) IC50 values of siTIG3(19+2) and siTIG3(16+3A). See the legend of Fig. S1 for details. (B) IC50 values of siSurvivin(19+2), siSurvivin(16+3A), and siSurvivin antisense strand. (C) IC50 values of siLamin(19+2) and siLamin(16+3A). (D) IC50 values of siIntegrin(19+2) and siIntegrin(16+3A).Figure S6. Strucures and activities of diverse asiRNAs. (A) Structures of 19+2 siRNAs that target diverse mRNAs. (B) Structures of 16+5A siRNAs that target diverse mRNAs. (C) Activities of siTIG3 variants. (D) Activities of siSurvivin variants. (E) Activities of siHIF1α-01 variants. (F) Activities of siCalcineurin variants. (G) Activities of siDBP variants. (H) Activities of siTEF variants. (I) Activities of siATF6 variants. (J) Activities of siHIF1α-02 variants. (K) Activities of siNF-kB variants. For (C)-(K), see the legend of Fig. S1 for details. All data in the graph represent mean ± SD values of three independent experiments.Figure S7. asiRNAs efficiently knockdown the protein levels of target genes. Control (reagent only) or siRNAs were transfected into HeLa cells using Lipofectamine 2000, and protein levels were analyzed by Western blot analysis 48 h following transfection. (A) Reduction of Survivin protein levels by 19+2 and 16+3A siRNAs targeting survivin gene. (B) Reduction of NF-κB protein levels by 19+2 and 16+5A siRNAs targeting NF-κB gene.Figure S8. siSurvivin 16+3A induces same phenotype with siSurvivn 19+2. (A) Nuclear abnormalities in siSurvivin-treated HeLa cells. HeLa cells were treated with transfection reagent only (Mock), siSurvivin(19+2), or siSurvivin(16+3A). DAPI staining of HeLa cells were performed 48 h after transfection. (B) The effects of Survivin siRNAs on cell cycle distribution. 48 h after the transfection of HeLa cells with transfection reagent only (Mock), siSurvivin(19+2), or siSurvivin(16+3A), cell cycle distribution was analyzed by flow cytometry. 2N, percentage of diploid cells; >4N, percentage of polyploid cells.Figure S9. AsiRNAs trigger gene silencing in Ago2-dependent manner. (A) Reduction of Ago2 mRNA levels by siAgo2 in HeLa cells. See the legend of Fig. S1 for details. (B) Reduction of Ago2 protein level by siAgo2 in HeLa cells. Control (reagent only) or siAgo2 were transfected into HeLa cells using Lipofectamine 2000, and protein levels were analyzed by western blot analysis 48 h following transfection. (C) Effect of Ago2 knockdown on the activity of siTIG3(19+2) and siTIG3(16+3A). 24 h following the pre-treatment of 0nM (reagent only) or siAgo2, siTIG3(19+2) or siTIG3(16+3A) were transfected into pre-treated HeLa cells. 24 h after siTIG3 transfection, TIG3 mRNA levels were analyzed by quantitative real-time RT-PCR. (D) Effect of Ago2 knockdown on the activity of siSurvivin(19+2) and siSurvivin(16+3A). (E) Effect of Ago2 knockdown on the activity of siLamin(19+2) and siLamin(16+3A). (F) Effect of Ago2 knockdown on the activity of siIntegrin(19+2) and siIntegrin(16+3A) For (D)-(F), see the legend of (C) for details. All data in the graph represent mean ± SD values of three independent experiments.Figure S10. Serum stability of 19+2 and 16+3A siRNAs. Each siRNA was incubated in 10% FBS solution for indicated periods. The amount of siRNA remaining was analyzed on a 15 % native gel by EtBr staining and quantified. The values in the graph represent mean ± SD values of at least two independent experiments. (A) Serum stability of siTIG3(19+2) and siTIG3(16+3A). (B) Serum stability of siSurvivin(19+2) and siSurvivin(16+3A).Figure S11. Activities of asymmetric and symmetric siRNA variants for their antisense and sense targets. (A) Structures of symmetric and asymmetric siRNA variants. (B) Gene silencing activities of sense and antisense strands of siSurvivin variants. (C) Gene silencing activities of sense and antisense strands of siTIG3 variants. See the legend of Fig. 3 for details.Figure S12. Comparison of competition potency and sense strand–mediated gene-silencing activity between the 16+3A and a 5'-NH2-sense 19+2 siRNAs. (A) Structures of siTIG3(19+2), siTIG3(16+3A) and 5'-NH2-sense siTIG3(19+2). (B) Antisense and sense strand activities of siTIG3(19+2), siTIG3(16+3A), and 5'-NH2-sense siTIG3(19+2). Each siRNA was transfected into HeLa cells with a luciferase vector containing sense or antisense target sequences of siTIG3. Luciferase activity was measured 48 h after transfection. (C) Competition potency of siTIG3(19+2), siTIG3(16+3A), and 5'-NH2-sense siTIG3(19+2). See the legend of Fig. 4a for details.
Supplementary Material
Structures and activities of blunt-ended, shorter-duplex siRNAs targeting TIG3 mRNA in HeLa cells. siRNAs were transfected into HeLa cells using Lipofectamine 2000, and TIG3 mRNA levels were analyzed by quantitative real-time RT-PCR 24 h after transfection. The “TIG3 mRNA levels” on the y-axis in the figure were calculated as the TIG3 mRNA level divided by the GAPDH (control) mRNA level. All data in the graph represent mean ± SD values of three independent experiments.
Activities of asiRNAs targeting TIG3 mRNA in T98G cells. siRNAs were transfected into T98G cells using Lipofectamine 2000, and TIG3 mRNA levels were analyzed by quantitative real-time RT-PCR 48 h after transfection. The “TIG3 mRNA levels” on the y-axis in the figure were calculated as the TIG3 mRNA level divided by the GAPDH (control) mRNA level. All data in the graph represent mean ± SD values of three independent experiments.
Structures and activities of siTIG3 variants with 15-bp duplex regions. (A) Structures of siRNAs that have 15-bp duplex region but different overhang structures. 15-4A refers to a 15 bp duplex with 4 nt 5'-overhang at the end of antisense strand. (B) Gene silencing activities of siTIG3 variants with different overhangs. See the legend of Fig. S1 for details. All data in the graph represent mean ± SD values of three independent experiments.
Structures of other asiRNAs. (A) Structures of asiRNAs that target Survivin mRNA. (B) Structures of asiRNAs that target LaminA/C mRNA. (C) Structures of asiRNAs that target Integrin αv mRNA.
IC50 values of 19+2 and 16+3A siRNAs. (A) IC50 values of siTIG3(19+2) and siTIG3(16+3A). See the legend of Fig. S1 for details. (B) IC50 values of siSurvivin(19+2), siSurvivin(16+3A), and siSurvivin antisense strand. (C) IC50 values of siLamin(19+2) and siLamin(16+3A). (D) IC50 values of siIntegrin(19+2) and siIntegrin(16+3A).
Strucures and activities of diverse asiRNAs. (A) Structures of 19+2 siRNAs that target diverse mRNAs. (B) Structures of 16+5A siRNAs that target diverse mRNAs. (C) Activities of siTIG3 variants. (D) Activities of siSurvivin variants. (E) Activities of siHIF1α-01 variants. (F) Activities of siCalcineurin variants. (G) Activities of siDBP variants. (H) Activities of siTEF variants. (I) Activities of siATF6 variants. (J) Activities of siHIF1α-02 variants. (K) Activities of siNF-kB variants. For (C)-(K), see the legend of Fig. S1 for details. All data in the graph represent mean ± SD values of three independent experiments.
asiRNAs efficiently knockdown the protein levels of target genes. Control (reagent only) or siRNAs were transfected into HeLa cells using Lipofectamine 2000, and protein levels were analyzed by Western blot analysis 48 h following transfection. (A) Reduction of Survivin protein levels by 19+2 and 16+3A siRNAs targeting survivin gene. (B) Reduction of NF-κB protein levels by 19+2 and 16+5A siRNAs targeting NF-κB gene.
siSurvivin 16+3A induces same phenotype with siSurvivn 19+2. (A) Nuclear abnormalities in siSurvivin-treated HeLa cells. HeLa cells were treated with transfection reagent only (Mock), siSurvivin(19+2), or siSurvivin(16+3A). DAPI staining of HeLa cells were performed 48 h after transfection. (B) The effects of Survivin siRNAs on cell cycle distribution. 48 h after the transfection of HeLa cells with transfection reagent only (Mock), siSurvivin(19+2), or siSurvivin(16+3A), cell cycle distribution was analyzed by flow cytometry. 2N, percentage of diploid cells; >4N, percentage of polyploid cells.
AsiRNAs trigger gene silencing in Ago2-dependent manner. (A) Reduction of Ago2 mRNA levels by siAgo2 in HeLa cells. See the legend of Fig. S1 for details. (B) Reduction of Ago2 protein level by siAgo2 in HeLa cells. Control (reagent only) or siAgo2 were transfected into HeLa cells using Lipofectamine 2000, and protein levels were analyzed by western blot analysis 48 h following transfection. (C) Effect of Ago2 knockdown on the activity of siTIG3(19+2) and siTIG3(16+3A). 24 h following the pre-treatment of 0nM (reagent only) or siAgo2, siTIG3(19+2) or siTIG3(16+3A) were transfected into pre-treated HeLa cells. 24 h after siTIG3 transfection, TIG3 mRNA levels were analyzed by quantitative real-time RT-PCR. (D) Effect of Ago2 knockdown on the activity of siSurvivin(19+2) and siSurvivin(16+3A). (E) Effect of Ago2 knockdown on the activity of siLamin(19+2) and siLamin(16+3A). (F) Effect of Ago2 knockdown on the activity of siIntegrin(19+2) and siIntegrin(16+3A) For (D)-(F), see the legend of (C) for details. All data in the graph represent mean ± SD values of three independent experiments.
Serum stability of 19+2 and 16+3A siRNAs. Each siRNA was incubated in 10% FBS solution for indicated periods. The amount of siRNA remaining was analyzed on a 15 % native gel by EtBr staining and quantified. The values in the graph represent mean ± SD values of at least two independent experiments. (A) Serum stability of siTIG3(19+2) and siTIG3(16+3A). (B) Serum stability of siSurvivin(19+2) and siSurvivin(16+3A).
Activities of asymmetric and symmetric siRNA variants for their antisense and sense targets. (A) Structures of symmetric and asymmetric siRNA variants. (B) Gene silencing activities of sense and antisense strands of siSurvivin variants. (C) Gene silencing activities of sense and antisense strands of siTIG3 variants. See the legend of Fig. 3 for details.
Comparison of competition potency and sense strand–mediated gene-silencing activity between the 16+3A and a 5'-NH2-sense 19+2 siRNAs. (A) Structures of siTIG3(19+2), siTIG3(16+3A) and 5'-NH2-sense siTIG3(19+2). (B) Antisense and sense strand activities of siTIG3(19+2), siTIG3(16+3A), and 5'-NH2-sense siTIG3(19+2). Each siRNA was transfected into HeLa cells with a luciferase vector containing sense or antisense target sequences of siTIG3. Luciferase activity was measured 48 h after transfection. (C) Competition potency of siTIG3(19+2), siTIG3(16+3A), and 5'-NH2-sense siTIG3(19+2). See the legend of Fig. 4a for details.
Acknowledgments
D.-k. L. was supported by grants from the Global Research Laboratory program by KICOS (grant no. 2008-00582), the Basic Research Program of KOSEF (grant no. R01-2005-000-10266-0), the National R&D Program for Cancer Control (grant no. 0520200-3), and SKKU start-up grant. S. K. was supported by the National Research Laboratory grant from KOSEF.
REFERENCES
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K., and , Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W., and , Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL., and , Linsley PS. Noise amidst the silence: off-target effects of siRNAs. Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Clark PR, Pober JS., and , Kluger MS. Knockdown of TNFR1 by the sense strand of an ICAM-1 siRNA: dissection of an off-target effect. Nucleic Acids Res. 2008;36:1081–1097. doi: 10.1093/nar/gkm630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S. RNAi in moderation. Nat Biotechnol. 2006;24:796–797. doi: 10.1038/nbt0706-796. [DOI] [PubMed] [Google Scholar]
- Yoo JW, Kim S., and , Lee DK. Competition potency of siRNA is specified by the 5'-half sequence of the guide strand. Biochem Biophys Res Commun. 2007;367:78–83. doi: 10.1016/j.bbrc.2007.12.099. [DOI] [PubMed] [Google Scholar]
- Koller E, Propp S, Murray H, Lima W, Bhat B, Prakash TP, et al. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers TA, Lima WF, Nichols JG., and , Crooke ST. Reduced levels of Ago2 expression result in increased siRNA competition in mammalian cells. Nucleic Acids Res. 2007;35:6598–6610. doi: 10.1093/nar/gkm663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Schlee M, Hornung V., and , Hartmann G. siRNA and isRNA: two edges of one sword. Mol Ther. 2006;14:463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K., and , MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Yoo JW, Hong SW, Kim S., and , Lee DK. Inflammatory cytokine induction by siRNAs is cell type- and transfection reagent-specific. Biochem Biophys Res Commun. 2006;347:1053–1058. doi: 10.1016/j.bbrc.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Zhu X., and , Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M, Furset G., and , Cekaite L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2'-modified RNAs. Biochem Biophys Res Commun. 2007;361:122–126. doi: 10.1016/j.bbrc.2007.06.177. [DOI] [PubMed] [Google Scholar]
- Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, et al. Strand-specific 5'-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, Hong SW, Kim S., and , Lee DK. A structure-activity relationship study of siRNAs with structural variations. Biochem Biophys Res Commun. 2007;359:997–1003. doi: 10.1016/j.bbrc.2007.06.004. [DOI] [PubMed] [Google Scholar]
- McNamara JO 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Jiang M, Arzumanov AA, Gait MJ., and , Milner J. A bi-functional siRNA construct induces RNA interference and also primes PCR amplification for its own quantification. Nucleic Acids Res. 2005;33:e151. doi: 10.1093/nar/gni144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Behlke MA, Rose SD, Chang MS, Choi S., and , Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Choung S, Kim YJ, Kim S, Park HO., and , Choi YC. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Biophys Res Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Zhou X, Murphy FR, Gehdu N, Zhang J, Iredale JP., and , Benyon RC. Engagement of alphavbeta3 integrin regulates proliferation and apoptosis of hepatic stellate cells. J Biol Chem. 2004;279:23996–24006. doi: 10.1074/jbc.M311668200. [DOI] [PubMed] [Google Scholar]
- Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- Yang D, Welm A., and , Bishop JM. Cell division and cell survival in the absence of survivin. Proc Natl Acad Sci USA. 2004;101:15100–15105. doi: 10.1073/pnas.0406665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R., and , Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N., and , Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A., and , Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, et al. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen T, Amarzguioui M, Babaie E., and , Prydz H. Similar behaviour of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 2003;31:2401–2407. doi: 10.1093/nar/gkg338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY., and , Rana TM. Potent RNAi by short RNA triggers. RNA. 2008;14:1714–1719. doi: 10.1261/rna.1161908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxman DJ, Livingstone LR, Zhang J, Conti BJ, Iocca HA, Williams KL, et al. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 2006;6:7. doi: 10.1186/1472-6750-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J., and , Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structures and activities of blunt-ended, shorter-duplex siRNAs targeting TIG3 mRNA in HeLa cells. siRNAs were transfected into HeLa cells using Lipofectamine 2000, and TIG3 mRNA levels were analyzed by quantitative real-time RT-PCR 24 h after transfection. The “TIG3 mRNA levels” on the y-axis in the figure were calculated as the TIG3 mRNA level divided by the GAPDH (control) mRNA level. All data in the graph represent mean ± SD values of three independent experiments.
Activities of asiRNAs targeting TIG3 mRNA in T98G cells. siRNAs were transfected into T98G cells using Lipofectamine 2000, and TIG3 mRNA levels were analyzed by quantitative real-time RT-PCR 48 h after transfection. The “TIG3 mRNA levels” on the y-axis in the figure were calculated as the TIG3 mRNA level divided by the GAPDH (control) mRNA level. All data in the graph represent mean ± SD values of three independent experiments.
Structures and activities of siTIG3 variants with 15-bp duplex regions. (A) Structures of siRNAs that have 15-bp duplex region but different overhang structures. 15-4A refers to a 15 bp duplex with 4 nt 5'-overhang at the end of antisense strand. (B) Gene silencing activities of siTIG3 variants with different overhangs. See the legend of Fig. S1 for details. All data in the graph represent mean ± SD values of three independent experiments.
Structures of other asiRNAs. (A) Structures of asiRNAs that target Survivin mRNA. (B) Structures of asiRNAs that target LaminA/C mRNA. (C) Structures of asiRNAs that target Integrin αv mRNA.
IC50 values of 19+2 and 16+3A siRNAs. (A) IC50 values of siTIG3(19+2) and siTIG3(16+3A). See the legend of Fig. S1 for details. (B) IC50 values of siSurvivin(19+2), siSurvivin(16+3A), and siSurvivin antisense strand. (C) IC50 values of siLamin(19+2) and siLamin(16+3A). (D) IC50 values of siIntegrin(19+2) and siIntegrin(16+3A).
Strucures and activities of diverse asiRNAs. (A) Structures of 19+2 siRNAs that target diverse mRNAs. (B) Structures of 16+5A siRNAs that target diverse mRNAs. (C) Activities of siTIG3 variants. (D) Activities of siSurvivin variants. (E) Activities of siHIF1α-01 variants. (F) Activities of siCalcineurin variants. (G) Activities of siDBP variants. (H) Activities of siTEF variants. (I) Activities of siATF6 variants. (J) Activities of siHIF1α-02 variants. (K) Activities of siNF-kB variants. For (C)-(K), see the legend of Fig. S1 for details. All data in the graph represent mean ± SD values of three independent experiments.
asiRNAs efficiently knockdown the protein levels of target genes. Control (reagent only) or siRNAs were transfected into HeLa cells using Lipofectamine 2000, and protein levels were analyzed by Western blot analysis 48 h following transfection. (A) Reduction of Survivin protein levels by 19+2 and 16+3A siRNAs targeting survivin gene. (B) Reduction of NF-κB protein levels by 19+2 and 16+5A siRNAs targeting NF-κB gene.
siSurvivin 16+3A induces same phenotype with siSurvivn 19+2. (A) Nuclear abnormalities in siSurvivin-treated HeLa cells. HeLa cells were treated with transfection reagent only (Mock), siSurvivin(19+2), or siSurvivin(16+3A). DAPI staining of HeLa cells were performed 48 h after transfection. (B) The effects of Survivin siRNAs on cell cycle distribution. 48 h after the transfection of HeLa cells with transfection reagent only (Mock), siSurvivin(19+2), or siSurvivin(16+3A), cell cycle distribution was analyzed by flow cytometry. 2N, percentage of diploid cells; >4N, percentage of polyploid cells.
AsiRNAs trigger gene silencing in Ago2-dependent manner. (A) Reduction of Ago2 mRNA levels by siAgo2 in HeLa cells. See the legend of Fig. S1 for details. (B) Reduction of Ago2 protein level by siAgo2 in HeLa cells. Control (reagent only) or siAgo2 were transfected into HeLa cells using Lipofectamine 2000, and protein levels were analyzed by western blot analysis 48 h following transfection. (C) Effect of Ago2 knockdown on the activity of siTIG3(19+2) and siTIG3(16+3A). 24 h following the pre-treatment of 0nM (reagent only) or siAgo2, siTIG3(19+2) or siTIG3(16+3A) were transfected into pre-treated HeLa cells. 24 h after siTIG3 transfection, TIG3 mRNA levels were analyzed by quantitative real-time RT-PCR. (D) Effect of Ago2 knockdown on the activity of siSurvivin(19+2) and siSurvivin(16+3A). (E) Effect of Ago2 knockdown on the activity of siLamin(19+2) and siLamin(16+3A). (F) Effect of Ago2 knockdown on the activity of siIntegrin(19+2) and siIntegrin(16+3A) For (D)-(F), see the legend of (C) for details. All data in the graph represent mean ± SD values of three independent experiments.
Serum stability of 19+2 and 16+3A siRNAs. Each siRNA was incubated in 10% FBS solution for indicated periods. The amount of siRNA remaining was analyzed on a 15 % native gel by EtBr staining and quantified. The values in the graph represent mean ± SD values of at least two independent experiments. (A) Serum stability of siTIG3(19+2) and siTIG3(16+3A). (B) Serum stability of siSurvivin(19+2) and siSurvivin(16+3A).
Activities of asymmetric and symmetric siRNA variants for their antisense and sense targets. (A) Structures of symmetric and asymmetric siRNA variants. (B) Gene silencing activities of sense and antisense strands of siSurvivin variants. (C) Gene silencing activities of sense and antisense strands of siTIG3 variants. See the legend of Fig. 3 for details.
Comparison of competition potency and sense strand–mediated gene-silencing activity between the 16+3A and a 5'-NH2-sense 19+2 siRNAs. (A) Structures of siTIG3(19+2), siTIG3(16+3A) and 5'-NH2-sense siTIG3(19+2). (B) Antisense and sense strand activities of siTIG3(19+2), siTIG3(16+3A), and 5'-NH2-sense siTIG3(19+2). Each siRNA was transfected into HeLa cells with a luciferase vector containing sense or antisense target sequences of siTIG3. Luciferase activity was measured 48 h after transfection. (C) Competition potency of siTIG3(19+2), siTIG3(16+3A), and 5'-NH2-sense siTIG3(19+2). See the legend of Fig. 4a for details.