Abstract
For subsets of Duchenne muscular dystrophy (DMD) mutations, antisense oligoribonucleotide (AON)-mediated exon skipping has proven to be efficacious in restoring the expression of dystrophin protein. In the mdx murine model systemic delivery of AON, recognizing the splice donor of dystrophin exon 23, has shown proof of concept. Here, we show that using cationic polymethylmethacrylate (PMMA) (marked as T1) nanoparticles loaded with a low dose of 2′-O-methyl-phosphorothioate (2′OMePS) AON delivered by weekly intraperitoneal (IP) injection (0.9 mg/kg/week), could restore dystrophin expression in body-wide striated muscles. Delivery of an identical dose of naked AON did not result in detectable dystrophin expression. Transcription, western, and immunohistochemical analysis showed increased levels of dystrophin transcript and protein, and correct localization at the sarcolemma. This study shows that T1 nanoparticles have the capacity to bind and convoy AONs in body-wide muscle tissues and to reduce the dose required for dystrophin rescue. By immunofluorescence and electron microscopy studies, we highlighted the diffusion pathways of this compound. This nonviral approach may valuably improve the therapeutic usage of AONs in DMD as well as the delivery of RNA molecules with many implications in both basic research and medicine.
Introduction
Duchenne muscular dystrophy (DMD) is an X-linked inherited muscle degenerative disorder mainly caused by frame-disrupting mutations due to large rearrangements in the dystrophin gene.1 DMD boys are affected by both severe skeletal muscle wasting and dilated cardiomyopathy. The milder allelic form of the disease, Becker muscular dystrophy, is due to in frame mutations, which preserve a shortened but functional protein. This observation has suggested that converting a DMD phenotype to a less severe condition may represent a possible therapeutic approach for DMD. In this context, restoration of dystrophin synthesis by exon skipping during pre-mRNA splicing by means of antisense oligoribonucleotide (AON) could potentially be applicable for up to 90% of DMD patients.2 Indeed, this approach has been successfully applied to the mdx mouse model, both in vitro and in vivo, to normal and DMD patients' myogenic cells, to the transgenic humanized DMD mouse, and to the canine model of DMD. These results have therefore generated optimism about the feasibility of developing effective therapy approaches for DMD.3,4,5 Indeed, it was recently shown that intramuscular injection of 2′-O-methyl-phosphorothioate (2′OMePS) AONs in four DMD patients was associated with relevant local rescue of dystrophin synthesis.6 Notwithstanding these encouraging results, the systemic administration of AONs is limited by their cellular uptake and nontoxic effective doses. The main challenge in this field is therefore to improve the delivery of AONs to all tissues affected by the disease, including skeletal and cardiac muscles, and to reduce the effective therapeutic dose in view of life-long treatment.6,7,8 In particular, to obtain exon skipping in the heart is of importance, because cardiomyopathy is currently the leading cause of death in DMD patients. Very recently two independent studies described a high yield of dystrophin protein in the heart by systemic delivery of morpholino oligomers conjugated with cell-penetrating peptide containing arginine.9,10 However, the nondegradable nature of phosphorodiamidate morpholino oligomer still raises concerns over their safety after extended systemic applications.11 Experimental evidence in the mdx model indicates that engineered AONs can efficiently reach the cardiac muscle also when cloned into recombinant adeno-associated virus vectors.12 However, the intrinsic immunogenicity of virus-associated proteins may impair the efficacy of viral-mediated gene inoculations preventing prolonged transgene expression. Indeed, whereas mice may be more tolerant to initial exposure to AAV, recent data demonstrated immune-mediated loss of recombinant adeno-associated virus persistence in the DMD dog model and in patients participating to a phase 1/2 clinical trial of β-hemophilia.13,14
The use of synthetic nonviral vectors based on polymeric systems is a versatile and safe approach for the in vivo delivery of bioactive molecules, including plasmid DNA, oligonucleotides, and peptides. They increase their stability and shelf life in biological fluids, improving their efficacy. Over the past decade, several polymeric delivery systems, such as liposomes, copolymers, nano-, and micro-spheres, have been developed.15 The compounds are encapsulated inside the polymeric matrix and released in vivo by a combination of diffusion and polymer degradation. However, following encapsulation and release, labile drugs, such as DNA and proteins, may undergo significant degradation accompanied by a reduction in drug activity.16 Moreover intracellular drug release from the polymeric matrix may be too slow to be effective. In fact, particles could be removed from the intracellular environment before much of the payload has been released.17
To achieve an effective binding, cationic micro- and nanospheres consisting of biodegradable polymers (poly(lactic-co-glycolic acid)) were therefore obtained in which cationic surfactants are able to adsorb drug onto particles' surface (i.e., cetyltrimethylammonium bromide) or cationic polymers (i.e., polyethyleneimine).18,19 However, this approach suffers from an inherent surface instability due to the occurrence of cationic surfactant desorption, thus ultimately leading to reproducibility problems and even toxic side effects. To overcome these problems, we designed and prepared a novel type of cationic core–shell nanospheres (T1), made up of a core of polymethylmethacrylate (PMMA), surrounded by a shell bearing cationic groups. PMMA T1 nanoparticles have cationic groups, ideal for AONs' binding, covalently bound to the particles thus avoiding desorption and instability problems. PMMA-based nanoparticles were already described as in vivo drug-delivery systems for the delivery of both DNA oligonucleotides and peptides.20,21
Anionic and cationic PMMA–based nanoparticles similar to the T1 sample used in this study, were already shown to be very promising delivery systems for protein and DNA vaccines or for modified peptide nucleic acids as the particle/bioactive molecules are readily taken up by the cells where they efficiently release the delivered drug, are safe in mice and nonhuman primates, even after multiple administration of high doses, and slowly biodegradable.22,23,24 This knowledge prompted us to evaluate T1 nanoparticles as alternative vehicles to deliver charged RNA-like AONs and to induce dystrophin rescue with improved efficiency and/or with more durable effect in mdx mice. We indeed demonstrate that T1 nanoparticles bind 2′OMePS oligoribonucleotides and have a body-wide distribution following IP administration. This was accompanied with dystrophin restoration both in skeletal muscles and in the heart. This rescue persisted up to 6 weeks after the last injection. Using T1 nanoparticles, the effective dose of AON was highly reduced (2.7 mg/kg) when compared to those used in previous studies on naked AONs delivery (120–240 mg/kg).25,26 Our results encourage further studies on T1 or other novel nanoparticles to evaluate applicable therapeutic employment for AON delivery in DMD.
Results
T1 nanoparticles and AON loading experiments
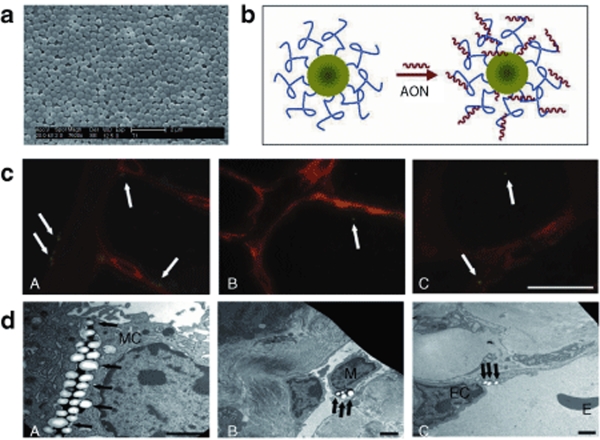
T1 nanoparticles (diameter measured by scanning electron microscope 417 nm, ζ-potential +46.5 mV, surface charge density 155 µmol/g) were obtained by emulsion polymerization (Figure 1a). They were made of a core of PMMA, surrounded by a shell bearing cationic groups. Loading experiments to test loading capacity, depending on the physico-chemical properties of the T1 nanoparticles, such as surface charge density (155 µmol/g) and surface area, showed that T1 nanoparticles (1 mg/ml) adsorbed onto their surface 2′OMePS M23D oligoribonucleotide in the concentration range of 10–100 µg/ml. The hydrophobic nature of the 2′OMePS oligoribonucleotide is responsible for its strong lipophilic interaction with the surface quaternary ammonium groups and with the matrix of the nanoparticles (Figure 1b). M23D adsorption on T1 nanospheres was a highly reproducible process with a loading efficiency, representing the percentage of bound versus unbound oligonucleotides, ranging from 20 to 100%. In particular, the adsorption efficiency was high at low AON concentration and it slightly decreased at high AON concentration, leading to the assembling of AON/nanoparticles complexes with 18 µg/mg of loading value (data not shown).
Figure 1.
T1 nanoparticles characteristics and biodistribution. (a) T1 nanoparticle scanning electron microscope image showing the nanoparticles size (diameter 417 nm). (b) Schematic representation of the lipophilic interaction between antisense oligoribonucleotide (AON) molecules and the surface quaternary ammonium groups onto nanoparticles. (See Supplementary Materials and Methods). (c) Biodistribution of T1-Fluo nanoparticles by fluorescence microscope analysis in group 3 mdx mice. A, diaphragm; B, skeletal muscle (gastrocnemius); C, heart. Tissue sections were labeled with antinidogen antibody (red staining). Isolated nanoparticles are detected inside the cytoplasm or close to the basement membrane of the muscle fibers (arrows). Bar = 20 μm. (d) Biodistribution of T1 nanoparticles by electron microscope analysis. The diaphragm shows the presence of several nanoparticles (A) between two mesothelial cells of the peritoneal side, and inside the cytoplasm of both (B) macrophages in lymphatic vessels and (C) endothelial cells in blood vessels (arrows). EC, endothelial cell; M, macrophage; MC, mesothelial cell.
Nanoparticles body-distribution
In order to monitor body distribution, three mdx mice (group 3 in Table 1) were treated via IP injections with fluorescent AON-free T1 nanoparticles and analyzed 1 and 6 weeks after last injection, obtaining similar results. Fluorescence analysis was performed on spleen, liver, heart, gastrocnemius, diaphragm, and quadriceps. In diaphragm, nanoparticles were detected close to the mesothelium (Figure 1c, A). Single particles were found intracellular in several myofibers of gastrocnemius and in the heart (Figure 1c, B and C). The number of particles/mm2 was higher in diaphragm when compared to gastrocnemius and quadriceps (about 10 and 2 particles/mm2, respectively). Transmission electron microscope examination confirmed the presence of nanoparticles in all tissues examined (Figure 1d). T1 nanoparticles appeared as electron-translucent round structures with an expected size of 500 nm. Nanoparticles were found both in the cytoplasm of circulating macrophages in lymphatic vessels and inside endothelial cells of blood vessels (Figure 1d, B and C).
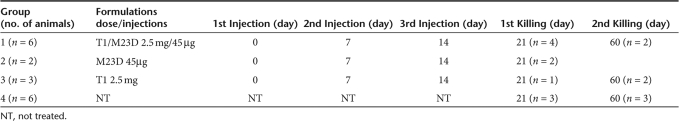
Table 1.
Mdx experiments schedule
Immunohistochemical analysis of dystrophin
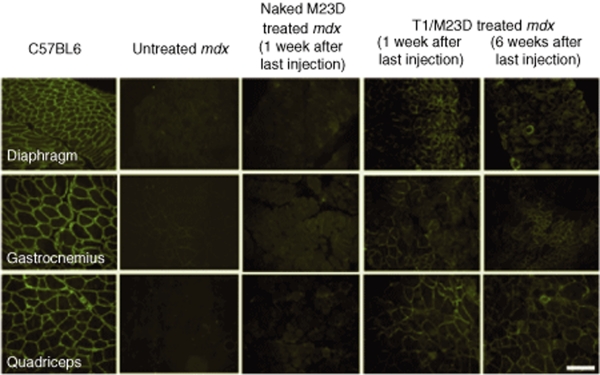
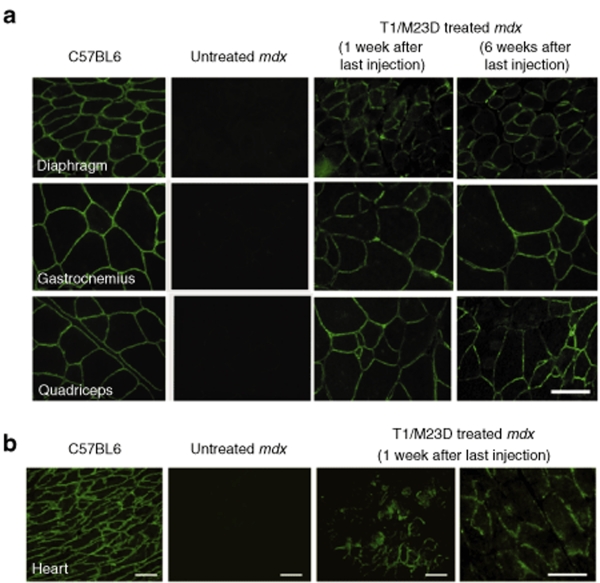
In all skeletal muscles from mice treated with the T1/M23D complexes, dystrophin expression was restored in a significant number of fibers. The immunolabeling pattern was characterized by clusters of dystrophin-expressing fibers (Figure 2). Restored dystrophin localized correctly at the sarcolemma, and the intensity of labeling was comparable to the wild type (WT) muscle fibers (Figure 3a). However, in some groups of fibers the labeling appeared heterogeneous. We found an average of 40, 40.27, and 45% of dystrophin-expressing fibers with a labeling covering 90–100% of the perimeter, in diaphragm, gastrocnemius, and quadriceps, respectively; the percentage of myofibers with a labeling ranging from 50 to 90%, was 44.2% in diaphragm, 55.3% in gastrocnemius, and 45.5% in quadriceps. Moreover, 10% in diaphragm, 3% in gastrocnemius, and 4% in quadriceps of myofibers showed a discontinuous pattern or a labeling that covered <50% of the perimeter. Immunohistochemical analysis of dystrophin in cardiac muscle of all T1/M23D-treated mdx mice examined 1 week after last injection revealed the presence of groups of dystrophin-expressing cardiomyocytes in different areas of the heart (Figure 3b). Dystrophin was absent in the heart of T1/M23D-treated mice killed 6 weeks after last injection (data not shown) and in control mdx mice (Figure 3b).
Figure 2.
Immunohistochemical findings in mdx skeletal muscles. Dystrophin immunolabeling in muscle fibers. Representative fields of cross sections from C57BL6 wild type, untreated mdx, naked M23D-treated mdx, and T1/M23D complexes treated mdx mice, diaphragm, gastrocnemius, and quadriceps skeletal muscle labeled with a polyclonal antidystrophin antibody, demonstrating absence of dystrophin in mdx mice untreated and treated with M23D naked AON and restoration of immunolabeling in several groups of muscle fibers after treatment with T1/M23D complexes analyzed 1 and 6 weeks after the last injection. The total injected amount of AON per animal, both naked or nanoparticles-combined, was 135 µg. Bar = 40 µm.
Figure 3.
Immunohistochemical findings in mdx skeletal muscles and heart. (a) Dystrophin appears correctly expressed at the membrane of muscle fibers of T1/M23D-treated mdx mice both 1 and 6 weeks after last treatment. The intensity of labeling in rescued dystrophin-positive muscle fibers is reminiscent of normal muscle. No dystrophin labeling is detected at the sarcolemma of untreated mdx myofibers. Bar = 20 µm. (b) Immunohistochemical analysis of dystrophin in heart from C57BL6, untreated mdx, and T1/M23D-treated mdx mice. Restoration of dystrophin immunolabeling is detected in focal areas of cardiac muscle of mice treated with T1/M23D complexes, 1 week after last injection. When compared with normal cardiac muscle, the dystrophin immunolabeling appears discontinuous at the membrane of treated cardiomyocytes. Bar = 50 µm.
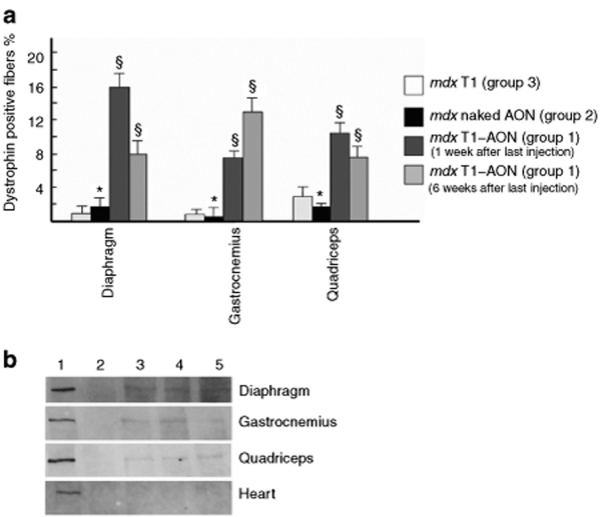
Quantitative analysis of dystrophin-positive fibers was performed in skeletal muscle of mdx mice treated with T1/M23D complexes, while cardiac muscle was not evaluated; the count of dystrophin-positive fibers in skeletal muscle excluded myofibers with a labeling that covered <50% of the perimeter or with a discontinuous pattern. The average of positive-muscle fibers varied between different skeletal muscles (Figure 4a). The number of dystrophin-positive fibers found in mdx mice treated with T1/M23D complexes was significantly increased when compared to mdx mice treated with the same dose of naked M23D or to control mdx mice, untreated or treated with T1-Fluo AON-free nanoparticles. mdx mice examined 1 week after last injection showed the highest number of dystrophin-positive fibers in diaphragm and quadriceps and the lowest in gastrocnemius. In mdx mice examined 6 weeks after the last injection, the highest number of dystrophin-positive fibers was detected in gastrocnemius and the lowest in diaphragm and quadriceps.
Figure 4.
Positive fibers counting and western blotting. (a) The statistical analysis shows a significant increase of dystrophin-positive fibers in diaphragm, quadriceps and gastrocnemius of mdx mice treated with T1/M23D complexes (group 1) if compared to mdx mice treated with M23D naked AON (group 2) or with T1 nanoparticles (group 3; see Table 1). The average of dystrophin-positive myofibers varies among different skeletal muscles and the mice examined 1 and 6 weeks after the last injection. Data represent mean ± SD. §P < 0.05. *P > 0.05. (b) Dystrophin immunoblot using DYS2 antibody shows restored expression of the protein in diaphragm, gastrocnemius, and quadriceps muscle from T1/M23D complexes treated mdx (lanes 3 and 4: 1 week after last injection, lane 5: 6 weeks after last injection, group 1 in Table 1), while no protein is detected in mdx untreated control (lane 2). Dystrophin protein is undetectable in heart from both untreated and treated mdx mice. Diaphragm, gastrocnemius, quadriceps, and heart from normal mice are used as positive controls, 1/30th of protein loaded in the other lanes (lane 1). AON, antisense oligoribonucleotide; T1, cationic polymethylmethacrylate.
Western blot
By western blot analysis the expression of dystrophin was clearly detected in gastrocnemius, quadriceps, and diaphragm from all mdx mice groups treated with T1/M23D complexes, examined both 1 and 6 weeks after last injection, but undetectable in the heart as well as in mdx mice treated with naked M23D or untreated (Figure 4b). Two different antibodies against dystrophin produced similar results. The size of the protein was indistinguishable from the full-length dystrophin of the normal muscle, as expected for a protein translated by a single exon 23 skipped transcript. No other lower molecular weight products were seen.
Transcription studies
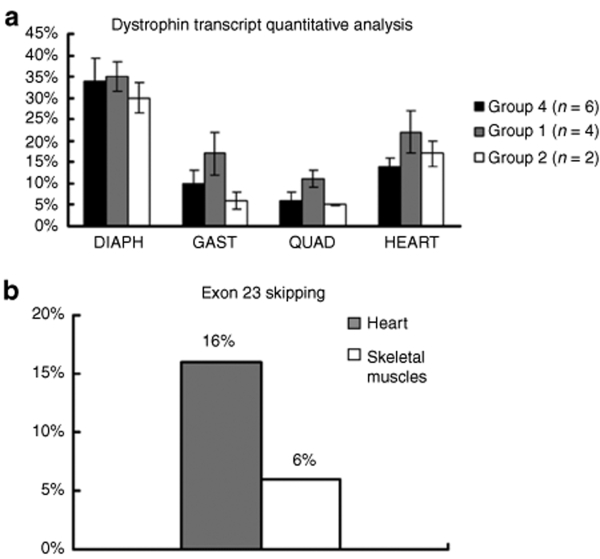
Quantitative evaluation of T1/M23D treatment on general dystrophin transcriptional level. In order to quantify differences in the amount of dystrophin transcript both in skeletal and cardiac muscle from untreated, T1/M23D- and naked M23D-treated animals, we performed a quantitative analysis using exon specific real-time assays (ESRAs). This demonstrated that untreated mdx mice showed a generally lower transcription level in respect to the amount of the transcript (considered as 100%) detected in the WT mice, therefore used as control. In addition, the transcription level in mdx follows a muscle-specific pattern, being the higher level of messenger in diaphragm (34% of the WT) and heart (14%), whereas lower levels were seen in gastrocnemius (10%) and quadriceps (6%) (Figure 5a). The mdx mice group 1 (Table 1), treated with T1/M23D compound and killed 1 week after last injection, showed a significant increase of dystrophin transcription levels in the heart (80% more transcript), gastrocnemius, and quadriceps (70% and 50% more, respectively), although at different extent among treated mice (Figure 5a). No effect was observed in the diaphragm, where transcription level remained comparable in treated and untreated animals. The mdx mice group 2, treated with naked M23D, did not show significant difference in the dystrophin transcription level in comparison with untreated mdx mice (Figure 5a).
Figure 5.
Transcript and exon skipping quantification. (a) Quantification of dystrophin transcript in skeletal and cardiac muscles from mdx treated and untreated animals in respect to wild type mice (considered as 100%). Dystrophin transcript expression was evaluated by relative-quantitative Real-Time PCR using β-actin as endogenous control. Histograms represent dystrophin transcript level calculated by the mean of exons 7, 22 and 25, 2−ΔΔCT values (expressed as percentage) in different skeletal muscles (diaphragm, quadriceps, and gastrocnemius) and heart. A total of 26 experiments has been performed (column represents the mean values). The analysis was performed on mdx mice from three different groups killed 1 week after the last injection: group 1 (n = 4; grey column), group 2 (n = 2; white column), and group 4 (n = 6; black column). (b) Percentage of exon 23 skipping determined by Real-time PCR in cardiac and skeletal muscles from mdx treated animals in comparison with untreated mice. The analysis was performed on group 1 mice (n = 4) in respect to group 4 mice (n = 3), all killed 1 week after the last injection. The percentage of exon 23 skipped transcript (2−ΔΔCT values) calibrated on exon 7, was evaluated in three different experiments and appeared to be clearly assessable in the cardiac muscle (16%; grey column) as well as in the skeletal muscles, represented by gastrocnemius and quadriceps (6%; white column).
Exon 23 skipped transcript quantification. The percentage of exon 23 skipped transcript, calibrated to exon 7 and taking as endogenous control mouse β-actin gene, appeared to be clearly assessable in the cardiac muscle (16%) as well as in the skeletal muscles (6%) of T1/M23D-treated mdx mice analyzed 1 week after last injection (Figure 5b). In addition, the correct exon 22–exon 24 junction was assessed by sequencing the nested PCR product (See Figure S2). Quantitative analyses were also performed on the treated mdx mice killed 6 weeks after last injection; however, the transcription levels were comparable to those of untreated mdx mice, and the exon 23 skipped transcript was undetectable.
Discussion
In this work, we describe the first application of cationic core–shell nanoparticles as effective nonviral vehicles for the delivery of charged AONs in vivo. We demonstrated that the T1 nanoparticles can efficiently bind 2′OMePS RNA-like oligonucleotides. We also demonstrated that T1 nanoparticles are widely distributed in various tissues/organs including heart and skeletal muscle. We also showed that IP administration of the M23D AON adsorbed onto T1 nanoparticles induced the restoration of dystrophin protein expression in skeletal muscles and, although at lower levels, in the heart of mdx mice. The dystrophin rescue is associated with increased dystrophin transcript and expression of high molecular weight dystrophin protein, as shown by western blot analysis. The novel dystrophin correctly localized at the sarcolemma, as detected by immunohistochemical analysis. These results demonstrate the effectiveness of this approach both in terms of body-wide distribution and protein synthesis restoration.
Moreover, dystrophin rescue was obtained using a very low dose of AON, corresponding to 1/50th to 1/80th of the routine dosage described in the literature for systemic treatments of mdx mice.25,26 Dystrophin synthesis is clearly induced by the T1/M23D complexes because the injection of the same dose of naked M23D did not produce any effect. The combination of slow release and depot effects, together with the protection from degradation/sequestration, afforded by this delivery system could be responsible of the very low amount of AON required for producing a functional effect. The above suggestion is supported by the well-known hydrophobic nature of phosphorothioates,27 which is responsible for the strong lipophilic interactions of oligonucleotides with quaternary ammonium groups and matrix of the nanoparticles.
Because a moderate toxicity is documented for PS-AONs, even in view of chronic treatment, the possibility of obtaining sustainable effects by using a very low dose is certainly realistic.
The second interesting result is the dystrophin rescue obtained in the heart, as shown in treated mdx mice killed 1 week after the last injection. Although we are aware that the amount of dystrophin rescue in the heart is low, we should consider that it has been obtained by 1/80th of the dose previously administered for systemic delivery of 2′OMePS.25 It is also known that other AON molecules as phosphorodiamidate morpholino oligomer-AONs do not reach the heart spontaneously but only when conjugated with cell-penetrating peptides.9,10,26 Therefore, despite the low amount of rescue as well as the lack of its persistence after 6 weeks, the T1/M23D complex represents the first conjugate able to improve the 2′OMePS oligomer uptake by systemic administration.
Although delivery of 2′OMePS was demonstrated to be improved by using synthetic cationic copolymers of polyethyleneimine and poly(ethylene glycol), this delivery was performed only locally because systemic administration of these copolymers may present toxic effects.3,28,29,30
We have also set up a novel quantitative analysis (ESRA) of dystrophin transcripts that provided the identification of a muscle-specific pattern of transcription in untreated mdx mice in comparison with WT mice. We used both β-actin and adjacent dystrophin exons as reference transcribed regions. The ESRA analysis allowed us both to identify a wide increase of transcription levels after treatment in the different muscles, including heart, and to quantify the dystrophin exon 23 skipped transcript in the heart (16%) and in skeletal muscles (6%). The increased amount of dystrophin transcript in T1/M23D-treated mdx mice could be due to an enhanced stability of skipped transcript and/or to the inhibition of nonsense mediated decay. The difference between heart and skeletal muscle exon 23 skipped transcript amounts is remarkable. Furthermore, it is also evident that there is a discrepancy between transcript and protein rescue, being inversely proportional in heart versus skeletal muscle. Although explaining this peculiar but relevant finding requires further studies, there are data showing that both the transcription and translation behavior varies between heart and skeletal muscle. This may be due to cis/trans acting tissue specific factors able to destabilize or stabilize the transcripts or more generally to the different transcription and translation efficiency in both tissues.31,32
Histological analysis showed no apparent tissue damage in any of the organs examined, such as liver, kidney, and spleen (Figure S1).
A possible disadvantage in using PMMA nanoparticles is related to their slow biodegradability, possibly causing adverse effects due to accumulation in chronic treatments. Furthermore, T1 nanoparticles might not be appropriate for intravenous usage, because the possibility to form small aggregates, expected by using >2.5 mg of nanoparticles.24 For these reasons and in order to improve the potential for such a novel approach, we are currently testing different nanoparticles, varying in size, RNA binding affinity, and bioerosion propensities.
In summary, our results show that IP administration of a low dose of specific AON–nanoparticle complexes can effectively restore dystrophin synthesis in both skeletal and cardiac muscle. Nanoparticles are therefore eligible as candidate delivery systems for RNA molecules supporting further investigations in particular for DMD AON-mediated therapeutics.
Materials and Methods
AON synthesis. M23D(+07-18) (5′-GGCCAAACCUCGGCUUACCUGAAAU-3′) AON against the boundary sequences of the exon and intron 23 of mouse dystrophin gene, contains 2′-O-methyl modified RNA and full-length phosphorothioate backbone.33 Oligonucleotide synthesis was carried out on an ÄKTA oligopilot plus 10 DNA/RNA synthesizer (GE Healthcare, Milano, Italy) using its trityl-on mode. The sequence was synthesized on a 2-µmol scale using Primer Support 200 loaded at 80 µmol/g (Amersham Biosciences, Milano, Italy). Commercial 2′-O-methyl phosphoroamidites (Proligo, Boulder, CO) were dissolved to a nominal concentration of 50 mmol/l in anhydrous acetonitrile (CH3CN) and activated with a 0.25 mol/l solution of 5-(bis-3,5-trifluoromethylphenyl)-1H-tetrazole (Proligo) in CH3CN. The final detritylation was achieved using a 0.1 mol/l aqueous solution of NaOAc (pH 3.0). Crude dimethyltryptamine protected and detritylated oligonucleotide were purified by an ÄKTAbasic ultra physical contact liquid chromatography system using an Amersham Biosciences Resource RPC 3 ml column eluted under a gradient of CH3CN in 0.1 mol/l triethylammonium acetate (pH 8).
The final oligonucleotide was dissolved in water and filtered through a short column of Dowex 50WX8 (Na+ form, 100–200 mesh) to obtain after lyophilization 0.8 µmol (40%) of target compound. The purity of the full-length desired product was evaluated by MALDI-TOF MS, 31P-NMR and RP-HPLC analyses.
T1 nanoparticles loading experiments. For phosphorothioate modified RNA-like oligonucleotide (2′OMePS-AON) adsorption experiments, 1.0 mg of freeze-dried nanospheres were suspended in 1.0 ml of 20 mmol/l sodium phosphate buffer (pH 7.4) and sonicated for 15 minutes. The appropriate amount of a concentrated aqueous solution of 2′OMePS-AON was then added to reach the final concentration (10 ÷ 100 µg/ml). The experiments were run in triplicate (SD ≤ 10%). The suspensions were continuously stirred at 25 °C for 2 hours. After microfuge centrifugation at about 18,000 rpm for 15 minutes, quantitative sedimentation of the AON–nanoparticle complexes was obtained and aliquots (10 ÷ 50 µl) of the supernatant were withdrawn, filtered on a Millex GV4 filter unit and diluted with sodium phosphate buffer. Finally, UV absorbance at Λ = 260 nm was measured. Adsorption efficiency (%) is calculated as 100 × (administered AON) − (unbound AON)/(administered AON).
Animals. All experiments were performed on male mdx mice (C57BL/ 10ScSn–Dmdmdx/J) or age-matched WT male mice (C57BL/10SnJ). All procedures were approved by the Animal Experimentation Ethics Committee. Mice were housed in temperature-controlled rooms (22 °C) with a humidity of 50% and a 12:12 hour light–dark cycle. Mice were purchased from the Jackson Laboratory (Bar Harbor, ME).
IP injections of T1–AON complexes, T1-Fluo-AON free, and naked AON. T1 nanoparticles toxicity tests were performed both in mice and in murine/human cells using up to 5 mg/injection (IP) in animals and 10 mg/ml in cells. No toxicity was observed both in in vivo and in in vitro systems (A. Caputo, Department of Histology, Microbiology, and Medical Biotechnology, personal communication, September 2006).
mdx mice (8–10 weeks of age) were IP injected with 250 µl of T1/M23D complex, containing 45 µg of M23D AON and 2.5 mg of T1 nanoparticles (thought to represent a well-tolerated dose both for multiple IP and for IV administration) dissolved in sterile unpreserved saline solution (0.9% sodium chloride), or 250 µl containing 45 µg of M23D dissolved in unpreserved saline solution, and monitored according to approved NIH and University guidelines. The complex suspension (250 µl) was slowly injected through the abdominal skin into the peritoneal cavity using an insulin syringe fitted with a 28-gauge needle. One group of mdx mice (n = 6) (group 1) received three identical injections of T1/M23D complex at days 0, 7, and 14. One group of mdx mice (n = 2) (group 2) received three identical injections of M23D naked AON at the same times. Controls were age-matched mdx mice injected at days 0, 7, and 14 with fluorescent T1 AON-free nanoparticles (n = 3) (group 3) and untreated mdx mice (n = 6) (group 4) as negative controls. The total amount of the administered M23D AON was 135 µg/animal. Table 1 summarizes treatments and killing of mice analyzed in the present work.
Harvest of tissues. At 1 week after the third injection, four mdx mice of group 1, two mdx mice of group 2, one mdx mouse of group 3 and three mdx mice of group 4, were killed and diaphragm, gastrocnemius, quadriceps, and cardiac muscles were isolated, blotted dry, trimmed of external tendon, pinned to Parafilm-covered cork, snap frozen in liquid N2-cooled isopentane, and stored at −80 °C until further processing. Muscles from the two remaining mdx mice of group 1 together with the control mdx mice, two from group 3 and three from group 4, were harvested 6 weeks after the last injection (16–18 weeks of age). Liver, kidney, and spleen were also harvested.
Fluorescent microscope analysis of nanoparticles distribution. Seven-micrometer thick frozen sections of liver, spleen, heart, diaphragm, quadriceps, and gastrocnemius skeletal muscles from mdx mice of group 3 (Table 1) were labeled with a rabbit antinidogen antibody (Calbiochem, San Diego, CA), selected as marker of basal lamina, incubated with an antirabbit tetramethyl rhodamine iso-thiocyanate–conjugated secondary antibody (DAKO, Glostrup, Denmark), washed several times with phosphate-buffered saline and observed with a Nikon Eclipse 80i fluorescence microscope (Nikon Instruments, Firenze, Italy). Serial sections, obtained from three different levels of each organ at 100-µm intervals were observed at 100× and images recorded with a Nikon digital camera.
Transmission electron microscope study of nanoparticles distribution. Liver, spleen, kidney, diaphragm, gastrocnemius and quadriceps skeletal muscles, and heart were fixed with 2.5% glutaraldehyde in 0.1 mol/l phosphate buffer for 3 hours and with 1% osmium tetroxide in Veronal buffer for 2 hours, dehydrated with ethanol and embedded in Epon E812.
Ultrathin sections were stained with lead citrate and uranyl acetate and observed in a Philips EM 400 transmission electron microscope (FEI, Hillsboro, OR), operated at 100 kV.
Immunohistochemical analysis of dystrophin. Seven-micrometer thick frozen sections of heart, diaphragm, quadriceps, and gastrocnemius skeletal muscle fragments from mdx treated with M23D AON T1-conjugated complex (group 1), M23D naked AON (group 2), T1-Fluo AON-free (group 3), and untreated mdx mice (group 4), were labeled with a polyclonal antidystrophin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:100, incubated with an antirabbit tetramethyl rhodamine iso-thiocyanate–conjugated secondary antibody (DAKO), washed several times and observed with a Nikon Eclipse 80i fluorescence microscope.
Evaluation and quantification of dystrophin-positive fibers. Dystrophin immunolabeled transverse sections from WT, treated, and untreated mdx mice were analyzed with a Nikon Eclipse 80i fluorescence microscope at 20× and images were taken with a fixed exposure time (0.5 seconds) using an high-resolution CCD camera (Nikon). Images were analyzed by NIS-Element BR2.20 (Nikon) imaging program and the threshold intensity for each tissue was determined on sections of WT mice. Because in T1/MD23 treated mdx mice groups of fibers showed a heterogeneous pattern, the percentage of the perimeter truly labeled by the antibody was determined for each fiber by using the AnalySIS program (Soft Imaging System, Muenster, Germany). The count of dystrophin-positive fibers excluded myofibers with a labeling that covered <50% of the perimeter or with a discontinuous pattern. The number of dystrophin-positive fibers was evaluated on three different levels, 300 µm apart from each other, in the diaphragm, gastrocnemius, quadriceps, and on eight serial sections, at 100-µm intervals, of the heart. At least 3,000 muscle fibers from the diaphragm and gastrocnemius and 5,000 from the quadriceps muscles, respectively, obtained from three different levels of tissue blocks for each sample, were studied for statistical evaluation; data were analyzed according to Mann–Whitney test, and the criterion for statistical significance was P < 0.05.
The counts were performed blind to sample identity and the relative patterns were confirmed by an independent observer in order to unbias the observation.
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis and immunoblotting analysis. Twenty-micrometer thick frozen muscle sections were homogenized with a lysis buffer containing 7 mol/l urea, 2 mol/l thiourea, 1% amidosulfobetaine-14, and 0.3% dithioerythritol, and then centrifuged at 1,500g for 10 minutes. Protein concentration was determined in the supernatants with the Bradford method. Aliquots of proteins from normal C57BL/10 mice (10 µg) and from muscles of treated or untreated mdx mice (300 µg) were loaded onto a 6% T sodium doedecyl sulphate–polyacrylamide gel and separated by electrophoresis. Samples were transferred to a nitrocellulose membrane overnight at 75 V. The membrane was blocked with nonfat dried milk for 60 minutes at room temperature and incubated overnight at 4 °C with the specific antibody DYS2 (a mouse monoclonal antibody to the carboxy terminal region of dystrophin, 1:100, NovoCastra, Newcastle, UK) or H-300 (a rabbit polyclonal antibody to the internal region of dystrophin, 1:200, Santa Cruz Biotechnology). After intervening washes, the membrane was incubated with horseradish peroxidase–conjugated goat antimouse or antirabbit IgG diluted 1:40,000 or 1:30,000, respectively. Immunocomplexes were visualized with the ECL Advance Western Blotting Detection Kit (Amersham Pharmacia Biotech, Buckinghamshire, UK).
RNA studies. Total RNA was purified from muscle biopsies of WT (n = 2), untreated (n = 6), and treated mdx mice (n = 8), by using RNeasy kit (Qiagen, La Jolla, CA) and reverse transcribed into cDNA using the high capacity cDNA reverse transcription kit (Applied Biosystems, Frankfurt, Germany). Six novel ESRAs detecting mdx exons 7, 8, 22, 23, 25, and 56, were specifically developed for this study. These exons were chosen because they do not undergo spontaneous alternative splicing. ESRA on exons 7, 22, 23, and 25 were used to quantify the percentage of exon 23 skipping in treated mice. Dystrophin transcript quantification was performed by comparison with the β-actin gene on each isolated muscle from both treated and control mdx mice. All these ESRAs are based on TaqMan technology and have been designed by PrimerExpress Applied Biosystems software (Applied Biosystems). Primers and probes sequences are available upon request. The amount of target sequences in respect to appropriate endogenous control (β-actin gene) was evaluated by the comparative CT method with respect to the endogenous β-actin control (ΔΔCt Method) (Applied Biosystems User Bulletin no. 2). Nested RT-PCR was performed as described in ref. 25, skipped transcript was analyzed by sequencing (ABI PRISM 3130 Automated Sequencer; Applied Biosystems).
Supplementary MaterialFigure S1. Morphological analysis of liver, kidney, and spleen from mdx mice treated with T1/M23D complex. analysed 1 week after last injection. Semithin sections stained with toluidine blue do not reveal any chronic inflammatory reaction in all tissues studied. Original magnification, 100xFigure S2. Nested RT-PCR analysis of dystrophin mRNA in muscle tissues of wild type, untreated, and T1/M23D-treated mdx mice. Primary PCR was performed by using 20F-26R primers spanning exons 20–26 of the mouse dystrophin gene, while nested amplification was carried out across exon 20-25 with a forward primer mapping downstream in respect to 20F (expected product: full-length 874bp, skipped 661bp). The muscles analysed were diaphragm, quadriceps, gastrocnemius and heart in wild type (lanes 1,4,7,10), untreated mdx (lanes 2,5,8,11) and T1/M23D treated mdx mice (lanes 3,6,9,12). Full-length dystrophin transcript was detected in all cDNA samples, and a shorter transcript originated from AON-induced exon 23 skipping was observed in quadriceps (lane 6), gastrocnemius (lane 9) and heart (lane 12) of T1/M23D treated mdx mice, analysed 1 week after last injection. Arrows indicate the fragment size of the DNA Molecular Weight Marker IX (Roche).Materials and Methods.
Supplementary Material
Morphological analysis of liver, kidney, and spleen from mdx mice treated with T1/M23D complex. analysed 1 week after last injection. Semithin sections stained with toluidine blue do not reveal any chronic inflammatory reaction in all tissues studied. Original magnification, 100x
Nested RT-PCR analysis of dystrophin mRNA in muscle tissues of wild type, untreated, and T1/M23D-treated mdx mice. Primary PCR was performed by using 20F-26R primers spanning exons 20–26 of the mouse dystrophin gene, while nested amplification was carried out across exon 20-25 with a forward primer mapping downstream in respect to 20F (expected product: full-length 874bp, skipped 661bp). The muscles analysed were diaphragm, quadriceps, gastrocnemius and heart in wild type (lanes 1,4,7,10), untreated mdx (lanes 2,5,8,11) and T1/M23D treated mdx mice (lanes 3,6,9,12). Full-length dystrophin transcript was detected in all cDNA samples, and a shorter transcript originated from AON-induced exon 23 skipping was observed in quadriceps (lane 6), gastrocnemius (lane 9) and heart (lane 12) of T1/M23D treated mdx mice, analysed 1 week after last injection. Arrows indicate the fragment size of the DNA Molecular Weight Marker IX (Roche).
Acknowledgments
The Telethon Italy grants GGP05115 and GUP07011 (both to A.F.) are acknowledged. Thanks are also due to the Industria Chimica Emiliana (ICE Reggio Emilia) Grant (to A.F.), to FAR2007 (University of Ferrara, to A.F.), to TREAT-NMD Network of Excellence of EU FP7 no. 036825 (to L.M. and Telethon, Italy). We are also grateful to the ISS National AIDS Programme grants to A.C., L.T., and M.L. supporting the nanoparticle technology platform. We wish to thank Judith van Deutekom (Prosensa–LUMC University, Leiden, NL) for the helpful suggestions and critical reading of the manuscript.
REFERENCES
- Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ., and , Den Dunnen JT.Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule Muscle Nerve 200634135–144.Review [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, van Ommen GJ, den Dunnen JT, et al. Antisense-induced multiexon skipping for Duchenne muscular dystrophy makes more sense. Am J Hum Genet. 2004;74:83–92. doi: 10.1086/381039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremmer-Bout M, Aartsma-Rus A, de Meijer EJ, Kaman WE, Janson AA, Vossen RH, et al. Targeted exon skipping in transgenic hDMD mice: a model for direct preclinical screening of human-specific antisense oligonucleotides. Mol Ther. 2004;10:232–240. doi: 10.1016/j.ymthe.2004.05.031. [DOI] [PubMed] [Google Scholar]
- McClorey G, Moulton HM, Iversen PL, Fletcher S., and , Wilton SD. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13:1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A., and , van Ommen GJ.Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications RNA 2007131609–1624.Review [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 't Hoen PA, van der Wees CG, Aartsma-Rus A, Turk R, Goyenvalle A, Danos O, et al. Gene expression profiling to monitor therapeutic and adverse effects of antisense therapies for Duchenne muscular dystrophy. Pharmacogenomics. 2006;7:281–297. doi: 10.2217/14622416.7.3.281. [DOI] [PubMed] [Google Scholar]
- Hoffman EP. Skipping toward personalized molecular medicine. N Engl J Med. 2007;357:2719–2722. doi: 10.1056/NEJMe0707795. [DOI] [PubMed] [Google Scholar]
- Jearawiriyapaisarn N, Moulton HM, Buckley B, Roberts J, Sazani P, Fucharoen S, et al. Sustained dystrophin expression induced by peptide-conjugated morpholino oligomers in the muscles of mdx mice. Mol Ther. 2008;16:1624–1629. doi: 10.1038/mt.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Moulton HM, Seow Y, Boyd C, Boutilier J, Iverson P, et al. Cell-penetrating peptide-conjugated antisense oligonucleotides restore systemic muscle and cardiac dystrophin expression and function. Hum Mol Genet. 2008;17:3909–3918. doi: 10.1093/hmg/ddn293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JH, Schray RC, Sirsi SR., and , Lutz GJ. Nanopolymers improve delivery of exon skipping oligonucleotides and concomitant dystrophin expression in skeletal muscle of mdx mice. BMC Biotechnol. 2008;8:35. doi: 10.1186/1472-6750-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denti MA, Rosa A, D'Antona G, Sthandier O, De Angelis FG, Nicoletti C, et al. Body-wide gene therapy of Duchenne muscular dystrophy in the mdx mouse model. Proc Natl Acad Sci USA. 2006;103:3758–3763. doi: 10.1073/pnas.0508917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Allen JM, Riddell SR, Gregorevic P, Storb R, Tapscott SJ, et al. Immunity to adeno-associated virus-mediated gene transfer in a random-bred canine model of Duchenne muscular dystrophy. Hum Gene Ther. 2007;18:18–26. doi: 10.1089/hum.2006.093. [DOI] [PubMed] [Google Scholar]
- Manno CS, Pierce GF, Arruda VR, Glader B, Ragni M, Rasko JJ, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- Li SD., and , Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13:1313–1319. doi: 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- Tamber H, Johansen P, Merkle HP., and , Gander B. Formulation aspects of biodegradable polymeric microspheres for antigen delivery. Adv Drug Deliv Rev. 2005;57:357–376. doi: 10.1016/j.addr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Little R., and , Kohane DS. Polymers for intracellular delivery of nucleic acids. J Mater Chem. 2008;18:832–841. [Google Scholar]
- O'Hagan D, Singh M, Ugozzoli M, Wild C, Barnett S, Chen MC, et al. Induction of potent immune responses by cationic microparticles with adsorbed human immunodeficiency virus DNA vaccines. J Virol. 2001;75:9037–9043. doi: 10.1128/JVI.75.19.9037-9043.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster CG, Kim N, Grode L, Barbu-Tudoran L, Schaper AK, Kaufmann SHE, et al. Cationic microparticles consisting of poly(lactide-co-glycolide) and polyethyleneimine as carriers systems for parental DNA vaccination. J Control Release. 2005;104:359–377. doi: 10.1016/j.jconrel.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Kreuter J. Evaluation of nanoparticles as drug-delivery systems. III: materials, stability, toxicity, possibilities of targeting, and use. Pharm Acta Helv. 1983;58:196–209. [PubMed] [Google Scholar]
- Kreuter J., and , Speiser PP. New adjuvants on a polymethylmethacrylate base. Infect Immun. 1976;13:204–210. doi: 10.1128/iai.13.1.204-210.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieneker F, Kreuter J., and , Löwer J. High antibody titres in mice with polymethylmethacrylate nanoparticles as adjuvant for HIV vaccines. AIDS. 1991;5:431–435. doi: 10.1097/00002030-199104000-00012. [DOI] [PubMed] [Google Scholar]
- Caputo A, Brocca-Cofano E, Castaldello A, De Michele R, Altavilla G, Marchisio M, et al. Novel biocompatible anionic polymeric microspheres for the delivery of the HIV-1 Tat protein for vaccine application. Vaccine. 2004;22:2910–2924. doi: 10.1016/j.vaccine.2003.12.025. [DOI] [PubMed] [Google Scholar]
- Castaldello A, Brocca-Cofano E, Voltan R, Triulzi C, Altavilla G, Laus M, et al. DNA prime and protein boost immunization with innovative polymeric cationic core-shell nanoparticles elicits broad immune responses and strongly enhance cellular responses of HIV-1 tat DNA vaccination. Vaccine. 2006;24:5655–5669. doi: 10.1016/j.vaccine.2006.05.058. [DOI] [PubMed] [Google Scholar]
- Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J, et al. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc Natl Acad Sci USA. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter J, Lou F, Rabinowitz A, Yin H, Rosenfeld J, Wilton SD, et al. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- Metelev V., and , Agrawal S. Ion-exchange high-performance liquid chromatography analysis of oligodeoxyribonucleotide phosphorothioates. Anal Biochem. 1992;200:342–346. doi: 10.1016/0003-2697(92)90476-n. [DOI] [PubMed] [Google Scholar]
- Sirsi SR, Schray RC, Guan X, Lykens NM, Williams JH, Erney ML, et al. Functionalized PEG-PEI copolymers complexed to exon-skipping oligonucleotides improve dystrophin expression in mdx mice. Hum Gene Ther. 2008;19:795–806. doi: 10.1089/hum.2007.129. [DOI] [PubMed] [Google Scholar]
- Nguyen HK, Lemieux P, Vinogradov SV, Gebhart CL, Guerin N, Paradis G, et al. Evaluation of polyetherpolyethyleneimine graft copolymers as gene transfer agents. Gene Ther. 2000;7:126–138. doi: 10.1038/sj.gt.3301052. [DOI] [PubMed] [Google Scholar]
- Grosse S, Aron Y, Honoré I, Thévenot G, Danel C, Roche AC, et al. Lactosylated polyethylenimine for gene transfer into airway epithelial cells: role of the sugar moiety in cell delivery and intracellular trafficking of the complexes. J Gene Med. 2004;6:345–356. doi: 10.1002/jgm.515. [DOI] [PubMed] [Google Scholar]
- Misquitta CM, Iyer VR, Werstiuk ES., and , Grover AK. The role of 3′-untranslated region (3′-UTR) mediated mRNA stability in cardiovascular pathophysiology. Mol Cell Biochem. 2001;224:53–67. doi: 10.1023/a:1011982932645. [DOI] [PubMed] [Google Scholar]
- Park SK., and , Prolla TA.Gene expression profiling studies of aging in cardiac and skeletal muscles Cardiovasc Res 200566205–212.Review [DOI] [PubMed] [Google Scholar]
- Gebski BL, Mann CJ, Fletcher S., and , Wilton SD. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet. 2003;12:1801–1811. doi: 10.1093/hmg/ddg196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphological analysis of liver, kidney, and spleen from mdx mice treated with T1/M23D complex. analysed 1 week after last injection. Semithin sections stained with toluidine blue do not reveal any chronic inflammatory reaction in all tissues studied. Original magnification, 100x
Nested RT-PCR analysis of dystrophin mRNA in muscle tissues of wild type, untreated, and T1/M23D-treated mdx mice. Primary PCR was performed by using 20F-26R primers spanning exons 20–26 of the mouse dystrophin gene, while nested amplification was carried out across exon 20-25 with a forward primer mapping downstream in respect to 20F (expected product: full-length 874bp, skipped 661bp). The muscles analysed were diaphragm, quadriceps, gastrocnemius and heart in wild type (lanes 1,4,7,10), untreated mdx (lanes 2,5,8,11) and T1/M23D treated mdx mice (lanes 3,6,9,12). Full-length dystrophin transcript was detected in all cDNA samples, and a shorter transcript originated from AON-induced exon 23 skipping was observed in quadriceps (lane 6), gastrocnemius (lane 9) and heart (lane 12) of T1/M23D treated mdx mice, analysed 1 week after last injection. Arrows indicate the fragment size of the DNA Molecular Weight Marker IX (Roche).