Abstract
The development of techniques to efficiently deliver genes using nonviral approaches can broaden the application of gene delivery in medical applications without the safety concerns associated with viral vectors. Here, we designed a clustered integrin-binding platform to enhance the efficiency and targetability of nonviral gene transfer to HeLa cells with low and high densities of αvβ3 integrin receptors. Arg-Gly-Asp (RGD) nanoclusters were formed using gold nanoparticles functionalized with RGD peptides and used to modify the surface of DNA/poly(ethylene imine) (PEI) polyplexes. DNA/PEI polyplexes with attached RGD nanoclusters resulted in either 5.4- or 35-fold increase in gene transfer efficiency over unmodified polyplexes for HeLa cells with low- or high-integrin surface density, respectively. The transfection efficiency obtained with the commercially available vector jetPEI-RGD was used for comparison as a vector without clustered binding. JetPEI-RGD exhibited a 1.2-fold enhancement compared to unmodified jetPEI in cells with high densities of αvβ3 integrin receptors. The data presented here emphasize the importance of the RGD conformational arrangement on the surface of the polyplex to achieve efficient targeting and gene transfer, and provide an approach to introduce clustering to a wide variety of nanoparticles for gene delivery.
Introduction
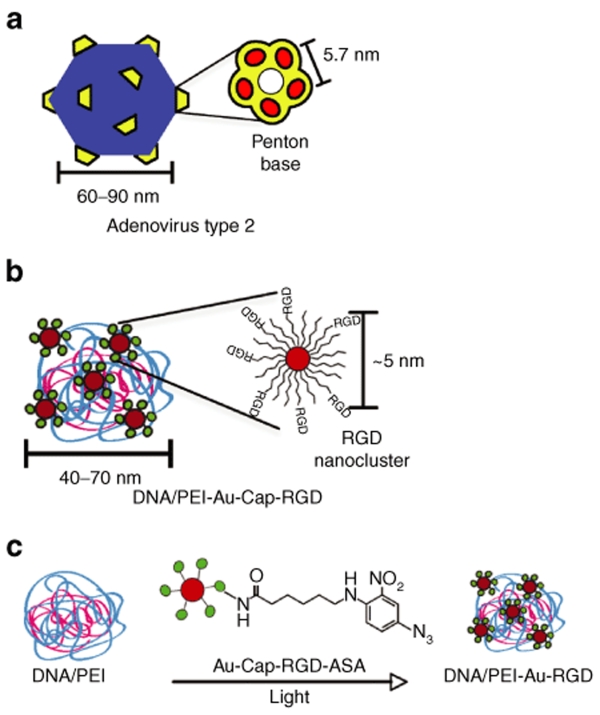
RGD ligand clustering has been implicated in the modulation of cell adhesion, cell migration, cell spreading, and nonviral gene transfer.1,2,3,4,5 For example, the maximum distance RGD ligands can be spaced for cell attachment and migration to occur on a flat surface is 58 nm (ref. 6) and it has been shown that integrin occupancy, conformation, and aggregation regulate integrin-signal transduction.7,8 Interestingly, four different adenovirus stereotypes9 have evolved to take advantage of clustered RGD ligands to gain access into cells through binding multiple integrin receptors simultaneously.10 The clustered RGD ligands are displayed on five penton-base proteins located at each of the 12 vertices of the virus surface. The penton-base protein protrudes five RGD peptide sequences located 5.7 nm apart,9,11 which have been found to be critical for viral cell entry.12 Inhibition of the penton-base protein using antibodies, RGD peptides, or by mutation results in decreased adenoviral internalization and overall transduction efficiency.11,13,14,15
The introduction of multivalent ligand binding to drug delivery carriers has been investigated as a method to enhance delivery of small molecular drugs16 or tumor-labeling agents17 and has been demonstrated to have increased effects over monovalent binding.18,19,20,21 However, the introduction of clustered ligand binding to nonviral gene delivery vectors has not been investigated. Nevertheless, ligands have been introduced to the surface of nonviral vectors to enhance targetability and overall gene transfer efficiency. These ligands include small molecules (e.g., folate and galactose), proteins (e.g., transferrin and antibodies)22 as well as RGD peptides.23,24 Although the direct RGD conjugation to DNA/poly(ethylene imine) (PEI) polyplexes increases the transfection efficiency in vitro and in vivo,23,24,25 the effect of RGD ligand clustering or the clustering of any other ligand on the efficiency and targetability of DNA/PEI polyplexes has not been investigated to date.
The cationic polymer PEI is one of the most widely used nonviral gene delivery vehicles for DNA, being used successfully both in vitro and in vivo.26 It condenses with DNA through its positively charged amines, protecting DNA from degradation and forming particles (polyplexes) that can enter the cell and result in transgene expression.26 The amines in PEI also serve as functional groups for chemical modification to introduce domains such as poly(ethylene glycol) to increase biocompatibility,27,28 and ligands and peptides that enhance targeting, internalization and trafficking.29
In this report, we investigated the effect of clustered RGD ligands on the transfection efficiency of DNA/PEI polyplexes using the penton base of Adenovirus type 2 as a design guide (Figure 1). Clustered RGD ligands were introduced to DNA/PEI polyplexes through attaching RGD peptide–modified gold nanoparticles to the polyplex surface. Gold nanoparticles were stabilized through the formation of a peptide monolayer-protected cluster (MPC) using a β-strand-forming peptide,30 which resulted in stable particles in high-salt solutions. RGD nanocluster-modified polyplexes were used to transfect high- and low-αvβ3-integrin density cells to determine whether our vector could transfect cells with high-integrin densities more efficiently. We demonstrated that the presence of clustered RGD ligands positively affects gene transfer to both high- and low-αvβ3-integrin density cells compared to unmodified polyplexes or polyplexes modified with RGD peptides that were not in a clustered conformation. Furthermore, the level of enhancement was shown to depend on the density of integrin receptors on the cell surface.
Figure 1.
Schematic comparing Adenovirus type 2 and DNA/PEI– Au-Cap-RGD-ASA polyplex. (a) Type 2 Adenovirus with penton-base proteins (reproduced from ref. 9). (b) DNA/PEI-Au-Cap-RGD-ASA polyplex with RGD nanoclusters. (c) Au-Cap-RGD-ASA nanoparticles are conjugated to preformed DNA/PEI polyplexes when exposed to UV light.
Results
RGD nanocluster formation and characterization
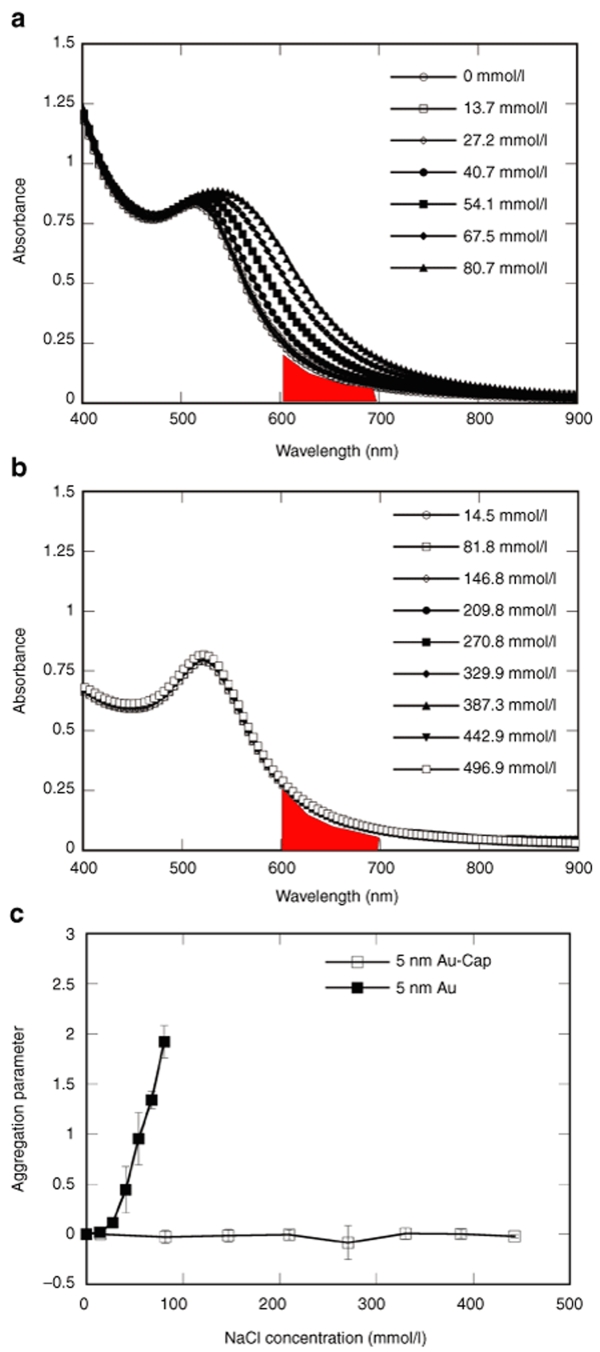
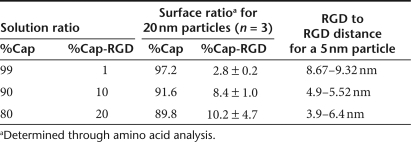
Stable RGD-modified gold nanoparticles (Au-Cap-RGD, RGD nanoclusters) and stabilized gold nanoparticles (Au-Cap) were synthesized through the modification of 5 nm gold nanoparticles with the self-assembling peptides CCVVVT (Cap), CCVVVT-RGD (Cap-RGD), CCVVVT-RGD-Azidosalicylic Acid (Cap-RGD-ASA), and CCVVVT-ASA (Cap-ASA) via thiol/gold chemisorption. Gold nanoparticle modification with the self-assembling peptides to form MPCs was confirmed through absorbance scans and dynamic light scattering (DLS). Using an absorption scan, the Au-Cap nanoparticles were shown to be stable in up to 500 mmol/l NaCl compared to only 15 mmol/l for unmodified gold nanoparticles, indicating the modification of the gold nanoparticles (Figure 2). The size of the gold nanoparticles increased by ~4 nm after Cap peptide modification, as expected, based on the length of the peptide (Figure 3a). To introduce bioactivity into the Au nanoparticles, MPC were formed with mixtures of Cap and Cap-RGD peptides and subjected to amino acid analysis to determine whether the ratio of peptides used in solution was conserved after attachment (Table 1). The ratio on the surface for 99:1 and 90:10 Cap to Cap-RGD-modified 20 nm nanoparticles were found to closely resemble the solution mixture as expected. However, when increased to 80:20, the ratio found on the surface was 89.8:10.2, suggesting that Cap-RGD peptides could only be packed efficiently up to 90 to 10 concentration ratio.
Figure 2.
Salt stability of modified and unmodified nanoparticles measured using UV-Vis absorbance spectroscopy. Salt concentration was increased using 2 µl for unmodified nanoparticles and 15 µl of 4.11 mol/l NaCl every 15 minutes. Wavelength scans of (a) 5 nm unmodified Au nanoparticles and (b) Au-Cap nanoparticles in solution with increasing NaCl concentration. Shaded area under the curves in a and b were used to calculate aggregation parameter. (c) Aggregation parameter plotted with mean ± SD for n = 3 for increasing NaCl concentration for 5 nm unmodified Au and Au-Cap. Aggregation parameter of unmodified Au show a large increase with small increases in NaCl concentration.
Figure 3.
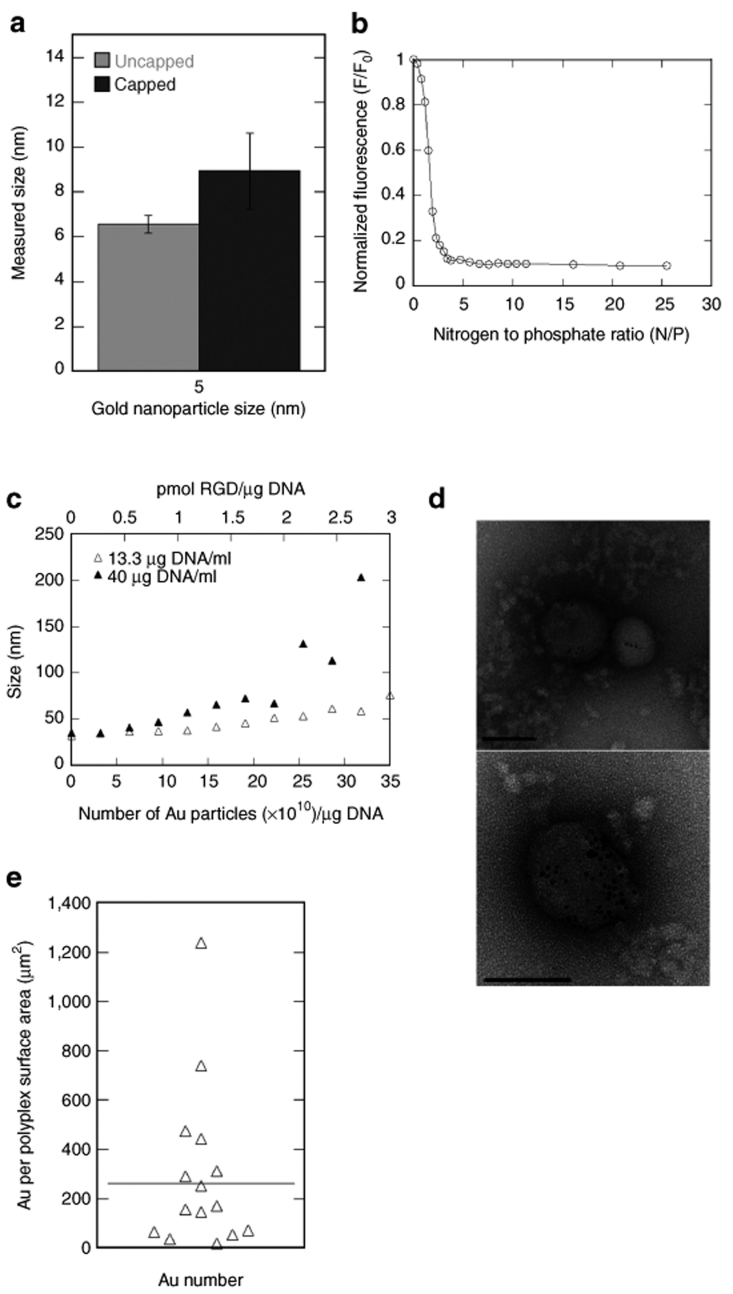
Characterization of DNA/PEI and DNA/PEI-Au-Cap-RGD-ASA polyplexes. (a) Mean size ± SD (n = 3) of 5 nm unmodified Au and Au-Cap measured using dynamic light scattering (DLS). (b) Ethidium bromide (EtBr) competition assay for DNA/PEI polyplex formation. Plasmid DNA (26.6 µg/ml in Tris–EDTA buffer) and EtBr (1.1 µg/ml) were added to a fluorimeter vial to a final volume of 150 µl and read before and after the addition of each aliquot of PEI (0.5 µl of 0.1 mg/ml). (c) Size of DNA/PEI polyplexes modified with 5 nm Au-Cap-RGD nanoparticles were measured using DLS for low (13.3 µg DNA/ml) and high concentration (40 µg DNA/ml). (d) Electron micrographs of negatively stained DNA/PEI-Au-Cap-RGD-ASA. All the polyplexes were observed to contain Au particles. Bar = 100 nm. (e) Number distribution of the amount of Au nanoparticles per surface area of polyplex (n = 17).
Table 1.
RGD to RGD distance approximation for 5 nm Au-Cap-RGD
DNA/PEI-Au-Cap-RGD polyplex characterization
To determine the nitrogen to phosphate ratio (N/P) that resulted in compacted particles at the lowest N/P ratio, an ethidium bromide (EtBr) exclusion assay was used where the amount of EtBr release is quantified as the polyplex forms. At an N/P ratio of 5, the EtBr exclusion curve (Figure 3b) began to plateau indicating full DNA/PEI complexation had been reached. Au-Cap-RGD-ASA nanoparticles were attached to DNA/PEI polyplexes (N/P of 10) using the photoreactive group ASA (Figure 1c), which reacts with amines when exposed to UV light. The sizes and degree of modification of the resulting DNA/PEI-Au-Cap-RGD polyplexes were measured as a function of Au-Cap-RGD-ASA nanoparticle concentration using DLS and visualized through transmission electron microscope (TEM). The size increased as Au-Cap-RGD-ASA nanoparticles were added for polyplexes formed with low (13.3 µg/ml) and high (40 µg/ml) DNA concentrations. Low concentration polyplexes showed a gradual increase in size from 32 to 76 nm for 0 to 35 × 1010 gold nanoparticles/µg DNA (Figure 3c). High-concentration polyplexes showed a gradual increase in size from 35 to 72 nm for 0 to 22 × 1010 nanoparticles/µg DNA. However, when higher amounts of Au-Cap-RGD-ASA nanoparticles were added (22 to 31 × 1010 nanoparticles/µg DNA) the size of the polyplexes increased from 72 to 203 nm (Figure 3c). Transmission electron micrographs of negatively stained DNA/PEI-Au-Cap-RGD polyplexes showed similar size distribution results to the DLS (Figure 3d). A maximum of 49 clusters of Au nanoparticles were observed in one polyplex structure. A distribution of polyplexes was seen containing different amounts of Au nanoparticles (Figure 3e) with an average of 262 nanoparticles/µm2 of polyplex surface area (~10 Au nanoparticles/polyplex). Gold nanoparticles were clearly visible, indicating attachment to the surface of the polyplex suggesting that the Au-Cap-RGD nanoclusters will be available for interaction with cell surface receptors. All polyplexes were observed to contain nanoparticles.
Toxicity of modified Au and DNA/ PEI-Au-Cap-RGD-ASA conjugates
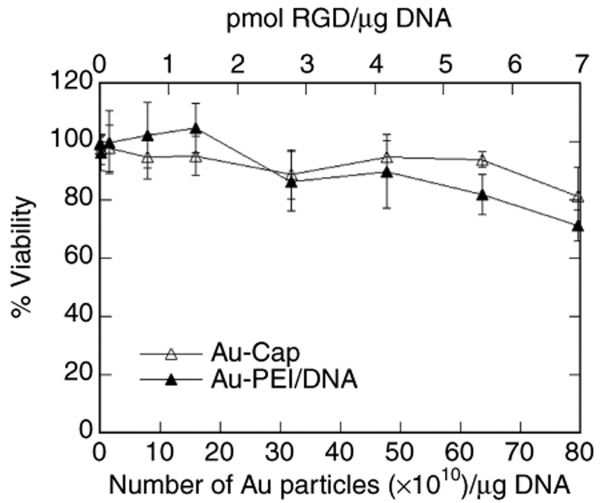
To determine whether 5 nm Au-Cap or DNA/PEI polyplexes modified with 5 nm Au-Cap-RGD-ASA were toxic to cells, the proliferation rate of HeLa cells exposed to Au-Cap or DNA/PEI-Au-Cap-RGD-ASA was compared to that of untreated cells (Figure 4). Au-Cap nanoparticles were not toxic to cells for all the concentrations tested up to 17.9 × 1010 particles/well (equivalent to 80 × 1010 particles/µg DNA). DNA/PEI polyplexes were conjugated with increasing amounts of Au-Cap-RGD-ASA (0.224 µg DNA/well) and it was observed that the addition of nanoparticles did not increase the toxicity of polyplexes for all concentrations tested except at the highest concentration of particles (80 × 1010 particles/µg DNA), where the proliferation rate decreased to 72%.
Figure 4.
Cell toxicity (proliferation) assay for Au-Cap nanoparticles and DNA/PEI-Au polyplexes. HeLa cells were plated in 96-well plates (7,000 cells/well) for 12 hours before exposing the cells to Au-Cap nanoparticles or DNA/PEI-Au-Cap-RGD-ASA polyplexes at varying concentrations of Au nanoparticles (1.7 µl/well) and testing for cell proliferation. The data were normalized to the absorbance of untreated samples to calculate the percent viability for each sample (n = 3).
Integrin expression
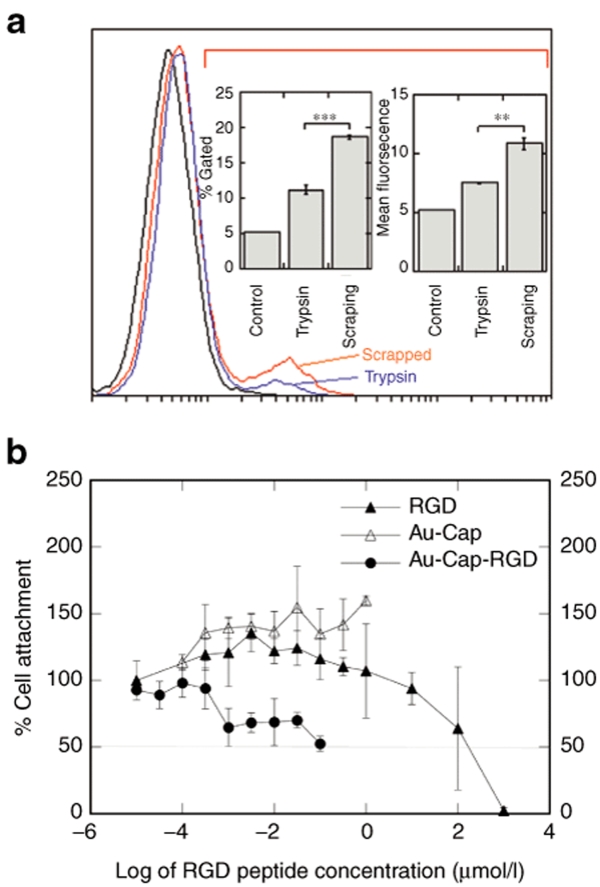
To generate HeLa cells with high- and low-αvβ3-integrin density, HeLa cells were passaged using trypsin or scrapping. The αvβ3 integrin expression profile of HeLa cells was determined using flow cytometry 12 hours after plating (Figure 5a). Cells that were detached via scrapping resulted in significantly (P < 0.01) higher densities of αvβ3 integrins on the cell surface than cells detached using trypsin. These cells will be referred to as high-αvβ3 density and low-αvβ3 density for the scrapped and trypsinized cells respectively.
Figure 5.
Effects of multivalent nanoparticles on HeLa cells. (a) Flow cytometry of αvβ3 integrin stained of HeLa cells passaged by scrapping or trypsinization measured 12 hours after plating. A gate that included 5% of cells in the unstained control was used in determining the percent of cells that were αvβ3 integrin positive. The amount of trypsin and scrapped αvβ3 integrin-positive cells are plotted with ***P < 0.0001 using a two-tailed t-test (n = 3). Mean fluorescence of all trypsin or scrapped cells were compared. **P < 0.001 using a two-tailed t-test (n = 3). (b) Antiadhesion assay of RGD peptide, Au-Cap, and Au-Cap-RGD nanoparticles with HeLa cells (n = 3). The multivalent Au-Cap-RGD nanoparticles required less of the equivalent amount of RGD than the monovalent free RGD peptide.
Antiadhesion assay
To determine the effectiveness of the RGD nanoclusters to bind to αvβ3 integrins, an antiadhesion assay was used (Figure 5b). The IC50 concentration at which 50% of cell adhesion is inhibited for free RGD peptide was ~102 µmol/l, which is comparable to that previously observed by others.18 In contrast, Au-RGD nanoclusters required an equivalent RGD peptide of ~10−1 µmol/l to achieve the IC50 level. Au-Cap did not affect cell attachment, indicating that the gold nanoparticle was not responsible for the observed results.
Transfection using RGD nanoclusters conjugated to DNA/PEI polyplexes
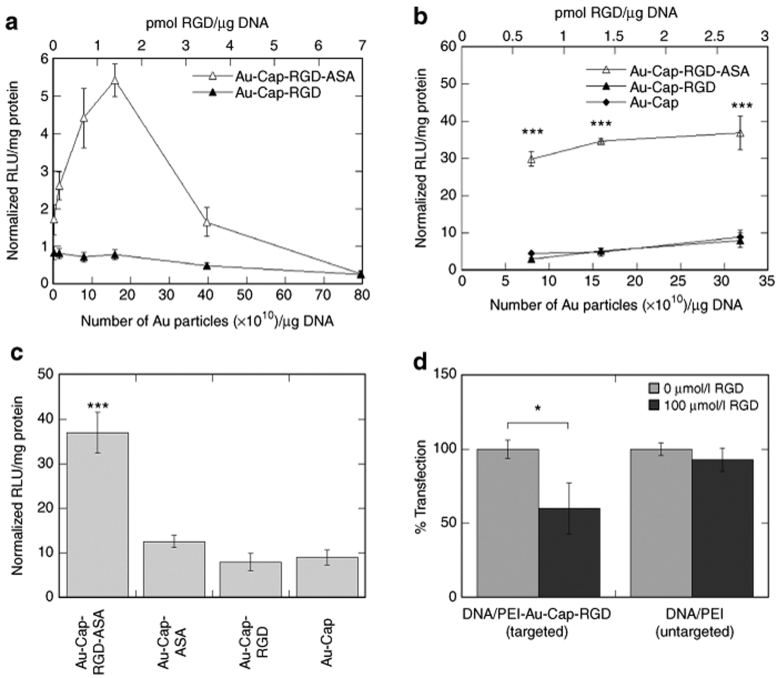
The effect of RGD nanoclusters on the transfection efficiency of DNA/PEI polyplexes was studied as a function of RGD nanoclusters bound to the polyplex and the density of integrins at the cell surface. DNA/PEI polyplexes were modified with RGD nanoclusters at concentrations ranging from 0.1 × 1010 to 80 × 1010 particles/µg DNA. For low-αvβ3-density cells maximal expression was observed for 16 × 1010 particles/µg DNA, showing a 5.4-fold increase in expression when compared to unmodified polyplexes (Figure 6a). The effect of RGD nanoclusters on the transfection efficiency of DNA/PEI polyplexes to cells with high-αvβ3-integrin density was studied with the range of RGD nanoclusters that resulted in the highest gene transfer for low-αvβ3-density cells, 8 × 1010 to 32 × 1010 particles/µg DNA (Figure 6b). Transfection of high-αvβ3 density cells resulted in a 35-fold enhancement in transfection efficiency compared to unmodified polyplexes. As controls for the transfections in both low and high-αvβ3-integrin density cells, polyplexes were mixed with Au-Cap-RGD nanoparticles that did not contain the ASA functional group and thus could not covalently bind to the polyplex. These transfections showed no increase in transgene expression for low-αvβ3-integrin density cells and an eightfold increase for high-αvβ3-integrin density cells compared to unmodified polyplexes. To further investigate the eightfold increase in transgene expression and to determine whether the gold nanoparticles themselves were responsive for the observed enhancement in gene transfer, polyplexes were mixed with Au-Cap or Au-Cap-ASA (16 × 1010 particles/µg DNA). Transfections with Au-Cap and Au-Cap-ASA-modified polyplexes resulted in an increase in transgene expression that was not statistically different from that observed for the Au-Cap-RGD (Figure 6c).
Figure 6.
Transfection efficiency in high- and low-αvβ3 integrin-expressing HeLa cells measured using a luciferase reporter gene. (a) Luciferase expression in low-αvβ3-integrin-expressing HeLa cells transfected with DNA/PEI-Au-Cap-RGD-ASA (with covalent linker) and DNA/PEI-Au-Cap-RGD (without covalent linker) (n = 3). (b) Luciferase expression in high-αvβ3-integrin-expressing HeLa cells transfected with DNA/PEI-Au-Cap-RGD-ASA (with covalent linker), DNA/PEI-Au-Cap-RGD (without covalent linker), and DNA/PEI-Au-Cap (n = 3). The symbol *** represents statistical significance to the level of P < 0.001 for DNA/PEI-Au-Cap-RGD-ASA compared to DNA/PEI-Au-Cap-RGD and DNA/PEI-Au-Cap using the Tukey test. (c) Luciferase expression in high-αvβ3-integrin-expressing HeLa cells transfected with DNA/PEI-Au-Cap-RGD-ASA (with covalent linker), DNA/PEI-Au-Cap-ASA (with covalent linker but no RGD), DNA/PEI-Au-Cap-RGD (without covalent linker), and DNA/PEI-Au-Cap (without covalent linker) (n = 3). The symbol *** represents statistical significance to the level of P < 0.001 for DNA/PEI-Au-Cap-RGD-ASA compared to DNA/PEI-Au-Cap-RGD, DNA/PEI-Au-Cap, and DNA/PEI-Au-Cap-ASA using the Tukey test. (d) Competitive binding of free RGD peptide for transfection of high-αvβ3-integrin-expressing HeLa cells with DNA/PEI-Au-Cap-RGD-ASA (n = 3). The symbol * represents statistical significance to the level of P < 0.05.
To ensure that the enhancement in gene transfer was due to the RGD nanoclusters binding to integrin receptors at the cell surface, a competitive binding using free RGD peptide was conducted (Figure 6d). DNA/PEI–Au-Cap-RGD-ASA polyplex gene transfer decreased by 40% when free RGD peptide was added to the transfection mixture. The addition of free RGD peptide to untargeted polyplexes had no effect on gene transfer.
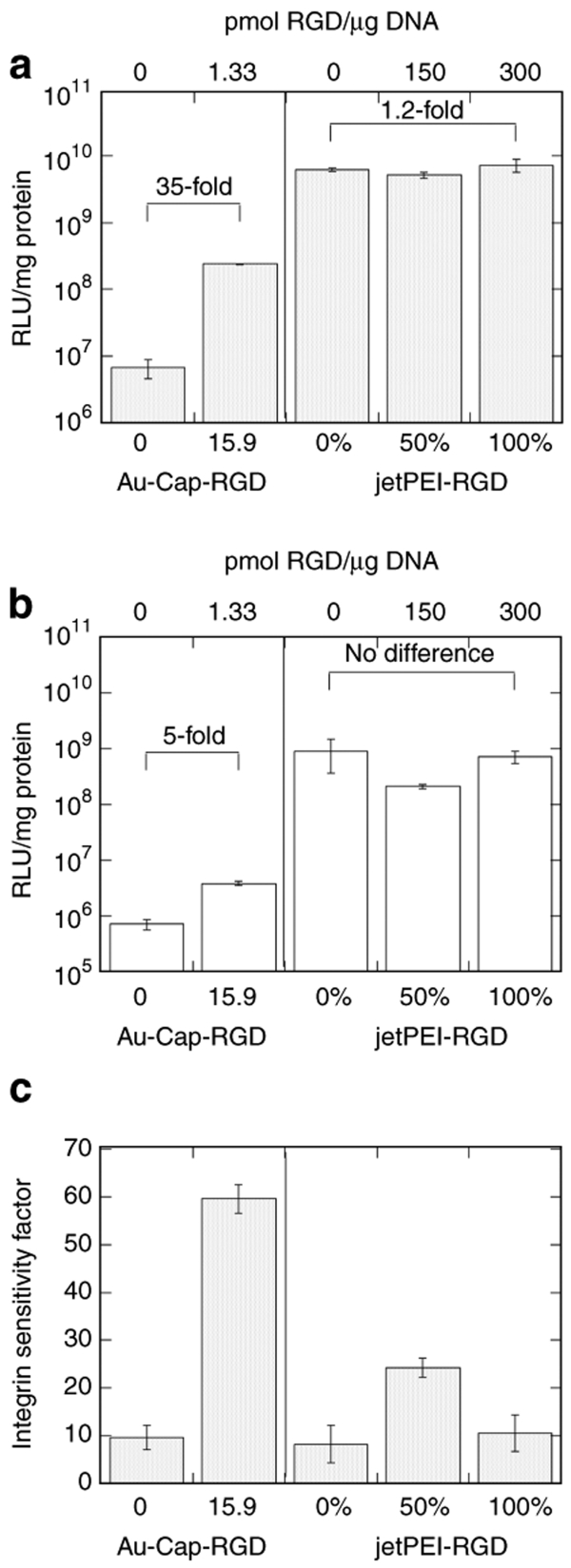
A commercially available RGD-modified PEI, jetPEI-RGD, was used for comparison of a delivery vector without clustered RGD ligands. High-αvβ3-density cells were transfected with jetPEI, jetPEI-RGD, or a 50:50 mixture (Figure 7a). A 1.2-fold increase in transfection efficiency was observed when jetPEI-RGD was used compared to a 35-fold increase when RGD nanocluster–modified polyplexes were used (Figure 7a). Transfection of low-αvβ3-density cells resulted in a 5.4-fold increase in expression for polyplexes modified with RGD nanoclusters, but no increase for jetPEI-RGD was observed (Figure 7b). To further compare the results obtained for the low- and high-αvβ3-density cells, we defined an integrin-sensitivity factor (ISF), a ratio of the transfection level achieved with high- and low-αvβ3-density cells (Figure 7c). Data from Figure 7a,b are used to generate Figure 7c. An ISF value of 60 for RGD nanoclusters-modified polyplexes was observed whereas for jetPEI-RGD the ISF was at most 20. Interestingly, cell trypsinization resulted in lower transgene expression for all of the vectors tested by at least an order of magnitude even for untargeted polyplexes.
Figure 7.
Transfection efficiency using vectors with clustered and homogeneous ligands. Luciferase transgene expression of DNA/PEI and DNA/PEI-Au-Cap-RGD-ASA compared to DNA/jetPEI and DNA/jetPEI-RGD for (a) high- and (b) low-αvβ3-integrin-expressing HeLa cells (n = 3). Sensitivity of the vector to integrin density can be compared using the transfection efficiency of high- and low-integrin-expressing cells by dividing the RLU/mg of the high-expressing cells to the RLU/mg of the low-expressing cells (integrin sensitivity factor). (c) integrin sensitivity factor comparison of HeLa cells for DNA/PEI, DNA/PEI-Au-Cap-RGD-ASA, DNA/jetPEI, and DNA/jetPEI-RGD (n = 3).
Discussion
Nonviral vectors are often modified with ligands to enhance targeting and increase their overall gene transfer efficiency. Although PEI has been previously modified with ligands23,24,31,32,33,34,35,36,37 to target corresponding receptors, the chemistries typically take advantage of the amine groups on the PEI backbone, resulting in ligands being unspecific and randomly distributed over the DNA/PEI polyplex. We explore an approach to modify PEI so that the ligands are clustered together and spatially constrained on the polyplex surface, allowing multiple ligands to bind to cell surface receptors simultaneously. In our approach, ligand-modified gold nanoparticles are attached to the surface of DNA/PEI polyplexes so they are presented as ligand clusters. Here we use spatially constrained RGD peptides as an example to test our ligand clustering platform and examined the effect that clustered RGD ligands has on nonviral gene transfer to HeLa cells.
We used the physical characteristics of the penton-base protein of Adenovirus type 2 to design the size and peptide-to-peptide distance of the RGD nanoclusters used to modify the surface of DNA/PEI polyplexes (Figure 1). The penton-base protein is ~16 nm in diameter and has an RGD to RGD distance of 5.7 nm.9 To achieve a similar size and RGD to RGD distance for the RGD nanoclusters, 5 nm gold nanoparticles were modified with two self-assembling peptides, CCVVVT (Cap) and CCVVVT-RGD (Cap-RGD). The Cap peptide was chosen for its ability to prevent gold nanoparticle aggregation in salt-containing solutions (Figure 2).30 The modification of gold nanoparticles with Cap peptide results in particles which were ~10 nm in diameter by DLS (Figure 3a). Control over the spacing of RGD peptides is achieved through mixing the Cap and Cap-RGD peptides in the solution mixture during gold nanoparticle modification. The resulting RGD-modified gold nanoparticles are assumed to have a uniform distribution of RGD peptides and based on the solution ratio and the number of peptides bound per gold nanoparticle an RGD to RGD distance can be estimated (Table 1, see Materials and Methods for a sample calculation). Mixing multiple types of alkane thiols has been widely investigated for the formation of mixed self-assembled monolayers on gold-coated substrates38 and gold nanoparticles,39,40 with the solution ratio corresponding to the surface ratio. To determine whether the solution mixture of Cap to Cap-RGD is also conserved on the nanoparticle surface, gold nanoparticles were modified with different ratios of Cap to Cap-RGD and the surface concentration of the peptides was determined through amino acid analysis.
For Au-Cap-RGD nanoparticles modified with up to 90 to 10 Cap to Cap-RGD ratio, the solution and surface ratios of Cap to Cap-RGD ratio were similar, indicating that simply mixing the two peptides in the reaction mixture results in similar ratios on the nanoparticle surface as those found in solution (Table 1). However, when the ratio of Cap to Cap-RGD peptide was increased to 80 to 20 the experimentally obtained ratio was 89.7:10.3, which suggest that there is an upper limit to how much RGD peptides can be introduced to the surface of the nanoparticle possibly due to a crowding effect of the RGD peptide on the nanoparticle surface. Five nanometer gold nanoparticles modified with 90:10 Cap to Cap-RGD ratio were explored to introduce clustered ligand binding to DNA/PEI polyplexes due to their similarities to the penton-base protein of Adenovirus 2 (Table 1).
Although we did not test gold nanoparticles with other RGD surface densities, based on literature reports on the importance of RGD spacing on multivalent targeting, we expect that the density of RGDs on the surface of the gold nanoparticle to play a role on efficient targeting. For example, the effect that RGD density has on multivalent targeting has been investigated using cyclic decapeptide RAFTs41 and RGD-modified iron dextran nanoparticles.18 In both examples, density of RGD peptides affected its multivalent events, with too high or too low amounts of RGD abolishing the multivalent effects observed with the optimal RGD concentrations.18,41
A cell adhesion assay was used to determine the effectiveness of our RGD nanoclusters to bind multiple integrin receptors simultaneously as previously done by others18,21 (Figure 4). Au-RGD nanoclusters required three orders of magnitude less of the equivalent RGD peptide compared to free RGD peptide to achieve the antiadhesion IC50. Similar clustering effects are seen for the attachment of HUVEC cells to vitronectin-coated plates using RGD-modified iron nanoparticles compared to free peptide.18,42 These results suggest that synthesized RGD nanoclusters are able to bind to multiple integrin receptors simultaneously.
RGD nanoclusters were introduced to the surface of polyplexes by mixing DNA/PEI polyplexes with RGD nanoclusters that contain the photoreactive group ASA (Figure 1) and were characterized using DLS and TEM (Figure 3b,c). DLS analysis showed a steady increase in the size of DNA/PEI polyplexes as gold nanoparticles were added (Figure 3b); however, the overall sizes of the RGD nanocluster-modified polyplexes remained <200 nm. Size is an important design parameter to ensure efficient cellular internalization since polyplexes <200 nm in diameter are internalized through clathrin-mediated endocytosis while large polyplexes (>200 nm) are internalized by caveole-mediated endocytosis.43 Polyplexes internalized through clathrin-mediated endocytosis have been show to internalize more quickly than larger particles.44
DNA/PEI polyplexes that were covalently modified with RGD nanoclusters resulted in enhanced nonviral gene transfer to HeLa cells, with either a 5.4- (low-integrin-density cells) or 35-fold (high-integrin-density cells) increase in expression (Figure 6). Polyplexes modified with RGD nanoclusters without the covalent crosslinker (no ASA) did not show an enhancement in expression, indicating covalent binding is necessary for the enhancement to occur and that electrostatic interactions are insufficient. To ensure that the enhancement in gene transfer was not a result of the attachment of gold nanoparticles to the polyplex surface, polyplexes were modified with Au-Cap-ASA and their effect on high-αvβ3-integrin density cells studied. Au-Cap-ASA-modified polyplexes resulted in transgene expression that was similar to that observed for electrostatically modified Au-Cap and Au-Cap-RGD polyplexes, indicating that the 35-fold enhancement in gene transfer observed for Au-Cap-RGD-ASA-modified polyplexes is not due to the gold nanoparticle alone. Although the DNA/PEI-Au polyplexes do not completely settle out of solution (observed from DLS and visual inspection) the increase in transfection seen in these controls can be explained by an increase in the density of the polyplexes, which has been implicated to increase the physical concentration of DNA at the cell surface.45 The specificity of RGD nanocluster–modified polyplexes to bind αvβ3 integrin receptors was shown using a competitive binding assay with free RGD peptide in solution. In the presence of free RGD a 40% decrease in transfection was observed, demonstrating that the proposed vector binds to cells through αvβ3 integrins (Figure 6d). A complete abolishment of transfection was not observed, however, indicating that either the vector binds also through other nonspecific interactions with the cell surface or the RGD peptide itself cannot fend off the RGD nanoclusters completely.
To better understand the effect that RGD nanoclusters have on nonviral gene transfer, we compared the transfection efficiency achieved using vectors with and without clustered ligands transfecting high- and low-αvβ3 density cells. An ISF was defined that compares how a delivery vector responds to the density of integrin receptors at the cell surface. DNA/PEI-Au-RGD-ASA shows an ISF of 60, whereas jetPEI-RGD shows an ISF of 20 at most (Figure 7c). This indicates that the presence of clustered RGD ligands on the polyplex surface renders the delivery vector more sensitive to the density of receptors at the cell surface and that the local arrangement of RGD ligands on the surface of the polyplex is an important design characteristic for targeting. Although the RGD nanoclusters have a higher binding affinity with the cells surface as seen in the cell adhesion assay, it cannot be concluded at this time whether the enhancement in gene transfer is a result of integrin clustering or simply to higher affinity of the vector for the cell surface.
Trypsin is generally used for cellular detachment of adherent cells during cell culture. We found that the common protocol of trypsinization, replating into culture dishes and transfecting 12–14 hours later results in much lower transfection rates than cells that were scrapped during splitting. At least one order of magnitude lower transfection efficiency was observed for all the vectors tested (Figure 7a,b), signifying a general effect that is not influenced by targeting or the presence of RGD nanoclusters. Although the full effect of trypsin on the cell surface was not studied, the density of αvβ3 integrin receptors is greatly reduced after trypsin treatment and did not fully recover 12 hours after plating (Figure 5a). It is likely that the densities of other receptors at the cell surface are also reduced and that these receptors/proteins also play a role in nonviral gene transfer. For example, syndecans have been previously implicated in PEI-mediated gene transfer to HeLa cells46 and surface nucleolin has shown to serve as a receptor for poly(ethylene glycol)-polylysine vectors in HeLa cells.47 Proteoglycans have also been known to play a role in cation-mediated gene transfer to HeLa and CHO cells.48
The toxicity of RGD nanoclusters and DNA/PEI polyplexes was determined using a cell proliferation assay and normalized to untreated cells (Figure 4). The results indicate that higher concentrations of nanoparticles are slightly toxic to cells. Similarly, gold nanoparticle toxicity has been previously reported for both 11-thioundecanoic acid and ammonium thiol–modified gold nanoparticles at high concentration due to the concentration-dependant charge interaction of the nanoparticles with the cell membrane.49
In summary, we report a strategy to introduce clustering to nonviral vectors and other nanoparticles of interest that mimics the physical structure of Adenovirus type 2 and that utilizes peptide-modified gold nanoparticles. We show that the presence of RGD nanoclusters on DNA/PEI polyplex surfaces enhances gene transfer and that the enhancement is dependent on the density of αvβ3 integrins on the cell surface, with higher αvβ3 densities resulting in higher luciferase transgene expression. Our approach to introduce clustering is versatile and can be used to introduce alternative ligands to target other receptors by simply modifying the Cap peptide with the ligand of interest, which we believe will be an ideal approach to enhance targeting and gene transfer efficiency of nonviral vectors.
Materials and Methods
Materials. Plasmid DNA was purified from bacteria culture using Qiagen (Santa Clara, CA) reagents and stored in Tris–EDTA buffer solution at −20 °C. The plasmid pEGFP-Luc was purchased from Clontech (Palo Alto, CA). All other reagents were purchased from Fisher (Chino, CA) unless specified.
Synthesis of CAP, CAP-RGD, CAP-RGD-ASA, Cap-ASA peptides. The peptides CCVVVT-COOH (Cap) and Ac-CCVVVTGRGDSPSSK-COOH (Cap-RGD) were purchased from Genscript (Piscataway, NJ) at 98.5% and >95% purity respectively. Cap-RGD-ASA was synthesized by reaction of Cap-RGD peptide with N-Hydroxysuccinimidyl-4-azidosalicylic acid (NHS-ASA; Pierce, Rockford, IL). Cap-RGD peptide (1 µmol) was made to react with 1.5 mg of NHS-ASA in 10% DMSO/0.1 mol/l carbonate buffer (pH 8.3) for 1 hour. The final solution was dialyzed in milliQ water and lyophilized. Cap-ASA was synthesized by acetylation of the N-terminus and reaction of Cap peptide with p-azidobenzoyl hydrazide (Pierce, Rockford, IL). The photoreactive group on Cap-ASA is not exactly the same as the one present on Cap-RGD-ASA (varies by an extra OH group), but the chemistry they undergo is the same; thus, they were given the same abbreviation. Cap peptide (1 µmol) was dissolved in 6% DMSO in 0.1 mol/l MES (pH 6.0), and 10 µl of 300 mmol/l acetic anhydride in THF was added and vortexed for 3 minutes. Acetic anhydride addition was repeated twice. The solution was dialyzed in milliQ water and lyophilized. The lyophilized peptide was dissolved in 500 µl of 0.1 mol/l MES (pH 6.0) and reacted with 0.2 mg EDC (Advanced ChemTech, Louisville, KY) and 0.3 mg NHS (Pierce, Rockford, IL) for 15 minutes. Azidobenzoyl hydrazide was added to a final concentration of 5 mmol/l and diluted to a final volume of 1 ml using phosphate-buffered saline (PBS) and reacted for 2 hours. The solution was dialyzed in milliQ water and lyophilized.
Formation of Au-Cap-RGD nanoparticles (RGD nanoclusters). Au-Cap-RGD nanoparticles of different degrees of RGD modification were formed using a mixture of Cap and Cap-RGD or Cap-RGD-ASA peptides to the desired ratio. A total of 200 nmol of peptide with varying mixtures of Cap peptide, dissolved in 6% DMSO/PBS, and Cap-RGD or Cap-RGD-ASA peptide dissolved in PBS was added to Au nanoparticles purchased from Ted Pella (Redding, CA) and was reacted for 24 hours. A 1/10 volume ratio of 10× phosphate buffer (pH = 7.4) was added and incubated for an additional 24 hours. Purification of Au-Cap was achieved through dialysis using a 8 k MWCO Float-a-lyzer (Spectrapor, Rancho Dominguez, CA) in phosphate buffer (3 mmol/l KCl, 8 mmol/l NaHPO4, 1 mmol/l KH2PO4, pH = 7.4). Concentration of the nanoparticles was achieved by dehydration using Spectra/Gel Absorbent (Spectrapor, Rancho Dominguez, CA). The number of peptides per particle was determined using amino acid analysis at the UCLA Biopolymer Lab. The concentration of Valines was used to calculate the amount of total peptides (Cap and Cap-RGD) reacted with the Au nanoparticles. The concentration of Arginine was used to calculate the amount of Cap-RGD peptides. The ratio of Cap to Cap-RGD peptides was determined using the calculated values. The sizes of Au-Cap and uncapped nanoparticles were measured using DLS with a Malvern Zetasizer nano ZS.
RGD spacing determination. Distances between RGD peptides on a particle surface can be calculated using surface area of the particle and theoretical peptide length. Assuming 1.93 peptides/nm2 (ref. 30), a 5 nm particle would contain 151 peptides. The theoretical distance of the RGD sequence from the surface (~2.7 nm) would increase effective particle diameter to 10.4 nm. Using the Cap-RGD peptides to Cap peptides ratio, we calculated the number of RGD peptides per particle. For example, for a 90:10 Cap to Cap-RGD ratio we obtained 15 Cap-RGD peptides per particle, which would equally share surface area of 23 nm2. This resulted in a theoretically calculated spacing of ~5 nm between RGD peptides.
Salt stability of Au-Cap. Peptide-modified (Au-Cap) and unmodified particles were tested to check their salt stability. Au and Au-Cap nanoparticles were added to semi-micro cuvettes and serial additions of NaCl solution were added every 15 minutes followed by a wavelength scan from 400–900 nm (Beckman DU 730, Fullerton, CA). Aggregation parameter (AP) used to quantify aggregation was calculated using the equation AP = (A − A0)/A0. A is the integral of absorbance from 600 to 700 nm, and A0 is the integral of absorbance from 600 to 700 nm of the initial solution without salt added (see Figure 2 area under the curve).
Size characterization of DNA/PEI-Au polyplexes. DNA/PEI polyplexes were formed by mixing equal volumes of plasmid DNA (4.4 µg) with 25 kDa–branched PEI (1 mg/ml, Sigma, St Louis, MO) to get an N/P of 10 in a final volume of 330 µl of milliQ water. PEI was added to the DNA solution, vortexed for 10 seconds, and incubated at room temperature for 15 minutes. Specified concentrations of Au-Cap, Au-Cap-RGD or Au-Cap-RGD-ASA nanoparticles (10 µl) were added to the DNA/PEI polyplexes and exposed to ambient light for 15 minutes. The optimal N/P ratio for DNA/PEI complexes was determined using an ethidium bromide (EtBr) exclusion assay by measuring fluorescence using a Modulus 20/20 Fluorimeter/Luminometer (Turner Biosystems, Sunnyvale, CA). The size of DNA/PEI-Au-Cap-RGD-ASA polyplexes was determined using DLS and TEM. DNA/PEI polyplexes for DLS size measurements were formed at both normal (13.3 µg DNA/ml) and high concentrations (40 µg DNA/ml) at an N/P of 10. Normal concentration DNA/PEI polyplexes were formed as previously described. High concentration DNA/PEI polyplexes were formed as previously described in a final volume of 110 µl of milliQ water. Au-Cap-RGD-ASA nanoparticles were added (2 µl per increment, 7 × 1013 particles/ml) to the DNA/PEI polyplexes, vortexed for 10 seconds, and incubated at room temperature for 2 minutes. Size readings were taken between each increment using a Malvern Zetasizer nano ZS (Malvern Instruments, Worcestershire, UK). High concentration DNA/PEI polyplexes were formed for TEM measurements and Au-Cap-RGD-ASA nanoparticles were added (3.95 µl, 3.5 × 1014 particles/ml), vortexed for 10 seconds, and incubated at room temperature for 15 minutes. A droplet of 3 µl DNA/PEI-Au-RGD solution was applied to a glow-discharged continuous carbon film–coated copper grid. The solution was suspended on the grid for 10 seconds before removing the excess liquid by blotting with Whatman filter paper. The grid was then washed twice with 5 µl of distilled water and incubated for 15 seconds with 2% uranyl-acetate solution for negative staining. The air-dried grid was then observed with a JEM-1230 TEM. The micrograph images were recorded under a magnification of 50,000 on a TIVPS 2k × 2k CCD camera.50
Integrin quantification with flow cytometry. HeLa cells were detached either through scrapping or using 0.25% Trypsin-EDTA during passaging, plated on 6-well plates (200,000 cells/well), and incubated for 12 hours. The cells were detached from the wells by scrapping and stained using Alexa488 conjugated anti-αvβ3 monoclonal antibody (MAb 23C6, Santa Cruz Biotechnology, Santa Cruz, CA) following the Santa Cruz Biotechnology flow cytometry protocol.
Antiadhesion assay. HeLa cells (ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (DMEM) medium (Invitrogen, Carlsbad, CA) supplemented with 10% bovine growth serum (Hyclone, Logan, UT). The cells were cultured at 37 °C, with 98% humidity and 5% CO2. Cells were regularly subcultured using trypsin. Next, 96-well plates were coated with human vitronectin by incubating a vitronectin solution (50 µl of 3 µg/ml, Invitrogen, Carlsbad, CA) in the well for 12 hours at 37 °C. The wells were washed three times with PBS and blocked with bovine serum albumin (50 µl of 2% bovine serum albumin in PBS) for 30 minutes at 37 °C. The wells were washed three times with PBS and 100 µl/well of serum-free DMEM was added before chilling on ice. HeLa cells were harvested by scrapping using cell scrapers (USA Scientific, Ocala, FL), pelleted, and resuspended in ice-cold serum-free DMEM to 400,000 cells/ml. 200 µl of serum-free DMEM was added to 50 µl of Au-Cap-RGD solution and vortexed before 250 µl of cell solution was added to the Au-Cap-RGD solution and was subsequently placed in ice for 15 minutes. 100 µl/well of the mixture was added and allowed to incubate at 37 °C for 50 minutes. The plate was submerged in PBS in a Ziploc bag and centrifuged upside down for 10 minutes at 250g. The plate was washed gently with additional PBS and removed. Remaining liquid was removed from the wells. The cells were fixed using 4% paraformaldehyde solution for 15 minutes and washed three times with PBS. Crystal violet solution was added to each well and incubated for 1 hour. The wells were then washed with milliQ water until no crystal violet was visible in solution. Crystal violet was solubilized using 10% acetic acid plate for 15 minutes and the absorbance at 570 nm was taken using the PowerWave microplate reader (BioTek, Winooski, VT).
Bolus transfection with polyplexes. HeLa cells were transfected using DNA/PEI-Au-Cap-RGD-ASA polyplexes. HeLa cells were either removed with 0.25% Trypsin-EDTA or scrapped and then plated 40,000 cells/well in DMEM supplemented with 10% bovine growth serum on a 24 well TC-plate (USA scientific, Ocala, FL). Cells were incubated for 12 hours (50% confluency) at 37 °C and 5% CO2. 1.33 µg of DNA from low-concentration DNA/PEI-Au polyplex solution, formed as previously described, was added per well. Immediately following addition, 1.5 mol/l NaCl solution was added to each well (10% of polyplex volume). JetPEI and jetPEI-RGD polyplexes were formed and added according to manufacturers protocols at an N/P of 5. Cells and polyplexes were allowed to incubate for 48 hours. Cells were lysed and a luciferase assay was performed as described in the Promega luciferase assay kit and measured using a Luminometer (Turner Biosystems, Modulus 20/20, Sunnyvale, CA). Protein concentration was measured using the Bradford reagent, Coomassie Plus (Pierce, Rockford, IL), following manufactures protocols.
Bolus transfection with RGD competition. HeLa cells were transfected using DNA/PEI-Au-Cap-RGD-ASA polyplexes in the presence of RGD peptide in solution. Bolus transfection using DNA/PEI-Au-Cap-RGD-ASA and DNA/PEI polyplexes was performed similarly as described above except RGD peptide (Ac-GCGYGRGDSPSSK-NH2, Standford PAN, Palo Alto, CA) dissolved in PBS was added to each well to a final concentration of 100 µmol/l and incubated at 37 °C for 15 minutes prior to addition of polyplexes. Cells and polyplexes were allowed to incubate for 4 hours before replacing with fresh media and incubating for an additional 44 hours. Cells were lysed and assayed as described previously.
Statistics. All statistical analyses were performed using the computer program Instat (GraphPad, San Diego, CA). Experiments were statistically analyzed using a one-way analysis of variance using the Tukey test, which compares all pairs of columns, using a 95% confidence interval. When only two groups were compared, the Student's t- test was used as indicated in the figure legends.
Acknowledgments
Flow cytometry was performed at the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards CA-16042 and AI-28697, and by the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA. Amino acid analysis was performed in the Biopolymer Laboratory at the UCLA Neuroscience Research Building. Electron microscopy imaging was partly sponsored by a Discovery grant and the National Institutes of Health HIVRAD and nanomedicine roadmap programs. Special thanks to Sean Anderson, Anandkia Dhaliwal, Leo Lei, and Talar Tokatlian for insightful discussions and Kaajal Baheti for technical assistance.
REFERENCES
- Arnold M, Cavalcanti-Adam EA, Glass R, Blummel J, Eck W, Kantlehner M, et al. Activation of integrin function by nanopatterned adhesive interfaces. Chemphyschem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA., and , Griffith LG. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J Cell Sci. 2002;115:1423–1433. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- Maheshwari G, Brown G, Lauffenburger DA, Wells A., and , Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113:1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- Kong HJ, Polte TR, Alsberg E., and , Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc Natl Acad Sci USA. 2005;102:4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HJ, Hsiong S., and , Mooney DJ. Nanoscale cell adhesion ligand presentation regulates nonviral gene delivery and expression. Nano Lett. 2007;7:161–166. doi: 10.1021/nl062485g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcanti-Adam EA, Micoulet A, Blummel J, Auernheimer J, Kessler H., and , Spatz JP. Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly. Eur J Cell Biol. 2006;85:219–224. doi: 10.1016/j.ejcb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Hughes PE., and , Pfaff M. Integrin affinity modulation. Trends Cell Biol. 1998;8:359–364. doi: 10.1016/s0962-8924(98)01339-7. [DOI] [PubMed] [Google Scholar]
- Humphries MJ. Integrin activation: the link between ligand binding and signal transduction. Curr Opin Cell Biol. 1996;8:632–640. doi: 10.1016/s0955-0674(96)80104-9. [DOI] [PubMed] [Google Scholar]
- Stewart PL, Chiu CY, Huang S, Muir T, Zhao Y, Chait B, et al. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 1997;16:1189–1198. doi: 10.1093/emboj/16.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Kauwe LK. Endocytosis of adenovirus and adenovirus capsid proteins. Adv Drug Delivery Rev. 2003;55:1485–1496. doi: 10.1016/j.addr.2003.07.010. [DOI] [PubMed] [Google Scholar]
- Chiu CY, Mathias P, Nemerow GR., and , Stewart PL. Structure of Adenovirus Complexed with Its Internalization Receptor, alpha vbeta 5 Integrin. J Virol. 1999;73:6759–6768. doi: 10.1128/jvi.73.8.6759-6768.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MJ., and , Wilson JM. Expression of alpha v beta 5 integrin is necessary for efficient adenovirus-mediated gene transfer in the human airway. J Virol. 1995;69:5951. doi: 10.1128/jvi.69.10.5951-5958.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi H, Koizumi N, Hosono T, Ishii-Watabe A, Uchida E, Utoguchi N, et al. CAR-or alpha v integrin-binding ablated adenovirus vectors, but not fiber-modified vectors containing RGD peptide, do not change the systemic gene transfer properties in mice. Gene Ther. 2002;9:769–776. doi: 10.1038/sj.gt.3301701. [DOI] [PubMed] [Google Scholar]
- Mathias P, Wickham T, Moore M., and , Nemerow G. Multiple adenovirus serotypes use alpha v integrins for infection. J Virol. 1994;68:6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham TJ, Mathias P, Cheresh DA., and , Nemerow GR. Integrins [alpha]v[beta]3 and [alpha]v[beta]5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- Schiffelers RM, Koning GA, ten Hagen TL, Fens MH, Schraa AJ, Janssen AP, et al. Anti-tumor efficacy of tumor vasculature-targeted liposomal doxorubicin. J Controlled Release. 2003;91:115–122. doi: 10.1016/s0168-3659(03)00240-2. [DOI] [PubMed] [Google Scholar]
- McCarthy JR., and , Weissleder R. Multifunctional magnetic nanoparticles for targeted imaging and therapy. Adv Drug Delivery Rev. 2008;60:1241–1251. doi: 10.1016/j.addr.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montet X, Funovics M, Montet-Abou K, Weissleder R., and , Josephson L. Multivalent effects of RGD peptides obtained by nanoparticle display. J Med Chem. 2006;49:6087–6093. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
- Dijkgraaf I, Kruijtzer J, Liu S, Soede A, Oyen W, Corstens F, et al. Improved targeting of the αvβ3 integrin by multimerisation of RGD peptides. Eur J Nucl Med Mol Imaging. 2007;34:267–273. doi: 10.1007/s00259-006-0180-9. [DOI] [PubMed] [Google Scholar]
- Kiessling LL, Gestwicki JE., and , Strong LE. Synthetic multivalent ligands as probes of signal transduction. Angew Chem. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok RJ, Schraa AJ, Bos EJ, Moorlag HE, Asgeirsdottir SA, Everts M, et al. Preparation and Functional Evaluation of RGD-Modified Proteins as AVB3 Integrin Directed Therapeutics. Bioconjug Chem. 2002;13:128–135. doi: 10.1021/bc015561+. [DOI] [PubMed] [Google Scholar]
- Davis ME. Non-viral gene delivery systems. Curr Opin Biotechnol. 2002;13:128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- Erbacher P, Remy JS., and , Behr JP. Gene transfer with synthetic virus-like particles via the integrin-mediated endocytosis pathway. Gene Ther. 1999;6:138–145. doi: 10.1038/sj.gt.3300783. [DOI] [PubMed] [Google Scholar]
- Kunath K, Merdan T, Hegener O, Haberlein H., and , Kissel T. Integrin targeting using RGD-PEI conjugates for in vitro gene transfer. J Gene Med. 2003;5:588–599. doi: 10.1002/jgm.382. [DOI] [PubMed] [Google Scholar]
- Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu M, Fischer D., and , Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J Gene Med. 2005;7:992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- Blessing T, Kursa M, Holzhauser R, Kircheis R., and , Wagner E. Different strategies for formation of pegylated EGF-conjugated PEI/DNA complexes for targeted gene delivery. Bioconjug Chem. 2001;12:529–537. doi: 10.1021/bc0001488. [DOI] [PubMed] [Google Scholar]
- Ogris M, Brunner S, Schuller S, Kircheis R., and , Wagner E. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6:595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- Kichler A.Gene transfer with modified polyethylenimines J Gene Med 20046S3–S10.Suppl. 1 [DOI] [PubMed] [Google Scholar]
- Lévy R, Thanh NT, Doty RC, Hussain I, Nichols RJ, Schiffrin DJ, et al. Rational and combinatorial design of peptide capping ligands for gold nanoparticles. J Am Chem Soc. 2004;126:10076–10084. doi: 10.1021/ja0487269. [DOI] [PubMed] [Google Scholar]
- Zanta MA, Boussif O, Adib A., and , Behr JP. In vitro gene delivery to hepatocytes with galactosylated polyethylenimine. Bioconjug Chem. 1997;8:839–844. doi: 10.1021/bc970098f. [DOI] [PubMed] [Google Scholar]
- Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R., and , Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 1998;5:1425–1433. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- Guo W., and , Lee RL. Receptor-targeted gene delivery viafolate-conjugated polyethylenimine. AAPS PharmSci. 1999;11:19. doi: 10.1208/ps010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Kursa M, Wagner E, Cotten M., and , Zenke M. Mannose polyethylenimine conjugates for targeted DNA delivery into dendritic cells. J Biol Chem. 1999;274:19087–19094. doi: 10.1074/jbc.274.27.19087. [DOI] [PubMed] [Google Scholar]
- Bettinger T, Remy JS., and , Erbacher P. Size reduction of galactosylated PEI/DNA complexes improves lectin-mediated gene transfer into hepatocytes. Bioconjug Chem. 1999;10:558–561. doi: 10.1021/bc990006h. [DOI] [PubMed] [Google Scholar]
- Chiu SJ, Ueno NT., and , Lee RJ. Tumor-targeted gene delivery via anti-HER2 antibody (trastuzumab, Herceptin) conjugated polyethylenimine. J Control Release. 2004;97:357–369. doi: 10.1016/j.jconrel.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Blessing T, Kursa M, Holzhauser R, Kircheis R., and , Wagner E. Different strategies for formation of PEGylated EGF-conjugated PEI/DNA complexes for targeted gene delivery. Bioconjug Chem. 2001;12:529–537. doi: 10.1021/bc0001488. [DOI] [PubMed] [Google Scholar]
- Daniel MC., and , Astruc D. Gold nanoparticles: assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- Ingram RS, Hostetler MJ., and , Murray RW. Poly-hetero-w;-functionalized alkanethiolate-stabilized gold cluster compounds. J Am Chem Soc. 1997;119:9175–9178. [Google Scholar]
- Wang Z, Lévy R, Fernig DG., and , Brust M. The peptide route to multifunctional gold nanoparticles. Bioconjug Chem. 2005;16:497–500. doi: 10.1021/bc050047f. [DOI] [PubMed] [Google Scholar]
- Garanger E, Boturyn D, Jin Z, Dumy P, Favrot MC., and , Coll JL. New multifunctional molecular conjugate vector for targeting, imaging, and therapy of tumors. Mol Ther. 2005;12:1168–1175. doi: 10.1016/j.ymthe.2005.06.095. [DOI] [PubMed] [Google Scholar]
- Carlson CB, Mowery P, Owen RM, Dykhuizen EC., and , Kiessling LL. Selective tumor cell targeting using low-affinity, multivalent interactions. ACS Chem Biol. 2007;2:119–127. doi: 10.1021/cb6003788. [DOI] [PubMed] [Google Scholar]
- Rejman J, Conese M., and , Hoekstra D. Gene transfer by means of lipo- and polyplexes: role of clathrin and caveolae-mediated endocytosis. J Liposome Res. 2006;16:237–247. doi: 10.1080/08982100600848819. [DOI] [PubMed] [Google Scholar]
- Ogris M, Steinlein P, Carotta S, Brunner S., and , Wagner E. DNA/polyethylenimine transfection particles: influence of ligands, polymer size, and PEGylation on internalization and gene expression. AAPS PharmSci. 2001;3:E21. doi: 10.1208/ps030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., and , Saltzman WM. Enhancement of transfection by physical concentration of DNA at the cell surface. Nat Biotech. 2000;18:893–895. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- Kopatz I, Remy JS., and , Behr JP. A model for non-viral gene delivery: through syndecan adhesion molecules and powered by actin. J Gene Med. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- Chen X, Kube DM, Cooper MJ., and , Davis PB. Cell surface nucleolin serves as receptor for DNA nanoparticles composed of pegylated polylysine and DNA. Mol Ther. 2008;16:333–342. doi: 10.1038/sj.mt.6300365. [DOI] [PubMed] [Google Scholar]
- Mislick KA., and , Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci USA. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CM, McCusker CD, Yilmaz T., and , Rotello VM. Toxicity of gold nanoparticles functionalized with cationic and anionic side chains. Bioconjug Chem. 2004;15:897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]
- Srivastava IK, Kan E, Sun Y, Sharma VA, Cisto J, Burke B, et al. Comparative evaluation of trimeric envelope glycoproteins derived from subtype C and B HIV-1 R5 isolates. Virology. 2008;372:273–290. doi: 10.1016/j.virol.2007.10.022. [DOI] [PubMed] [Google Scholar]