Abstract
In our earlier work we showed that either spermidine or spermine could support the growth of spe2Δ or spe3Δ polyamine-requiring mutants, but it was unclear whether the cells had a specific requirement for either of these amines. In the current work, we demonstrate that spermidine is specifically required for the growth of Saccharomyces cerevisiae. We were able to show this specificity by using a spe3Δ fms1Δ mutant that lacked both spermidine synthase and the FMS1-encoded amine oxidase that oxidizes spermine to spermidine. The polyamine requirement for the growth of this double mutant could only be satisfied by spermidine; i.e., spermine was not effective because it cannot be oxidized to spermidine in the absence of the FMS1 gene. We also showed that at least one of the reasons for the absolute requirement for spermidine for growth is the specificity of its function as a necessary substrate for the hypusine modification of eIF5A. Spermine itself cannot be used for the hypusine modification, unless it is oxidized to spermidine by the Fms1 amine oxidase. We have quantified the conversion of spermine in vivo and have shown that this conversion is markedly increased in a strain overexpressing the Fms1 protein. We have also shown this conversion in enzymatic studies by using the purified amine oxidase from yeast.
Keywords: eIF5A, polyamine, polyamine oxidase
Polyamines, such as putrescine, spermidine, and spermine, are ubiquitous cellular polycationic compounds. Their essential nature has been demonstrated by many studies with inhibitors and, in particular, with mutants in the biosynthetic pathway (1–3).
In many studies some of the phenotypic effects of spermidine and spermine have been interchangeable. This has also been true of our recent studies with Saccharomyces cerevisiae. Thus, strains unable to synthesize spermidine because of deletions (spe2Δ or spe3Δ) in the biosynthetic pathway† did not grow in amine-free medium unless spermidine or spermine was added to the growth medium (4, 5). However, because either spermidine or spermine permitted growth, it was unclear whether the cells had any specific requirement for spermidine that could not be satisfied by spermine. In the present work we show that S. cerevisiae cells do indeed have an absolute requirement for spermidine and that this requirement cannot be met by spermine unless the spermine is first oxidized to spermidine by an amine oxidase.
We also show that at least one of the reasons for the absolute requirement for spermidine (and not spermine) for growth is the specificity of its function as a necessary substrate for the hypusine modification of eIF5A. eIF5A is a eukaryote protein synthesis initiation factor that is essential for growth (6–10) and may be involved in mRNA metabolism, translation, and ribosome biogenesis (11, 12). In many cell types hypusine modification has been shown to occur by the transfer of the 4-aminobutyl moiety of spermidine to the ε-amino group of a specific lysine residue of precursor eIF5A (13, 14). Studies with yeast mutants have shown that this hypusine modification is essential for growth of S. cerevisiae; i.e., mutants defective in hypusine synthesis are lethal (6, 8, 15, 16). Although earlier work with S. cerevisiae had shown that in wild-type cells label from spermidine could be incorporated into eIF5A (17), these data did not indicate whether either spermidine or spermine could serve as the direct source for the hypusine modification. In our current studies we show that in S. cerevisiae, as in other cell types (13, 18–20), only spermidine (and not spermine) can be used for this modification.
Materials and Methods
Yeast strains and plasmids used are listed in Table 1.
Table 1. Yeast strains and plasmids.
| Strain | Genotype | Source or ref. |
|---|---|---|
| Y505 | Mat α his6 leu2 ura3 spe3Δ::URA3 | 5 |
| Y537 (ATCC 4005488) | Mat a his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 spe3Δ::KANMX | S. cerevisiae gene deletion bank (21) BY4741-5488 |
| Y547 (ATCC 400595) | Mat a his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 fms1Δ::KANMX | S. cerevisiae gene deletion bank (21) BY4741-0595 |
| Y548 | Mat a his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 fms1Δ::KANMX | This study; diploid of the cross of Y505 and Y547 |
| Mat α his6 leu2 ura3 spe3Δ::URA3 | ||
| Y549 | Mat α met15 leu2 ura3 spe3Δ::URA3 fms1Δ::KANMX | Sporulation of Y548 |
| Y551 | Mat α met15 leu2 ura3 spe3Δ::URA3 | Sporulation of Y548 |
| Y559 | Mat a his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 spe3Δ::KANMX/pYFMS1 | This study; Y537-pYES 2.1/PGAL1-FMS1/6XHis |
| Y560 | Mat a his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 fms1Δ::KANMX/pYFMS1 | This study; Y547-pYES 2.1/PGAL1-FMS1/6XHis |
Materials. N1-Acetylspermine trihydrochloride was from Fluka; spermine, spermidine, and putrescine as their respective hydrochlorides were purchased from Sigma. Commercially available spermine tetrahydrochloride was purified on a Dowex 50 column and was eluted with a HCl gradient. N1-Acetylspermidine was prepared by addition of 4-bromobutyronitrile to monoacetyldiaminopropane and reduction of the product with Pt(H2). Restriction enzymes were obtained from New England Biolabs. Custom gene-specific oligonucleotides for homologous cloning were synthesized by Invitrogen. The vector (modified pYES 2.1-PGAL1/V5-His/lacZ, as described below) for overexpression of the Fms1 protein was purchased from Invitrogen. Precast 12% Bis-Tris NuPAGE gel and related buffers and reagents were also obtained from Invitrogen. Y-PER 6XHis fusion protein purification kit was purchased from Pierce. [14C]Spermine tetrahydrochloride (113 mCi/mmol, 50 μCi/ml; catalog no. CFA511) and [14C]spermidine trihydrochloride (112 mCi/mmol, 50 μCi/ml; catalog no. CFA512) were from Amersham Pharmacia; both amines were labeled in the diaminobutane moiety.
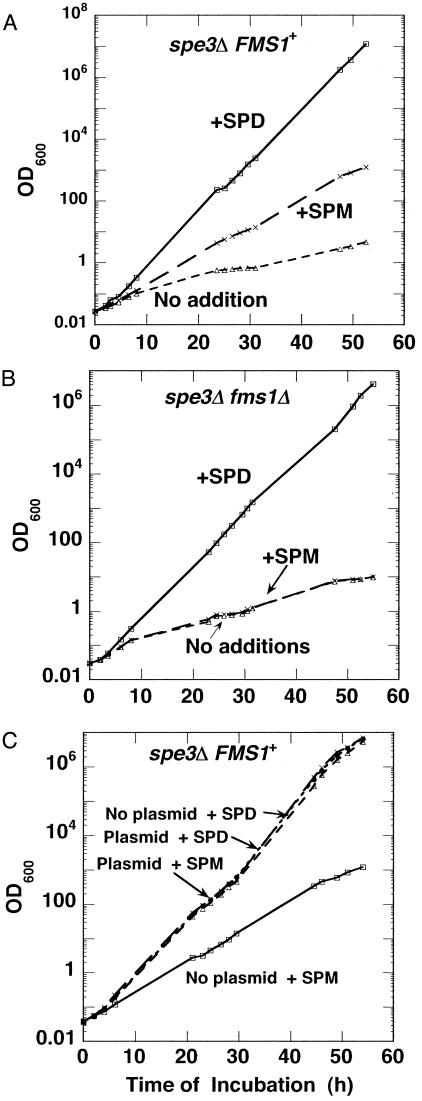
Culture Conditions. Colonies were inoculated from YPAD (yeast extract, peptone, dextrose, adenine) plates into synthetic dropout (SD) medium‡ plus 10–8 M spermidine and grown for 20–25 generations. The cultures were then diluted with SD medium to an OD600 of ≈0.03, and either spermidine or spermine was added to aliquots as indicated in the legend to Fig. 3. The cultures were incubated at 30°C with shaking, and the optical density (OD600) was followed. Whenever the optical density reached ≈1.0 OD600, the cultures were diluted into fresh prewarmed medium, and the incubations were continued. The data presented in Fig. 3 has been corrected for these dilutions.
Fig. 3.
Spermine-to-spermidine conversion by FMS1 encoded amine oxidase is necessary for growth of spe3Δ cells. Cultures of spe3Δ FMS1+ (Y551) (A) and spe3Δ fms1Δ (Y549) (B) were grown in SD medium containing 10–8 M spermidine as described in Materials and Methods and, at zero time, diluted in amine-free medium to an OD600 of 0.03 with the addition of either spermidine (10–5 M) or spermine (10–5 M, purified), or without any addition. Optical densities were noted at different time points. Cultures were diluted in the same medium when the OD600 value reached ≈1. The OD values in the figure were corrected for these dilutions. (C) The cultures were grown and diluted in glucose-containing medium in a way similar to above, but experiments were carried out in spe3Δ FMS1+ cells with (Y559) and without (Y537) the pYFMS1 plasmid in the presence of either 10–5 M spermidine or 10–5 M spermine. SPD, spermidine; SPM, spermine.
Radioactive Experiments. For the experiments testing the conversion of radioactive spermine to spermidine, the cultures were grown as described above in SD medium containing 10–8 M spermidine, diluted to an OD600 of 0.01 into fresh SD medium containing [14C]spermine (final concentration 10–6 M spermine, 0.1μCi/ml culture), and incubated overnight (18–20 h) at 30°C with shaking. The cells were harvested and washed twice with SD medium; the cell pellet was extracted with 5 volumes of cold 5% trichloroacetic acid. Aliquots were spotted on a cation-exchange thin-layer chromatographic sheet (Polygram Ionex-25, Macherey-Nagel, Düren, Germany) and developed with 2 M KCl (5). After drying, the plate was exposed to a Fuji BAS-MS imaging plate for several days. The imaging plate was scanned by a Fuji FLA-3000 scanner and spots were quantified by image gauge software (Fuji).
For the experiments testing whether [14C]spermidine or [14C]spermine is incorporated into eIF5A, the cultures were grown in a way similar to the experiment above. The trichloroacetic acid precipitate was washed with 5% cold trichloroacetic acid containing added nonradioactive spermidine or spermine followed by washing with 100% ethanol to remove the trichloroacetic acid. The precipitate was resuspended in 0.05 M Tris·Cl, pH 9.0. An aliquot was mixed with 4× lithium dodecyl sulfate (LDS) sample buffer (obtained from Invitrogen) plus DTT and run on their 12% Bis-Tris NuPAGE gel with Mes-SDS gel running buffer together with standards. The gel was stained, dried, and exposed to the Fuji BAS-MS imaging plate before scanning.
Polyamine and Protein Estimation. The polyamine content of the cells grown with and without spermine was estimated by HPLC analysis of the 5% trichloroacetic acid extracts as described (5) with modification of the buffer to a 70:30 ratio of KCl/NaCl. Protein was estimated by a protein-dye-binding assay (22), with use of BSA as a standard.
Insertion of FMS1 Gene into a Galactose-Inducible Vector. Because previous work had shown that yeast contains an amine oxidase that is encoded by the FMS1§ gene, we inserted this gene into a galactose-inducible vector for studies on the effect of the overexpression of this enzyme in vivo and for the purification of the enzyme for in vitro assays. The FMS1 gene (YMR020W) was first amplified by PCR from yeast genomic DNA by using the primers 5′-ATGAATACAGTTTCACCAGCC (upstream) and 5′-TTTCAGTAAGTCAGAGATTCG (downstream). For cloning the gene into our yeast strains (Y537 and Y547) by homologous recombination (26), a second PCR amplification was carried out, with the product of the first PCR as a template (after purification) and two primers 5′-TAATATACCTCTATACTTTAACGTCAAGGAGAAAAAACCCCGGATATGAATACAGTTTCA (upstream) and 5′-TACATGATGCGGCCCTCTAGAAAC.
TCAATGGTGATGGTGATGATGTTTCAGTAAGTCAGA (downstream) that contained the necessary vector and gene sequences (the gene sequences in bold) plus the codons (in italics) needed for a C-terminal 6XHis-tag. This PCR product and BamHI–BstEII-digested vector DNA (pYES2.1-PGAL1/V5-His/lacZ, Invitrogen) were used to transform Y537 (spe3Δ FMS1+) and Y547 (SPE3+ fms1Δ) cells (Table 1) by using the lithium acetate method (27). Ura+ transformants were selected on SD minus uracil plates. Confirmation of the FMS1+ genotype was carried out by PCR.
Partial Purification of the FMS1-Encoded Amine Oxidase. A single colony of Y560 (see Table 1 for genotype) was inoculated into 200 ml of SD minus uracil medium (2% glucose). The culture was incubated at 30°C with shaking to 1.8–2.0 OD600 and then was diluted to 0.3–0.4 OD600 in 1,000 ml of SD medium containing galactose (2%) plus raffinose (1%) instead of glucose; incubation was continued until the OD was 0.8. The cells were harvested by centrifugation. Extraction and purification through a Ni column (1 ml) was carried out with the Y-PER 6XHis fusion protein purification kit obtained from Pierce. The column was eluted with the buffer supplied (containing 200 mM imidazole). The fractions (0.5 ml) were assayed by electrophoresis on 4–12% NuPAGE/SDS gel, and immunodetection with anti-His C-terminal antibody (Invitrogen) (data not shown). Fractions 8 and 9, which contained the purest protein (≈80% pure) were dialyzed, concentrated, and used for the assays. We estimate that this preparation was purified 120-fold compared with the crude extract.¶
The assay mixture, containing 1 mM substrate, 0.1 M Tris·Cl (pH 8) buffer, and 6 μg of enzyme in a 200-μl volume, was incubated at 37°C. At 30 min, aliquots were treated with an equal volume of 10% cold trichloroacetic acid, and the extract was assayed for appearance of product by HPLC chromatography.
Results
Spermine Cannot Be Converted to Spermidine in Vivo in a fms1Δ Mutant. In our previous work we reported that [14C]spermine could be converted into [14C]spermidine in vivo in cells of S. cerevisiae (5). To test whether the Fms1 protein carried out this conversion in vivo, we compared this conversion in a spe3Δ FMS1+ strain and a spe3Δ fms1Δ strain. As shown in Fig. 1, the spermine-to-spermidine conversion occurs in a spe3Δ FMS1+ strain (Y551), but not in a spe3Δ fms1Δ strain (Y549), indicating that the FMS1-encoded amine oxidase is required for the in vivo conversion of spermine to spermidine.
Fig. 1.
Fms1-encoded amine oxidase is necessary for spermine-to-spermidine conversion in vivo.[14C]Spermine (113 mCi/mmol, 50 μCi/ml) was added at 10–6 M concentration to the partially amine-depleted spe3Δ FMS1+ (Y551) and spe3Δ fms1Δ (Y549) cultures as mentioned in the text. Cells were harvested, and polyamines were extracted and run on a Polygram cation-exchange chromatographic plate with 2 M KCl. Results were obtained by scanning a Fuji BAS-MS imaging plate as mentioned in Materials and Methods. The scanned values are expressed in percent of [14C]spermine-to-[14C]spermidine conversion and are shown below each figure. The right-hand lane contains standard [14C]spermine. (The calculated values were corrected for a 0.3% contamination of radioactive spermidine in commercial [14C]spermine.) [Rf values: spermine (SPM), 0.17; spermidine (SPD), 0.33.]
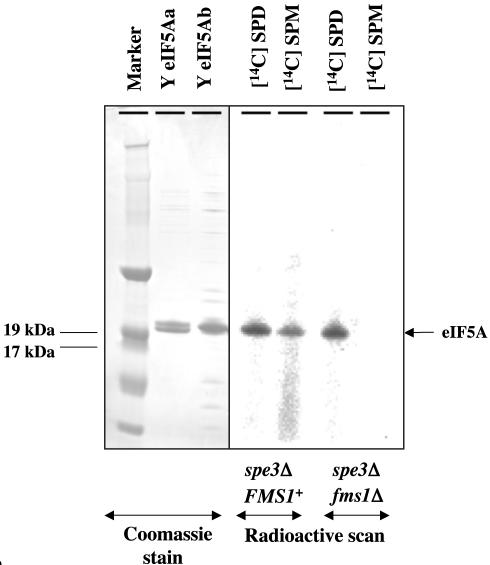
Spermine Cannot Be Used for Hypusine Biosynthesis in S. cerevisiae in the Absence of FMS1 Gene. Cultures of spe3Δ fms1Δ and spe3Δ FMS1+ were grown in the presence of 10–6 M [14C]spermine (113 mCi/mmol, 0.1μCi/ml culture) or [14C]spermidine (112 mCi/mmol, 0.1μCi/ml culture) for 20 h. Cells were harvested, and the protein was precipitated with 5% cold trichloroacetic acid. The pellet was solubilized and run on SDS/PAGE as described in Materials and Methods. In the spe3Δ FMS1+ strain either [14C]spermidine or [14C]spermine was incorporated into a single protein that migrated together with known yeast eIF5A (≈20–21 kDa) (28, 29), whereas only [14C]spermidine was incorporated into this protein in the spe3Δ fms1Δ double mutant (Fig. 2). To confirm that the labeled protein was modified eIF5A, we hydrolyzed the labeled band in 6 N HCl (109°C, overnight) and found that the radioactivity eluted in the same position as authentic hypusine by ion-exchange chromatography (13) (data not shown). These results indicate that the radiolabeled band was hypusine-modified yeast eIF5A, and spermine cannot be used for hypusine modification of eIF5A in vivo until it is oxidized to spermidine by the Fms1 amine oxidase.
Fig. 2.
Spermine cannot be used for hypusine or eIF5A modification unless it is converted to spermidine. The cultures were grown as shown in Fig. 1, except that either [14C]spermidine (112 mCi/mmol, 50 μCi/ml) or [14C]spermine (113 mCi/mmol, 50 μCi/ml) was added in separate but comparable cultures. The cells were harvested, and the cellular precipitates after 5% trichloroacetic acid extraction from both [14C]spermidine- and [14C]spermine-grown cultures were washed, dried, and resuspended as detailed in Materials and Methods. Part of the suspension (15 μl) was mixed with lithium dodecyl sulfate sample buffer and run on a 12% Bis-Tris NuPAGE gel together with protein molecular weight markers (Invitrogen) and yeast eIF5Aa and eIF5Ab. After running, the gel was stained, dried, and exposed to Fuji BAS-MS imaging plate for 3–5 days. Part of the Coomassie-stained gel is included to show the positions of yeast eIF5As run in parallel with the radioactive samples. SPD, spermidine; SPM, spermine.
Spermine Does Not Support Growth of a spe3Δ Mutant in the Absence of the FMS1 Gene. Previous studies have shown that mature eIF5A (i.e., after the hypusine modification of precursor eIF5A) is essential for yeast growth (6, 8, 15, 16). Therefore, because the above-described experiments showed that spermine cannot be converted to spermidine in the absence of the FMS1 gene, and that spermine cannot be used as a substrate for the formation of hypusine and for the maturation of eIF5A, we expected that the spe3Δ fms1Δ double mutant should have an absolute requirement for spermidine for growth. As shown in Fig. 3B, this requirement is indeed the case. Either spermidine or spermine will support the growth of a spe3Δ FMS1+ strain, but spermine will not support the growth of the spe3Δ fms1Δ double mutant, because it cannot be converted to spermidine.
The experiments above show that the amine oxidase encoded by the FMS1 gene is required for the conversion of spermine to spermidine. However, as shown in Fig. 3A, even in the spe3Δ FMS1+ strain the growth rate with spermine was much slower (nearly 50%) than that obtained with addition of a similar concentration of spermidine (10–5 M). To test whether the slow growth with spermine was due to a limitation in the conversion of spermine to spermidine in this strain (i.e., with only the chromosomal copy of the FMS1 gene) or to some other factor such as spermine uptake, we repeated the growth experiments with a comparable strain (spe3Δ background) containing a plasmid (pYFMS1) with the FMS1+ gene (Y559). As shown in Fig. 3C, the growth rate with spermine addition in the plasmid-containing cells was nearly the same as the growth rate with spermidine addition. Thus, even though these growth experiments were carried out in glucose-containing medium (i.e., without galactose induction), sufficient overexpression of the Fms1 protein (confirmed by anti-His antibody, data not shown) occurred to permit enough conversion of spermine to spermidine to permit optimum growth. Indeed, we found by HPLC analysis that the intracellular spermine-to-spermidine conversion increased (30% in the plasmid-containing cells compared with 3–8% in the cells without the pYFMS1 plasmid) (data not shown), albeit much less than in the culture grown in galactose (shown below).
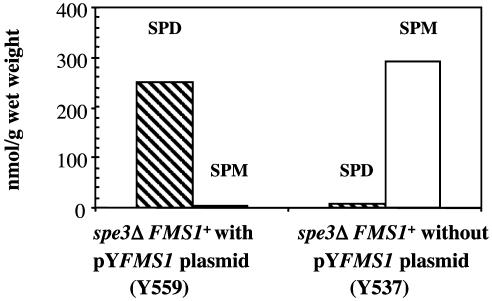
Spermine-to-Spermidine Conversion in Vivo Is Markedly Increased after Induction of the FMS1 Gene. To confirm the ability of the FMS1-encoded amine oxidase to convert spermine to spermidine in vivo, we grew a spe3Δ FMS1+ (Y537) strain and a spe3Δ FMS1+/pYFMS1 (Y559) harboring the FMS1 gene under a galactose-inducible promoter in a galactose-containing medium with 10–5 M spermine. The spe3Δ FMS1+ (Y537) cells contained 290 nmol of spermine per gram of wet weight and 6 nmol of spermidine (Fig. 4). On the other hand, the spe3Δ FMS1+/pYFMS1 cells (Y559) contained nearly undetectable spermine, but a markedly increased spermidine content (249 nmol/g wet weight).
Fig. 4.
Marked increase in in vivo oxidation of spermine to spermidine in a strain overexpressing Fms1 protein. HPLC analysis showed complete conversion of intracellular spermine to spermidine after a 20-h incubation in galactose-containing medium in spe3Δ FMS1+/pYFMS1 cells (Y559) but not in spe3Δ FMS1+ cells without the pYFMS1 plasmid (Y537). The cells were grown in 10–5 M spermine (see Materials and Methods). SPD, spermidine; SPM, spermine.
In Vitro Conversion of Spermine to Spermidine by the Purified Fms1 Amine Oxidase. We have purified the enzyme from a yeast strain overxpressing FMS1 gene (Y560) after galactose induction to study the in vitro substrate specificity of the enzyme. The assay results (Table 2) confirmed the recent data obtained by Landry and Sternglanz, who used a recombinant yeast protein from E. coli (25), showing that the Fms1-encoded amine oxidase converts spermine and N1-acetylspermine to spermidine in vitro. Our purified protein from yeast showed absorbance peaks at 375 and 455 nm (data not shown), similar to the peaks of FAD found by Landry and Sternglanz in their preparation (25). The enzyme has greater activity with N1-acetylspermine as the substrate than with spermine (Table 2). The purified enzyme also uses N1-acetylspermidine as a substrate. As in their experiments (25) the enzyme does not oxidize spermidine or putrescine.
Table 2. In vitro assay showing substrate specificity.
| Substrate | Product assayed | Polyamine oxidase activity, nmol per mg of protein per min |
|---|---|---|
| Spermine | Spermidine | 390 |
| N1-Acetylspermine | Spermidine | 800 |
| N1-Acetylspermidine | Putrescine | 270 |
Substrates (1 mM) were incubated in the presence of partially purified (≈80%) His-tagged Fms1 protein as mentioned in Materials and Methods, and, after 30 min, reactions were stopped. Activity was measured on the basis of amine conversion (appearance of amine product) as determined by HPLC analysis. No oxidation was observed by using spermidine or putrescine as substrate (data not shown).
Discussion
Our current studies show that spermidine is specifically required for the growth of S. cerevisiae. Although we had previously shown that either spermidine or spermine can satisfy the polyamine requirements of a spe2Δ or a spe3Δ mutant of S. cerevisiae (4, 5), these results were complicated by the ability of S. cerevisiae to convert spermine to spermidine in vivo.
In the current work we show that spermidine is specifically required for S. cerevisiae in a strain that is deleted in both SPE3 and FMS1 (spe3Δ fms1Δ, Table 1). We show that only spermidine, and not spermine, will satisfy the growth requirement of the spe3Δ fms1Δ strain (Fig. 3B). These experiments were stimulated by the findings of White et al. (24, 30) that the gene FMS1 codes for an amine oxidase that converts spermine to 3-aminopropionaldehyde, a necessary precursor of the β-alanine moiety of pantothenate. More recently, while our work was in progress, Landry and Sternglanz (25) extended the studies of White et al. (24) and showed in vitro that the purified Fms1 protein produces both spermidine and 3-aminopropanal when the substrate is spermine. Our studies show that this reaction also occurs in vivo and is due to the action of the Fms1 protein.
One reason for the specific requirement of S. cerevisiae for spermidine is its importance as a substrate for hypusine biosynthesis and formation of mature eIF5A. This use is particularly important because hypusine-modified eIF5A (not the unmodified precursor) is essential for growth of S. cerevisiae (6, 8, 15, 16). Thus, we have shown that the spe3Δ fms1Δ mutant is able to incorporate [14C]spermidine but not [14C]spermine in vivo into the eIF5A protein (Fig. 2). These in vivo results are the first demonstration that in S. cerevisiae, as in other cell types (13, 18–20), only spermidine can be used as a substrate for the hypusine modification; spermine cannot be used unless it is first oxidized to spermidine by the amine oxidase.
White et al. (24) showed that spermine, and not spermidine, is specifically required for the biosynthesis of β-alanine and panthothenate. Thus, our experiments showing a specific requirement for spermidine, together with the findings of White et al. (24), demonstrate that spermidine and spermine have separate and specific functions in the yeast cell; namely, hypusine and pantothenate synthesis, respectively. Further work is needed, with use of the spe3Δ fms1Δ mutant (or a spe2Δ fms1Δmutant), to test whether this specificity also applies to any of the other functions of the polyamines that others and we have described (1–3).
As discussed above, both in vivo and in vitro, the FMS1-encoded amine oxidase carries out the reaction:
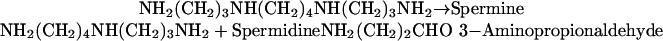 |
However our results, and those of Landy and Sternglanz (25), showed that the enzyme also oxidizes N1-acetylspermine and N1-acetylspermidine. Indeed, in our results the rate of oxidation with acetylspermine was 2.5-fold greater than that found with spermine as the substrate. We also found that this difference was true in vivo; namely, when cells were grown with N1-acetylspermine, the cells contained more spermidine than when the cells were grown with added spermine (data not shown). These results were of particular interest because of the many experiments with the mammalian polyamine oxidase/N-acetyltransferase system, showing that spermine was usually acetylated before oxidation (31–33). This result brings up the question of whether the physiological substrate for the S. cerevisiae enzyme in vivo is spermine or whether the spermine is first converted to acetylspermine. If the spermine first had to be acetylated before oxidation to spermidine, one would expect the accumulation of [14C]acetylspermine in the strain lacking the FMS1 gene (Y549) after incubation with [14C]spermine. However, in the experiments reported in Fig. 1, no [14C]acetylspermine was detected in the spe3Δ fms1Δ culture, indicating that no detectable acetylation had occurred. In addition, until now no homologue of the mammalian spermidine/spermine N1-acetyltransferase gene has been found in the S. cerevisiae genome data bank. Thus, the in vivo oxidation of spermine in yeast may be different from the polyamine oxidase/N-acetyltransferase system described in mammalian tissues (33–36). However, polyamine oxidation involving both acetyl and nonacetyl pathways has been reported in other fungal systems (37).
It is unclear as to what the physiological role of this oxidation pathway is in wild-type yeast cells growing with adequate pantothenate in the yeast medium. Such cells have enough intracellular spermidine. Thus, the fms1Δ SPE3+ cells grow normally in the presence of pantothenate in the medium without addition of spermine. Even though the Fms1 amine oxidase can oxidize spermine to spermidine for hypusine modification and 3-aminopropionaldehyde (for pantothenate biosynthesis), demonstration of the functional importance of the spermine-to-spermidine conversion in vivo can only be observed by using either the double mutants (such as spe3Δ fms1Δ) or by using the strains overexpressing Fms1 protein as we described above.
Acknowledgments
We thank Drs. David Balasundaram and Nobuko Hamasaki for their critical comments and valuable suggestions, Dr. Tibor B. Roberts for his generous help and useful discussions and for his protocol for plasmid construction by homologous recombination, and Drs. Edith Wolff and M. H. Park for performing the hypusine assay by chromatography of the 14C-labeled protein hydrolysate and for providing recombinant yeast eIF5Aa and eIF5Ab.
Abbreviation: SD, synthetic dropout.
Footnotes
Polyamine biosynthetic pathway.
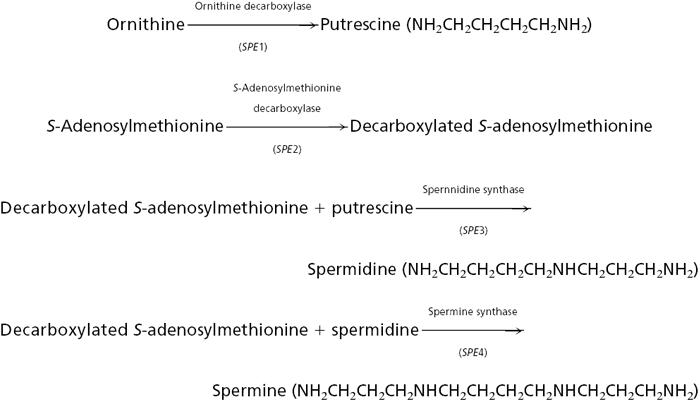 |
The SD medium was made from Bio 101 yeast nitrogen base minimal medium without vitamins (catalog no. 4030-612). To each liter were added 10 ml of 1 M K2HPO4 to bring the pH to 6.7, 10 ml of 0.1% calcium pantothenate, 1 ml of vitamin mixture, and 20 g of glucose and necessary supplements. The medium was sterilized by filtration.
FMS1 was first identified as Fenpropiomorph Multicopy Suppressor gene1 by Joets et al. (23) and shown to have some degree of homology to the mammalian gene for monoamine oxidase. The FMS1 gene is located on the right arm of yeast chromosome XIII. Subsequently, the Fms1 protein was identified as an amine oxidase involved in the metabolism of spermine (24, 25).
As described above, purification of the amine oxidase was made from an overexpressed yeast strain. While this work was in progress, Landry and Sternglanz (25) reported a similar purification of the yeast His-tagged Fms1 protein by using an overexpression system in Escherichia coli.
References
- 1.Tabor, C. W. & Tabor, H. (1984) Annu. Rev. Biochem. 53, 749–790. [DOI] [PubMed] [Google Scholar]
- 2.Pegg, A. E. (1986) Biochem. J. 234, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, S. S. (1998) A Guide to the Polyamines (Oxford Univ. Press, New York).
- 4.Balasundaram, D., Tabor, C. W. & Tabor, H. (1991) Proc. Natl. Acad. Sci. USA 88, 5872–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamasaki-Katagiri, N., Tabor, C. W. & Tabor, H. (1997) Gene 187, 35–43. [DOI] [PubMed] [Google Scholar]
- 6.Schnier, J., Schwelberger, H. G., Smit-McBride, Z., Kang, H. A. & Hershey, J. W. (1991) Mol. Cell. Biol. 11, 3105–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park, M. H., Wolff, E. C. & Folk, J. E. (1993) Trends Biochem. Sci. 18, 475–479. [DOI] [PubMed] [Google Scholar]
- 8.Kang, H. A. & Hershey, J. W. (1994) J. Biol. Chem. 269, 3934–3940. [PubMed] [Google Scholar]
- 9.Park, M. H., Lee, Y. B. & Joe, Y. A. (1997) Biol. Signals 6, 115–123. [DOI] [PubMed] [Google Scholar]
- 10.Chen, K. Y. & Liu, A. Y. (1997) Biol. Signals 6, 105–109. [DOI] [PubMed] [Google Scholar]
- 11.Zuk, D. & Jacobson, A. (1998) EMBO J. 17, 2914–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valentini, S. R., Casolari, J. M., Oliveira, C. C., Silver, P. A. & McBride, A. E. (2002) Genetics 160, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park, M. H., Cooper, H. L. & Folk, J. E. (1981) Proc. Natl. Acad. Sci. USA 78, 2869–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park, M. H., Liu, T. Y., Neece, S. H. & Swiggard, W. J. (1986) J. Biol. Chem. 261, 14515–14519. [PubMed] [Google Scholar]
- 15.Sasaki, K., Abid, M. R. & Miyazaki, M. (1996) FEBS Lett. 384, 151–154. [DOI] [PubMed] [Google Scholar]
- 16.Park, M. H., Joe, Y. A. & Kang, K. R. (1998) J. Biol. Chem. 273, 1677–1683. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, E. D., Mora, R., Meredith, S. C., Lee, C. & Lindquist, S. L. (1987) J. Biol. Chem. 262, 16585–16589. [PubMed] [Google Scholar]
- 18.Gerner, E. W., Mamont, P. S., Bernhardt, A. & Siat, M. (1986) Biochem. J. 239, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byers, T. L., Lakanen, J. R., Coward, J. K. & Pegg, A. E. (1994) Biochem. J. 303, 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, C. H. & Park, M. H. (2000) Biochem. J. 352, 851–857. [PMC free article] [PubMed] [Google Scholar]
- 21.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 22.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 23.Joets, J., Pousset, D., Marcireau, C. & Karst, F. (1996) Curr. Genet. 30, 115–120. [DOI] [PubMed] [Google Scholar]
- 24.White, W. H., Gunyuzlu, P. L. & Toyn, J. H. (2001) J. Biol. Chem. 276, 10794–10800. [DOI] [PubMed] [Google Scholar]
- 25.Landry, J. & Sternglanz, R. (2003) Biochem. Biophys. Res. Commun. 303, 771–776. [DOI] [PubMed] [Google Scholar]
- 26.Ma, H., Kunes, S., Schatz, P. J. & Botstein, D. (1987) Gene 58, 201–216. [DOI] [PubMed] [Google Scholar]
- 27.Gietz, D., St. Jean, A., Woods, R. A. & Schiestl, R. H. (1992) Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwelberger, H. G., Kang, H. A. & Hershey, J. W. (1993) J. Biol. Chem. 268, 14018–14025. [PubMed] [Google Scholar]
- 29.Kang, K. R., Wolff, E. C., Park, M. H., Folk, J. E. & Chung, S. I. (1995) J. Biol. Chem. 270, 18408–18412. [DOI] [PubMed] [Google Scholar]
- 30.White, W. H., Skatrud, P. L., Xue, Z. & Toyn, J. H. (2003) Genetics 163, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolkenius, F. N. & Seiler, N. (1987) Biochim. Biophys. Acta 923, 125–135. [DOI] [PubMed] [Google Scholar]
- 32.Seiler, N. (1995) Prog. Brain Res. 106, 333–344. [DOI] [PubMed] [Google Scholar]
- 33.Casero, R. A. J. & Pegg, A. E. (1993) FASEB J. 7, 653–661. [PubMed] [Google Scholar]
- 34.Murray-Stewart, T., Wang, Y., Devereux, W. & Casero, R. A. J. (2002) Biochem. J. 368, 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Y., Murray-Stewart, T., Devereux, W., Hacker, A., Frydman, B., Woster, P. M. & Casero, R. A. J. (2003) Biochem. Biophys. Res. Commun. 304, 605–611. [DOI] [PubMed] [Google Scholar]
- 36.Vujcic, S., Liang, P., Diegelman, P., Kramer, D. L. & Porter, C. W. (2003) Biochem. J. 370, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Large, P. J. (1992) FEMS Microbiol. Rev. 8, 249–262. [DOI] [PubMed] [Google Scholar]