Abstract
Lentiviral vector-based gene therapy has been used to target the human immunodeficiency virus (HIV) using an antisense env payload. We have analyzed lentiviral-vector integration sites from three treated individuals. We compared integration sites from the ex vivo vector-transduced CD4+ cell products to sites from cells recovered at several times after infusion. Integration sites were analyzed using 454 pyrosequencing, yielding a total of 7,782 unique integration sites from the ex vivo product and 237 unique sites from cells recovered after infusion. Integrated vector copies in both data sets were found to be strongly enriched within active genes and near epigenetic marks associated with active transcription units. Analysis of integration relative to nucleosome structure on target DNA indicated favoring of integration in outward facing DNA major grooves on the nucleosome surface. There was no indication that growth of transduced cells after infusion resulted in enrichment for integration sites near proto-oncogene 5′-ends or within tumor suppressor genes. Thus, this first look at the longitudinal evolution of cells transduced with a lentiviral vector after infusion of gene modified CD4+ cells provided no evidence for abnormal expansions of cells due to vector-mediated insertional activation of proto-oncogenes.
Introduction
The first unambiguously successful human gene therapy involved use of γ-retroviral vectors to restore the mutant genes in hematopoeitic stem cells from patients with inherited immunodeficiencies.1,2 However, five leukemias have now been reported among the 20 patients successfully treated in these trials, in which the vector integrated upstream of a proto-oncogene and increased its rate of transcription.3,4,5,6,7 Proliferating cells accumulated further genetic lesions, ultimately resulting in clonal expansion and leukemia.
Although many factors can potentially contribute to adverse events (possible oncogenicity of transgenes, multiplicities of infection, etc.),8,9,10 many in the field have suggested that use of lentiviral vectors may lead to reduced genotoxicity. (i) Lentiviruses have a different distribution of favored integration sites in the human genome than is seen with γ-retroviruses—lentiviruses favor integration within active transcription units, while γ-retroviruses favor integration near transcription start sites and associated features such as CpG islands and DNAseI cleavage sites.11,12,13,14,15,16,17,18,19 The association of γ-retroviral integration with gene 5′-ends15 may be part of the reason insertional activation is seen with these vectors. (ii) human immunodeficiency virus (HIV) infection is not associated with insertional activation of proto-oncogenes and transformation by this mechanism. (iii) Lentiviral vectors can infect cell types not easily accessible with γ-retroviral vectors. (iv) Murine models have been devised to quantify genotoxicity of retroviral vectors, and these consistently show that lentiviral vectors are less toxic than γ-retroviral vectors.10,16
Levine et al. carried out a first in human study in which a lentiviral vector was introduced into five HIV-infected subjects who had failed at least two prior antiretroviral drug regimens.17 The vector, VRX496, expressed an antisense HIV env-gene that had been shown to inhibit HIV replication in a cell culture model.20 Peripheral blood CD4+ T cells were harvested from each subject by apheresis, depleted of CD8+ cells and monocytes, transduced with the lentiviral vector ex vivo, activated via CD3 and CD28 costimulation, and expanded before being cryopreserved. Following quality control testing, the cells were reinfused at a dose of 10 billion cells per subject. The participants have been followed for a median of ~4 years and no leukemias or other serious adverse events associated with study treatment have been detected (to be reported elsewhere), strengthening the idea that lentiviral vectors are safe for use in gene therapy. The transduced cells diminished in number after infusion but were detectable at 2 years in three of five subjects. All patients had detectable viral loads throughout the experiment. In some patients a transient reduction in viral load was observed, though it is uncertain whether this was a consequence of the gene therapy treatment. There was no evidence of abnormal proliferation of cells after infusion. We note that expression of the env antisense RNA was directed by the HIV long-terminal repeat (LTR) and so required Tat protein for efficient transcription, which was not supplied by the vector and would only be available upon super-infection with wild-type HIV.
In order to evaluate the safety of the long-term use of lentiviral vectors, it remains essential to investigate whether those cells that have persisted longest contained integration sites near genes involved in growth control or transformation. Such an evolutionary bias was seen in a recent trial treating chronic granulomatous disease using stem cells modified with a γ-retroviral vector, in which the cells that persisted were enriched for integration events near the proto-oncogenes MDS1-EVI1, PRDM16, or SETBP1.21
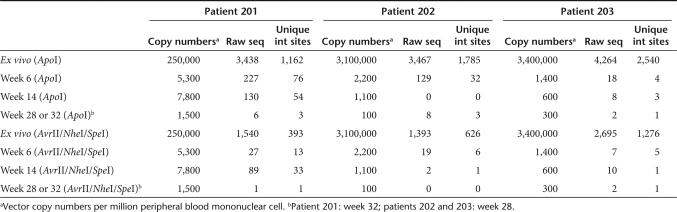
A follow-on Phase I/II clinical trial was initiated in early stage HIV-infected subjects who have well-controlled viral loads. This study has evaluated the safety of up to 6 infusions of 10 billion CD4+ T cells modified with a lentiviral vector expressing a segment of the env gene (VRX496) and corresponding antiviral efficacy during interruption of highly active antiretroviral therapy. In the studies presented here, we analyze the distribution of integrated vector copies in the ex vivo transduced CD4+ T cells (before infusion), and in cells from the first three subjects recovered at 6, 14, and 28 or 32 weeks after reinfusion (Table 1). In a previous study, a small number of integration sites (192) were recovered from the ex vivo transduced cell population from the first VRX496 trial and characterized.17 Here we characterize integration sites from the follow-on Phase I/II trial, using DNA bar coding and pyrosequencing to characterize 7,782 sites from the ex vivo transduced product and 237 sites recovered from three patients after infusion. In cells recovered from study subjects, we found no significant enrichment of vector copies in or near known proto-oncogenes or tumor suppressors, arguing against the idea that activation of proliferative pathways by vector-mediated insertional mutagenesis has contributed to cellular persistence.
Table 1.
Subjects and time points studied
Results
Recovery and analysis of host–vector DNA junctions
We analyzed integration site distributions in the first three study subjects out of 15 treated in a Phase I/II clinical trial of multiple infusions of the VRX496 vector-transduced CD4+ T cells. These subjects were selected because they were the first treated and so allowed longer term follow-up. We sampled the initially transduced population of T cells before reinfusion, and peripheral blood mononuclear cell (PBMC) from three time points after infusion (Table 1).
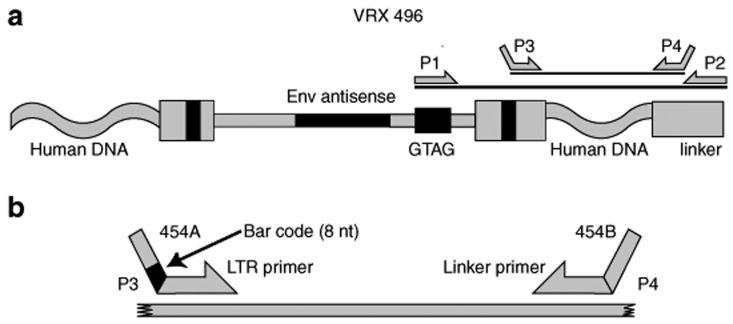
To isolate integration sites from the VRX496-transduced cells, we needed a way to distinguish the HIV-based vector from the HIV strains circulating in the patients. For this we took advantage of the short unique sequence (GTAG) engineered in the VRX496 vector (Figure 1). We isolated genomic DNA from cells of the ex vivo transduced product or PBMC recovered from study subjects, cleaved with restriction enzymes (either ApoI or a cocktail of AvrII + SpeI + NheI), then ligated DNA linkers on the restriction-cut DNA ends. We then amplified using one primer complementary to the GTAG sequence and the other complementary to the DNA linker (Figure 1a). A nested PCR step was then used, with the second round primers binding to the HIV LTR sequence within the vector, and the second again binding to the DNA linker.
Figure 1.
Diagram of integration site recovery strategy. (a) Use of a GTAG primer for first round amplification, followed by nested PCR for second round amplification. A detailed description of the VRX496 vector can be found in ref. 20. (b) Use of DNA bar coding to index amplification products during the second round PCR. LTR, long-terminal repeat.
We wished to use the 454/Roche method for deep pyrosequencing of integration sites,22 but we also wanted to sequence multiple samples simultaneously to maximize efficiency and minimize costs. For this reason, we bar coded23,24,25,26 our second round amplification primers (Figure 1b). The pyrosequence read begins at the 3′-end of the sequence marked “454 A ” in the diagram. We introduced eight base recognition sites abutting this in the primer, which indexed each amplification product by patient and sample type. Thus all the amplification products could be sequenced as a pool, and integration site sequences separated subsequently using the linked DNA bar code.
After pyrosequencing, we recovered a total of 17,482 sequence reads which after dereplication yielded 8,019 unique integration sites. Of these, the great majority were from the ex vivo cell product harvested before infusion (7,782), likely because the concentration and number of transduced cells were considerably lower after infusion (Table 1). A total of 237 unique integration sites were recovered from PBMC postinfusion. This number is relatively low but sufficient for statistical analysis of strong trends in the data.
Correct recovery of junctions between the vector DNA and flanking host sequences could be verified in two ways. In one, negative controls during the PCR amplification showed that the GTAG amplification step was required to form amplification products in the second round, indicating that proviruses formed from the circulating HIV did not give rise to products. In the second approach to verification, we required a perfect match between the vector LTR sequence and the isolated recovered sequence, so that any sequences derived from the pre-existing HIV infection that differed from the VRX496 vector were excluded. No high-abundance sequence polymorphisms were detected over the LTR region analyzed, supporting the idea that only integrated VRX 496 vectors gave rise to the recovered sequences.
Primary sequence features at integration sites
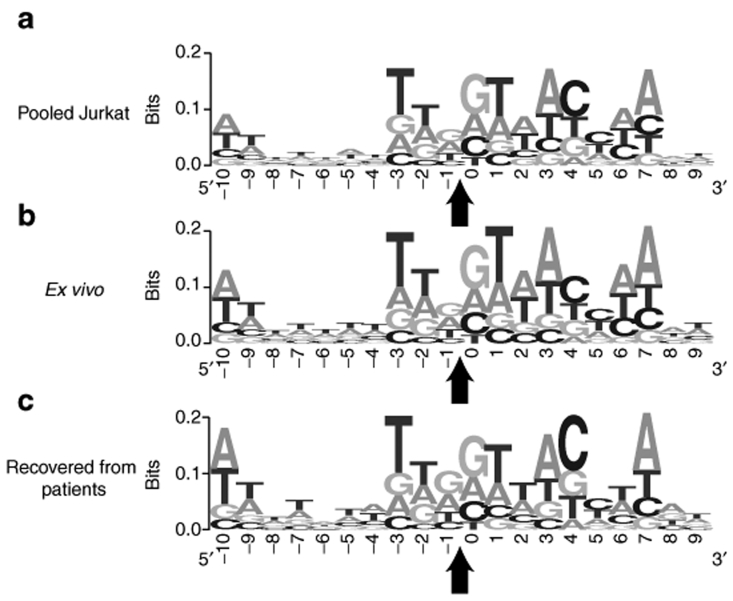
We first asked whether the weakly conserved favored primary sequences characteristic of HIV integration sites were present in VRX 496 data sets. Previous work has shown that a favored palindromic sequence can be detected when many HIV integration sites are aligned, the inverted repeat structure likely originating from the symmetry of the IN–DNA complexes responsible for covalent DNA joining at each end. This primary sequence has been seen in all previously analyzed HIV integration site data sets, and serves as a quality control check here.14,27,28,29,30 The ex vivo and patient-derived data sets were analyzed together with a previously published data sets of 40,000 HIV integration sites generated by infection of the Jurkat T-cell line (Figure 2).18 A comparison of the information content at integration sites for the VRX496 samples and the Jurkat data sets18 showed highly similar base frequencies at each position (Figure 2).
Figure 2.
Analysis of the information content in the local sequence at HIV integration sites. HIV integration sites in each data set were aligned and conserved bases identified. The y-axes indicate bits of information at each base; perfect conservation of a base would score as two bits. (a) Sequences from HIV infection of Jurkat cells.18 (b) Sequences from the VRX496 ex vivo samples. (c) Sequences from the VRX496 samples recovered from patients. The arrow indicates the location of the host virus DNA junction after integration. HIV, human immunodeficiency virus.
Provirus accumulation on the human chromosomes
We next analyzed the integration site data sets for enrichment or depletion relative to identifiable chromosomal features. For comparison in the statistical analysis, we also generated sets of matched random controls. For this, a large library of random sites was generated, and then the distances to restriction enzyme recognition sites scored. Each experimental site was matched with 10 control sites that were positioned the same number of bases from a restriction site as for the experimental site. That is, if an integration site was isolated after cleavage with ApoI, and the distance from the ApoI site to the edge of the VRX496 sequence was 80 bp, then 10 random control sites were drawn from the pool that were also 80 bp from an ApoI site. In the following statistical analysis integration sites were compared to their paired matched random controls. This helps control for the severe isolation bias resulting from preferred recovery of integration sites optimally positioned near restriction sites.19 In addition, we used two different restriction enzyme cocktails (either ApoI or AvrII + SpeI + NheI) to increase the recoverable numbers of integration sites.
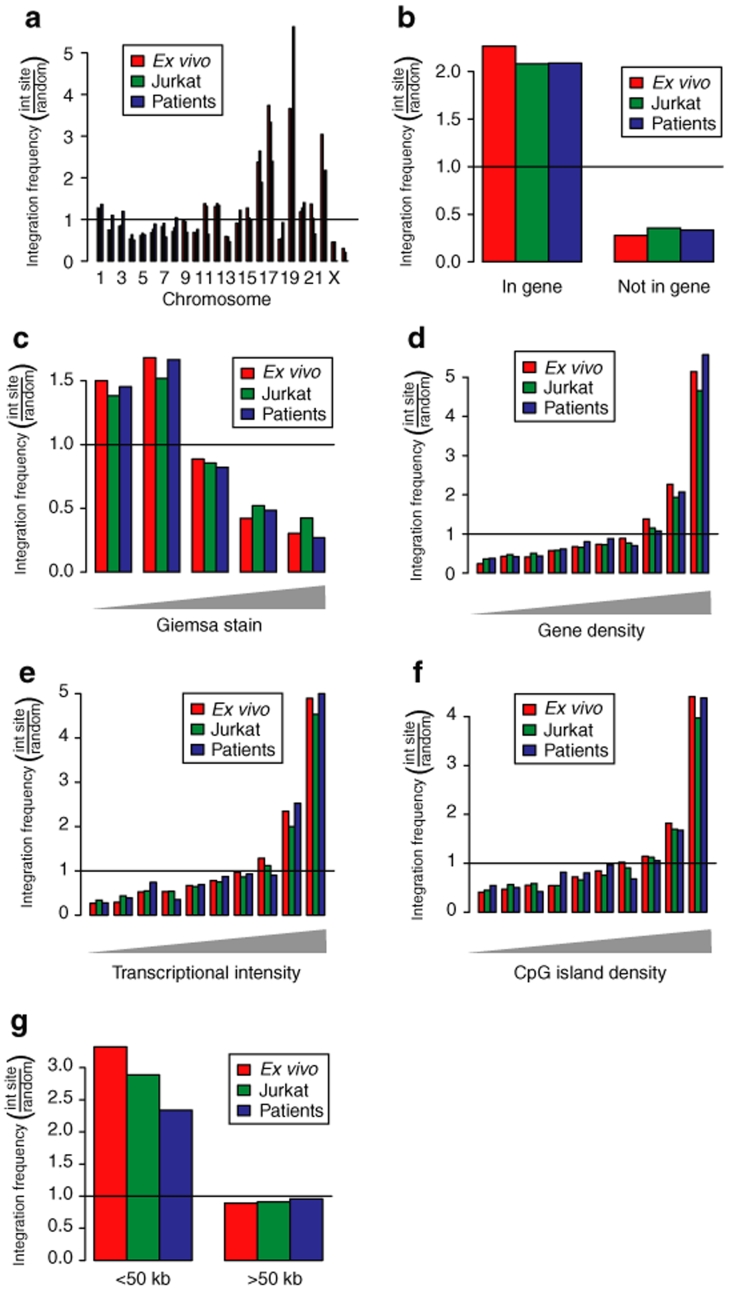
A comparison of the chromosomal distribution of integration sites for the ex vivo, patient derived, and Jurkat data sets is shown in Figure 3a. For each comparison, the observed proportion of integration sites in each chromosome was divided by the frequency in the matched random controls. Thus values above one indicate enrichment compared to random, while values below one indicate depletion. We found that the gene rich chromosomes 16, 17, 19, and 22 were favored for integration in all data sets, while the gene sparse chromosomes 4, 13, and Y were disfavored. These observations parallel findings for previously analyzed HIV integration site data sets.11,12,13,18,31
Figure 3.
Comparison of in vivo and ex vivo lentiviral vector integration sites to the integration sites from control infections of Jurkat cells—analysis of proximity to genomic features. (a) Chromosomal distribution of integration sites in the ex vivo sample, the patient-derived sample, and the control infections of Jurkat cells.18 Random integration would correspond to the line at one. Favored integration is indicated by the bars above the line, disfavored by the bars below. Only every other chromosome is numbered. The right-most chromosome is Y. (b) Frequency of integration in RefGenes. (c) Frequency of integration in Giemsa dark and light bands. The Giemsa dark regions (left side) are higher in gene density. (d) Integration frequency in gene rich regions (scored over 8-Mb intervals surrounding integration sites). (e) Integration frequency in regions of differing transcriptional intensity (scored over 8-Mb intervals surrounding integration sites). The transcriptional intensity measure is similar to the gene density measure, but only genes scored as active using Affymetrix microarrays are counted. (f) Integration frequency in regions of differing CpG island density (scored over 8-Mb intervals surrounding integration sites). (g) Frequency of integration near proto-oncogene 5′-ends. Random integration would have bar heights of one on the y-axis.
Provirus accumulation near chromosomal features
A series of studies were then carried out analyzing the density of proviruses near identifiable genomic landmarks. Figure 3b compares integration frequency in transcription units (as scored by the RefGenes database). As above, the observed proportion of integration sites in RefGenes was divided by the proportion in the matched random controls, so that values above one indicate enrichment. All three data sets showed enriched provirus accumulation within transcription units, as has been seen in previous studies of lentiviral DNA integration. There were no major differences among data sets, though a slight enrichment was observed in the ex vivo data set (P < 0.001 versus Jurkat data, P = 0.033 versus patient-derived data, χ2). Similarly, integration was favored for all three data sets in chromosomal regions annotated as Giemsa dark, which are regions of high gene density (Figure 3c).
Integration in chromosomal intervals was then quantified, comparing integration frequency to gene density, density of expressed genes, and density of CpG islands (Figure 3d–f). For each study, the density of each feature was quantified over an 8-Mb region surrounding each integration site. To quantify the density of expressed genes, transcriptional profiling data were used to annotate those genes in the most highly expressed 50% of all genes queried on the microarrays used, then the numbers of these genes in each interval scored. Each data set was compared to the matched random control. In all three data sets, integration was favored at increasing density of the indicated feature. In the human genome, gene dense regions are Giemsa dark, enriched in highly expressed genes, and enriched in CpG islands (which are commonly associated with gene regulatory regions). Although HIV integration is disfavored very close to CpG islands (i.e., <1 kb from a CpG island center), over much longer 8-Mb intervals, the density of CpG islands serves as another marker for gene density. Thus, integration in all three data sets was favored in regions dense in genes and associated features.
Integration near proto-oncogenes or within tumor suppressor genes
Our main reason for analyzing the patient integration sites was to monitor for possible genotoxicity, for example insertional activation of oncogenes or inactivation of tumor suppressor genes. During routine clinical monitoring of patients, there has been no evidence for abnormal expansion of cell clones harboring the VRX496 vector, nor any other indication of serious adverse events due to the transfer of gene modified CD4+ T cells (to be reported elsewhere). However, even in the absence of adverse events, it remained possible that cells with altered growth properties due to insertional mutagenesis might persist preferentially. To investigate this, we analyzed the distribution of VRX496 integration sites relative to known proto-oncogenes and tumor suppressor genes as compiled in our Cancergenes database (available at http://microb230.med.upenn.edu/protocols/cancergenes.html).
Each integration site was annotated for whether or not it was within 50 kb of the 5′ end of a known proto-oncogene or tumor suppressor gene (Figure 3g). The proportions in each data set were divided by the proportions in the matched random control for comparison. Integration sites from all three data sets are more commonly found near the 5′-ends of proto-oncogenes than expected by chance. However, there was no enrichment for proto-oncogenes in the patient-derived integration site data set compared to the control data sets, and indeed the association of the in vivo set was the lowest of the three evaluated.
We also compared the patient-derived integration sites to a specialized list of known proto-oncogenes implicated as important in lymphoid cells (prepared by Marina Cavazzana-Calvo, Salima Hacein-Bey-Abina, Alain Fischer, and their colleagues; see http://microb230.med.upenn.edu/protocols/cancergenes.html for the list). None of the 237 integration sites recovered from VRX496-treated patients were within 50 kb of any of these genes.
We also investigated the behavior of cell clones over time by asking whether any of the integration sites found in patients could be detected at more than one time point. No overlaps were found. Thus there has been no evidence for expansion of any specific transduced cell clones to date.
Provirus accumulation near sites of histone post-translational modification and bound chromosomal proteins
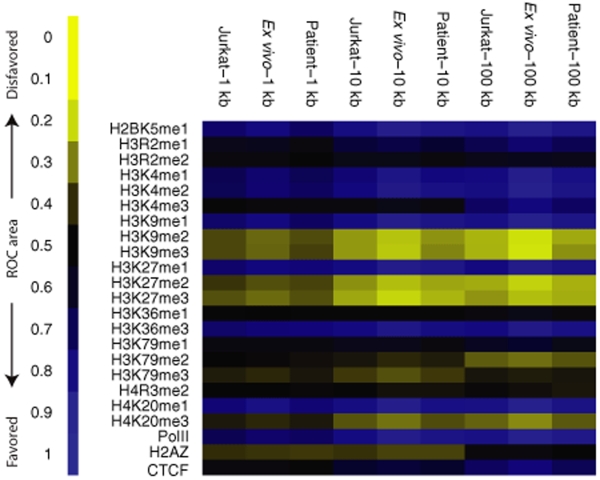
A previous study of the Jurkat data set suggested that lentiviral integration frequency correlated with several types of epigenetic marks in the human genome,18 so we asked whether significant differences could be detected among the three data sets. For this we used the data of Barski et al.,32 who used chromatin-immunoprecipitation and Solexa sequencing to map 23 types of histone modification or bound chromatin proteins genome-wide in human T cells. For each type of annotation, between ~1 and 16 million sequence tags were mapped. We investigated whether these epigenetic marks correlated with integration frequency, and again compared the ex vivo, patient derived, and Jurkat data sets. The direction of correlations and their strengths were quantified using receiver operating characteristic area methods as described in Berry et al.,14 allowing the observations to be summarized as heat maps.
Figure 4 shows the relationship of integration frequency to the density of epigenetic marks. Comparisons were carried out versus the matched random controls over chromosomal segments of three lengths (1, 10, and 100 kb) since Berry et al. found that some correlations between integration frequency and genomic annotation were dependent on the interval size studied. Many correlations were strongest over longer genomic intervals, potentially because the larger numbers of sequence tags in the larger intervals allow finer discrimination.
Figure 4.
Comparison of in vivo and ex vivo lentiviral vector integration sites to the integration sites from control infections of Jurkat cells—analysis of proximity to sites of histone methylation and bound DNA binding proteins. Values for each data set were compared to matched random controls. The direction and strength of each trend is quantified using the ROC area method described in ref. 14, the key to the left of the figure indicates the scale. Each row in the plot corresponds to a different form of histone post-translational modification or bound protein as scored in the Barski et al. “ChIP-seq ” data.32 Each column corresponds to an integration site data set. Comparisons were carried out over 1, 10, or 100 kb genomic intervals. ROC, receiver operating characteristic.
HIV provirus accumulation in all three data sets was positively correlated with histone methylation patterns characteristic of active transcription units. These included H2BK5me1, H3K4me1 and me2, H3K9me1, H3K27me1, H3K36me3, H4K20me1. RNA polymerase II was also positively associated. Provirus accumulation was generally negatively associated with marks linked to repression of transcription, including H3K9me2 and me3, H3K27me2 and me3, and H4K20me3. The H3K9me2 and me3 marks are associated with peri-centromeric heterochromatin, and centromeric heterochromatin has been reported to be negatively associated with integration frequency.11,28 Over shorter intervals the histone H2 variant H2AZ was negatively associated with integration, probably because it is found in promoter regions which are disfavored for HIV integration. Over longer intervals it is neither positively nor negatively associated, likely reflecting a balance between the above negative effects and enrichment due to favoring of integration in gene rich regions, which drive the associations in comparisons over longer genomic intervals. Thus integration sites were positively correlated with epigenetic marks associated with active transcription and negatively correlated with marks associated with transcriptional repression.
Provirus integration on nucleosome-bound DNA
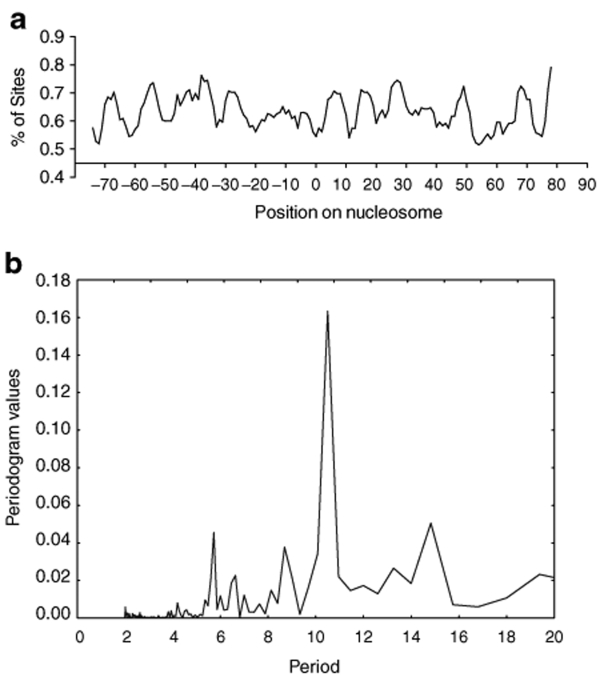
A previous study of HIV integration in the Jurkat T-cell line suggested that lentiviral integration occurs on nucleosome-bound DNA,18 and earlier studies suggested favored integration on nucleosome-bound episomes and in vitro.33,34,35,36,37 It was thus of interest to ask whether nucleosome-bound chromosomal DNA was also the integration target in primary CD4+ T cells in the ex vivo data set. We took advantage of the nucleosome prediction algorithm devised by Segal et al.38 to map histone positions in the 5 kb of genomic DNA sequence surrounding each integration site. This allowed us to map the position of each integration site relative to the center of symmetry of the nucleosomes bound to target DNA. Figure 5a shows a strong periodic pattern for integration frequency relative to the histone center of symmetry. Consistent with the previous report,18 alignment of the periodic pattern relative to the nucleosome axis of dyad symmetry indicated that integration was favored in the outward-facing major grooves of DNA bound on nucleosomes. A Fourier transformation analysis of this plot revealed a peak at ~10.5 bp (Figure 5b), matching the periodicity of the DNA helix. These data indicate that lentiviral integration favors the outwardly facing major grooves of nucleosome-bound DNA in human primary cells that are natural targets of HIV infection.
Figure 5.
Ex vivo lentiviral vector integration favors the major grooves of DNA bound on nucleosomes. Positions of lentiviral integration sites on nucleosomes were predicted using the nucleosome prediction algorithm developed by Segal et al. (a) The percentage of total ex vivo lentiviral vector integration sites at each base pair (y-axis) is plotted relative to the dyad axis of nucleosome symmetry (position 0; the scale is in base pairs). (b) Fourier transformation of the data from a, showing the ~10.5 bp periodicity of integration frequency.
Comparing integration site populations from ex vivo infection of cells from late stage versus stably suppressed HIV patients
In the initial test of the VRX496 vector,17 the subjects were late stage HIV patients who had failed multiple ART regimens. In the subjects from the multiple dose clinical trial studied here, the subjects had well suppressed HIV RNA viral load and were otherwise healthy. It was thus of interest to ask whether the differences in immune status of the cell donors in each trial resulted in alterations in the distribution of vector integration sites.
Integration site sequences for the ex vivo products from the two trials were compared over multiple measures of integration in transcription units, near CpG islands, within gene rich regions, near gene 5′-ends, G/C content, and Giemsa stained regions (data not shown). No statistically significant differences were detected. We conclude that the differences between the subjects in the two trials did not result in detectable differences in vector integration targeting.
Discussion
In summary, the analysis of VXR496 integration sites from cells transduced ex vivo or recovered from study subjects during 28–32 weeks after gene transfer yielded no evidence for the preferential survival or expansion of cells with integration sites near proto-oncogenes or tumor suppressors. However, some caution is warranted in interpreting integration site surveys in the context of gene therapy trials, because the integration site recovery methods using restriction enzymes are severely biased,19 though use of multiple restriction enzymes for each sample improves recovery somewhat. Thus, this study and all previous studies represent surveys of easily recoverable sites. Nevertheless, in the first study of lentiviral vector integration sites recovered from study subjects presented here, no adverse trends were detectable in the samples studied. This is in contrast, for example, to the recent chronic granulomatous disease study, where worrisome integration events were documented during early follow-up after infusion.21 It is not clear from this study alone whether lentiviral vectors are in fact safer than γ-retroviral vectors, or whether different modifications of the lentiviral vector such as introduction of strong internal promoters might result in detectable clonal skewing. We note that the chronic granulomatous disease study and SCID-X1 studies targeted CD34+ stem cells, although the study reported here targeted mature T cells—thus, it remains to be seen whether adverse trends become apparent after lentiviral gene therapy of stem cells. Nevertheless, our data showed no sign of preferential expansion of T cells with potentially adverse lentiviral integration sites.
This study presents the largest sample of lentiviral vector integration sites in a natural target cell for HIV infection yet reported, and the analysis provides a rich catalog of the genomic landmarks dictating HIV integration frequency. HIV integration was found to be strongly favored near a collection of epigenetic marks associated with active transcription, and disfavored near marks associated with transcriptional repression. Some of the patterns are readily interpretable. For example, the finding that RNA polymerase II is positively associated with HIV integration simply reflects the finding that integration is favored in active genes. The finding of favored integration near sites of H3K36 trimethylation likely reflects the association of this mark with transcription units of active genes. The large number of sites also allowed the demonstration that integration shows a periodic pattern relative to the underlying positions of nucleosomes, providing the first data that HIV-based lentiviral vectors favor integration on nucleosomal target DNA in primary T cells. Looking ahead, the availability of large integration site data sets, together with increasingly detailed genome-wide annotation, offers many new routes to understanding the mechanisms responsible for lentiviral vector integration targeting.
Materials and Methods
Clinical samples. PBMC from patient blood or apheresis samples were obtained under an IRB approved protocol at the University of Pennsylvania. The protocol (0407-667) was reviewed and approved by the NIH RAC/OBA. A detailed report on the clinical trial will be published elsewhere.
Integration site recovery, sequencing, and analysis. Integration sites were recovered and sequenced using the 454 pyrosequencing technology as described in refs. 18,19. Briefly, genomic DNA was extracted from ex vivo transduced CD4+ cells or PBMC derived from patients using the DNeasy tissue kit (Qiagen, Valencia, CA). For each genomic DNA sample, digestions with two different cocktails of restriction enzymes (AvrII/SpeI/NheI, or ApoI) were performed. The digested DNA samples were ligated to linkers, then amplified by nested PCR as previously described in refs. 18,19. To selectively amplify host–vector DNA junctions resulting from vector integration (rather than provirus integration from the circulating HIV), a first round PCR primer specific for the engineered short unique sequence (GTAG) in the VRX496 vector was used. Each second round lentiviral specific primer contains a unique 8 nt barcode which indexes the amplification products (Figure 1). The primers used for the nested PCR were as in ref. 18. The PCR products were gel purified, pooled, and sequenced using the 454 pyrosequencing platform. Integration sites were determined to be authentic if the sequences began within 3 bp of vector LTR ends, had a >98% sequence match to the human genome, and had a unique best hit when aligned to the draft human genome (hg18) using BLAT. All integration site sequences are available in GenBank.
Bioinformatic analysis. A 20-bp target DNA sequence surrounding each integration site was extracted from the draft human genome (hg18), and aligned using WebLogo (http://weblogo.Berkeley.edu/logo.cgi). Detailed bioinformatic methods for analysis of association with chromosomal features are described in Berry et al.14 For analysis of association with epigenetic modifications and bound chromatin proteins, the data of Barski et al.32 were used. The methods for generating heat maps based in receiver operating characteristic curves are as described in ref. 14. The placement of nucleosomes on chromosomal regions hosting vector integration events was determined using the nucleosome positioning prediction tool developed by Segal et al., which was available at http://genie.weizmann.ac.il/pubs/nucleosomes06/index.html. For this analysis, 5 kb of human DNA sequence surrounding each integration site was extracted and analyzed. The positions of integration sites on nucleosomes were smoothed using a 5-bp moving window. The periodicity of integration frequency relative to nucleosome positioning was determined by Fourier transformation analysis using Statistica (StatSoft, Tulsa, OK).
Acknowledgments
We are grateful to members of the Bushman laboratory for help and suggestions. This work was supported by NIH grants AI52845 and AI66290 and an ACTG grant to the University of Pennsylvania. G.P.W. was supported by NIH NIAID T32 AI07634 (Training Grant in Infectious Diseases) and the University of Pennsylvania SOM Department of Medicine Measey Basic Science Fellowship Award.
References
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods NB, Bottero V, Schmidt M, von Kalle C., and , Verma IM. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440:1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- Thrasher AJ, Gaspar HB, Baum C, Modlich U, Schambach A, Candotti F, et al. Gene therapy: X-SCID transgene leukaemogenicity Nature 2006443E5, E6–6.discussion [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR., and , Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR., and , Bushman FD. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Mol Ther. 2006;14:218–225. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J., and , Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and , Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- De Palma M, Montini E, Santoni de Sio FR, Benedicenti F, Gentile A, Medico E, et al. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoetic cells. Blood. 2005;105:2307–2315. doi: 10.1182/blood-2004-03-0798. [DOI] [PubMed] [Google Scholar]
- Levine BL, Humeau LM, Boyer J, MacGregor RR, Rebello T, Lu X, et al. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc Natl Acad Sci USA. 2006;103:17372–17377. doi: 10.1073/pnas.0608138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC., and , Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Yu Q, Binder GK, Chen Z, Slepushkina T, Rossi J, et al. Antisense-mediated inhibition of human immunodeficiency virus (HIV) replication by use of an HIV type 1-based vector results in severely attenuated mutants incapable of developing resistance. J Virol. 2004;78:7079–7088. doi: 10.1128/JVI.78.13.7079-7088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Hoffmann C, Ronen K, Malani N, Minkah N, Rose HM, et al. Massively parallel pyrosequencing in HIV research. AIDS. 2008;22:1411–1415. doi: 10.1097/QAD.0b013e3282fc972e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ., and , Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SW., and , Griffith JD. Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. J Virol. 1996;70:6459–6462. doi: 10.1128/jvi.70.9.6459-6462.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S, Hoffmann C., and , Bushman FD. Chromosome structure and HIV-1 cDNA integration: centromeric alphoid repeats are a disfavored target. J Virol. 1998;72:4005–4014. doi: 10.1128/jvi.72.5.4005-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman AG., and , Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci USA. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM., and , Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffi A, Mitchell RS, Hoffmann C, Leipzig J, Shinn P, Ecker JR, et al. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol Ther. 2006;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Pryciak PM., and , Varmus HE. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- Pryciak PM, Sil A., and , Varmus HE. Retroviral integration into minichromosomes in vitro. EMBO J. 1992;11:291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak PM, Müller HP., and , Varmus HE. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc Natl Acad Sci USA. 1992;89:9237–9241. doi: 10.1073/pnas.89.19.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss D, Bushman FD., and , Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss D, Reeves R, Bushman FD., and , Wolffe AP. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J Biol Chem. 1994;269:25031–25041. [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]