Abstract
Human extracellular superoxide dismutase (EC-SOD; EC 1.15.1.1) is a scavenger of superoxide anions in the extracellular space. The amino acid sequence is homologous to the intracellular counterpart, Cu/Zn superoxide dismutase (Cu/Zn-SOD), apart from N- and C-terminal extensions. Cu/Zn-SOD is a homodimer containing four cysteine residues within each subunit, and EC-SOD is a tetramer composed of two disulfide-bonded dimers in which each subunit contains six cysteines. The amino acid sequences of all EC-SOD subunits are identical. It is known that Cys-219 is involved in an interchain disulfide. To account for the remaining five cysteine residues we purified human EC-SOD and determined the disulfide bridge pattern. The results show that human EC-SOD exists in two forms, each with a unique disulfide bridge pattern. One form (active EC-SOD) is enzymatically active and contains a disulfide bridge pattern similar to Cu/Zn-SOD. The other form (inactive EC-SOD) has a different disulfide bridge pattern and is enzymatically inactive. The EC-SOD polypeptide chain apparently folds in two different ways, most likely resulting in different three-dimensional structures. Our study shows that one gene may produce proteins with different disulfide bridge arrangements and, thus, by definition, different primary structures. This observation adds another dimension to the functional annotation of the proteome.
Two isoforms of Cu/Zn-containing superoxide dismutase (SOD) enzymes exist in mammals (1, 2). Cu/Zn-SOD is found in the intracellular space, and extracellular SOD (EC-SOD) predominantly is found in the extracellular matrix of most tissues (3). Both enzymes dismutate the superoxide anion into hydrogen peroxide and oxygen with diffusion-limited rate constants (>109 M–1 sec–1), and both are inhibited by cyanide and azide (4, 5). Human Cu/Zn-SOD is a homodimer with a molecular mass of 32 kDa, and human EC-SOD is a tetramer of ≈135 kDa (4). The subunit of each isoform contains one Cu(II) and one Zn(II) atom. The central region of EC-SOD (His-96 to Gly-193) is homologous to human Cu/Zn-SOD and contains all of the ligands essential for the coordination of the active site Cu(II) and Zn(II) ions (6, 7). The N-terminal region of EC-SOD is important for the formation of tetramers (8–10), and the C-terminal region (Val-194–Ala-222) encompasses a heparin-binding region, which is responsible for the immobilization of EC-SOD in the extracellular matrix (11, 12). The heparin-binding region of EC-SOD can be removed by an intracellular proteolytic event before secretion (13, 14). Consequently, EC-SOD tetramers with no (type A), intermediate (type B), or high (type C) affinity for the extracellular matrix are produced (11, 12). These tetramers are composed of cleaved polypeptides (type A), intact polypeptides (type C), or a mixture (type B). In addition, the C-terminal region supports the formation of disulfide-linked dimers via Cys-219 (15, 16).
The x-ray structure of Cu/Zn-SOD shows that the active-site channel is maintained by an intrapolypeptide disulfide bond between two highly conserved cysteines (6). This disulfide bond is essential for enzymatic activity, and the analogous cysteines are found at positions 107 and 189 in human EC-SOD (7). The conservation of these cysteine residues and other residues important for the active-site geometry, as well as a similar sensitivity to inhibitors, suggest that the central region of EC-SOD has similar structural characteristics. Despite numerous efforts, the crystal structure of human EC-SOD has not been determined. The lack of suitable crystals could be due to heterogeneous posttranslational modifications (17) or other properties of human EC-SOD distinct from Cu/Zn-SOD.
In this study we demonstrate that the human EC-SOD polypeptide folds in two distinct ways with different disulfide bridge patterns. This imposes structural differences between the two folded polypeptides, as demonstrated by the absence of SOD activity in one form. Our data show that the cellular machinery of protein folding may produce two different structures from polypeptides with identical amino acid sequence.
Materials and Methods
Purification of EC-SOD from Human Aorta. EC-SOD was purified from human aorta as described (15), except that the cation exchange chromatography step was omitted.
Polyacrylamide Gel Electrophoresis and Western Blotting. Proteins were separated by SDS/PAGE in 5–15% polyacrylamide gels (18). Proteins analyzed under nonreducing conditions were boiled in the presence of iodoacetamide (IAA) before loading. For Western blotting, proteins were electrophoretically transferred to a polyvinylidene difluroride membrane in 10 mM 3-(cyclohexylamino)-1-propane sulfonic acid/10% methanol, pH 11 (19). The membrane was blocked with 5% (wt/vol) skimmed milk in 20 mM Tris-HCl/150 mM NaCl, pH 7.4. EC-SOD was subsequently detected by using a rabbit anti-human EC-SOD antiserum and peroxidase-conjugated goat anti-rabbit Ig (Sigma).
Separation of Monomeric and Dimeric EC-SOD. The monomers and disulfide-linked dimers of EC-SOD were separated by reversed-phase HPLC. Approximately 100 μg of purified EC-SOD in Tris-HCl and NaCl was acidified by the addition of trifluoroacetic acid (TFA) and applied to an Aquapore RP-300 C8 reversed-phase HPLC column (2.1 mm × 220 mm; Brownlee Lab). Bound proteins were eluted by a segmented linear gradient increasing the concentration of solvent B (90% acetonitrile/0.08% TFA) in solvent A (0.1% TFA) from 0% to 30% in 5 min (6% B min–1) followed by 0.5% B min–1 from 30% to 60%. The column was operated at 23°C at a flow rate of 200 μl min–1. Protein was detected at 220 and 280 nm, and fractions were collected manually.
Alkylation and Separation of EC-SOD Monomers. The collected fraction containing monomeric EC-SOD was lyophilized and redissolved in 30 mM Hepes, pH 8.3, containing 5 M guanidinium hydrochloride and 25 mM IAA. The reaction was performed at 23°C for 30 min. The material was subsequently acidified by the addition of TFA, and the alkylated monomers were separated by reversed-phase HPLC as described in Separation of Monomeric and Dimeric EC-SOD.
Reconstitution of EC-SOD Activity. Alkylated and separated EC-SOD monomers were sequentially dialyzed against 50 mM acetate/10 mM EDTA, pH 3.8; 50 mM acetate/100 mM NaCl, pH 3.8; 100 mM acetate, pH 3.8; 100 mM acetate, pH 5.5; 100 mM acetate/50 μM CuSO4/20 μM ZnSO4, pH 5.5; and 10 mM Tris-HCl, pH 7.4 as described (20, 21).
SOD Activity Stain. Samples were separated by SDS/PAGE as described above, omitting boiling before analysis. EC-SOD retains activity in SDS/PAGE loading buffer if not heated. The gel was then equilibrated in 20 mM Tris-HCl, pH 7.4, three times for 10 min each, and SOD activity was detected with nitroblue tetrazolium, riboflavin, and N,N,N′,N′-tetramethylethylenediamine as described (15). The activity was also determined in fluid phase by using the xanthine oxidase/cytochrome c assay (20). The protein concentration used to calculate specific activity was estimated by SDS/PAGE and Coomassie blue staining.
Peptide Map of Separated EC-SOD Monomers. Collected fractions from reversed-phase HPLC (≈300 μl) were added to 130 μl of 0.5 M Hepes, pH 8.3, and 2.5 μg of porcine trypsin (Promega) and placed at 37°C overnight. The digested material was concentrated to 100 μl with a SpeedVac centrifuge (Sorvall, New-town, CT) and acidified by the addition of TFA. The tryptic peptides were separated by reversed-phase HPLC as described by using an Aquapore RP300 column and a 60-min linear gradient of 1% B min–1. Collected peptides destined for N-terminal amino acid analyses were applied to a Biobrene pre-cycled glass-fiber filter (Applied Biosystems) and subjected to automated Edman degradation as described (15).
Mass Spectrometric Analysis of Tryptic Peptides. Peptides were analyzed by matrix-assisted laser desorption/ionization (MALDI)-MS by using a quadrupole/time-of-flight (Q-TOF) Ultima Global mass spectrometer (Micromass, Manchester, U.K.) with alpha-cyano-4-hydroxycinnamic acid (Sigma) as the matrix. Selected peptides were also analyzed by nanoelectrospray MS by using a Q-TOF mass spectrometer (Micromass). Before analysis, peptides were purified by using a Poros 50 R2 (PerSeptive Biosystems, Framingham, MA) micropurification column (22). The expected mass of peptides and proteins was calculated by using general protein/mass analysis for windows software (http://welcome.to/gpmaw).
Limited Reduction of EC-SOD Double Peptide. Aliquots (20 μl) of the fraction containing the EC-SOD double peptide were lyophilized and rehydrated in 50 mM Hepes, pH 8.3, containing 0, 0.01, or 0.1 mM DTT. The tubes were placed at 23°C in the dark for 30 min to allow for partial reduction of disulfide bonds. The material was then added to 5 μl of 100 mM IAA in 50 mM Hepes, pH 8.3, to alkylate free sulfhydryl groups. The peptide of interest was subsequently analyzed by electrospray ionization (ESI) tandem MS to identify the alkylated cysteine residue.
Results
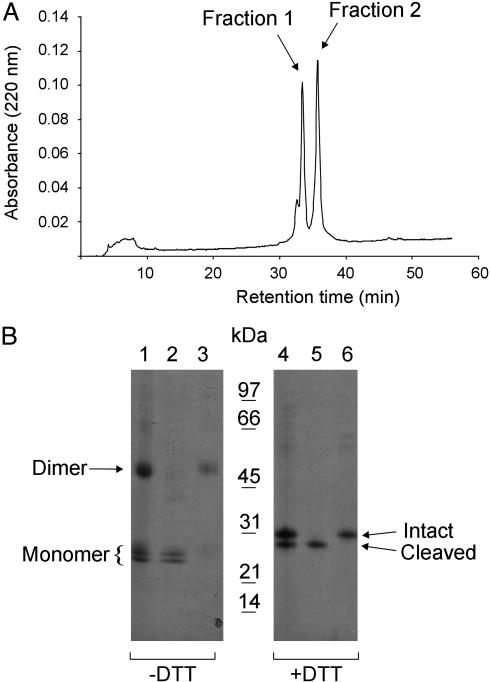
Separation of Human EC-SOD Monomer and Dimer. Reversed-phase HPLC analysis of purified EC-SOD (Fig. 1B, lanes 1 and 4) resulted in two distinct peaks (Fig. 1 A). Nonreducing SDS/PAGE analysis revealed that the material collected in fraction 1 migrated as a closely spaced doublet, corresponding to the size of a monomer (Fig. 1B, lane 2). Fraction 2 contained the disulfide-linked EC-SOD dimer (Fig. 1B, lane 3). When fraction 1 was analyzed in the reduced state, a single band of 28 kDa was detected (Fig. 1B, lane 5). This band represents EC-SOD lacking the C-terminal heparin-binding domain. The material in fraction 2 with a mass of 31 kDa corresponded to the intact EC-SOD (Fig. 1B, lane 6). These analyses show that reversed-phase HPLC may be used to separate the EC-SOD monomer from the disulfide-linked dimer.
Fig. 1.
Separation of monomeric and dimeric human EC-SOD. (A) Purified EC-SOD was subjected to reversed-phase HPLC by using a C8 column and a shallow gradient of acetonitrile in TFA (0.5% solvent B min–1). The absorption profile at 220 nm is shown, with the two collected peaks indicated. (B) The separated material was analyzed by SDS/PAGE and Coomassie blue staining under nonreducing (–DTT) and reducing (+DTT) conditions as indicated. Lanes 1 and 4 represent purified EC-SOD used for the reversed-phase HPLC separation. Lanes 2 and 5 and lanes 3 and 6 represent the material collected in fractions 1 and 2, respectively. The positions of the disulfide-linked dimer and the monomer (Left) and the intact and cleaved forms under nonreducing conditions (Right) are indicated. A molecular mass marker is shown in the center. The analysis shows that EC-SOD can be separated into monomer and dimer by reversed-phase HPLC.
Separation of Monomeric EC-SOD into Two Distinct Forms. The reversed-phase HPLC isolated monomeric EC-SOD (Fig. 1 A, fraction 1) was recovered in 0.1% TFA and acetonitrile. Under these conditions the pH is ≈1 and EC-SOD is likely to be somewhat unfolded. When the pH was raised to 8.3 we observed some dimerization due to disulfide bridge exchange caused by free cysteine residues. This dimerization was completely eliminated by the addition of IAA before the increase of the pH. In general, thorough alkylation using IAA was used throughout this study to eliminate unwanted disulfide exchange (23).
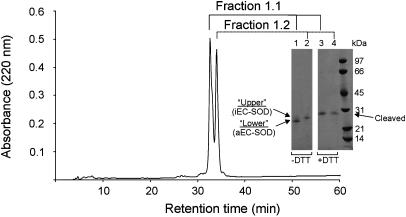
After alkylation of the monomeric EC-SOD, the sample separated into two peaks when analyzed by reversed-phase HPLC by using the same protocol as described above (Fig. 2). Nonreducing SDS/PAGE analysis showed that fraction 1.1 contained the lower band of the doublet (Fig. 2, lane 1) and fraction 1.2 contained the upper band (lane 2). Under reducing conditions each fraction ran as a 28-kDa band corresponding to cleaved monomeric EC-SOD (Fig. 2, lanes 3 and 4). The observed difference in migration under nonreducing conditions could be caused by disulfide-dependent variations in shape. This is emphasized by the finding that the two forms migrate the same in SDS/PAGE after reduction.
Fig. 2.
Separation of EC-SOD monomers. The material collected in fraction 1 (Fig. 1 A) was alkylated with IAA and rechromatographed by reversed-phase HPLC by using the same conditions as in Fig. 1. The alkylated material separated into two peaks, as detected by absorption at 220 nm. Collected fractions were denoted 1.1 and 1.2 as shown. The separated material was analyzed by SDS/PAGE and Coomassie blue staining under nonreducing (–DTT) and reducing (+DTT) conditions (Inset). Lanes 1 and 3 and lanes 2 and 4 contain material collected in fractions 1.1 and 1.2, respectively. The two closely spaced bands under nonreducing conditions are denoted upper and lower band, as indicated. An arrow indicates the position of cleaved EC-SOD under reducing conditions. A molecular mass marker is included on the right.
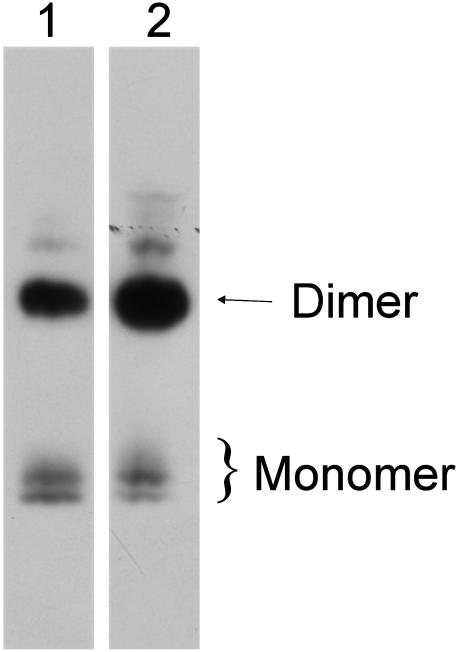
Both Forms of EC-SOD Are Present in Freshly Homogenized Tissue. To investigate whether both forms of EC-SOD were present in tissue, we analyzed freshly homogenized human aorta by nonreducing SDS/PAGE and Western blotting (Fig. 3). Both purified EC-SOD (Fig. 3, lane 1) and aorta homogenate (lane 2) contained the doublet of EC-SOD monomers and the dimer. We conclude that the two monomeric EC-SOD species are present in the tissue and that the upper and lower bands are not generated during protein purification.
Fig. 3.
Both EC-SOD monomers are present in human aorta. Purified EC-SOD (lane 1) and human aorta homogenate (lane 2) were analyzed by nonreducing SDS/PAGE and Western blotting. The analysis shows that the two bands corresponding to monomeric EC-SOD are detected both in purified EC-SOD (lane 1) and in the aorta homogenate (lane 2). The two distinct bands are thus present in tissue and are not a result of manipulation during purification.
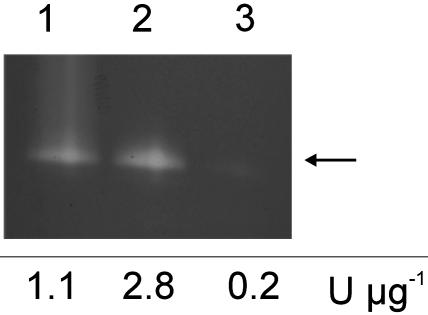
SOD Activity of the Two EC-SOD Forms. The reversed-phase HPLC-separated EC-SOD monomers (Fig. 2) were reconstituted with Cu(II) and Zn(II) by sequential dialysis (20, 21). The activity of the material was subsequently analyzed by SDS/PAGE followed by staining for SOD activity (Fig. 4). Because the samples were not boiled before SDS/PAGE, both the upper and lower form of monomeric EC-SOD migrated at the same position, as indicated by an arrow. The analysis of the recharged forms showed that the lower band was active (Fig. 4, lane 2) and the upper band displayed no activity (lane 3). In addition, the activity of the recharged material was estimated by a spectrophotometric assay. The SOD activity of the lower band was >2-fold higher when compared with equal amounts of purified EC-SOD (Fig. 4). The upper band showed no activity. Because the activity of purified EC-SOD was approximately half of that of purified active EC-SOD (aEC-SOD), the population of EC-SOD molecules is likely composed of equal molar amounts of active and inactive monomers. We propose to annotate the lower band aEC-SOD and the upper band inactive EC-SOD (iEC-SOD).
Fig. 4.
Activity stain of separated EC-SOD monomers. The EC-SOD monomers were reconstituted with Cu(II) and Zn(II) and separated by SDS/PAGE. Three micrograms of purified EC-SOD was loaded in lane 1. Approximately 1 μg of recharged monomeric EC-SOD collected in fractions 1.1 and 1.2 (Fig. 2 A) was loaded in lanes 2 and 3, respectively. The arrow indicates the position of both monomeric forms on SDS/PAGE analysis of unboiled EC-SOD. No SOD activity can be detected in lane 3. The specific activity determined by the xanthine oxidase/cytochrome c assay is shown below the gel.
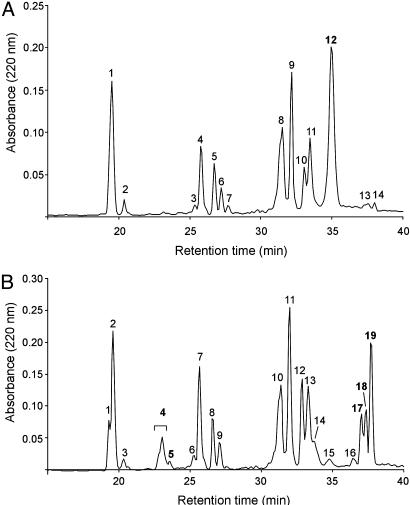
The Unreduced Peptide Maps of aEC-SOD and iEC-SOD Are Not Identical. To investigate the structural basis for the difference in SOD activity, we generated tryptic peptides of both forms. The peptides were separated by reversed-phase HPLC, and a comparison of the traces revealed several differences (Fig. 5). The peptide map of aEC-SOD (Fig. 5A) contained a major peak eluting after 35 min. This peak was absent in the iEC-SOD trace, and several other distinct peaks with retention times of 23, 33, and 37–39 min were observed (Fig. 5B). After reduction of aEC-SOD and iEC-SOD, the tryptic peptide maps were identical (data not shown). These data are consistent with differences in the disulfide pattern of aEC-SOD and iEC-SOD (see below).
Fig. 5.
Tryptic peptide map of EC-SOD monomers. EC-SOD collected in fractions 1.1 and 1.2 (Fig. 2) were digested with trypsin. The generated peptides were separated by reversed-phase HPLC, and the 220-nm traces of aEC-SOD (fraction 1.1) (A) and iEC-SOD (fraction 1.2) (B) are shown. All collected peptides were analyzed by Edman degradation and MALDI-MS. The represented differences between the traces are numbered in bold and are presented in Table 1.
Disulfide Bonding of Separated Tryptic Peptides. The distribution of Lys and Arg in EC-SOD generates three cysteine-containing peptides. One of these peptides (Leu-187–Arg-202) contains the three cysteine residues at positions 189, 190, and 195 (C189C190VVGVC195). Moreover, the tryptic peptide containing Cys-45 includes a Thr/Ala polymorphism at position 40 (24). This generates two Cys-45 containing peptides with a 30-Da mass difference (Table 1). The polymorphism was not linked to the disulfide isoforms. Fractions collected from the reversed-phase HPLC separation (Fig. 5) were analyzed by Edman degradation, MALDI, or ESI-MS (Table 1).
Table 1. Detection of Cys-containing EC-SOD peptides in fractions from tryptic digest.
| Fraction no. | Cys45* | Cys107 | Cys189/190/195 | Unreduced mass (m/z)† |
|---|---|---|---|---|
| aEC-SOD | ||||
| F12 | Arg-35—Arg-59 | Ala-94—Arg-134 | Leu-187—Arg-202 (+IAA)‡ | 8,789.94 (8,789.73) |
| Arg-186—Arg-202 (+IAA) | 8,945.89 (8,945.92) | |||
| iEC-SOD | ||||
| F4 | Asp-36—Arg-59 (+IAA) | N.D. | N.D. | 2,537.04 (2,537.17) |
| Arg-35—Arg-59 (+IAA) | 2,693.12 (2,693.27) | |||
| F5 | Asp-36—Arg-59 (+IAA) | N.D. | N.D. | 2,507.02 (2,507.16) |
| Arg-35—Arg-59 (+IAA) | 2,663.09 (2,663.26) | |||
| F17 | N.D. | Ala-94—Arg-134 | Arg-186—Arg-202 | 6,251.57 (6,252.08) |
| Leu-187—Arg-202 | 6,095.70 (6,095.89) | |||
| F18 | N.D. | Ala-94—Arg-134 | Arg-186—Arg-202 | 6,251.55 (6,252.08) |
| F19 | N.D. | Ala-94—Arg-134 | Leu-187—Arg-202 | 6,095.19 (6,095.89) |
Fractions indicated in Fig. 5 were reduced and analyzed by MALDI-MS. The detected peptides containing the indicated Cys residues are listed. Incomplete digestion by trypsin is indicated in italics. N.D., Not detected.
The tryptic peptide containing Cys-45 encompasses a Thr/Ala polymorphism at position 40.
The mass of unreduced material was determined by ESI-MS. Calculated mass is given in parentheses.
Mass increase due to alkylation (+57 Da) is indicated by +IAA in parentheses.
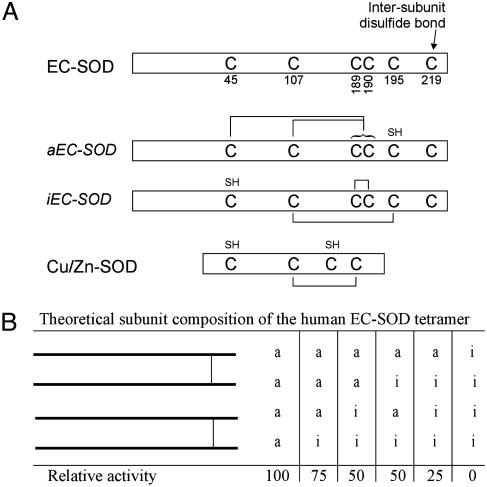
Disulfide Bridges of aEC-SOD. Fraction 12 includes all cysteine-containing peptides (Table 1). The mass spectrometric analysis indicated that the peptides eluting in this fraction were connected by disulfide bridges (only the Thr-40 variant is shown in Table 1). In addition, the data indicated that one cysteine residue of the Cys-189/190/195-containing peptide was alkylated. To identify this cysteine, the peptide was reduced with DTT without subsequent alkylation and analyzed by ESI tandem MS (data not shown). The analysis demonstrated that Cys-195 was alkylated. Consequently, Cys-189 or Cys-190 is disulfide-linked to Cys-45. Cys-107 is disulfide-linked to Cys-189 or Cys-190 (whichever one is not in linkage to Cys-45). Because of the vicinal position of Cys-189/190, we were not able to assign these specifically to Cys-45 or Cys-107. However, a sequence alignment of EC-SOD and Cu/Zn-SOD predicts that a Cys-107–Cys-189 disulfide exists. In addition, this disulfide is essential for enzymatic activity (7). Taken together, these data show that Cys-195 is free in native aEC-SOD whereas the remaining Cys-189/190 are connected to Cys-45 or Cys-107 (see Fig. 6A).
Fig. 6.
Schematic showing the different EC-SOD forms. (A) Human EC-SOD (P08294) depicted with cysteine residues indicated (top). The two different forms of EC-SOD are shown in the middle with the determined cysteine connectivity indicated. Underivatized cysteine residues of the native protein are indicated by SH. Human Cu/Zn-SOD (P00441) is aligned at the bottom. (B) The theoretical composition of EC-SOD tetramers and the relative SOD activity of these forms. Two intact disulfide-linked dimers are shown.
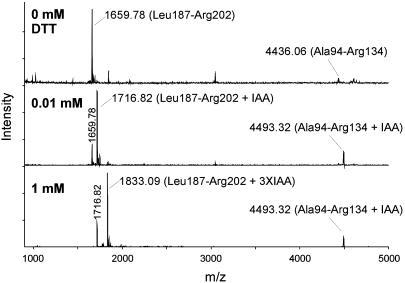
Disulfide Bridges of iEC-SOD. The peptide containing Cys-45 eluted in fractions 4 and 5 (Table 1). Due to a slight difference in hydrophobicity caused by the Thr/Ala polymorphism, the peptides eluted in two fractions. The cysteine residue was alkylated in both fractions, indicating that Cys-45 is free in native iEC-SOD. This was supported by the identical mass of reduced (data not shown) and unreduced (Table 1) material. The peptides containing Cys-107 and Cys-189/190/195 eluted in fractions 17–19. The three peaks were seen repeatedly in independent iEC-SOD digests and were in part caused by incomplete digestion of the Arg-185–Arg-186–Leu-187 peptide bonds. The mass of the unreduced material indicated that the two peptides were connected by a disulfide (Table 1). In addition, the data suggest that the Cys-189/190/195-containing peptide does not contain any free sulfhydryl groups due to the absence of carbamidomethyl cysteine. Because one of the three cysteines of this peptide forms an interpeptide disulfide bond with Cys-107, the remaining two cysteines must form an intrapeptide disulfide bond. To identify the cysteine involved in the interpeptide disulfide bond, we subjected the peptide to limited reduction (fraction 19 was used for this analysis). Generated sulfhydryl groups were subsequently alkylated by the addition of IAA, and the material was analyzed by MALDI-MS (Fig. 7). In the absence of a reducing agent, the two individual peptides could be detected (Fig. 7 Top). This is caused by dissociation of the disulfide bond during the MALDI-MS analysis (25). When the peptide was reduced by 0.01 mM DTT, masses corresponding to the singly alkylated Leu-187–Arg-202 peptide (1,716.82 Da) and the alkylated Ala-94–Arg-134 peptide (4,493.32 Da) were detected (Fig. 7 Middle). Apparently, the interpeptide disulfide bridge was reduced by 0.01 mM DTT. However, this concentration was not sufficient to reduce the intrapeptide disulfide bridge, because only one carbamidomethyl cysteine in the Leu-187–Arg-202 peptide was generated. If the peptide was reduced by 1 mM DTT, the triply alkylated Leu-187–Arg-202 peptide (1,833.09 Da) was detected, indicating that all disulfide bridges had been reduced (Fig. 7 Bottom). These data suggest that the interpeptide disulfide is particularly sensitive to reduction. This allowed us to distinguish between the intra- and interdisulfide bonds by limited reduction and alkylation. To assign the interpeptide disulfide bridge, we subjected the partially reduced and alkylated Leu-187–Arg-202 peptide (1,716.82) (Fig. 7 Middle) to ESI tandem MS. The analysis of the data indicated that the alkylated cysteine was found at position 195 (data not shown). The two peptides are thus connected by a disulfide bridge between Cys-107 and Cys-195, and the vicinal Cys-189/190 forms the intrapeptide disulfide bond (see Fig. 6A). The disulfide pattern of aEC-SOD is likely to force a more constrained structure of EC-SOD under denaturing conditions, because the N- and C-terminal regions are joined by the Cys-45–Cys-189/190 disulfide. This could explain the difference in migration of aEC-SOD and iEC-SOD when analyzed by nonreducing SDS/PAGE (Fig. 2).
Fig. 7.
Limited reduction of EC-SOD double peptide. To determine the location of the interpeptide disulfide bond between the two connected peptides detected in iEC-SOD F19 (Table 1), we subjected the double peptide to limited reduction. The peptides were analyzed by MALDI-MS. Without reduction (0 mM DTT; Top), the two connected peptides can be detected individually. This is due to disruption of the disulfide bridge during MALDI analysis. The interpeptide disulfide bond can be reduced by 0.01 mM DTT (Middle). The cysteines involved in this bond are alkylated (+IAA). When fully reduced (1 mM DTT, Bottom), all cysteines of the Leu-187–Arg-202 peptide are alkylated (plus 3× IAA). The singly alkylated Leu-187–Arg-202 generated by 0.01 mM DTT was subjected to ESI tandem MS for further analysis.
Discussion
We observed that, when purified human EC-SOD was analyzed by nonreducing SDS/PAGE, monomeric EC-SOD migrated as a closely spaced doublet. After reduction of disulfides, the two forms had the same apparent mass. This led us to speculate that the difference in migration under nonreducing conditions was caused by variations in the disulfide bridge pattern. We were aware of possible artifacts caused by disulfide exchange at neutral pH (23). However, the two bands were similarly present in freshly homogenized human aorta. We used three different preparations of EC-SOD purified from 10–20 pooled aortas and observed the same distribution of aEC-SOD and iEC-SOD. In addition, if the difference in disulfide bridge patterns were the result of disulfide exchange, we would have expected to see several forms and not just two. We also note that the rearrangement of the disulfide bonds of iEC-SOD to aEC-SOD or vice versa requires the breakage and reformation of two disulfides (see Fig. 6A). These results substantiate the conclusion that the two forms are not generated by disulfide scrambling during protein purification.
Limited reduction established that iEC-SOD contained a disulfide between the two vicinal cysteines (see Fig. 6A). This linkage is unusual but has been reported in both prokaryotic and eukaryotic proteins (26–28). Vicinal cysteine disulfides have been implicated in electron transfer or function as regulatory switches (29, 30). Whether this structure in iEC-SOD has any biological role is not known at present. EC-SOD from mouse (31), rat (32), and rabbit (33) contains only five cysteines lacking the cysteine homologous to Cys-195 in human EC-SOD. This fact prevents these animals from establishing the Cys-107–Cys-195 disulfide to produce iEC-SOD (Fig. 6A). Therefore, it is likely that only aEC-SOD exists in these animals. Consequently, the putative biological role of iEC-SOD in humans is absent in mouse, rat, and rabbit, a fact that will have to be considered when using these animals for in vivo EC-SOD studies.
Our data suggest that aEC-SOD and iEC-SOD are present in equimolar ratios, indicating that the generation of these two forms is a regulated event. Interestingly, structural data show that Cu/Zn-SOD forms a heterodimeric intermediate with its copper chaperone (CCS) (34). During complex formation a transient intersubunit disulfide bond between Cys-229 of CCS and Cys-57 of Cu/Zn-SOD is formed. Because Cys-57 participates in the disulfide bond essential for enzymatic activity, this disulfide must be reestablished after copper insertion and dissociation of the Cu/Zn-SOD–CCS complex. This implies that a series of redox processes takes place. A similar interaction between human EC-SOD and (metallo)chaperones may be responsible for the generation of transient intersubunit disulfides and the formation of aEC-SOD or iEC-SOD. Likewise, redox-regulated chaperone activity could regulate the ratio of aEC-SOD and iEC-SOD to shift or “fine-tune” the extracellular antioxidant level, e.g., during episodes of oxidative stress. One example of such a redox-regulated chaperone is Hsp33, which is active only under oxidizing conditions (35, 36). However, it is also possible that the formation of iEC-SOD and aEC-SOD is a regulated extracellular event.
If the assembly of the EC-SOD tetramer during the biosynthesis is a random event, six possible combinations of tetramers exist: aa–aa, aa–ai, aa–ii, ai–ai, ai–ii, and ii–ii (Fig. 6B). These tetramers will express decreasing levels of SOD activity depending on how many iEC-SOD monomers the EC-SOD molecule contains. It is theoretically possible to produce EC-SOD with no SOD activity at all. The existence of EC-SOD tetramers with different levels of enzymatic activity is likely to have profound implications on the biological functions of the protein.
The thermodynamic hypothesis of protein folding implies that a given protein adopts its three-dimensional shape as a result of peptide bond constraints and chemical and physical properties of the amino acid residues. Furthermore, molecular chaperones and other proteins assist the folding of proteins in vivo (for reviews, see refs. 37 and 38). The data presented in this study suggest that this intracellular folding machinery may be capable of folding polypeptide chains with the same sequence to produce proteins with distinct disulfide bridge patterns and enzymatic activity. However, our data do not exclude the possibility that the formation of aEC-SOD and iEC-SOD is an extracellular event. Nonetheless, this finding implies that the number of products from the human genome may be higher than anticipated (39).
Acknowledgments
This work was supported by the Carlsberg Foundation (S.V.P.), the Danish Research Council (J.J.E. and P.H.), and National Institutes of Health Grants R01 HL63700 (to T.D.O.), P01 HL31992E (to J.D.C.), and RO1 EY12712 (to J.J.E.).
Abbreviations: SOD, superoxide dismutase; EC-SOD, extracellular SOD; aEC-SOD, active EC-SOD; iEC-SOD, inactive EC-SOD; IAA, iodoacetamide; TFA, trifluoroacetic acid; ESI, electrospray ionization; MALDI, matrix-assisted laser desorption/ionization.
References
- 1.Fridovich, I. (1995) Annu. Rev. Biochem. 64, 97–112. [DOI] [PubMed] [Google Scholar]
- 2.Zelko, I. N., Mariani, T. J. & Folz, R. J. (2002) Free. Radical Biol. Med. 33, 337–349. [DOI] [PubMed] [Google Scholar]
- 3.Fattman, C. L., Schaefer, L. M. & Oury, T. D. (2003) Free. Radical Biol. Med. 35, 236–256. [DOI] [PubMed] [Google Scholar]
- 4.Marklund, S. L. (1982) Proc. Natl. Acad. Sci. USA 79, 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marklund, S. L. (1984) Biochem. J. 220, 269–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tainer, J. A., Getzoff, E. D., Beem, K. M., Richardson, J. S. & Richardson, D. C. (1982) J. Mol. Biol. 160, 181–217. [DOI] [PubMed] [Google Scholar]
- 7.Hjalmarsson, K., Marklund, S. L., Engstrom, A. & Edlund, T. (1987) Proc. Natl. Acad. Sci. USA 84, 6340–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlsson, L. M., Marklund, S. L. & Edlund, T. (1996) Proc. Natl. Acad. Sci. USA 93, 5219–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tibell, L. A., Skarfstad, E. & Jonsson, B. H. (1996) Biochim. Biophys. Acta 1292, 47–52. [DOI] [PubMed] [Google Scholar]
- 10.Stenlund, P., Andersson, D. & Tibell, L. A. (1997) Protein Sci. 6, 2350–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adachi, T., Kodera, T., Ohta, H., Hayashi, K. & Hirano, K. (1992) Arch. Biochem. Biophys. 297, 155–161. [DOI] [PubMed] [Google Scholar]
- 12.Sandstrom, J., Carlsson, L., Marklund, S. L. & Edlund, T. (1992) J. Biol. Chem. 267, 18205–18209. [PubMed] [Google Scholar]
- 13.Enghild, J. J., Thogersen, I. B., Oury, T. D., Valnickova, Z., Hojrup, P. & Crapo, J. D. (1999) J. Biol. Chem. 274, 14818–14822. [DOI] [PubMed] [Google Scholar]
- 14.Bowler, R. P., Nicks, M., Olsen, D. A., Thogersen, I. B., Valnickova, Z., Hojrup, P., Franzusoff, A., Enghild, J. J. & Crapo, J. D. (2002) J. Biol. Chem. 277, 16505–16511. [DOI] [PubMed] [Google Scholar]
- 15.Oury, T. D., Crapo, J. D., Valnickova, Z. & Enghild, J. J. (1996) Biochem. J. 317, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fattman, C. L., Enghild, J. J., Crapo, J. D., Schaefer, L. M., Valnickova, Z. & Oury, T. D. (2000) Biochem. Biophys. Res. Commun. 275, 542–548. [DOI] [PubMed] [Google Scholar]
- 17.Stenlund, P. & Tibell, L. A. (1999) Protein Eng. 12, 319–325. [DOI] [PubMed] [Google Scholar]
- 18.Bury, A. F. (1981) J. Chromatogr. 213, 491–500. [Google Scholar]
- 19.Matsudaira, P. (1987) J. Biol. Chem. 262, 10035–10038. [PubMed] [Google Scholar]
- 20.McCord, J. M. & Fridovich, I. (1969) J. Biol. Chem. 244, 6049–6055. [PubMed] [Google Scholar]
- 21.Boissinot, M., Karnas, S., Lepock, J. R., Cabelli, D. E., Tainer, J. A., Getzoff, E. D. & Hallewell, R. A. (1997) EMBO J. 16, 2171–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kussmann, M., Nordhoff, E., Rahbek-Nielsen, H., Haebel, S., Rossel-Larsen, M., Jakobsen, L., Gobom, J., Mirgorodskaya, E., Kroll-Kristensen, A., Palm, L. & Roepstorff, P. (1997) J. Mass. Spectrom. 32, 593–601. [Google Scholar]
- 23.Ryle, A. P. & Sanger, F. (1955) Biochem. J. 60, 535–556. [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada, H., Yamada, Y., Adachi, T., Goto, H., Ogasawara, N., Futenma, A., Kitano, M., Miyai, H., Fukatsu, A., Hirano, K. & Kakumu, S. (1997) Jpn. J. Hum. Genet. 42, 353–356. [DOI] [PubMed] [Google Scholar]
- 25.Patterson, S. D. & Katta, V. (1994) Anal. Chem. 66, 3727–3732. [DOI] [PubMed] [Google Scholar]
- 26.Schiering, N., Kabsch, W., Moore, M. J., Distefano, M. D., Walsh, C. T. & Pai, E. F. (1991) Nature 352, 168–172. [DOI] [PubMed] [Google Scholar]
- 27.Blake, C. C., Ghosh, M., Harlos, K., Avezoux, A. & Anthony, C. (1994) Nat. Struct. Biol. 1, 102–105. [DOI] [PubMed] [Google Scholar]
- 28.Olsen, E. H., Rahbek-Nielsen, H., Thogersen, I. B., Roepstorff, P. & Enghild, J. J. (1998) Biochemistry 37, 408–416. [DOI] [PubMed] [Google Scholar]
- 29.White, S., Boyd, G., Mathews, F. S., Xia, Z. X., Dai, W. W., Zhang, Y. F. & Davidson, V. L. (1993) Biochemistry 32, 12955–12958. [DOI] [PubMed] [Google Scholar]
- 30.Kao, P. N. & Karlin, A. (1986) J. Biol. Chem. 261, 8085–8088. [PubMed] [Google Scholar]
- 31.Folz, R. J., Guan, J., Seldin, M. F., Oury, T. D., Enghild, J. J. & Crapo, J. D. (1997) Am. J. Respir. Cell Mol. Biol. 17, 393–403. [DOI] [PubMed] [Google Scholar]
- 32.Willems, J., Zwijsen, A., Slegers, H., Nicolai, S., Bettadapura, J., Raymackers, J. & Scarcez, T. (1993) J. Biol. Chem. 268, 24614–24621. [PubMed] [Google Scholar]
- 33.Laukkanen, M. O., Lehtolainen, P., Turunen, P., Aittomaki, S., Oikari, P., Marklund, S. L. & Yla-Herttuala, S. (2000) Gene 254, 173–179. [DOI] [PubMed] [Google Scholar]
- 34.Lamb, A. L., Torres, A. S., O'Halloran, T. V. & Rosenzweig, A. C. (2001) Nat. Struct. Biol. 8, 751–755. [DOI] [PubMed] [Google Scholar]
- 35.Graf, P. C. & Jakob, U. (2002) Cell. Mol. Life. Sci. 59, 1624–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aslund, F. & Beckwith, J. (1999) Cell 96, 751–753. [DOI] [PubMed] [Google Scholar]
- 37.Stevens, F. J. & Argon, Y. (1999) Semin. Cell Dev. Biol. 10, 443–454. [DOI] [PubMed] [Google Scholar]
- 38.Frand, A. R., Cuozzo, J. W. & Kaiser, C. A. (2000) Trends Cell. Biol. 10, 203–210. [DOI] [PubMed] [Google Scholar]
- 39.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]