Abstract
The multiprotein Mediator complex has been shown to interact with gene-specific regulatory proteins and RNA polymerase II in vitro. Here, we use chromatin immunoprecipitation to analyze the recruitment of Mediator to GAL genes of yeast in vivo. We find that Mediator associates exclusively with transcriptionally active and not inactive GAL genes. This association maps to the upstream activating sequence, rather than the core promoter, and is independent of RNA polymerase II, general transcription factors, and core promoter sequences. These findings support the idea of Mediator as a primary conduit of regulatory information from enhancers to promoters in eukaryotic cells.
The activation of transcription is one of the major mechanisms of gene regulation in eukaryotes. It entails the binding of gene-specific activator proteins to enhancers [termed upstream activating sequences (UASs) in yeast] and the recruitment of the RNA polymerase II (pol II) transcription machinery. Activator proteins are presumed to target one or more components of the transcription machinery, and two main targets have been proposed: the TATA-binding protein (TBP)-associated factors (TAFs), which are components of the multisubunit TFIID complex (1, 2), and the 20-subunit Mediator complex, which associates with pol II to form a holoenzyme (3–5). The TAF hypothesis was challenged by genetic studies in yeast showing the destruction of TAFs without concomitant loss of transcriptional activation (6, 7), and by results of chromatin immunoprecipitation (ChIP) showing a lack of interaction of TAFs with many transcriptionally active yeast promoters (8, 9). A recent systematic analysis showed that no TAF is universally required for transcription (10). The Mediator hypothesis, on the other hand, has been supported by genetic studies; 13 of the 20 subunits of isolated Mediator are products of genes identified in screens for mutations affecting transcriptional regulation in yeast (11–13). Moreover, at least two subunits (Srb4 and Srb6) are required for all pol II transcription (13, 14).
The purpose of the present work was to investigate Mediator interaction with yeast promoters and to compare the results with those obtained previously for TAFs. Our initial ChIP experiments were modeled after those on TAFs, which interact through TBP and the TFIID complex with core promoter elements (TATA box and transcription start site) (8, 9). As Mediator forms a complex with pol II, we expected it would similarly interact with core promoter elements through pol II. We found, however, for GAL genes, responsive to the Gal4 activator protein (15), that Mediator preferentially associates with UASs and not with core promoter elements. Pursuing this result, we analyzed the association of Mediator with GAL genes in cells defective in assembly of the pol II transcription machinery, and with a truncated GAL1 gene lacking a TATA box. The results showed that Mediator associates with GAL UASs independently of the pol II transcription machinery and in the absence of a functional core promoter.
Materials and Methods
Strains and Plasmid Construction. Strains used in this study derive from W303-1A (MATa ade2 ura3 his3 leu2 trp1) and were obtained through transformation or genetic crosses by using standard procedures. TAP-tagged Mediator subunits and hemagglutinin (HA)-tagged TBP were generated as described (16, 17). Strains containing the temperature-sensitive TFIIB allele sua7-1 and their isogenic WT counterparts were constructed as follows. (i) SUA7 (encoding TFIIB) was disrupted by introducing a sua7Δ::LEU2 fragment lacking most of SUA7 ORF (nucleotides 207–972) and containing the LEU2 marker (18) into diploid strains homozygous for RGR1::RGR1-TAP-TRP1 or SRB6::SRB6-TAP-TRP1 and HA3-TBP, ade2, ura3, his3, leu2, trp1. Disruption was confirmed by PCR and tetrad analysis. (ii) The disrupted strains were transformed with plasmids pRS413-SUA7 or pRS413-sua7-1 (kindly provided by S. Bhaumik and M. Green), and haploid cells containing a disrupted SUA7 allele and plasmid pRS413-SUA7 or pRS413-sua7-1 were generated by sporulation and isolated by tetrad dissection. Strains containing pGAL1-xylE and pGAL1Δ-xylE chimeric genes were generated by one-step integration at the URA3 locus of two pRS306 (URA3) derivatives constructed by cloning into EcoRI/XbaI-digested pRS306, an EcoRI/BamHI PCR fragment containing xylE ORF from Pseudomonas putida together with a BamHI/XbaI PCR fragment containing either GAL1 whole promoter (pGAL1, spanning position –13 to –537) or GAL1 UAS (pGAL1Δ, panning position –225 to –537).
ChIP. Crosslinked chromatin was prepared essentially as described (17). Heat-shocked cell cultures were cooled from 37°C to 28°C and crosslinked for 10–15 min with 1.4% formaldehyde. After extraction, crosslinked chromatin was collected by centrifugation at 17,400 × g for 20 min and solubilized by sonication (size of DNA fragments ranging from 100 to 1,000 bp with an average of 400 bp); the remaining debris was eliminated by centrifugation at 12,100 × g for 15 min. TAP-tagged proteins were immunoprecipitated with rabbit IgG-agarose (Sigma) for 60 min at room temperature. Immune complexes were washed twice in FA lysis buffer containing 1·M NaCl, once in 10 mM Tris·HCl (pH 8.0), 0.25 M LiCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% Na-deoxycholate, once in 10 mM Tris·HCl (pH 8.0), 1 mM EDTA, 0.150 M urea, and once in 10 mM Tris·HCl (pH 8.0), 1 mM EDTA. HA-tagged TBP and RNA pol II were immunoprecipitated with anti-HA (F7, Santa Cruz Biotechnology) and anti-RNA pol II (8WG16, Covance, Denver, PA) mouse monoclonal antibodies coupled to protein A-Sepharose beads (Amersham Pharmacia). Gal4 was immunoprecipitated with rabbit polyclonal antibodies against the Gal4 DNA-binding domain (Santa Cruz Biotechnology). Immune complexes were washed as described (17). Immunoprecipitated chromatin was eluted by heating for 20 min at 65°C in 25 mM Tris·HCl, pH 7.5, 5 mM EDTA, 0.5% SDS. Formaldehyde crosslinkings were reversed by heating the eluate over night at 65°C in the presence of1mg/ml Pronase (Roche Biochemicals). DNA was purified by using QIAquick DNA cleanup system (Qiagen, Valencia, CA) and analyzed by quantitative PCR as described (17). Reactions were carried out in 15 μl containing 1 μM of each primer, 0.2 mM of each dNTP, and 1 μCi of [α-32P]dATP (specific activity, 3,000 Ci/mmol). PCR products were separated in an 8% TBE polyacrylamide gel and quantified by using a PhosphorImager with imagequant software (Molecular Dynamics). The IP/Total ratio was calculated by dividing the amount of PCR product obtained with the IP DNA by the amount obtained with the total DNA, using for each time at least two dilutions confirmed to be in the linear range of the PCR (standard error <15%). For each immunoprecipitation, the highest value obtained was set to 100, and other values were expressed relative to this maximum.
Analysis of mRNA Levels. Total RNA was isolated by extraction with hot acidic phenol (19). Samples were digested with RNase-free DNase I (Sigma), and cDNA was generated by StrataScript reverse transcriptase (Stratagene) by using p(dN)6 random hexamer (Roche Biochemicals). A primer specific for U4 small nuclear RNA (sequence, 5′-CACCGAATTGACCATGAG-3′) was incorporated in the reaction to serve as internal reference. The cDNAs were quantified by real-time PCR by using the LightCycler instrument and LightCycler–FastStart DNA Master SYBR Green I reaction mix (Roche Biochemicals). Data were analyzed with the LightCycler data analysis software.
Results
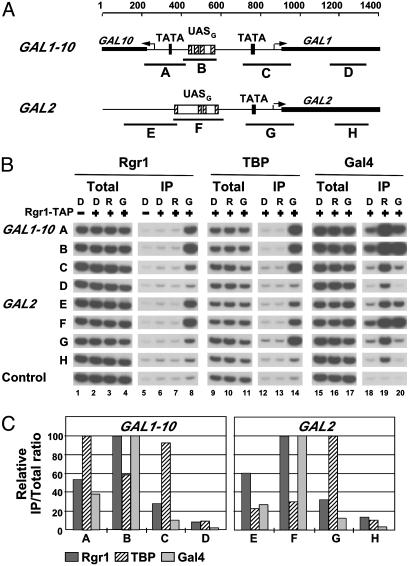
Association of Mediator with UASs. We began by analyzing the association of Rgr1, an integral component of the Mediator complex, essential for cell growth (20), with GAL1, GAL2, and GAL10 genes. ChIP analysis was performed with yeast cells expressing a TAP-tagged form of Rgr1. Cells were grown under activating conditions, in the presence of galactose, or under nonactivating conditions, in the presence of raffinose or glucose. The cells were crosslinked with formaldehyde, and chromatin was isolated and fragmented by sonication. Fragments bearing Rgr1 were precipitated with IgG-agarose, and DNA was detected in the precipitate by quantitative PCR with primer pairs specific for the enhancer (UASG), the core promoter (TATA box and transcription start site), or the ORF (Fig. 1A). For comparison, ChIP analysis was performed in parallel on Gal4, the activator protein which binds to UASG and activates the transcription of GAL promoters in the presence of galactose (15, 21), and also on TBP, whose binding to the TATA box correlates with transcriptional activity (17, 22). Gal4 protein was precipitated with anti-Gal4 antibody, whereas TBP was expressed with an HA tag and precipitated with anti-HA antibody.
Fig. 1.
Rgr1 association with GAL genes. (A) Schematic of GAL1–10 and GAL2 promoters and regulatory regions showing location of primer pairs used in ChIP analysis. For GAL1–10:A, –690/–481; B, –491/–322; C, –142/+81; and D, +252/+439 (coordinates are given relative to GAL1 ATG). For GAL2: E, –794/–524; F, –591/–538; G, –179/+70; and H, +285/+444 (coordinates are given relative to GAL2 ATG). The arrows indicate the position of transcription start sites. (B) Chromatin from cells expressing TAP-tagged (+) or untagged (–) Rgr1 and HA3-tagged TBP grown in YP medium containing glucose (D), raffinose (R), or galactose (G) was immunoprecipitated with IgG-agarose (Rgr1), anti-HA (TBP), or anti-Gal4 (Gal4) antibodies. Immunoprecipitated (IP) and input (Total) DNA were analyzed by radioactive PCR by using primers shown in A and primers located in a transcriptionally inactive region (Control). PCR products were separated on an 8% TBE polyacrylamide gel and visualized by autoradiography. Signals shown in Total or IP derive from identical dilutions and were confirmed to be in the linear range of PCR. (C) Quantitation of the ChIP data obtained from the cells grown in galactose. Amounts of PCR products were quantified by PhosphorImager analysis, and IP/Total ratios were calculated by dividing the amount of PCR product obtained with IP DNA by the amount obtained with the total DNA, using each time at least two dilutions confirmed to be in the linear range of the PCR (standard error <15%, data not shown). The highest value obtained in each immunoprecipitation was set to 100, and other values were expressed relative to this maximum.
The results for Gal4 showed a strong association with all GAL promoters in the presence of galactose and raffinose, and a 4- to 5-fold weaker association in the presence of glucose (Fig. 1B, lanes 18–20). This result is in agreement with previous studies showing that Gal4 is expressed in the absence of galactose, although its expression is diminished in glucose due to the general glucose-dependent repression system in yeast (15, 21). This result also confirms that Gal4 binds to UASG in vivo in both the presence and the absence of galactose (15, 21). In contrast, TBP was associated with GAL promoters only in cells grown in galactose, where activation takes place, but not in cells grown in raffinose or glucose, where activity of DNA-bound Gal4 is inhibited by Gal80 (Fig. 1B, lanes 12–14). The results for Rgr1 showed a strong association with all GAL promoters in cells grown in galactose (Fig. 1B, lanes 5–8). PCR signals were as much as 50-fold greater for the TAP-tagged Rgr1 strain than for the untagged strain (Fig. 1B, compare lanes 5 and 8). In contrast, no association was observed in cells grown in raffinose or glucose. Thus, Rgr1 specifically associates with transcriptionally active GAL promoters but not with transcriptionally inactive ones.
Comparison of the ChIP results for Gal4 and TBP shows that the analysis could distinguish between association with UASG and core promoter elements, despite their close proximity in some cases (only 150 bp apart for GAL10). For instance, the PCR signals for Gal4 association with the GAL1–10 promoter and intergenic region formed a single peak centered on UASG, whereas those for TBP formed two peaks over the core promoters, with a trough over UASG (Fig. 1C). Similarly, the PCR signals for Gal4 and TBP association with the GAL2 regulatory region formed two distinct peaks centered on UASG and the core promoter, respectively. In all cases, the Rgr1 distribution was very similar to that of Gal4 but strikingly different from that of TBP (Fig. 1C). For Rgr1 as well as for Gal4, the PCR signals from UAS regions were always at least 2-fold greater than those from the core promoter regions, whereas for TBP, the PCR signals from UAS regions were always lower than those from the adjacent core promoter regions. Evidently, the series of overlapping DNA fragments produced by sonication, ranging in size from 100 to 1,000 bp, with an average of ≈400 bp, is capable of resolving associations with sequences separated by as few as 150 bp. We conclude that the Rgr1 subunit of Mediator preferentially associates with the UAS of transcriptionally active GAL genes.
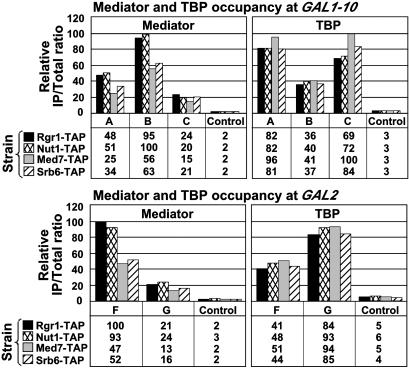
The same experimental approach was applied to yeast strains expressing TAP-tagged forms of the Mediator subunits Srb6, Med7, and Nut1. Electron microscopy and image processing has revealed a division of Mediator in three modules: head, which includes Srb6; middle, which includes Med7 and Nut1; and tail, with Rgr1 apparently straddling the junction between middle and tail (23). The ChIP results for Med7, Nut1, and Srb6 were essentially the same as those for Rgr1 (Fig. 2). First, all four Mediator subunits were associated with GAL promoters in the presence of galactose and not in the presence of raffinose or glucose (Fig. 2 and data not shown). Differences in PCR signals among the Mediator subunits were most likely due to differences in crosslinking efficiency and not to differences in gene activity among the four tagged strains. Indeed, TBP occupancy at GAL1, GAL2, and GAL10 was the same in the four strains, indicating that GAL genes were equally expressed in all of the strains. Second, all four subunits were preferentially associated with UAS regions, whereas TBP was preferentially associated with core promoter regions (Fig. 2).
Fig. 2.
Mediator association with GAL genes. Equivalent amounts of crosslinked chromatin extracts prepared from cells grown in YP galactose medium and expressing Rgr1-TAP, Nut1-TAP, Med7-TAP, or Srb6-TAP and HA3-TBP were immunoprecipitated in parallel with IgG-agarose (Mediator) or anti-HA antibodies (TBP). Immunoprecipitated (IP) and input (Total) DNA were analyzed with primer pairs described in Fig. 1.
We conclude that the entire Mediator complex is associated with UASG under activating conditions. In the absence of any reported Mediator–DNA interaction, we presume that Mediator subunits are crosslinked by formaldehyde to UAS DNA through Gal4 protein. The PCR signals obtained for Mediator subunits, and thus apparent crosslinking efficiencies, were severalfold lower than those for Gal4 protein or for TBP, consistent with indirect crosslinking of Mediator subunits to the DNA (data not shown). Moreover, Mediator was shown to interact with Gal4 in vitro (24).
It is noteworthy that Mediator is not associated with UASG under nonactivating conditions. Genetic studies have demonstrated a role of Mediator in repression as well as activation of transcription of many promoters (11), so Mediator might have been found at GAL genes in the presence of raffinose or glucose. Indeed, Gal4 is bound to UASG and therefore in a position to recruit Mediator under these conditions. Instead, we detect Mediator at GAL genes only in cells grown in galactose, suggesting that repression in raffinose or glucose does not require Mediator.
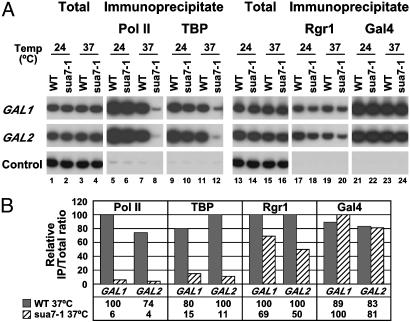
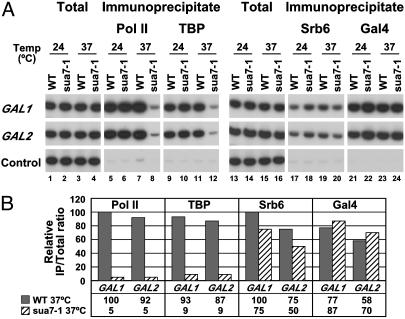
Mediator–UAS Interaction Is Independent of the pol II Transcription Machinery. The strongly preferential association of Mediator with UAS regions was unexpected because Mediator is known to interact with pol II (5, 23, 25), which is found associated with the core promoters and ORFs of transcribed genes. It has been suggested that Mediator is recruited to promoters as part of an even larger complex, or holoenzyme, including not only pol II but also general transcription factors (12, 26). To investigate such possibilities, we performed ChIP analysis on a yeast strain bearing a temperature-sensitive allele (sua7-1) of the general transcription factor TFIIB, which prevents assembly of the pol II machinery at restrictive temperatures (22). WT and sua7-1 cells expressing TAP-tagged Rgr1 or TAP-tagged Srb6 were grown in the presence of galactose at a permissive temperature (24°C) and then raised to a restrictive temperature (37°C) for 40 min. Immunoprecipitates containing pol II and TBP were analyzed with PCR primers specific for the core promoter region, and immunoprecipitates containing Mediator subunits and Gal4 were analyzed with primers for UASG (Figs. 3 and 4). The results showed an almost complete loss of pol II (20-fold decrease) and TBP (10-fold decrease) from GAL promoters in sua7-1 cells raised to a restrictive temperature, compared with WT cells (Figs. 3 and 4, lanes 5–12). These results confirm that inactivation of TFIIB in sua7-1 cells has a rapid and dramatic effect on assembly of the pol II machinery at promoters (22). In contrast, there was only a slight loss (<2-fold) of Rgr1 (Fig. 3, lanes 17–20) and Srb6 (Fig. 4, lanes 17–20), and no loss of Gal4 (Figs. 3 and 4, lanes 21–24). We conclude that Mediator is capable of association with transcriptionally active genes independently of pol II and general transcription factors.
Fig. 3.
Association of Rgr1 with GAL genes in TFIIB mutant cells. (A) TFIIB temperature-sensitive (sua7-1) and isogenic WT cells, both expressing Rgr1-TAP and HA3-TBP, were grown in YP galactose medium at permissive temperature (24°C) and shifted for 40 min to restrictive temperature (37°C). Chromatin was immunoprecipitated with anti-pol II (Pol II), anti-HA (TBP), IgG-agarose (Rgr1), and anti-Gal4 (Gal4) antibodies. Immunoprecipitated and input (Total) DNA were analyzed with primers for GAL1 and GAL2 (TATA element region for pol II and TBP and UASG region for Rgr1 and Gal4) and primers for a transcriptionally inactive region (Control). (B) Quantification by PhosphorImager analysis of the ChIP data obtained in the experiment presented in A. Quantification was carried out as described in Fig. 1.
Fig. 4.
Association of Srb6 with GAL genes in TFIIB mutant cells. Same as in Fig. 3 with cells expressing Srb6-TAP instead of Rgr1-TAP.
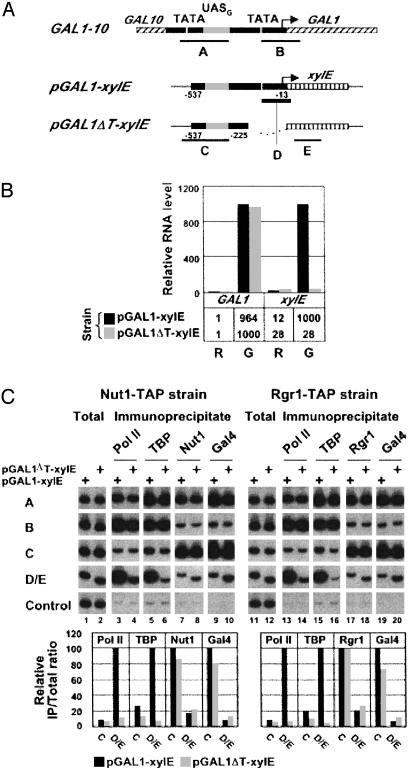
Mediator–UAS Interaction in the Absence of Core Promoter Elements. In the previous experiment, sua7-1 was inactivated while GAL genes were already undergoing transcription. It might therefore be argued that pol II, TBP, TFIIB, and other general transcription factors are not required for the retention of Mediator at the UAS but are nonetheless required for its recruitment upon the initiation of transcription. To investigate this possibility, we compared the association of Mediator with UASG in a chimera containing the entire GAL1 promoter and regulatory region fused to the bacterial xylE gene (pGAL1-xylE), and the association of Mediator with UASG in the same chimera lacking the TATA element (pGAL1ΔT-xylE) (Fig. 5A). Both chimeric genes were integrated at the URA3 chromosome locus, leaving intact the native GAL1 gene. Measurement of mRNA levels by RT-PCR showed that transcription from pGAL1-xylE was induced 100-fold upon addition of galactose to cells grown in raffinose, whereas removal of the TATA element prevented any induction at all (Fig. 5B). As a control, transcription of GAL1 was induced to the same extent in cells containing pGAL1-xylE and pGAL1ΔT-xylE.
Fig. 5.
Association of Mediator with an isolated UASG. (A) Schematic of GAL1–10 regulatory region and pGAL1-xylE and pGAL1ΔT-xylE chimeric genes showing location of primers used in the ChIP analysis. The arrow indicates the position of GAL1 transcription start site. The chimeric genes contain a fragment of GAL1 promoter spanning position –537 to –13 (pGAL1-xylE) and position –537 to –225 (pGAL1ΔT-xylE) fused to xylE ORF and are integrated at the URA3 locus. (B) Quantitation of GAL1 and xylE RNA levels by RT-PCR in pGAL1-xylE and pGAL1ΔT-xylE cells grown in raffinose (R) and induced for 90 min with galactose (G). GAL1 and xylE maximum RNA levels were arbitrarily set to 1,000. (C) ChIP analysis of galactose-induced pGAL1-xylE and pGAL1ΔT-xylE cells expressing either Nut1-TAP or Rgr1-TAP. Crosslinked chromatin was immunoprecipitated with anti-pol II (Pol II), anti-HA (TBP), IgG-agarose (Rgr1), and anti-Gal4 (Gal4) antibodies. Immunoprecipitated and input (Total) DNA were analyzed by radioactive PCR with primers shown in A and primers specific for a transcriptionally inactive region (Control). Pair D was used specifically for samples deriving from pGAL1-xylE cells, and pair E was specifically for samples deriving from pGAL1ΔT-xylE cells. PCR products were separated on an 8% TBE polyacrylamide gel and visualized by autoradiography (Upper). Quantification in Lower was carried out as described in Fig. 1.
Crosslinked chromatin was prepared from cells grown under activating conditions. Immunoprecipitates containing pol II, TBP, Nut1 or Rgr1, and Gal4 were analyzed with PCR primers specific for pGAL1-xylE and pGAL1ΔT-xylE (pairs C–E) and, as a control, with primer pairs specific for the native GAL1 gene (pairs A and B). The results showed that association of pol II and TBP with the core promoter was much diminished by removal of the TATA element (9.3- and 15.4-fold, respectively) (Fig. 5C, lanes 3–6 and 13–16; compare results with probes D/E for pGAL1-xylE and pGAL1ΔT-xylE; note that D and E are at the same distance from UASG). By contrast, no significant change in association of Nut1 and Rgr1 subunits of the Mediator with UASG was observed upon removal of the TATA element from pGAL1-xylE (Fig. 5C, lanes 7, 8, 17, and 18; compare results with primer pair C for pGAL1-xylE and pGAL1ΔT-xylE). We conclude that Mediator is recruited to GAL promoters in the absence of a functional core promoter and without simultaneous recruitment of polII and the general transcription factors. Therefore, Mediator recruitment to GAL promoters does not require Mediator to interact with the core promoter, nor does it require the presence of pol II and the general transcription factors. These results strongly suggest that Mediator is not recruited to GAL promoters or, in all likelihood, to other promoters, as part of a preassembled holoenzyme with the rest of the pol II transcription machinery.
Discussion
The chief finding of this study is the distinction between Mediator interaction with UASs and core promoter sequences. The distinction is supported by three lines of evidence. First, Mediator was localized by ChIP analysis to GAL UASs. Although the distance from the UAS to the core promoter was as short as 150 bp (for GAL10), ChIP analysis with DNA fragments of, on average, ≈400 bp was sufficient for localization to the UAS, as shown by the clear difference in ChIP profiles obtained for Mediator and TBP. Second, Mediator–UAS interaction was unaffected by a mutation preventing assembly of a transcription initiation complex at the core promoter, in contrast with pol II and TBP, whose interaction with the core promoter was abolished. Finally, Mediator interacted with a UAS in the absence of a key core promoter element, the TATA box, whereas pol II and TBP did not.
Our results have two major implications: they provide strong support for a central role of Mediator in transcriptional activation and they contradict the proposal that a pol II holoenzyme, comprising pol II, Mediator, and general transcription factors, is recruited en bloc to the promoters of active genes (12). The primary association of Mediator is evidently with transcriptional activators rather than with the pol II machinery. Mediator does interact with pol II, as shown by biochemical evidence for a Mediator–pol II complex (5, 27), and by structural analysis of this complex (23). However, the interaction with pol II is transient, occurring only during transcription initiation (28), and is therefore less readily detected by ChIP analysis than the interaction of Mediator with activators and UASs, which is persistent.
Our spatial discrimination between interactions of Mediator with GAL UAS and core promoter elements complements a temporal discrimination among transcription protein interactions with GAL promoters published while this manuscript was in preparation (29). It was shown that Mediator interacts with GAL1 promoter more rapidly than do pol II, TBP, TFIIE, or TFIIH following the induction of transcription. Both spatial and temporal discrimination between Mediator and pol II interactions were previously demonstrated for the HO endonuclease promoter (30, 31). The combined results from studies of four GAL promoters and the HO promoter points to the generality of the conclusions regarding Mediator interaction with UASs and the independent recruitment of Mediator and pol II.
The generality of the conclusions from studies in yeast extends to higher organisms. It was shown, at the resolution of indirect immunofluorescence from Drosophila polytene chromosomes, that heat shock transcription factor and Mediator colocalize to heat shock response elements rapidly following induction by heat shock (32). This colocalization occurred for heat shock response elements in the absence of associated core promoters. Neither TAFs nor pol II exhibited such behavior. The inference from this work, that Mediator interacts with heat shock transcription factor, is supported by biochemical studies of human transcription factors. For example, human thyroid hormone receptor protein was recovered from uninduced cells as an individual polypeptide, whereas it was isolated as a complex with human Mediator following hormonal induction (33). Vitamin D receptor, SREBP, and other inducible transcriptional activators were similarly isolated as tight complexes with Mediator from human cells grown under inducing conditions (34).
Our results thus contribute to the emerging picture of Mediator as the central molecule of transcriptional regulation. Mediator interacts with transcriptional activators and with enhancers of all genes so far analyzed, in contrast with TAFs, whose interactions are restricted to the core promoters of a limited number of genes. Mediator is in a position to orchestrate the entire series of events that ensues upon activator binding and culminates in the initiation of transcription.
Acknowledgments
We are especially grateful to Dominique Thomas and Yolande Surdin-Kerjan for their constant support. We thank Kevin Struhl for his role in initiating this collaboration, and Sukesh Bhaumik, Michael Green, and Michael Hampsey for providing plasmids and yeast strains. This work was supported by Centre National de la Recherche Scientifique and ACI Grant 5319 from the French Ministry of Research (to L.K.), postdoctoral fellowships from the Deutsche Forschungsgemeinschaft, the Schweizer Nationalfonds, and the Norvartis Foundation (to T.B.), and National Institutes of Health Grant GM36659 (to R.D.K.).
Abbreviations: ChIP, chromatin immunoprecipitation; pol II, RNA polymerase II; TBP, TATA-binding protein; TAF, TBP-associated factors; UAS, upstream activating sequence; HA, hemagglutinin.
References
- 1.Pugh, B. F. & Tjian, R. (1990) Cell 61, 1187–1197. [DOI] [PubMed] [Google Scholar]
- 2.Dynlacht, B. D., Hoey, T. & Tjian, R. (1991) Cell 66, 563–576. [DOI] [PubMed] [Google Scholar]
- 3.Kelleher, R. J., III, Flanagan, P. M. & Kornberg, R. D. (1990) Cell 61, 1209–1215. [DOI] [PubMed] [Google Scholar]
- 4.Flanagan, P. M., Kelleher, R. J., III, Sayre, M. H., Tschochner, H. & Kornberg, R. D. (1991) Nature 350, 436–438. [DOI] [PubMed] [Google Scholar]
- 5.Kim, Y. J., Bjorklund, S., Li, Y., Sayre, M. H. & Kornberg, R. D. (1994) Cell 77, 599–608. [DOI] [PubMed] [Google Scholar]
- 6.Moqtaderi, Z., Bai, Y., Poon, D., Weil, P. A. & Struhl, K. (1996) Nature 383, 188–191. [DOI] [PubMed] [Google Scholar]
- 7.Walker, S. S., Reese, J. C., Apone, L. M. & Green, M. R. (1996) Nature 383, 185–188. [DOI] [PubMed] [Google Scholar]
- 8.Kuras, L., Kosa, P., Mencia, M. & Struhl, K. (2000) Science 288, 1244–1248. [DOI] [PubMed] [Google Scholar]
- 9.Li, X. Y., Bhaumik, S. R. & Green, M. R. (2000) Science 288, 1242–1244. [DOI] [PubMed] [Google Scholar]
- 10.Shen, W. C., Bhaumik, S. R., Causton, H. C., Simon, I., Zhu, X., Jennings, E. G., Wang, T. H., Young, R. A. & Green, M. R. (2003) EMBO J. 22, 3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson, M. (1997) Annu. Rev. Cell Dev. Biol. 13, 1–23. [DOI] [PubMed] [Google Scholar]
- 12.Lee, T. I. & Young, R. A. (2000) Annu. Rev. Genet. 34, 77–137. [DOI] [PubMed] [Google Scholar]
- 13.Myers, L. C. & Kornberg, R. D. (2000) Annu. Rev. Biochem. 69, 729–749. [DOI] [PubMed] [Google Scholar]
- 14.Holstege, F. C., Jennings, E. G., Wyrick, J. J., Lee, T. I., Hengartner, C. J., Green, M. R., Golub, T. R., Lander, E. S. & Young, R. A. (1998) Cell 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 15.Johnston, M. (1987) Microbiol. Rev. 51, 458–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borggrefe, T., Davis, R., Bareket-Samish, A. & Kornberg, R. D. (2001) J. Biol. Chem. 276, 47150–47153. [DOI] [PubMed] [Google Scholar]
- 17.Kuras, L. & Struhl, K. (1999) Nature 399, 609–613. [DOI] [PubMed] [Google Scholar]
- 18.Pinto, I., Ware, D. E. & Hampsey, M. (1992) Cell 68, 977–988. [DOI] [PubMed] [Google Scholar]
- 19.Iyer, V. & Struhl, K. (1996) Proc. Natl. Acad. Sci. USA 93, 5208–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y., Bjorklund, S., Jiang, Y. W., Kim, Y. J., Lane, W. S., Stillman, D. J. & Kornberg, R. D. (1995) Proc. Natl. Acad. Sci. USA 92, 10864–10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bash, R. & Lohr, D. (2001) Prog. Nucleic Acid Res. Mol. Biol. 65, 197–259. [DOI] [PubMed] [Google Scholar]
- 22.Li, X. Y., Virbasius, A., Zhu, X. & Green, M. R. (1999) Nature 399, 605–609. [DOI] [PubMed] [Google Scholar]
- 23.Asturias, F. J., Jiang, Y. W., Myers, L. C., Gustafsson, C. M. & Kornberg, R. D. (1999) Science 283, 985–987. [DOI] [PubMed] [Google Scholar]
- 24.Jeong, C. J., Yang, S. H., Xie, Y., Zhang, L., Johnston, S. A. & Kodadek, T. (2001) Biochemistry 40, 9421–9427. [DOI] [PubMed] [Google Scholar]
- 25.Davis, J. A., Takagi, Y., Kornberg, R. D. & Asturias, F. A. (2002) Mol. Cell 10, 409–415. [DOI] [PubMed] [Google Scholar]
- 26.Ptashne, M. & Gann, A. (1997) Nature 386, 569–577. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, C. M., Koleske, A. J., Chao, D. M. & Young, R. A. (1993) Cell 73, 1361–1375. [DOI] [PubMed] [Google Scholar]
- 28.Svejstrup, J. Q., Li, Y., Fellows, J., Gnatt, A., Bjorklund, S. & Kornberg, R. D. (1997) Proc. Natl. Acad. Sci. USA 94, 6075–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryant, G. O. & Ptashne, M. (2003) Mol. Cell 11, 1301–1309. [DOI] [PubMed] [Google Scholar]
- 30.Bhoite, L. T., Yu, Y. & Stillman, D. J. (2001) Genes Dev. 15, 2457–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosma, M. P., Panizza, S. & Nasmyth, K. (2001) Mol. Cell 7, 1213–1220. [DOI] [PubMed] [Google Scholar]
- 32.Park, J. M., Werner, J., Kim, J. M., Lis, J. T. & Kim, Y. J. (2001) Mol. Cell 8, 9–19. [DOI] [PubMed] [Google Scholar]
- 33.Fondell, J. D., Ge, H. & Roeder, R. G. (1996) Proc. Natl. Acad. Sci. USA 93, 8329–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malik, S. & Roeder, R. G. (2000) Trends Biochem. Sci. 25, 277–283. [DOI] [PubMed] [Google Scholar]